Abstract
Objective
To determine the spectrum and prevalence of mutations in the RYR2-encoded the cardiac ryanodine receptor in cases with exertional syncope and normal QTc.
Background
Mutations in the RYR2 cause type 1 catecholaminergic polymorphic ventricular tachycardia (CPVT1), a cardiac channelopathy with increased propensity for lethal ventricular dysrhythmias. Most RYR2 mutational analyses target 3 canonical domains encoded by < 40% of the translated exons. The extent of CPVT1-associated mutations localizing outside of these domains remains unknown as RYR2 has not been examined comprehensively in most patient cohorts.
Methods
Mutational analysis of all RYR2 exons was performed using PCR, DHPLC, and DNA sequencing on 155 unrelated patients (49% females, 96% white, age at diagnosis 20 ± 15 years, mean QTc 428 ± 29 ms), with either clinical diagnosis of CPVT (n = 110) or an initial diagnosis of exercise-induced long QT syndrome (LQTS) but with QTc < 480 ms and a subsequent negative LQTS genetic test (n = 45).
Results
Sixty-three (34 novel) possible CPVT1-associated mutations, absent in 400 reference alleles, were detected in 73 unrelated patients (47%). Thirteen new mutation-containing exons were identified. Two thirds of the CPVT1-positive patients had mutations that localized to one of 16 exons.
Conclusions
Possible CPVT1 mutations in RYR2 were identified in nearly half of this cohort. 45 of the 105 translated exons are now known to host possible mutations. Considering that ~65% of CPVT1-positive cases would be discovered by selective analysis of 16 exons, a tiered targeting strategy for CPVT genetic testing should be considered.
Keywords: Ryanodine Receptor, Catecholaminergic Polymorphic Ventricular Tachycardia, Sudden Cardiac Death, Exertional Syncope
INTRODUCTION
Catecholaminergic polymorphic ventricular tachycardia (CPVT) is a potentially lethal, heritable arrhythmia syndrome often manifesting as exercise-induced ventricular arrhythmias, syncope or sudden death.1 With mortality rates of 30-50% by age 35 years, CPVT is one of the most malignant cardiac channelopathies expressed predominately in young patients with otherwise structurally normal hearts2. While the resting 12-lead electrocardiogram (ECG) is typically normal, the hallmark arrhythmia, bidirectional VT, is often present during exercise stress testing and has been considered pathognomonic for CPVT.1,3
CPVT stems from an alteration of intracellular calcium handling involving the critical calcium-induced calcium release mechanism of myocardial cells. At the molecular level, gain of function mutations in the cardiac ryanodine receptor encoded by RYR2 account for at least 50% of CPVT cases and is annotated as type 1 CPVT (CPVT1).3 Mutations in CASQ2-encoded calsequestrin are responsible for the very rare, autosomal recessive form known as type 2 CPVT (CPVT2).2,4
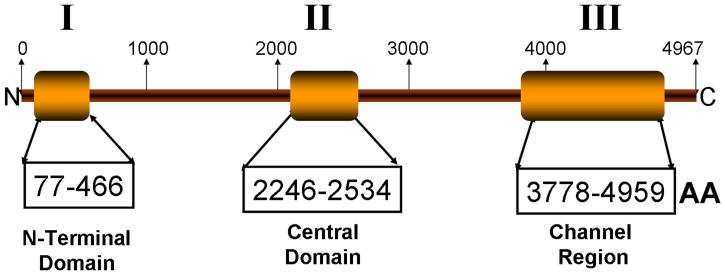
The cardiac ryanodine receptor (RyR2), encoded by the 105-exon-containing RYR2 gene, is one of the largest ion channel proteins comprised of 4967 amino acids; localizes to the sarcoplasmic reticulum, and controls intracellular calcium release and cardiac contraction. Since the sentinel discovery of a CPVT-causing RYR2 mutation5, a cluster distribution involving three discrete protein regions has been reported. Based in a potential physiological role for these “hot-spots”, these regions have been termed “domains” I, II and III (Figure 1)6,7. Similar mutation clustering is observed in the RYR1 gene which encodes the skeletal muscle RyR1 and is linked to malignant hyperthermia and central core disease8-10. However, since the majority of CPVT cases have not undergone the entire RYR2 scan, the prevalence of mutations residing outside these three canonical domains (i.e. ~61 exons that encode for 2570 amino-acids) remains unknown.
Figure 1. Mutation clustering in the cardiac ryanodine receptor (RyR2).
Mutations are distributed in three “hot-spots” regions, called domains I (N-terminal), II (central) and III (channel region)6,7. AA: amino-acid number estimated for each domain.
Currently, among research laboratories and clinical diagnostic laboratories, there is no consensus or clear definition of the “RYR2 targeted scan” resulting in enormous discrepancy in the number of exons studied by each research group or commercial company. This situation has an important impact in “gene-negative” definition, genotype-phenotype correlation and patient quality of care. In the present study, we sought to determine the prevalence of mutations throughout RYR2’s entire open reading frame in a large cohort of unrelated cases referred to 2 different institutions for exertional syncope and, using a combined analysis of the previous reported mutations and the novel mutations found in this cohort, we propose a novel, targeted “genetic approach” for CPVT1 genetic testing.
METHODS
Study Participants
We studied a cohort of 155 unrelated patients referred to either the Windland Smith Rice Sudden Death Genomics Laboratory at Mayo Clinic, Rochester, MN or the Department of Clinical Genetics, Academic Medical Center, University of Amsterdam, Netherlands for genetic testing between August 2001 and June 2008. A clinical diagnosis of CPVT was rendered in 110 patients by either one of the authors (MJA, AAMW) or the referring physician. Of these, 78 were classified as “strong CPVT phenotype” because of exertional syncope plus documentation of bidirectional or polymorphic ventricular tachycardia (BVT/PVT) while 32 were classified as “possible CPVT phenotype” based on the presence of exertional syncope and stress test induced ventricular ectopy but not BVT/PVT. In addition, 45 cases were referred as “possible/atypical long QT syndrome (LQTS)” because of exertional syncope and QTc values < 480 ms. All were genotype negative for the 12 known LQTS-susceptibility genes.
Following receipt of written consent for this Mayo Foundation Institutional Review Board and Amsterdam Academic Medical Center Medical Ethical Committee approved protocol, genomic DNA was extracted from peripheral blood lymphocytes using the Purgene DNA extraction kit (Gentra, Inc, Minneapolis, MN, USA). In cases with suspected mosaicism, additional DNA from saliva was isolated using the ORAgene kit (DNA Genotek, Ottawa, Ontario, Canada) and DNA from skin fibroblasts and hair-roots was isolated using the QIAamp DNA minikit (Qiagen, USA).
Mutational Analysis
Comprehensive open reading frame/splice site mutational analysis of all 105 RYR2 exons was performed using polymerase chain reaction (PCR), denaturing high performance liquid chromatography (DHPLC), and DNA sequencing as described previously.11 The flanking primers used for PCR were published previously or designed with Oligo software (Molecular Biology Insights, Inc., Cascade Colo.) and are available on request. We also searched for large genomic rearrangements affecting exon 3 as reported previously12.
All putative pathogenic variants must have been absent in 400 reference alleles (100 healthy white and 100 healthy black) obtained from the Human Genetic Cell Repository sponsored by the National Institute of General Medical Sciences and the Coriell Institute for Medical Research (Camden, New Jersey) in order to be considered as potentially disease-related.
Statistical Analysis
We used the JMP Statistical Software (JMP 6.0, 2005; SAS Institute Inc, Cary, NC). All continuous variables are reported as mean ± SD. Differences between continuous variables were evaluated using unpaired Student t tests, and nominal variables were analyzed using chi-square analysis. Statistical significance was considered at p < 0.05.
RESULTS
The demographic characteristics of the 155 unrelated patients are shown in Table 1. 96% were Caucasians, 49% were females, age at symptoms was 20 ± 15 yrs, and average QTc was 428 ± 29 ms. The mean age of onset of symptoms was significantly lower in RYR2 mutation positive subjects compared to those with a negative genetic test (16.7 ± 12.3 vs 23.8 ± 16.6 yrs respectively, p<0.004).
Table 1. Demographics Characteristics of the Cohort.
| CPVT Strong Phenotype |
CPVT Possible Phenotype |
Gene Negative LQTS |
Total | |
|---|---|---|---|---|
| No. of Patients | 78 | 32 | 45 | 155 |
| Age (yrs) mean ± SD | 20 ± 15 | 20 ± 16 | 22 ± 14 | 20 ± 15 |
| QTc (ms) mean ± SD | 415 ± 26 | 434 ± 30 | 434 ± 27 | 427 ± 29 |
| %Female | 47 | 44 | 57 | 49 |
| RYR2 Positives (%) | n=47 (60.2%) | n=12 (37.5%) | n=14 (31.1%) | n=73 (47.1%) |
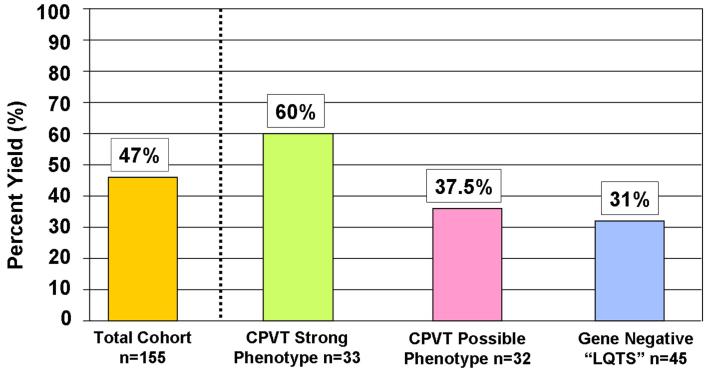
Overall, 77 (63 unique, 34 novel) putative disease causing mutations were identified in 73 cases (47%, Table 2, Figure 2). 41/73 mutation positive cases (56%) were females. Putative mutations were absent in 400 references alleles and most of the mutated residues exhibit highly conservation across species (Supplemental Table). The yield of the genetic test was significantly higher among the 78 cases classified clinically as “strong CPVT phenotype” compared to the 32 cases diagnosed as “possible CPVT phenotype” (60% vs 37.5%, p < 0.04). Notably, nearly one-third of the 45 “gene negative LQTS” cases had a rare missense mutation in RYR2 (Table 1, Figure 3). Four out of the 73 RYR2 mutation-positive cases hosted multiple mutations (5.5%). As expected, we observed a mutation clustering distribution across RYR2; nevertheless, ten mutations found in 11 cases resided outside the three canonical domains, specifically, between domain I and II; 8 of them exhibited a strong CPVT phenotype. Three large genomic rearrangements comprising exon 3 were detected in three unrelated cases involving a 3.6 kb deletion in one and a 1.1 kb deletion in two cases.
Table 2. Compendium of RYR2 mutations and polymorphisms reported to date.
Putative mutations are indicated in red, n=129 (including 2 large genomic rearrangements involving exon 3, not detectable by regular genetic scan), polymorphisms in blue n=15.
| No. | Mutation Number (Figure 1) |
Exon | Base Position | Amino-acid Change |
Location | Cases hosting the variant (n=108) |
AA hosting the variant (n=100) |
CC hostingthe variant (n=100) |
Reference |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 3 | 1.1kb deletion* | Exon 3 del | NT | 2 | Bhuiyan12 | ||
| 2 | 2 | 3 | 3.6kb deletion* | Exon 3 del | NT | 1 | Novel | ||
| 3 | 3 | 3 | 184 C>T | L62F | NT | 1 | Novel | ||
| 4 | 3 | 230 C>T | A77V | NT | d’Amati G27 | ||||
| 5 | 4 | 3 | 241 A>C | M81L | NT | 1 | Novel | ||
| 6 | 5 | 8 | 493 C>T | P164S | NT | 1 | Choi28 | ||
| 7 | 8 | 506 G>A | R169Q | NT | Hsueh29 | ||||
| 8 | 6 | 8 | 527 G>A | R176Q | NT | 1 | Tiso24 | ||
| 9 | 7 | 8 | 556 G>A | V186M | NT | 1 | Tiso24 | ||
| 10 | 8 | 8 | 567 A>T | E189D | NT | 1 | Davis30 | ||
| 11 | 10 | 6337 G>A | H240R | NT | Tester31 | ||||
| 12 | 9 | 10 | 727 G>A | E243K | NT | 1 | Novel | ||
| 13 | 10 | 12 | 985 T>C | F329L | NT | 1 | Novel | ||
| 14 | 11 | 12 | 994C>T | R332W | NT | 1 | Novel | ||
| 15 | 12 | 13 | 1072 G>A | G357S | NT | 1 | Novel | ||
| 16 | 13 | 13 | 1129 G>A | V377M | NT | 3 | Novel | ||
| 17 | 14 | 1240 C>T | R414C | NT | Tester32 | ||||
| 18 | 14 | 14 | 1241 G>T | R414L | NT | 1 | Choi28 | ||
| 19 | 15 | 14 | 1244 C>G | T415R | NT | 1 | Novel | ||
| 20 | 14 | 1255 A>T | I419F | NT | Choi28 | ||||
| 21 | 16 | 14 | 1258 C>T | R420W | NT | 3 | Bauce33 | ||
| 22 | 17 | 14 | 1259 G>A | R420Q | NT | 2 | Novel | ||
| 23 | 15 | 1298 T>C | L433P | NT | Tiso24 | ||||
| 24 | 18 | 15 | 1396 C>G | P466A | NT | 1 | Tester14 | ||
| 25 | 19 | 16 | 1519 G>A | V507I | NT | 5 | 4 | Novel | |
| 26 | 20 | 17 | 1646 C>T | A549V | NT | 1 | Novel | ||
| 27 | 19 | Not Reported | S616L | CL | Marjmaa21 | ||||
| 28 | 21 | 21 | 2216 G>A | R739H | CL | 1 | Novel | ||
| 29 | 22 | 26 | 3038 G>A | R1013Q | CL | 1 | Novel | ||
| 30 | 27 | Not Reported | R1051P | CL | Marjmaa21 | ||||
| 31 | 23 | 28 | 3320 C>T | T1107M | CL | 1 | Novel | ||
| 32 | 24 | 28 | 3407 C>T | A1136V | CL | 3 | 2 | Novel | |
| 33 | 25 | 37 | 5170 G>A | E1724K | CL | 2 | Postma34 | ||
| 34 | 26 | 37 | 5509 G>A | E1837K | CL | 1 | Novel | ||
| 35 | 27 | 37 | 5654 G>A | G1885E | CL | 2 | 6 | Milting26 | |
| 36 | 28 | 37 | 5656 G>A | G1886S | CL | 11 | 20 | 9 | Milting26 |
| 37 | 29 | 40 | 6137 A>G | E2045G | CL | 1 | Novel | ||
| 38 | 41 | 6337 G>A | V2113M | CL | 1 | Tester31 | |||
| 39 | 41 | Not Reported | G2145R | CL | Marjmaa35 | ||||
| 40 | 30 | 42 | 6467 A>G | Y2156C | CL | 1 | Novel | ||
| 41 | 31 | 42 | 6504 C>G | H2168Q | CL | 2 | Novel | ||
| 42 | 32 | 42 | 6548 A>T | E2183V | CL | 1 | Novel | ||
| 43 | 33 | 43 | D2216V | CL | 1 | Novel | |||
| 44 | 34 | 44 | 6740 C>T | S2246L | CL | 1 | Priori5 | ||
| 45 | 35 | 44 | 6761 C>T | A2254V | CL | 1 | Postma34 | ||
| 46 | 45 | 6800 G>A | R2267H | CL | Tester36 | ||||
| 47 | 36 | 45 | 6886 G>C | E2296Q | FKBP | 1 | Novel | ||
| 48 | 45 | 6916 G>A | V2306I | FKBP | Laitinen37 | ||||
| 49 | 45 | 6919 T>C | F2307L | FKBP | Berge38 | ||||
| 50 | 46 | 6933 G>A | E2311D | FKBP | Priori3 | ||||
| 51 | 46 | 6992 T>C | V2321M | FKBP | Nishio39 | ||||
| 52 | 46 | 6982 C>T | P2328S | FKBP | Laitinen37 | ||||
| 53 | 46 | 6992 T>C | F2331S | FKBP | Creighton40 | ||||
| 54 | 46 | 7076 G>A | R2359Q | FKBP | Aizawa41 | ||||
| 55 | 47 | 7157 A>T | N2386I | FKBP | Tiso24 | ||||
| 56 | 37 | 47 | 7158 G>A | A2387T | FKBP | 3 | Tester14 | ||
| 57 | 47 | 7158 G>A | A2387P | FKBP | Bagattin42 | ||||
| 58 | 38 | 47 | 7165 A>C | M2389L | FKBP | 1 | Tester43 | ||
| 59 | 47 | 7175 A>5 | Y2392C | FKBP | Bauce33 | ||||
| 60 | 47 | 7181 C>G | A2394G | FKBP | Postma34 | ||||
| 61 | 39 | 47 | 7202 G>A | R2401H | FKBP | 1 | Aizawa41 | ||
| 62 | 47 | 7207 G>T | R2401L | FKBP | Creighton40 | ||||
| 63 | 40 | 47 | 7207 G>A | A2403T | FKBP | 1 | Choi28 | ||
| 64 | 41 | 47 | 7210 C>A | R2404T | FKBP | 1 | Beckman44 | ||
| 65 | 42 | 48 | 7258 A>T | R2420W | FKBP | 1 | Novel | ||
| 66 | 49 | 7422 G>C | P2474S | FKBP | Priori3 | ||||
| 67 | 49 | 7423 G>T | V2475F | FKBP | Tester32 | ||||
| 68 | 49 | 7511 C>T | T2504M | FKBP | Tiso24 | ||||
| 69 | 49 | Not Reported | L2487I | FKBP | Tester43 | ||||
| 70 | 50 | 7528 T>C | T2510A | FKBP | Tester31 | ||||
| 71 | 50 | 7599 C>G | L2534V | FKBP | Hasdemir45 | ||||
| 72 | 43 | 61 | 8874 A>G | Q2958R | Cytosol | 40 | 10 | 36 | Tiso24 |
| 73 | 69 | Not Reported | N3308S | Cytosol | Marjamaa21 | ||||
| 74 | 75 | Not Reported | R3570W | Cytosol | Marjamaa35 | ||||
| 75 | 83 | 11332 C>T | L3778F | Cytosol | Priori3 | ||||
| 76 | 44 | 83 | 11399 G>T | C3800F | Cytosol | 1 | Tester14 | ||
| 77 | 86 | 11636 T>C | L3879P | Cytosol | Tester31 | ||||
| 78 | 87 | 11773 C>G | Q3925E | Cytosol | Tester31 | ||||
| 79 | 45 | 88 | 11814 C>A | S3938R | Cytosol | 1 | Tester14 | ||
| 80 | 88 | 11836 G>A | G3946S | Cytosol | Priori3 | ||||
| 81 | 88 | Not Reported | G3946A | Cytosol | Davis30 | ||||
| 82 | 88 | 11876 C>T | S3959L | Cytosol | Tester31 | ||||
| 83 | 46 | 89 | 11916 G>T | M3972I | Cytosol | 1 | Novel | ||
| 84 | 47 | 89 | 11917 G>C | D3973H | Cytosol | 1 | Novel | ||
| 85 | 48 | 89 | 11921 T>A | L3974Q | Cytosol | 1 | Novel | ||
| 86 | 49 | 90 | 11989 A>G | K3997E | Cytosol | 1 | Novel | ||
| 87 | 50 | 90 | 12028 G>A | V4010M | Cytosol | 1 | Tester43 | ||
| 88 | 90 | Not Reported | F4020L | Cytosol | Postma34 | ||||
| 89 | 51 | 90 | 12226 A>G | E4076K | Cytosol | 1 | Postma34 | ||
| 90 | 90 | 12290 A>G | N4097S | Cytosol | Tester46 | ||||
| 91 | 90 | 12311 A>T | N4104I | Cytosol | Postma34 | ||||
| 92 | 90 | 12312 C>G | N4104K | Cytosol | Priori3 | ||||
| 93 | 90 | Not Reported | L4105F | Cytosol | Hasdemir47 | ||||
| 94 | 52 | 90 | 12322 C>A | H4108N | Cytosol | 1 | Postma34 | ||
| 95 | 90 | Not Reported | H4108Q | Cytosol | Postma34 | ||||
| 96 | 53 | 90 | 12370 A>G | S4124G | Cytosol | 1 | Novel | ||
| 97 | 54 | 90 | 12371 G>C | S4124T | Cytosol | 1 | Tester14 | ||
| 99 | 90 | Not Reported | R4144C | Cytosol | Berge38 | ||||
| 100 | 90 | 12436 G>A | E4146K | Cytosol | Tester46 | ||||
| 101 | 55 | 90 | 12446 A>G | Y4149S† | Cytosol | 1 | Novel | ||
| 102 | 56 | 90 | 12470 G>A | R4157Q | Cytosol | 1 | Novel | ||
| 103 | 90 | 12472 A>C | T4158P | Cytosol | Tester46 | ||||
| 104 | 57 | 90 | 12476 A>G | Q4159P | Cytosol | 1 | Novel | ||
| 105 | 58 | 90 | 12533 A>G | N4178S | Cytosol | 3 | Novel | ||
| 106 | 59 | 90 | 12559 G>C | E4187Q | Cytosol | 1 | Novel | ||
| 107 | 60 | 90 | 12586 A>G | T4196A | Cytosol | 1 | Tester | ||
| 108 | 90 | 12601 C>A | Q4201R | Cytosol | Laitinen37 | ||||
| 109 | 61 | 90 | 12845 C>T | A4282V | Cytosol | 1 | Tester43 | ||
| 110 | 90 | 12919 C>T | R4307C | Cytosol | Callis48 | ||||
| 111 | 62 | 90 | 12944 G>A | G4315E | Cytosol | 1 | Novel | ||
| 112 | 91 | 13291 G>A | E4431K | Cytosol | Berge38 | ||||
| 113 | 93 | 13489 C>T | R4497C | TMD | Priori5 | ||||
| 114 | 63 | 93 | 13496 T>G | F4499C | TMD | 1 | Choi28 | ||
| 115 | 93 | 13512 G>A | M4504I | TMD | Bagattin42 | ||||
| 116 | 64 | 93 | 13528 G>A | A4510T | TMD | 2 | Choi28 | ||
| 117 | 93 | Not Reported | F4511L | TMD | Beckmann44 | ||||
| 118 | 65 | 94 | 13666 G>A | A4556T | TMD | 1 | Tester14 | ||
| 119 | 94 | 13695 C>A | S4565R | TMD | Tester36 | ||||
| 120 | 95 | 13819 G>C | A4607P | TMD | Bagattin42 | ||||
| 121 | 95 | 13831 G>A | E4611K | TMD | Berge38 | ||||
| 122 | 96 | Not Reported | W4645R | TMD | Beery49 | ||||
| 123 | 66 | 96 | 13948 A>G | K4650E | TMD | 1 | Novel | ||
| 124 | 96 | 13957 A>G | V4653F | TMD | Laitinen37 | ||||
| 125 | 67 | 97 | 13967-13972 Dup |
4657-4658 ins EY |
TMD | 1 | Tester14 | ||
| 126 | 97 | Not Reported | G4662S | TMD | Postma34 | ||||
| 127 | 68 | 97 | 14011 G>C | G4671R | TMD | 1 | Choi28 | ||
| 128 | 69 | 99 | 14205-14208 Del |
N4736 Del | TMD | 1 | Novel | ||
| 129 | 99 | 14285 A>C | H4762P | TMD | Postma34 | ||||
| 130 | 70 | 100 | 14311 G>A | V4771I | TMD | 2 | Priori3 | ||
| 131 | 71 | 100 | 14369 G>A | R4790Q | TMD | 1 | Novel | ||
| 132 | 72 | 100 | 14414 A>G | K4805R | TMD | 1 | Novel | ||
| 133 | 73 | 101 | 14465 G>A | R4822H | TMD | 1 | Novel | ||
| 134 | 74 | 101 | 14542 G>A | I4848V | TMD | 2 | Choi28 | ||
| 135 | 101 | 14552 T>G | F4851C | TMD | Aizawa41 | ||||
| 136 | 101 | 14579 C>G | A4860G | TMD | Priori3 | ||||
| 137 | 102 | 14601 T>G | I4867M | CT | Priori3 | ||||
| 138 | 102 | 14639 T>C | V4880A | CT | Bagattin42 | ||||
| 139 | 103 | 14683 A>G | N4895D | CT | Priori3,5 | ||||
| 140 | 103 | 14705 C>T | P4902L | CT | Laitinen37 | ||||
| 141 | 103 | Not Reported | P4902S | CT | Postma34 | ||||
| 142 | 104 | 14806 G>A | G4936R | CT | Tester31 | ||||
| 143 | 105 | 14848 G>A | E4950K | CT | Priori3 | ||||
| 144 | 75 | 105 | 14876 G>A | R4959Q | CT | 2 | Laitinen37 |
Predicted location: NT = Amino-Terminal, CL = Cytoplasmic Loop, FKBP = 12.6 (Calstabin) binding domain, TMD = Transmembrane domain, CT = C Terminal.
Large genomic rearrangement comprising intron 2-3 and intron 2-4 resulted in inframe deletion of exon 3. AA: African Americans Controls, CC: Caucasians Controls.
Mosaicism.
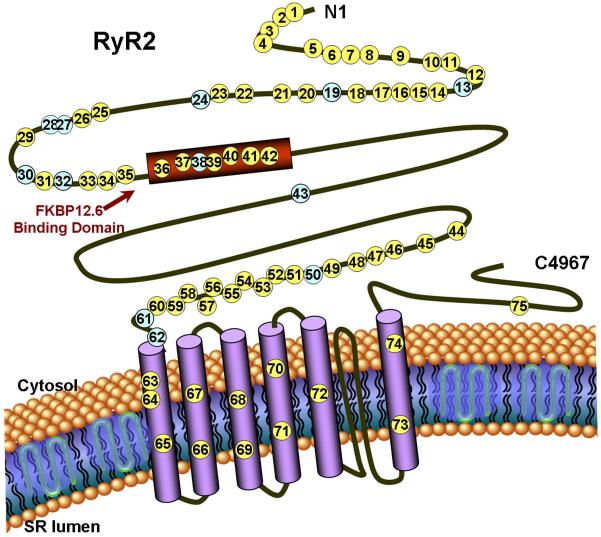
Figure 2. RyR2 channel topology and localization of mutations and polymorphisms.
Linear topology of the cardiac ryanodine receptor (RyR2); putative pathogenic mutations (yellow circles) and polymorphisms (blue circles) found on this study-cohort are shown in the approximate location. The number within the circle corresponds to the mutation # on Table 1.
Figure 3. Prevalence of RYR2 mutations by subgroups.
The yield from the entire RYR2 scan on this cohort is shown on the left. Bars on the right side show the sensitivity in the 3 different subgroups of this cohort.
One proband had a maternally inherited Y4149S (tyrosine, Y, at position 4149 mutated to serine, S) missense mutation. Although the proband’s mother was asymptomatic and had an unremarkable exercise ECG; germline mosaicism was suspected clinically because more than one offspring was affected. Accordingly, Y4149S mosaicism was detected in her being highest in the hair-roots (~25%), less in leucocytes (~20%) and in fibroblasts and buccal epithelium (~15-18%).
Twelve non-synonymous single nucleotide polymorphisms (6 novel) were also identified, 7 of them were seen only in controls and 5 in cases and controls (Table 2). Four novel polymorphisms localize between domain I and II. The most common polymorphism was Q2958R with an heterozygous prevalence of 34% in Caucasians and 10% in African-Americans; followed by G1886S with a prevalence of 20% (African Americans) and 9% (Caucasians). V377M was found only in African-Americans with a prevalence of 3%. Finally, Y2156C, E2183V, M2389L, V4010M, A4282V and G4315E are rare variants observed only once in different control subjects. Thus, within the exons hosting putative CPVT1-associated mutations, the background prevalence of rare amino acid substitutions among the 200 apparently healthy volunteers was 3% (3/100 Caucasians and 3/100 African Americans, Table 2).
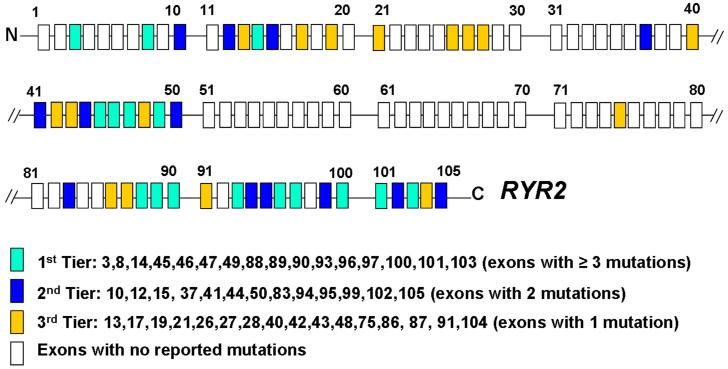
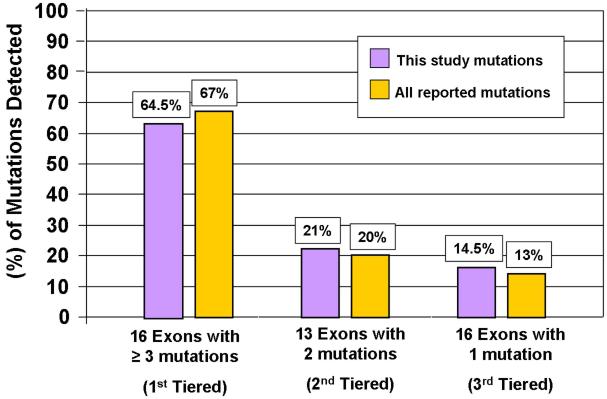
We evaluated the number of mutations in each exon reported to date in the literature (Table 2), excluding exons containing only polymorphisms. As such; 127 unique mutations were analyzed, including those found within this cohort. Sixteen exons hosted > 3 distinct CPVT1-associated mutations; 13 exons had at least 2 mutations reported while an additional 16 exons had, so far, only a single mutation reported (Figure 4). This mutation clustering phenomena might facilitate a tiered strategy that may yield a more cost-effective approach for CPVT genetic testing. If we consider that the average charge for the current RYR2 commercial tests available on the market is approximately $0.40 per coding nucleotide (http://pgxhealth.com, www.preventiongenetics.com), the estimated charge for the entire RYR2 coding region scan would be approximately $6000 per patient, meaning that the commercial charge to analyze this 155 patient cohort in its entirety would have approached $1 million US dollars. In comparison, the total charge to scan only the 45 mutation-hosting exons that have been reported to date exon-containing mutations reported to date would be about 50% less. Further, a reflex tiered strategy would reduce the cost significantly. As modeled here, using a 3-tiered reflex genetic test strategy based on Figure 4, the genetic scan of the first tier of exons in our cohort would cost $190,960.00 (~$1200 per case) and would detect nearly two-thirds of those CPVT cases that are due to mutations in RYR2. The charge to reflex to the second tier genetic scan would add < $1000 per case and combined, nearly 90% of the RYR2-mutation positive cases (CPVT1) would be identified. Reflexing to the third tier would capture the remaining RYR2-positive cases and the charge to do so would be ~$123,225 US dlls ($795.00 US dlls/case, Figure 5).
Figure 4. Possible tiered strategy for reflex genetic testing.
Schematic representation of the 105 coding exons of the RYR2 gene. Boxes in colors: all the exon-containing mutations reported to date. Boxes in white: exons free of reported mutations. The tiered strategy was built based on the number of mutations containing in each exon as shown by three different colors. The 1st tier included 16 exons, 2nd tier 13 exons and 3rd tier 16 exons. Exons containing control variants were not included.
Figure 5. Yield from RYR2 mutational analysis based on a tiered strategy.
Retrospective analysis of the mutations detected in our cohort and in the world-wide compendium of mutations reported to date. The percentage of mutations that would be detected using the tired strategy is shown.
DISCUSSION
Exertional Syncope: LQTS or CPVT?
It has been reported that nearly 30% of CPVT cases have been misdiagnosed as “LQTS with normal QT intervals” or “concealed LQTS”.13 Recently, we demonstrated that nearly 6% of 269 LQTS genotype negative patients hosted a putative CPVT1-causing RyR2 mutation14. Here, we included only referral cases of “atypical/possible LQTS” with a phenotype of exertional syncope and QTc < 480 ms. Herein, the yield of RYR2 mutations for these 45 cases was 31%; indicating the critical importance of properly distinguishing between CPVT and LQTS. CPVT-related arrhythmias can be easily reproduced during an exercise stress test, isoproterenol infusion or by other forms of adrenergic stimulation15,16. The induction of polymorphic ventricular tachycardia or bidirectional VT, characterized by 180° alternating QRS axis on a beat-to-beat basis, sets CPVT apart from “concealed” or “borderline” LQTS.
RYR2 genetic approach: Targeted scan and tiered strategy
Our results confirm that mutation clustering exists. The functional significance of mutation clustering remains unclear. It has been suggested, however, that a domain-domain interaction is crucial for channel function17-19 and a defective inter-molecular interaction may be crucial in disease phenotypes. Interestingly, in this study 11/64 (17%) of the putative mutations localize outside the considered canonical domains.
Based upon our results and after analyzing a large publicly available compendium of the 127 RYR2 putative mutations known to date (Table 2), we propose an expanded genetic approach for research/investigational laboratories. A reasonable RYR2 scan will include the analysis of at least 45 exons in total known to host all published mutations reported to date. Since some exons (19) imbibed in the hot-spot region remain free of mutations so far, a more ambitious and “comprehensive” RYR2 genetic test would include these exons as well resulting in a 64-exon scan (exons 3-28, 37-50, 75 and 83-105).
The mutation clustering phenomena might facilitate a tiered strategy that may yield a more cost-effective approach for CPVT genetic testing. Figure 4 summarizes this proposed tiered strategy. The approach was developed considering the number of mutations in each exon reported to date in the literature. The first tier comprises those exons (N=16) now known to host > 3 unique CPVT-associated mutations. The second tier includes 13 exons with at least 2 mutations reported while the third tier consists of the final 16 exons where, so far, only a single mutation within that exon has been reported. Considering that ~65% of the RYR2 mutation-positive cases might have a mutation in the first tier of 16 RYR2 exons, the charge of the genetic analysis in this group could be reduced by approximately half (predicted $1232.00 US dlls/case for the first tier of 16 exons vs $3019.00 US dlls/case for the entire sequencing of exons-containing reported mutation).
In case of negative results, we suggest that the pseudo-comprehensive (64 exon) RYR2 scan mentioned previously (exons 3-28, 37-50, 75 and 83-105) be performed. Additional “rare” although documented causes of CPVT should also be considered, like large RYR2 genomic rearrangements involving exon 3 and mutations in calsequestrin 2 (CASQ2) and Kir2.1 (KCNJ2)20. The area surrounding exon 3 is highly susceptible to large Alu-repeat-mediated genomic rearrangements; we documented 3 unrelated cases hosting large heterozygous deletions involving exon 3 that could not be detected by regular genetic screening using DHPLC or direct DNA sequencing. Validating this observation, exon 3 deletion was also reported recently in a different cohort where 2 unrelated cases (out of 33), hosted a 1.1kb deletion, including exon 321.
Polymorphisms in RYR2, not that rare and with potential functional effect
It has been considered that RYR2 is not a polymorphic gene. However, 15/142 (10.5%) missense variants reported to date were found in controls. We did not scan the entire RYR2 gene in control subjects. Instead, since we focused on the exon-containing mutations, the rate of non-synonymous genetic variation throughout all of RYR2 may be higher. Importantly however, among the exons now known to host possible CPVT1-associated missense mutations, similarly rare amino acid substitutions were found in only 6 of the 200 control subjects examined in this study. Although not a true case-control genetic epidemiologic study, if validated, this would suggest that among cases where CPVT is strongly suspected, there would be a 95% estimated probability that the identification of a rare missense mutation would likely represent the pathogenic basis for the patient’s CPVT rather than merely being only a rare amino acid substitution.
We have learned that common polymorphisms in other ion channels have the potential to modify the clinical phenotype22,23; polymorphisms in RYR2 may have the same potential. RyR2-Q2958R is the most common RYR2 polymorphism; was described for the first time 9 years ago24 and is particularly common in Caucasians (34%). The second most common polymorphism in RYR2 is G1886S (20% African Americans, 9% Caucasians) followed by G1885E (6% Caucasians). Interestingly, in vitro studies in heterologous systems have demonstrated that both G1885E and G1886S polymorphisms caused a significant increase in the cellular Ca(2+) oscillation activity compared with RyR2 wild-type channels. Further, when both polymorphisms were introduced in the same RyR2 subunit, the store-overload-induced calcium release activity was nearly completely abolished25. The clinical consequences of this “RyR2 loss of function” in vitro phenotype is not clear, however, compound heterozygosity involving these two polymorphisms has been reported in right ventricular dysplasia26. The potential functional effects of the 6 novel polymorphisms identified in this study are unknown.
It is important to remark that none of the novel mutations detected on this study have been functionally characterized to further bolster the contention of pathogenicity. However, less than 15% of the mutations reported to date in RYR2 have been studied in vitro, pathogenicity has been suspected based on co-segregation with the disease and absence in control subjects. Here, co-segregation with the disease data was not available for all cases. Instead, the prevalence of putative mutations amongst strong cases (~60%) was markedly higher than in controls (~3%) and all putative mutations were absent in 400 reference alleles. Thus, although the precise contribution of each discrete mutation to the phenotype remains to be determined, statistically, the estimated probability for pathogenicity for RYR2 mutations found in strong cases is quite high (~95%).
Mosaicism in RYR2
This is the first report involving RYR2 mosaicism which was transmitted to descendants, presumably causing sudden death in two children and full blown CPVT in one child from the age of 9 years. RYR2 mutations, in many circumstances (~20% in our cohort) are de novo in origin, but it could also be present in a mosaic form in the asymptomatic parents, which requires attention during genetic counseling as well as during genetic screening.
Clinical Significance
This study represents the first analysis of RYR2 mutation distribution in a large cohort of cases. Our results contribute to a better delineation of the “hot spot” region with important consequences in “gene negative” definition. The identification of novel common variants in control subjects will allow a better interpretation of the CPVT genetic test and the detection of RYR2 mosaicism and confirmation of exon 3 deletion in different patients-cohort, provide novel genetic possibilities in the pathogenesis of CPVT. Moreover, the possibility of a tiered strategy for RYR2 genetic scan may enable a more cost-effective genetic approach to analyzing one of the largest genes in the human genome. Finally, we emphasize the critical importance of properly distinguishing between CPVT and LQTS (including Andersen-Tawil syndrome), two different diseases with a similar clinical presentation but different clinical outcomes and different responsiveness to pharmacotherapy.
CONCLUSION
Although intimidating as one of the largest genes in the human genome, results from this comprehensive open reading frame analysis involving one of the largest cohorts of unrelated patients examined, combined with a detailed analysis of all published CPVT1-associated mutations indicate that to date, only 45 of RYR2’s 105 translated exons host a putative CPVT1-associated mutation thus far. Moreover, an initial targeting of only 16 exons would allow the identification of putative mutations in ~65% of the RYR2-mutation positive cases, though compound heterozygosity may be missed. Finally, given the present estimate of 3% frequency for rare missense mutations among controls, one must be cognizant of the possibility of a “false positive” especially as the pre-test probability of a CPVT diagnosis decreases. The ~33% yield that was observed among the “possible” cases of CPVT indicates that perhaps 90% of the mutations, identified among cases labeled as “possible CPVT” or so-called “atypical LQTS” with exercise-induced syncope and QTc < 480 ms, are pathogenic whereas 10% of those mutations may represent “false positives”.
Supplementary Material
ACKNOWLEDGEMENTS
Michael J. Ackerman and Argelia Medeiros-Domingo are supported by the Mayo Clinic Windland Smith Rice Comprehensive Sudden Cardiac Death and Leducq Fondation programs. Arthur A. M. Wilde is supported by The Interuniversity Cardiology Institute of the Netherlands (ICIN) project 27 and by a Leducq Fondation program grant “05CVD01, Alliance Against Sudden Cardiac Death.”
Footnotes
DISCLOSURES
Dr. Ackerman is a consultant for PGxHealth and chairs their FAMILION Medical/Scientific Advisory Board (approved by Mayo Clinic’s Medical-Industry Relations Office and Conflict of Interests Review Board). In addition, a license agreement pertaining to “mutations in the ryanodine receptor 2 gene and heart disease”, resulting in consideration and royalty payments, was established between PGxHealth and Mayo Clinic Health Solutions in 2007.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Leenhardt A, Lucet V, Denjoy I, et al. Catecholaminergic polymorphic ventricular tachycardia in children. A 7-year follow-up of 21 patients. Circulation. 1995;91:1512–1519. doi: 10.1161/01.cir.91.5.1512. [DOI] [PubMed] [Google Scholar]
- 2.Swan H, Piippo K, Viitasalo M, et al. Arrhythmic disorder mapped to chromosome 1q42-q43 causes malignant polymorphic ventricular tachycardia in structurally normal hearts. J Am Coll Cardiol. 1999;34:2035–2042. doi: 10.1016/s0735-1097(99)00461-1. [DOI] [PubMed] [Google Scholar]
- 3.Priori SG, Napolitano C, Memmi M, et al. Clinical and molecular characterization of patients with catecholaminergic polymorphic ventricular tachycardia. Circulation. 2002;106:69–74. doi: 10.1161/01.cir.0000020013.73106.d8. [DOI] [PubMed] [Google Scholar]
- 4.Postma AV, Denjoy I, Hoorntje TM, et al. Absence of calsequestrin 2 causes severe forms of catecholaminergic polymorphic ventricular tachycardia. Circ Res. 2002;91:e21–26. doi: 10.1161/01.res.0000038886.18992.6b. [DOI] [PubMed] [Google Scholar]
- 5.Priori SG, Napolitano C, Tiso N, et al. Mutations in the cardiac ryanodine receptor gene (hRyR2) underlie catecholaminergic polymorphic ventricular tachycardia. Circulation. 2001;103:196–200. doi: 10.1161/01.cir.103.2.196. [DOI] [PubMed] [Google Scholar]
- 6.Yano M, Yamamoto T, Ikeda Y, et al. Mechanisms of Disease: ryanodine receptor defects in heart failure and fatal arrhythmia. Nat Clin Pract Cardiovasc Med. 2006;3:43–52. doi: 10.1038/ncpcardio0419. [DOI] [PubMed] [Google Scholar]
- 7.George CH, Jundi H, Thomas NL, et al. Ryanodine receptors and ventricular arrhythmias: emerging trends in mutations, mechanisms and therapies. J Mol Cell Cardiol. 2007;42:34–50. doi: 10.1016/j.yjmcc.2006.08.115. [DOI] [PubMed] [Google Scholar]
- 8.Benkusky NA, Farrell EF, Valdivia HH. Ryanodine receptor channelopathies. Biochem Biophys Res Commun. 2004;322:1280–1285. doi: 10.1016/j.bbrc.2004.08.033. [DOI] [PubMed] [Google Scholar]
- 9.McCarthy TV, Quane KA, Lynch PJ. Ryanodine receptor mutations in malignant hyperthermia and central core disease. Hum Mutat. 2000;15:410–417. doi: 10.1002/(SICI)1098-1004(200005)15:5<410::AID-HUMU2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 10.McCarthy EJ. Malignant hyperthermia: pathophysiology, clinical presentation, and treatment. AACN Clin Issues. 2004;15:231–237. doi: 10.1097/00044067-200404000-00009. [DOI] [PubMed] [Google Scholar]
- 11.Ackerman MJ, Tester DJ, Jones GS, et al. Ethnic differences in cardiac potassium channel variants: Implications for genetic susceptibility to sudden cardiac death and genetic testing for congenital long QT syndrome. Mayo Clinic Proceedings. 2003;78:1479–1487. doi: 10.4065/78.12.1479. [DOI] [PubMed] [Google Scholar]
- 12.Bhuiyan ZA, van den Berg MP, van Tintelen JP, et al. Expanding spectrum of human RYR2-related disease: new electrocardiographic, structural, and genetic features. Circulation. 2007;116:1569–1576. doi: 10.1161/CIRCULATIONAHA.107.711606. [DOI] [PubMed] [Google Scholar]
- 13.Priori SG, Napolitano C, Schwartz PJ. Low penetrance in the long-QT syndrome: clinical impact. Circulation. 1999;99:529–533. doi: 10.1161/01.cir.99.4.529. [DOI] [PubMed] [Google Scholar]
- 14.Tester DJ, Kopplin LJ, Will ML, et al. Spectrum and prevalence of cardiac ryanodine receptor (RyR2) mutations in a cohort of unrelated patients referred explicitly for long QT syndrome genetic testing. Heart Rhythm. 2005;2:1099–1105. doi: 10.1016/j.hrthm.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 15.Sumitomo N, Harada K, Nagashima M, et al. Catecholaminergic polymorphic ventricular tachycardia: electrocardiographic characteristics and optimal therapeutic strategies to prevent sudden death. Heart. 2003;89:66–70. doi: 10.1136/heart.89.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vyas H, Hejlik J, Ackerman MJ. Epinephrine QT stress testing in the evaluation of congenital long-QT syndrome: diagnostic accuracy of the paradoxical QT response. Circulation. 2006;113:1385–1392. doi: 10.1161/CIRCULATIONAHA.105.600445. [DOI] [PubMed] [Google Scholar]
- 17.Wang R, Chen W, Cai S, et al. Localization of an NH(2)-terminal disease-causing mutation hot spot to the “clamp” region in the three-dimensional structure of the cardiac ryanodine receptor. J Biol Chem. 2007;282:17785–17793. doi: 10.1074/jbc.M700660200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z, Wang R, Zhang J, et al. Localization of a disease-associated mutation site in the three-dimensional structure of the cardiac muscle ryanodine receptor. J Biol Chem. 2005;280:37941–37947. doi: 10.1074/jbc.M505714200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.George CH, Yin CC, Lai FA. Toward a molecular understanding of the structure-function of ryanodine receptor Ca2+ release channels: perspectives from recombinant expression systems. Cell Biochem Biophys. 2005;42:197–222. doi: 10.1385/CBB:42:2:197. [DOI] [PubMed] [Google Scholar]
- 20.Tester DJ, Arya P, Will M, et al. Genotypic heterogeneity and phenotypic mimicry among unrelated patients referred for catecholaminergic polymorphic ventricular tachycardia genetic testing. Heart Rhythm. 2006;3:800–805. doi: 10.1016/j.hrthm.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 21.Marjamaa A, Laitinen-Forsblom P, Lahtinen AM, et al. Search for cardiac calcium cycling gene mutations in familial ventricular arrhythmias resembling catecholaminergic polymorphic ventricular tachycardia. BMC Med Genet. 2009;10:12. doi: 10.1186/1471-2350-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poelzing S, Forleo C, Samodell M, et al. SCN5A polymorphism restores trafficking of a Brugada syndrome mutation on a separate gene. Circulation. 2006;114:368–376. doi: 10.1161/CIRCULATIONAHA.105.601294. [DOI] [PubMed] [Google Scholar]
- 23.Makielski JC, Ye B, Valdivia CR, et al. A ubiquitous splice variant and a common polymorphism affect heterologous expression of recombinant human SCN5A heart sodium channels. Circ Res. 2003;93:821–828. doi: 10.1161/01.RES.0000096652.14509.96. [DOI] [PubMed] [Google Scholar]
- 24.Tiso N, Stephan DA, Nava A, et al. Identification of mutations in the cardiac ryanodine receptor gene in families affected with arrhythmogenic right ventricular cardiomyopathy type 2 (ARVD2) Hum Mol Genet. 2001;10:189–194. doi: 10.1093/hmg/10.3.189. [DOI] [PubMed] [Google Scholar]
- 25.Koop A, Goldmann P, Chen SR, et al. ARVC-related mutations in divergent region 3 alter functional properties of the cardiac ryanodine receptor. Biophys J. 2008;94:4668–4677. doi: 10.1529/biophysj.107.122382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milting H, Lukas N, Klauke B, et al. Composite polymorphisms in the ryanodine receptor 2 gene associated with arrhythmogenic right ventricular cardiomyopathy. Cardiovasc Res. 2006;71:496–505. doi: 10.1016/j.cardiores.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 27.d’Amati G, Bagattin A, Bauce B, et al. Juvenile sudden death in a family with polymorphic ventricular arrhythmias caused by a novel RyR2 gene mutation: evidence of specific morphological substrates. Hum Pathol. 2005;36:761–767. doi: 10.1016/j.humpath.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 28.Choi G, Kopplin LJ, Tester DJ, et al. Spectrum and frequency of cardiac channel defects in swimming-triggered arrhythmia syndromes. Circulation. 2004;110:2119–2124. doi: 10.1161/01.CIR.0000144471.98080.CA. [DOI] [PubMed] [Google Scholar]
- 29.Hsueh CH, Weng YC, Chen CY, et al. A novel mutation (Arg169Gln) of the cardiac ryanodine receptor gene causing exercise-induced bidirectional ventricular tachycardia. Int J Cardiol. 2006;108:276–278. doi: 10.1016/j.ijcard.2005.02.051. [DOI] [PubMed] [Google Scholar]
- 30.Davis D, Gow R, Birnie D, et al. Syncope While Swimming: Identification of Novel RyR2 Mutations. Heart Rhythm. 2006;3:P4–62. [Google Scholar]
- 31.Tester DJ, Medeiros-Domingo A, Ackerman MJ. Post-Mortem Cardiac Channel Genetic Testing for Autopsy Negative Sudden Unexplained Death. Heart Rhythm. 2009:6. doi: 10.1016/j.mayocp.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tester DJ, Kopplin LJ, Creighton W, et al. Pathogenesis of unexplained drowning: new insights from a molecular autopsy. Mayo Clin Proc. 2005;80:596–600. doi: 10.4065/80.5.596. [DOI] [PubMed] [Google Scholar]
- 33.Bauce B, Rampazzo A, Basso C, et al. Screening for ryanodine receptor type 2 mutations in families with effort-induced polymorphic ventricular arrhythmias and sudden death: early diagnosis of asymptomatic carriers. J Am Coll Cardiol. 2002;40:341–349. doi: 10.1016/s0735-1097(02)01946-0. [DOI] [PubMed] [Google Scholar]
- 34.Postma AV, Denjoy I, Kamblock J, et al. Catecholaminergic polymorphic ventricular tachycardia: RYR2 mutations, bradycardia, and follow up of the patients. J Med Genet. 2005;42:863–870. doi: 10.1136/jmg.2004.028993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marjamaa A, Laitinien-Forsblom P, Toivonen L, et al. Ryanodine Receptor (RYR2) Mutations in Sudden Unexplained Death: Studies in Extended Pedigrees and Phenotypic Characterization In Vitro. Circulation. 2007;116:607. doi: 10.1016/j.ijcard.2009.08.041. [DOI] [PubMed] [Google Scholar]
- 36.Tester DJ, Dura M, Carturan E, et al. A mechanism for sudden infant death syndrome (SIDS): stress-induced leak via ryanodine receptors. Heart Rhythm. 2007;4:733–739. doi: 10.1016/j.hrthm.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laitinen PJ, Brown KM, Piippo K, et al. Mutations of the cardiac ryanodine receptor (RyR2) gene in familial polymorphic ventricular tachycardia. Circulation. 2001;103:485–490. doi: 10.1161/01.cir.103.4.485. [DOI] [PubMed] [Google Scholar]
- 38.Berge KE, Haugaa KH, Fruh A, et al. Molecular genetic analysis of long QT syndrome in Norway indicating a high prevalence of heterozygous mutation carriers. Scand J Clin Lab Invest. 2008;68:362–368. doi: 10.1080/00365510701765643. [DOI] [PubMed] [Google Scholar]
- 39.Nishio H, Iwata M, Tamura A, et al. Identification of a novel mutation V2321M of the cardiac ryanodine receptor gene of sudden unexplained death and a phenotypic study of the gene mutations. Leg Med (Tokyo) 2008 doi: 10.1016/j.legalmed.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 40.Creighton W, Virmani R, Kutys R, et al. Identification of novel missense mutations of cardiac ryanodine receptor gene in exercise-induced sudden death at autopsy. J Mol Diagn. 2006;8:62–67. doi: 10.2353/jmoldx.2006.050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aizawa Y, Mitsuma W, Ikrar T, et al. Human cardiac ryanodine receptor mutations in ion channel disorders in Japan. Int J Cardiol. 2007;116:263–265. doi: 10.1016/j.ijcard.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 42.Bagattin A, Veronese C, Bauce B, et al. Denaturing HPLC-based approach for detecting RYR2 mutations involved in malignant arrhythmias. Clin Chem. 2004;50:1148–1155. doi: 10.1373/clinchem.2003.030734. [DOI] [PubMed] [Google Scholar]
- 43.Tester D, Salisbury B, Judson R, et al. Spectrum and prevalence of genetic variants in the RyR2-encoded cardiac ryanodine receptor-calcium release channel in healthy subjects. Circulation. 2005;11:516. [Google Scholar]
- 44.Beckmann BM, Wilde AA, Kaab S. Dual inheritance of sudden death from cardiovascular causes. N Engl J Med. 2008;358:2077–2078. doi: 10.1056/NEJMc0708596. [DOI] [PubMed] [Google Scholar]
- 45.Hasdemir C, Priori SG, Overholt E, et al. Catecholaminergic polymorphic ventricular tachycardia, recurrent syncope, and implantable loop recorder. J Cardiovasc Electrophysiol. 2004;15:729. doi: 10.1046/j.1540-8167.2004.03408.x. [DOI] [PubMed] [Google Scholar]
- 46.Tester DJ, Spoon DB, Valdivia HH, et al. Targeted mutational analysis of the RyR2-encoded cardiac ryanodine receptor in sudden unexplained death: a molecular autopsy of 49 medical examiner/coroner’s cases. Mayo Clin Proc. 2004;79:1380–1384. doi: 10.4065/79.11.1380. [DOI] [PubMed] [Google Scholar]
- 47.Hasdemir C, Aydin HH, Sahin S, et al. Catecholaminergic polymorphic ventricular tachycardia caused by a novel mutation in the cardiac ryanodine receptor. Anadolu Kardiyol Derg. 2008;8:E35–36. [PubMed] [Google Scholar]
- 48.Callis TE, Harris-Kerr C, Carr JL, et al. Case-Control genetic comparison of the cardiac ryanodine receptor in catecholaminergic polymorphic ventricular tachycardia. Heart Rhythm. 2009:6. [Google Scholar]
- 49.Beery T, Shah M, Benson D. Genetic Characterization of Familial CPVT after 30 Years. Heart Rhythm. 2008;5:AB2–2. doi: 10.1177/1099800409333369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.