Abstract
Many pathogenic bacteria use a regulatory process termed quorum sensing (QS) to produce and detect small diffusible molecules to synchronize gene expression within a population. In Gram-negative bacteria, the detection of, and response to, these molecules depends on transcriptional regulators belonging to the LuxR family. Such a system has been discovered in the intracellular pathogen Brucella melitensis, a Gram-negative bacterium responsible for brucellosis, a worldwide zoonosis that remains a serious public health concern in countries were the disease is endemic. Genes encoding two LuxR-type regulators, VjbR and BabR, have been identified in the genome of B. melitensis 16 M. A ΔvjbR mutant is highly attenuated in all experimental models of infection tested, suggesting a crucial role for QS in the virulence of Brucella. At present, no function has been attributed to BabR. The experiments described in this report indicate that 5% of the genes in the B. melitensis 16 M genome are regulated by VjbR and/or BabR, suggesting that QS is a global regulatory system in this bacterium. The overlap between BabR and VjbR targets suggest a cross-talk between these two regulators. Our results also demonstrate that VjbR and BabR regulate many genes and/or proteins involved in stress response, metabolism, and virulence, including those potentially involved in the adaptation of Brucella to the oxidative, pH, and nutritional stresses encountered within the host. These findings highlight the involvement of QS as a major regulatory system in Brucella and lead us to suggest that this regulatory system could participate in the spatial and sequential adaptation of Brucella strains to the host environment.
Keywords: Brucella, intracellular pathogen, Quorum sensing, LuxR-type regulator, adaptation, proteome, transcriptome, ChIP
Short abstract
Some pathogens use the regulatory process termed Quorum Sensing (QS) to synchronize gene expression within bacterial population. We report here the first genome scale study of the Quorum Sensing system of the intracellular pathogen Brucella melitensis. Our combined proteomic and transcriptomic data suggest that Quorum Sensing is involved in the spatial and sequential adaptation of B. melitensis to the host environment.
Introduction
Bacteria of the genus Brucella are the etiological agents of brucellosis, the most widespread zoonotic disease worldwide, resulting in more than 500 000 new reported human cases per year.(1) Animal brucellosis is a disease affecting wild and domestic animals, causing abortion and sterility and producing huge economic losses.(2) Several of the nine Brucella species can infect humans, causing a chronic, debilitating disease with severe and sometimes fatal outcomes. As a result, these bacteria represent a significant public health concern in endemic countries (predominantly in the Mediterranean region and areas of Asia, Africa and Latin America).1,3 Because of their potential use as weapons, B. melitensis, B. suis and B. abortus strains have been classified as select agents by the Center for Disease Control and Prevention in the U.S.A.(4)
Brucella strains are Gram-negative intracellular pathogens belonging to the α-2 proteobacteria group. The virulence of these bacteria is based on their capacity to infect professional and nonprofessional phagocytes.5−8 This remarkable adaptation to the intracellular environment and their ability to modulate the host innate immune response(9) allows the Brucellae to establish and maintain chronic infections. During host cell infection, Brucella containing vacuoles (BCVs) traffic along the endocytic pathway and fuse transiently with both late endosomes and lysosomes, and such interactions are required for further maturation of BCVs into an ER-derived replication-permissive organelle.(10) The virulence strategies of these bacteria seem to be based on poor stimulatory activity and toxicity for host cells,(9) resistance to intracellular killing,(11) adaptation to intracellular stresses12,13 and creation of the replication-permissive compartment in professional and nonprofessional phagocytes.8,14
During infection, Brucella spp. are confronted with very diverse environments and host defense mechanisms.12,15−17 Thus, completion of a successful infection cycle is crucially dependent on fine-tuning gene expression in response to environmental stimuli.(18) Among the systems that allow such regulations, quorum sensing (QS) is of particular interest because of its documented involvement in the virulence of Brucella(19) and other pathogens.20,21 QS is a communication system used by a large number of bacteria to synchronize gene expression within a population. This system involves the synthesis, release and subsequent detection of small diffusible molecules called autoinducers (commonly N-acyl-homoserine lactones or AHLs in Gram-negatives bacteria). When AHL concentrations reach a threshold level, they bind to LuxR-type transcriptional regulators and modify their activity (for review see ref (22)). Since QS was first discovered in V. fischeri in the late 1970s,(23) the conceptual role of this communication system in prokaryotic biology has evolved considerably. QS was first described as a system allowing bacteria to sense population density.(24) However, the autoinducer concentrations can be affected by numerous parameters like diffusion, spatial distribution, and degradation.25,26 These latter factors are particularly relevant given the intravacuolar localization of Brucella spp. in host cells.
Genes encoding two LuxR-type regulators have been identified in the B. melitensis 16 M genome,(27) the previously described VjbR regulator19,28 and BabR,(29) also known as BlxR.(30) While the virulence of a ΔvjbR strain is highly attenuated in all experimental model tested, BabR seems to play a minor, if any role, in B. melitensis 16 M virulence.(31) Despite the lack of a gene encoding a classical AHL synthase in the genome of B. melitensis, we have previously identified low amounts of C12−HSL in culture supernatants from these strains.(32) This autoinducer down-regulates the expression of flagellar genes,(19) and the expression of the virB operon encoding a Type four secretion system (T4SS),32,33 two virulence factors involved in the establishment of chronic infection(34) and the control of Brucella containing vacuole (BCV) maturation, respectively.(35) Experimental evidence suggests that VjbR mediates the effect of C12−HSL on virB transcription(28) by binding to a 18 bp palindromic motif in the virB promoter.(36) Moreover it was recently demonstrated that VjbR is involved in the regulation of exopolysaccharide (EPS) synthesis and/or export and the production of several outer membrane proteins (OMPs), some of which are involved in virulence, suggesting that this regulator plays a crucial role in the regulation of the surface properties of B. melitensis 16 M.(28)
The work described in this paper is the first attempt to identify the QS regulon of an intracellular pathogen. To accomplish this, we characterized ΔbabR and ΔvjbR mutants by 2D-DIGE and microarray analysis on the same samples. We identified 101 QS targets using the proteomic approach and 338 QS target genes by transcriptome analysis. To focus on the most confident targets, we focus only on those that were identified by both proteomic and microarray analysis and those from the microarray analysis that were confirmed by qRT-PCR, chromatin immunoprecipitation (chIP) or other biological validation experiments. This combinatorial screen allowed us to select 149 VjbR and BabR target genes representing 4.7% of the B. melitensis 16 M genome. Interestingly many of these targets were regulated by both VjbR and BabR, suggesting a cross-talk between these two LuxR type regulators. Our analysis revealed that the QS system of this intracellular bacterium is a global regulatory system because VjbR and BabR control (directly or not) genes and proteins involved in stress response, metabolic adaptation and virulence. In the light of these results, we therefore propose that the B. melitensis QS system may play a role in fine-tuning the spatiotemporal adaptation of the bacteria to their intracellular niche.
Experimental section
Bacterial Strains and Culture Conditions
Brucella melitensis strains were grown with shaking at 37 °C in 2YT medium (10% yeast extract, 10 g L−1 tryptone, 5 g L−1 NaCl) containing the appropriate antibiotics, from an initial optical density at 600 nm (OD600) of 0.05. For transcriptomic and proteomic analyses, 100 mL of 2YT without antibiotic were inoculated with wild-type strain, ΔvjbR or ΔbabR mutants to an OD600 of 0.05. Cultures were grown in triplicate, and incubated at 37 °C with shaking to an OD600 of 0.75. Ten milliliters of culture was used for protein preparation, and the rest was used for RNA extraction.
Nalidixic acid (Nal) and gentamycin (Gnt) were used at 25 μg mL−1 and 50 μg mL−1 respectively. Synthetic N-dodecanoyl-dl-homoserine lactone (C12−HSL; Fluka) was prepared in acetonitrile (ACN) and added to bacterial growth media at 5 μM final concentration. The same volume of ACN was used as a negative control.
Mutant Construction
The ΔbabR and ΔvjbR mutant strains were constructed by gene replacement employing a kanamycine resistance gene and previously described procedures.19,31
For ChIP experiments, the plasmid pSB502 harboring a C-terminal fusion between the flag tag and the vjbRHTH region coding for the HTH region of VjbR (amino acids 181 to 260) was designed as following. First, we constructed the Gateway destination vector pSB500 allowing C-terminal fusions of an ORF with the flag epitope under Plac control. The Gw-Flag cassette was excised from the pGEMT-Gw-FLAG Cter (from Geraldine Laloux) by an ApaI/SacI restriction. The resulting fragment was purified and ligated in the pBBR1MCS-5(37) plasmid restricted by the same enzymes to obtain the destination vector pSB500 (containing a Gnt resistance cassette). The entry clone pSB102 containing vjbRHTH(28) was used together with the destination vector pSB500 during Gateway LR reaction as described by Dricot and co-workers.(38) The resulting vector pSB502 and the pBBRmcs-5 plasmid (negative control) were introduced in Brucella melitensis ΔvjbR strain by mating.
Matings were performed by mixing 200 μL of E. coli S17−1 donor cells liquid culture (overnight culture) and 1 mL of the B. melitensis NalR recipient strain (overnight culture). Cells were centrifuged 2 min at 7000 rpm and washed two times with 2YT. The pellets were resuspended in 10 μL of 2YT and spotted on a 2YT plate for 4 h. Bacteria were then transferred onto a 2YT plate containing Gnt and Nal. After 3 days of incubation at 37 °C, the exconjugates were replicated on a 2YT plate containing Nal and Gnt.
Microarray Experiments
RNA Preparation
Total RNA was extracted from B. melitensis 16 M and the isogenic ΔvjbR and ΔbabR mutants (all cultured in triplicate) as follows: 45 mL of culture (OD600 of 0.75) were centrifuged at 3500 rpm for 15 min. Bacterial pellets were resuspended in 100 μL SDS 10% and 20 μL proteinase K (20 mg mL−1) and incubated at 37 °C with shaking for 1 h. Five milliliters of TRIzol Reagent (Invitrogen) were added and suspensions were vigorously shacken. After 10 min of incubation at 65 °C, 1 mL chloroform was added to the suspensions and the mixtures were shacken and incubated at room temperature for 5−10 min. Samples were then centrifuged at 14.000 rpm for 15 min at 4 °C. Then, 2.5 mL 2-propanol were added to the aqueous phases and samples were stored overnight at −20 °C. After centrifugation for 30 min at 14.000 rpm at 4 °C, pellets were washed with 75% (RNase free) ethanol. Supernatants were discarded and pellets were dried 15 min at room temperature. Total RNA samples were resuspended in 100 μL RNase free water, incubated 10 min at 55 °C and stored at −80 °C. The integrity of the RNA and the absence of DNA were checked by gel electrophoresis. RNA quantity was measured using a NanoDrop spectrophotometer (ND-1000, Thermo Fisher Scientific).
Microarray Analysis
Microarray design and analysis were made by NimbleGen Systems, Inc. from catalogue design for B. melitensis 16 M chromosomes I (NC_003317) and II (NC_003318) with 20 probes per gene (10 perfect matches and 10 mismatches). Each probe (24 mer) was replicated three times on a chip (design includes random GC probes). Triplicate RNA samples of each strain were mixed and one chip was analyzed per strain. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible for reviewers through GEO Series accession number GSE8844 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=jzmhfuoccmeugfi&acc=GSE8844).
All of the analysis was performed using the statistical program in the stats package.(39) Data obtained from the microarray analysis were preprocessed using the RMA algorithm,(40) as provided by NimbleGen Systems, Inc. Two pair wise comparisons were performed (Δvjbr vs wt) and (Δbabr vs wt). For each comparison, the fold change was computed as the ratio of intensity averages (mutant/wt). A Student t test was used for statistical analysis of overexpression and under-expression. Genes presenting both a fold change greater than 1.3 (or below 0.7) and statistical significance at the alpha level 0.005 were defined as being over- or under-expressed between the two strains being compared.
Two-Dimensional Difference in Gel Electrophoresis (2D-DIGE)
Samples Preparation and Electrophoresis
Proteins were extracted from 10 mL of B. melitensis 16 M and ΔvjbR and ΔbabR cultures (OD600 0.75) in triplicate. Cultures were centrifuged at 3500 rpm for 10 min. Bacterial pellets were washed three times with 20 mL PBS before resuspension in 2 mL chloroform. The mixtures were incubated at room temperature for 1 h and then centrifuged at 3500 rpm for 10 min at 4 °C. Pellets were resuspended in PBS to obtain an OD600 of 100 and the cell suspensions subjected to three freeze/thaw cycles. Protein concentration for the cell lysates were determined using the BCA Protein Assay (Pierce) and protein concentrations were adjusted to 5−10 μg μL−1. Samples were divided into 100 μg aliquots and one volume of 10% trichloroacetic acid (TCA) was added. The mixtures incubated for 5 min on ice and centrifuged at 14 000 rpm for 3 min at 4 °C. Pellets were resuspended in one volume of 5% TCA and the mixes were incubated 5 min on ice. Samples were centrifuged at 14 000 rpm for 3 min at 4 °C and pellets were washed with ice cold acetone. After centrifugation an additional centrifugation step, pellets were resuspended in a mix of 40 μL Buffer 1 (40 μM Tris HCl pH 8.5, 0.3% SDS) and 4 μL Buffer 2 (0.4 M Tris HCl pH 8.5, 1 mg mL−1 DNaseI, 0.25 mg mL−1 RNase A; 50 mM MgCl2).
We used the 2D-DIGE method to compare total protein extracts from wt and ΔvjbR strains and from wt and ΔbabR strains. For each comparison, two types of gels (pH 4−7 and pH 7−11 NL) were run in triplicate. Proteins were labeled with CyDye DIGE Fluor, minimal dyes (GE Healthcare) according to the manufacturer, which allows the detection of two prelabeled protein samples and an internal standard on the same 2-D electrophoresis gel. Two samples of 25 μg (wt and ΔvjbR or wt and ΔbabR) were labeled with Cy3 and Cy5, respectively, and analyzed on the same gel together with an internal standard labeled with Cy2 (25 μg). The internal standard was a pool that included an equal amount of proteins of all samples run on triplicate gels. Labeled proteins were first separated by isoelectric focusing in immobilized pH gradient (IPG) gels, linear pH 4−7 gradient or nonlinear pH 7−11 gradient, using IPGphor (GE Healthcare). IPG pH 4−7 gels were run for 3 h at 300 V, 6 h at 1000 V, 3 h at 8000 V and 50 000 Vh at 8000 V and nonlinear IPG pH 7−11 gels were run for 4 h at 500 V, 7 h at 1000 V, 3 h at 8000 V and 60 000 Vh at 8000 V. First-dimension gels were laid on the top of 10% polyacrylamide gels and run using the Ettan Dalt II System (GE Healthcare) at constant 1.5W per gel for 18 h overnight at 15 °C. Gels were scanned with the Typhoon 9600 laser scanner (GE Healthcare) and images were analyzed with the DeCyder Differential Analysis Software (GE Healthcare).
The differential in-gel analysis mode of the DeCyder software was used to merge the Cy2, Cy3, and Cy5 images for each gel, to detect spot limits for the calculation of normalized spot volumes/protein abundances and to determine abundance differences between samples run on the same gel. The biological variation analysis mode of DeCyder was then used to match all pairwise image comparisons from difference in-gel analyses for a comparative cross-gel statistical analysis. Comparison of normalized Cy3 and Cy5 spot volumes with the corresponding Cy2 standard spot volumes within each gel gave a standardized abundance. This value was compared across all gels for each matched spot and a statistical analysis was performed. The Biological Variation Analysis (BVA) provides the average ratios between B. melitensis 16 M and mutated strain, with a threshold at ±1.3 and a t test confidence of ≤0.05, generating a list of spots of interest. All selected spots were picked, digested and identified using LC−MS/MS.
Mass Spectrometry and Protein Identification
To identify selected spots, preparative gels including 300 μg of proteins (from B. melitensis 16 M, ΔvjbR and ΔbabR triplicate samples) were performed following the protocol described above except that they were post stained with ruthenium(II) tris(bathophenanthroline disulfonate) overnight (7 μL of ruthenium/1 L of 20% ethanol) after 6 h of fixation in 30% ethanol, 10% acetic acid and 3 × 30 min in 20% ethanol at 20 °C.(41)
Protein spots were excised from preparative gels by using the Ettan Spot Picker (GE Healthcare) and in-gel tryptic digestion performed as previously described.(42) The gel pieces were twice washed with distilled water and then treated with 100% acetonitrile. The proteolytic digestion was performed by the addition of 3 μL of modified trypsin (Promega) suspended in 50 mM NH4HCO3 cold buffer. Proteolysis was performed overnight at 37 °C. The supernatant was collected and combined with the eluate of a subsequent elution step with 5% formic acid.
MALDI-TOF Identification
Digested peptides digest were desalted using C18 Geloader pipet Tips (Proxeon Biosystems) and directly eluted on the target with a mix (1:1 v/v) of α-cyano-4-hydroxyciannamic acid (in 7:3 v/v acetonitrile/0.1% formic acid) and 2,5-dihydroxybenzoic acid (in 7:3 v/v acetonitrile/0.1% trifluoracetic acid). Peptide mass fingerprints were obtained using a MALDI-MX mass spectrometer (Waters, Mildorf, U.S.A.) piloted with MassLynx 4.0 software (Waters). ProteinLynx Global Server 2.2.5 (Waters) was used as the peaklist generating software. MALDI calibration was done with ADH digest and two lockmass calibrations were used. First, an external lockmass with ADH digest (m/z: 1618.84 Da) and finally we applied an internal lockmass based on the trypsin autodigestion peak at 2211.1046 Da. The background subtract threshold was fixed at 15% (polynomial 5, we combined all spectra). An in house Mascot 2.2 server was used as database search engine, PMF search was performed on the Proteobacteria subset of the National Center for Biotechnology Information nonredundant database (NCBInr; 1 391 518 sequences in October 2008). Parameters for peptide matching were a peptide tolerance of 100 ppm, a maximum of one missed cleavage, carbamidomethylation was allowed as a fixed modification and oxidation of methionine was allowed as a variable modification. For all protein identifications, a minimal individual score of 73 and expected value below 1 were used for the identification criteria. All MS/MS spectra can be found in the Supporting Information.
Q-TOF Identification
The digests were separated by reverse phase liquid chromatography using a 75 μm × 150 mm reverse phase NanoEase column (Waters) in a CapLC (Waters) liquid chromatography system. Q-TOF2 and CapLC systems were piloted by MassLynx 4.0 (Waters). Peak lists were created using Mascot Distiller 2.2 (Matrix Science). Enzyme specificity was set to trypsin and the maximum number of missed cleavages per peptide was set at 1. Carbamidomethylation was allowed as a fixed modification and oxidation of methionine was allowed as a variable modification. Mass tolerance for the monoisotopic precursor peptide window was set to 100 ppm and MS/MS tolerance window to ±0.3 Da. We also specified ESI-Q-TOF as the instrument. The peak lists were searched against the Proteobacteria subset of the National Center for Biotechnology Information nonredundant database (NCBInr; 1 391 518 sequences in October 2008). For all protein identifications, a minimal individual ions score of 45 (identity score) and expected value below 1 were used for the initial identification criteria. In the case of redundant protein identifications, the protein identification with the highest score was selected. Moreover, the correlation between theoretical pI and molecular mass of the protein with the position of the corresponding spot in the 2D gel was also taken into account. All MS/MS spectra can be found in the Supporting Information.
Quantitative Real-Time RT-PCR
Total RNA samples were prepared as described above on B. melitensis 16 M wild-type strain grown in 2YT with 5 μM final concentration C12−HSL or ACN at 37 °C with shaking to an OD600 of 0.75. DNA was removed from the samples using the DNA-free kit (Ambion) and reverse-transcription performed with SuperScript II Reverse Transcriptase (Invitrogen). cDNA samples were used as template in real-time PCR reactions. Primers were designed with the PrimerExpress 2.0 (Applied Biosystems; sequences are listed in Table 3, Supporting Information), PCR products ranged from 80 to 100 bp. Real-time PCR reactions were performed with SYBR Green Mix (Applied Biosystems) in 96-well Optical Reaction plates (Applied Biosystems). Ratios were calculated using the ΔΔCT method for each primer in an Applied Biosystems Step One Plus real-time PCR instrument. Results for each target mRNA was normalized to BMEI0861 mRNA and averaged.
Chromatin Immunoprecipitation Assay
ΔvjbR pSB502 (encoding vjbRHTH C-terminal flag fusion) and ΔvjbR pBBR1MCS-5 (negative control) strains were grown in 2YT at 37 °C to an OD600 of 0.75. ChIP experiments were performed essentially as described(43) using antiflag m2 monoclonal antibodies (Sigma). Briefly, after bacterial growth, formaldehyde (1%) was added to 10 mL of triplicate cultures and the cultures placed at room temperature for 10 min before quenching the reaction with glycine (125 mM) for 5 min. Bacteria were collected and washed with cold phosphate-buffered saline twice. The cells were lysed in 0.9 mL of lysis solution (10 mM Tris pH 8.0, 50 mM NaCl, 10 mM EDTA, 20% sucrose, 20 mg mL−1 lysozyme) and 0.9 mL of 2× RIPA solution (100 mM Tris pH 8.0, 300 mM NaCl, 2% Nonidet P-40, 1% sodium deoxycholate, 0.2% SDS). The cell extracts were sonicated to fragment DNA to an average size of 500 bp and centrifuged 30 min at 13 000 rpm 4 °C, supernatants were stored at −80 °C. Fifteen μL of the extract was removed for total DNA preparation. For immunoprecipitation of VjbR cross-linked DNA, a portion of the extracts (500 μL) was first cleared with 80 μL of Sepharose-Protein G beads (Sigma) for 1 h at 4 °C and then incubated with 4 μL of monoclonal antiflag m2 antibodies (Sigma) for 4 h at 4 °C. The beads were washed twice with 1× RIPA solution, then twice with LiCl/detergent solution (10 mM Tris pH 8.0, 250 mM LiCl, 1 mM EDTA, 0.5% Nonidet P-40, 0.5% sodium deoxycholate), and finally with TE buffer. The immunoprecipitated material was eluted with 130 μL of elution buffer (25 mM Tris pH 8.0, 5 mM EDTA, 0.5% SDS) for 20 min at 65 °C. Cross-linking of immunoprecipitated and total DNA was reversed by incubation at 65 °C overnight. After Pronase treatment, the immunoprecipitated and total DNA were purified using the PCRapace Kit (Invitek GmbH, Germany) according to the manufacturer.
Analysis of the immunoprecipitated DNA was performed using quantitative PCR with input and immunoprecipitated DNA samples as templates. All promoter-specific primers were designed with Primer Express 1.0 (Applied Biosystems, see supplementary Figure 3) for amplicon sizes and primer localization and sequences). PCR products ranged from 80 to 100 bp. Real-time PCR reactions were performed in 25 μL SYBR Green Mix (Applied Biosystems) in 96-well Optical Reaction plates (Applied Biosystems). Relative quantification using a standard curve method was performed for each primer in an Applied Biosystems 7900HT real-time PCR instrument (absolute quantification method). Input DNA values were used to normalize ChIP, which are presented as a percentage of precipitated DNA (IP)/total DNA (IN).
Assessment of B. melitensis Stress Responses
Alkaline and Acid Resistance
B. melitensis 16 M and isogenic ΔvjbR and ΔbabR strains were grown in 2YT up to an OD600 of 1.0 and diluted to an OD600 of 0.05 in 2YT adjusted to the required pH with HCl or NaOH. Cultures were incubated at 37 °C with shaking for 72 h, and OD600 were measured after 24, 48, and 72 h of incubation.
Resistance to Bile Salts
In vitro resistance of B. melitensis 16 M and the ΔvjbR and ΔbabR strains to bile salts was evaluated as follows. The wt, ΔvjbR and ΔbabR strains were grown in 2YT up to an OD600 of 1.0 and were diluted to an OD600 0.05 in 2YT or in 2YT containing 0.1% bile salts (Fluka). Cultures were then incubated at 37 °C with shaking for 18 h and serial dilutions were plated on 2YT medium for CFU counting.
Results and Discussion
Proteomic Analysis of Brucella QS Mutants
To define the QS regulon of B. melitensis 16 M, we compared both QS mutants (ΔvjbR and ΔbabR) to the parental (wt) strain by proteomic analysis. Knowing that VjbR, BabR and several virulence factors are expressed during midexponential growth phase, total proteins were extracted under these conditions. 2D-DIGE was then used to compare total protein extracts from three independent midexponential phase cultures of B. melitensis 16 M and isogenic ΔvjbR and ΔbabR mutants. For each comparison, two types of gels (pH 4−7 and pH 7−11 NL) were run. Two samples (wt/ΔvjbR or wt/ΔbabR) labeled with Cy3 and Cy5 respectively, were analyzed on the same gel, together with an internal standard labeled with Cy2 (see Material and Methods section). We defined a protein as being affected by the mutation of VjbR or BabR if a difference in abundance of a least 30% compared to the wt strain (Student t test p < of 0.05) was observed for that protein in all three gels (one gel for each independent culture). Selected proteins spots corresponding to the 101 different proteins listed in Table 1 were picked, digested and identified using LC−MS/MS. The production of 35 of these proteins is directly or indirectly regulated by VjbR and 66 by BabR. Interestingly, numerous identified proteins are predicted to be involved in metabolic pathways such as central metabolism or amino acid metabolism, respiration, transport of amino acids, sugars and other molecules, secretion and translation.
Table 1. Targets Identified by 2D-DIGE Analysisa.
| A | ||||||||
|---|---|---|---|---|---|---|---|---|
| cellular function | BMEnnnnn | identification | accession no. | F.C. | # peptides | C % | score | method |
| ΔbabR, pH 4−7 | ||||||||
| A.A metabolism | BMEI0231 | NAD specific glutamate dehydrogensase | AAL51413.1 | 0.37 | 2 | 1 | 194 | Q-TOF |
| BMEI0451 | 2-isopropyl malate synthase | AAL51632.1 | 0.21 | 3 | 5 | 183 | Q-TOF | |
| BMEI0811 | l-serine dehydratase | AAL51992.1 | 0.66 | 5 | 10 | 267 | Q-TOF | |
| BMEI0979 | Glutamine synthase | AAL52160.1 | 0.30 | 2 | 4 | 125 | Q-TOF | |
| BMEI1620 | Ornithine carbamoyltransferase | AAL52801.1 | 0.64 | 2 | 4 | 99 | Q-TOF | |
| BMEI1638 | Glutamate synthase | AAL52819.1 | 2.21 | 4 | 9 | 267 | Q-TOF | |
| BMEII0371 | β-alanine pyruvate transaminase | AAL53613.1 | 1.78 | 6 | 16 | 412 | Q-TOF | |
| BMEII0559 | Aminomethyltransferase | AAL53801.1 | 1.68 | 3 | 7 | 191 | Q-TOF | |
| Carbohydrate metabolism | BMEI0310 | Glycéraldehyde 3-phosphate deshydrogenase | AAL51491.1 | 1.44 | 3 | 9 | 187 | Q-TOF |
| BMEI1413 | GDP-mannose 4,6-dehydratase | AAL52594.1 | 0.68 | 7 | 16 | 412 | Q-TOF | |
| BMEI1779 | Fructokinase | AAL52960.1 | 1.61 | 4 | 13 | 246 | Q-TOF | |
| BMEII0358 | 2-dehydro-3-dehydro-phosphogalactonase aldolase | AAL53600.1 | 1.38 | 2 | 10 | 141 | Q-TOF | |
| Cell wall/envelope | BMEI0727 | d-alanine-d-alanine ligase A | AAL51908.1 | 1.71 | 6 | 13 | 376 | Q-TOF |
| Central metabolism | BMEI0138 | Succinyl coA synthetase beta chain | AAL51320.1 | 1.91 | 8 | 18 | 545 | Q-TOF |
| BMEI0161 | Succinate dehydrogenase | AAL51343.1 | 0.16 | 5 | 9 | 310 | Q-TOF | |
| BMEI0836 | Citrate synthase | AAL52017.1 | 0.56 | 2 | 4 | 120 | Q-TOF | |
| BMEI0851 | Enolase | AAL52032.1 | 1.34 | 10 | 20 | 614 | Q-TOF | |
| BMEII0248 | Phosphoglycerate mutase | AAL53489.1 | 1.74 | 4 | 19 | 228 | Q-TOF | |
| BMEII0511 | Phosphogluconate dehydratase | AAL53753.1 | 0.08 | 2 | 3 | 144 | Q-TOF | |
| Lipid metabolism | BMEI0543 | Choloylglycine hydrolase | AAL51724.1 | 0.11 | 6 | 13 | 375 | Q-TOF |
| BMEI1112 | 3-oxo-acyl-carrier protein synthase | AAL52293.1 | 1.65 | 5 | 12 | 327 | Q-TOF | |
| BMEI1196 | EnoylCoA hydratase | AAL52377.1 | 1.51 | 2 | 7 | 113 | Q-TOF | |
| BMEI1512 | Enoyl-(acyl-carrier protein) reductase | AAL52693.1 | 0.74 | 7 | 23 | 453 | Q-TOF | |
| Nucleotide metabolism | BMEI1643 | N-carbamoyl-l-amino acid amidohydrolase | AAL52824.1 | 1.62 | 3 | 7 | 210 | Q-TOF |
| Other metabolism | BMEI0176 | Porphobilinogene deaminase | AAL51358.1 | 0.13 | 3 | 10 | 203 | Q-TOF |
| BMEI0219 | Malonate semialdehyde dehydrogenase | AAL51401.1 | 0.51 | 3 | 7 | 184 | Q-TOF | |
| BMEI0712 | CBIG protein/precorrin-3B C17-methyltransferase | AAL51893.1 | 0.08 | 3 | 5 | 231 | Q-TOF | |
| BMEI1588 | Carboxynorspermidine dehydrogenase | AAL52769.1 | 0.72 | 3 | 8 | 204 | Q-TOF | |
| Protein synthesis | BMEI0481 | LSU Ribosomal Protein L25P | AAL51662.1 | 0.71 | 6 | 21 | 472 | Q-TOF |
| BMEI0742 | EF-Tu | AAL51923.1 | 1.98 | 14 | 37 | 1130 | Q-TOF | |
| BMEI1483 | 50S ribosomal Protein L9 | AAL52664.1 | 0.12 | 2 | 9 | 121 | Q-TOF | |
| BMEI1915 | SSU ribosoma protein S1P | AAL53096.1 | 1.77 | 2 | 3 | 155 | Q-TOF | |
| Regulation | BMEI0626 | Transriptional regulator GntR familly | AAL51807.1 | 2.64 | 2 | 5 | 110 | Q-TOF |
| BMEII0299 | IclR family transcriptional regulator | AAL53541.1 | 0.73 | 2 | 7 | 83 | Q-TOF | |
| BMEII1116 | LuxR regulator VjbR | AAL54358.1 | 0.55 | 2 | 9 | 164 | Q-TOF | |
| Replication/transcription | BMEI0588 | DNA repair protein RecN | AAL51769.1 | 0.21 | 4 | 8 | 344 | Q-TOF |
| BMEI0749 | DNA-directed RNA polymerase beta chain | AAL51930.1 | 0.30 | 7 | 4 | 458 | Q-TOF | |
| BMEI1823 | DNA gyrase B | AAL53004.1 | 0.60 | 7 | 8 | 417 | Q-TOF | |
| Respiration | BMEI0096 | Electron transfer flavoprotein beta subunit | AAL51278.1 | 1.54 | 6 | 27 | 435 | Q-TOF |
| BMEI0249 | ATP Synthase Alpha Chain | AAL51431.1 | 0.76 | 5 | 9 | 329 | Q-TOF | |
| BMEI0487 | ATP synthase beta subunit/transription termination factor rho | AAL51668.1 | 1.61 | 4 | 11 | 216 | Q-TOF | |
| Stress/chaperone | BMEI0123 | Peptidyl-prolyl cis−trans isomerase | AAL51278.1 | 1.52 | 7 | 21 | 426 | Q-TOF |
| BMEI0195 | ATP-Dependent Clp Protease, ATP-Binding Subunit ClpB | AAL51377.1 | 0.74 | 18 | 20 | 1275 | Q-TOF | |
| BMEI0613 | Protease DO | AAL51794.1 | 1.66 | 7 | 12 | 465 | Q-TOF | |
| BMEI2002 | DnaK | AAL53183.1 | 1.78 | 15 | 22 | 1049 | Q-TOF | |
| BMEII0401 | Thioredoxine | AAL53643.1 | 1.71 | 3 | 9 | 224 | Q-TOF | |
| BMEII1048 | GroEL | AAL54290.1 | 0.63 | 21 | 48 | 1727 | Q-TOF | |
| Transport/secretion | BMEI1716 | Trehalose maltose Binding Protein | AAL52897.1 | 1.62 | 5 | 11 | 328 | Q-TOF |
| BMEI1930 | Leucine-, isoleucine-, valine-, threonine-, and alanine-binding protein precursor | AAL53111.1 | 1.61 | 2 | 7 | 133 | Q-TOF | |
| BMEII0098 | High affiny branched chain amino acid transport ATP-binding protein livF | AAL53339.1 | 1.38 | 3 | 10 | 166 | Q-TOF | |
| BMEII0590 | Sugar binding protein | AAL53832.1 | 2.68 | 11 | 27 | 781 | Q-TOF | |
| BMEII0601 | Cystine binding periplasmic protein | AAL53843.1 | 1.38 | 4 | 13 | 303 | Q-TOF | |
| BMEII0734 | Periplasmic oligopeptide Binding protein precursor | AAL53976.1 | 1.76 | 8 | 16 | 589 | Q-TOF | |
| BMEII0923 | Spermidine/putrescine-binding protein | AAL54165.1 | 1.52 | 3 | 9 | 193 | Q-TOF | |
| Unassigned | BMEI1201 | Hypothetical cytosolic protein | AAL52382.1 | 2.64 | 6 | 17 | 477 | Q-TOF |
| BMEI1211 | General L-amino acid-binding periplasmic protein AAPJ precursor | AAL52392.1 | 2.07 | 2 | 5 | 151 | Q-TOF | |
| BMEI1747 | aldehyde dehydrogenase | AAL52928.1 | 0.66 | 5 | 9 | 360 | Q-TOF | |
| BMEI1819 | Alcohol dehydrogenase deshydrogenase | AAL53000.1 | 1.44 | 3 | 7 | 192 | Q-TOF | |
| ΔvjbR, pH 4−7 | ||||||||
| A.A. metabolism | BMEI0101 | Cysteine synthase A | AAL51283.1 | 0.68 | 2 | 7 | 147 | Q-TOF |
| BMEI0386 | Succinate semialdehyde dehydrogenase | AAL51567.1 | 1.72 | 5 | 11 | 352 | Q-TOF | |
| BMEI1925 | Acetyl-CoA Carboxylase Alpha Chain/Propionyl-CoA Carboxylase Alpha Chain | AAL53106.1 | 0.75 | 4 | 5 | 244 | Q-TOF | |
| Central metabolism | BMEI0851 | Enolase | AAL52032.1 | 0.56 | 5 | 11 | 311 | Q-TOF |
| Nucleotide metabolism | BMEI0522 | Carbamoyl Phosphate synthase large subunit | AAL51703.1 | 0.60 | 12 | 9 | 725 | Q-TOF |
| BMEI1127 | Phosphoribosylformylglycinamidine Synthase | AAL52308.1 | 0.83 | 7 | 8 | 429 | Q-TOF | |
| Protein synthesis | BMEI0837 | Glutamyl tRNA synthase | AAL52018.1 | 1.64 | 2 | 3 | 121 | Q-TOF |
| BMEI1047 | Tyrosyl tRNA synthase | AAL52228.1 | 0.68 | 5 | 10 | 332 | Q-TOF | |
| Regulation | BMEI0417 | PdhS | AAL51598.1 | 0.70 | 4 | 5 | 232 | Q-TOF |
| BMEI0558 | Transcriptional regulator ArsR | AAL51739.1 | 0.68 | 5 | 15 | 389 | Q-TOF | |
| Replication/transcription | BMEI0880 | Single strand binding protein | AAL52061.1 | 2.03 | 4 | 22 | 263 | Q-TOF |
| Transport/secretion | BMEII0105 | Iron regulated outer membrane protein FrpB | AAL53346.1 | 0.80 | 4 | 6 | 244 | Q-TOF |
| ΔbabR, pH 7−11 NL | ||||||||
| Cell wall/envelope | BMEI1404 | Mannosyltransferase | AAL52585.1 | 1.38 | 1119 | 35 | 151 | Maldi-TOF |
| Other metabolism | BMEI0859 | Lipoyl synthetase | AAL52040.1 | 0.52 | 14148 | 46 | 123 | Maldi-TOF |
| ΔvjbR, pH 7−11 NL | ||||||||
| A.a. metabolism | BMEI1970 | S-adenosylmethionine synthetase | AAL53151.1 | 1.7 | 4 | 8 | 268 | Q-TOF |
| Cell wall/envelope | BMEI0035 | d-alanyl-d-alanine carboxypeptidase | AAL51217.1 | 1.42 | 25158 | 75 | 214 | Maldi-TOF |
| BMEI0575 | UDP-N-acetylmuramoylalanyl-d-glutamyl-2,6-diaminopimelate-d-alanyl-d- alanyl ligase | AAL51756.1 | 3.26 | 11141 | 33 | 98 | Maldi-TOF | |
| BMEI1029 | Outer membrane protein TolC | AAL52210.1 | 4.22 | 16148 | 33 | 108 | Maldi-TOF | |
| BMEI1435 | Polysaccharide deacetylase | AAL52616.1 | 0.37 | 8115 | 45 | 103 | Maldi-TOF | |
| BMEII0374 | Alanine racemase | AAL53616.1 | 1.45 | 17121 | 50 | 110 | Maldi-TOF | |
| BMEII1028 | Tetraacyldisaccharide 4′-kinase | AAL54270.1 | 0.52 | 16127 | 62 | 147 | Maldi-TOF | |
| Protein synthesis | BMEI0741 | 23S rRNA methyltransferase | AAL51922.1 | 0.68 | 9144 | 55 | 73 | Maldi-TOF |
| BMEI0747 | LSU ribosomal protein L10P | AAL51928.1 | 2.37 | 614 | 37 | 78 | Maldi-TOF | |
| BMEI0753 | SSU ribosomal protein S7P | AAL51934.1 | 1.93 | 5112 | 44 | 58 | Maldi-TOF | |
| BMEI1169 | SSU ribosomal protein S9P | AAL52350.1 | 1.93 | 317 | 17 | 42 | Maldi-TOF | |
| BMEI1267 | Dimethyladenosine transferase | AAL52448.1 | 0.40 | 1017 | 51 | 155 | Maldi-TOF | |
| Regulation | BMEI0808 | Transcriptional Regulator, MerR Family | AAL51989.1 | 0.73 | 615 | 34 | 79 | Maldi-TOF |
| Replication/transcription | BMEI1035 | Atp-dependent rna helicase | AAL52216.1 | 1.5 | 4 | 9 | 268 | Q-TOF |
| Transport/secretion | BMEI0469 | Purine nucleoside permease | AAL51650.1 | 0.18 | 913 | 29 | 138 | Maldi-TOF |
| BMEII0032 | Channel protein VirB8 homologue | AAL53273.1 | 0.26 | 912 | 53 | 157 | Maldi-TOF | |
| BMEII0033 | Channel protein VirB9 homologue | AAL53274.1 | 0.49 | 611 | 36 | 100 | Maldi-TOF | |
| BMEII0593 | Glucose ABC transporter ATPase | AAL53835.1 | 2.92 | 11116 | 45 | 133 | Maldi-TOF | |
| BMEII0863 | Oligopeptide transport ATP-binding protein appD | AAL54105.1 | 1.45 | 17124 | 38 | 183 | Maldi-TOF | |
| Unassigned | BMEI1193 | Cell wall degradation protein | AAL52374.1 | 0.75 | 8117 | 18 | 69 | Maldi-TOF |
| BMEII0002 | Ribosomal-protein-serine acetyltransferase | AAL53243.1 | 1.4 | 817 | 45 | 118 | Maldi-TOF | |
| BMEII0431 | Oxidoreductase | AAL53673.1 | 2.47 | 10115 | 26 | 109 | Maldi-TOF | |
| B | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gel | cellular function | BMEnnnnn | identification | accession no. | F.C. | sequence | C % | score | m/z | charge | method |
| ΔbabR, pH 4−7 | |||||||||||
| AA metabolism | BMEI0516 | Aspartate aminotransferase | AAL51697.1 | 0.68 | QAAIAAINR | 2 | 51 | 464,2757 | 2+ | Q-TOF | |
| Central metabolism | BMEI0791 | Isocitrate deshydrogenase | AAL51972.1 | 0.68 | ASFNYGLKR | 2 | 49 | 528,2996 | 2+ | Q-TOF | |
| Central metabolism | BMEI1436 | pyruvate phosphate dikinase | AAL52617.1 | 0.47 | TPQNITEEAR | 1 | 68 | 579,815 | 2+ | Q-TOF | |
| Stress/chaperone | BMEI1367 | Superoxide Dismutase Mn | AAL52548.1 | 1.38 | LLEGSGLEGK | 4 | 48 | 501,78 | 2+ | Q-TOF | |
| Transport/secretion | BMEII0593 | ATP GDP Binding protein ABC transporter | AAL53835.1 | 1.93 | SVFFDSASQTR | 2 | 51 | 622,8208 | 2+ | Q-TOF | |
| Unassigned | BMEI1939 | D-3-phosphoglycerate dehydrogenase | AAL53120.1 | 0.60 | GSLQNEPDILAALDR | 4 | 121 | 806,4188 | 2+ | Q-TOF | |
| ΔvjbR, pH 7−11 NL | |||||||||||
| Cell wall/envelope | BMEI0223 | Membrane-bound lytic murein transglycosylase B | AAL51405.1 | 2.56 | YAQATINADR | 3 | 79 | 561,8065 | 2+ | Q-TOF |
A. Proteins identified in the 2D-DIGE analysis of babR and vjbR mutant strains. B. Proteins identified by one single peptide in the 2D-DIGE analysis of babR and vjbR mutant strains. BMEnnnnn: ORF number; F.C.: fold change compared with the wild type strain; # peptides: numbers of unique peptides identified (for MALDI identification: number of peaks that match to the tryptic peptides vs. number of peaks that do not match to the tryptic peptides); C %: percentage sequence coverage of the protein; Score: identity score; Method: method used for the identification of the protein.
Transcriptomic Analysis of Brucella QS Mutants
We chose to combine our 2D-DIGE analysis with a transcriptomic study of both QS mutants. Total RNA samples taken at the proteomic analysis step (from the same cultures) were used to maximize the correlation between these two complementary approaches. RNA samples were pooled, retro-transcribed and labeled before hybridization to a B. melitensis DNA microarray (Nimblegen).
The gene expression pattern of the ΔvjbR strain was compared to profiles generated from the B. melitensis 16 M strain. This analysis led to the identification of 296 coding sequences (CDS) (9.2% of the genome) differentially expressed in the vjbR mutant strain (see Supplementary Table 1, Supporting Information). Contrary to what was expected based on previous experiments examining the expression of the virB and fliF promoters,(19) a subset of the predicted VjbR regulon is overexpressed in the mutant strain.
The gene expression pattern of the ΔbabR strain was compared to profiles generated from the parental strain and revealed that BabR regulated the expression of 42 CDS in B. melitensis 16 M (1.3% of the genome, see Supplementary Table 1, Supporting Information).
Our analysis reveals that the regulation of a significant fraction of the B. melitensis 16 M genome is influenced by a mutation affecting the QS system. This is consistent with the proposition that QS could act as a global regulatory system in this intracellular pathogen. Similar observations have been previously made in Escherichia coli(44) and in the opportunistic pathogen Pseudomonas aeruginosa.45−47 However, we suspect that LuxR-type regulators may directly control only for a fraction of the identified target genes since the expression of genes encoding several transcriptional or post-transcriptional regulators is affected by the ΔvjbR and/or the ΔbabR mutations as can be seen Table 2F.
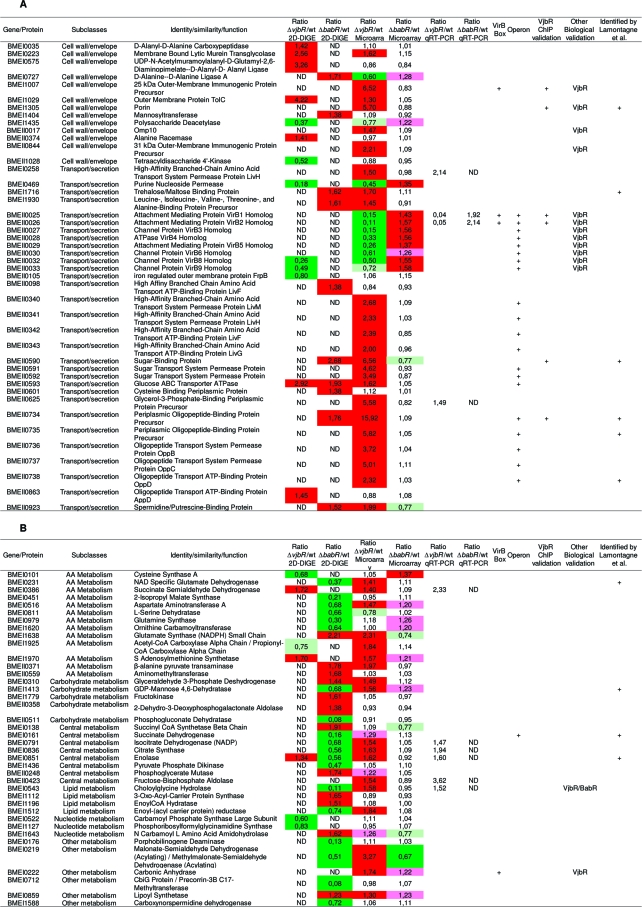
Table 2. Targets Identified in This Studya.
 |
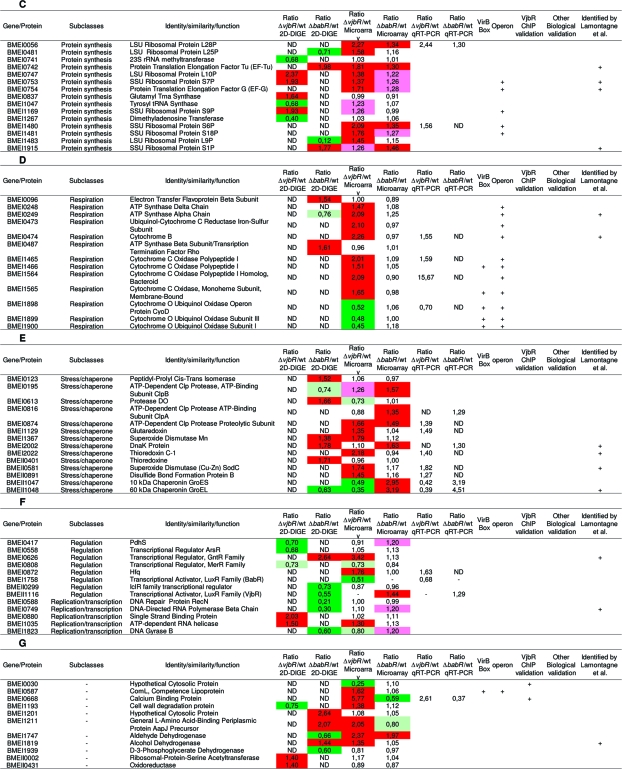 |
Summary table of targets genes identified in this study and connections with other published results. Each target is defined by a BMEnnnnn number (corresponding to the ORF number of the gene in Brucella melitensis 16 M genome), a functional class and a predicted function. A: Cell wall biogenesis and transport/secretion subclasses. B: Metabolism subclass. C: Translation subclass. D: Respiration process subclass. E: Stress response subclass. F: Regulation subclass. G: Unclassified targets. In the fold change column, colors represent the regulator’s effect: red when the regulator exerts a repressive role (fold change >1.3) and green when the regulator exerts an activation role (fold change <0.7). Light colors were used for genes with a lower fold change (pink: 1.3 > fold change >1.2; olive-green: 0.8 > fold change >0.7). Twenty-nine targets of interest were analyzed by qRT-PCR on new biological samples to validate microarray results. These results are listed in the “Ratio mutant/wt qRT-PCR” column. The “VirB Box” column indicates with a “+” genes containing in their promoter sequence the box identified by de Jong(35) for VjbR regulation. “Operon” column indicates genes which are predicted by BioCyc or KEGG DAS to be part of an operon. Positive results for VjbR ChIP experiments are labeled with a “+” in the “VjbR ChIP validation” column. When biological validations were available (such as Western blots, bile salts resistance test.. .), mutant strain’s name tested can been found in the “Biological validation” column. In the last column, genes identified by a “+” have been found by Lamontagne and coworkers(17) to be implicated in Brucella abortus intracellular adaptation. ND: not determined.
Notably, several previously known VjbR-regulated genes were identified in this transcriptomic study, (e.g., virB and omp genes) thus providing an “a priori” validation for the use of the microarray analysis.
Validation of Transcriptional Profiling Results by qRT-PCR
To further validate the results collected from the microarray analysis, we performed a reverse transcription experiment followed by quantitative PCR (qRT-PCR) on RNA samples prepared at exponential growth phase (same OD600nm as the transcriptomic experiments but harvested from new cultures). Total RNA was extracted from B. melitensis 16 M and isogenic ΔvjbR and ΔbabR mutants. We selected 29 CDS of particular interest (including CDS putatively involved in stress response, virulence and central metabolism) for this analysis. As shown in Table 2, for all the genes tested, the fold changes in transcription detected by qRT-PCR are similar to the fold changes detected by the microarray analysis. A negative control used for each qRT-PCR reaction showed that no genomic DNA contamination occurred in the RNA samples (data not shown).
Selection of the Most Confident B. melitensis QS Targets
We used both proteomic and transcriptomic analyses of vjbR and babR mutants to define the QS regulon of B. melitensis 16 M. In order to select the most confident targets, the results obtained with these two complementary methods were combined with previous data on genes regulated by VjbR and BabR.19,28,36 As the proteomic analysis was performed on three independent samples whereas the transcriptomic one was done on a pool of the corresponding RNA samples, we first based our selection on targets identified by the 2D-DIGE analysis (n = 99). We then added to the list CDS identified in the transcriptomic analysis only if they have been confirmed by qRT-PCR (n = 29), ChIP (n = 8) or a previous biological validation (n = 14). Finally, we added CDS predicted to belong to the same transcriptional unit as one of the above selected CDS (n = 38). Using this combinatorial analysis, we got a selection of 149 genes whose expression or the amount of gene products formed is affected (directly or indirectly) by VjbR and/or BabR, they are listed in Table 2.
Connections between the Two Brucella QS-Regulators
Analysis of the combined data led to the observation that 27 targets are regulated by both BabR and VjbR (Table 3). The two regulators act in an opposite way on 55% of the genes, including the virB genes, and genes encoding chaperones and transporters. These results strongly suggest a crosstalk between the two QS-regulators of Brucella. Two recent studies demonstrate that VjbR activates its own expression.30,36 One of these studies demonstrated a positive regulatory effects of both QS regulators on their own genes as well as the gene encoding the other regulator.(30) However, our transcriptomic analysis revealed that VjbR has a 2-fold activating effect on babR expression whereas BabR has a 1.5-fold repressing effect on vjbR (Table 2). This observation was confirmed by two different qRT-PCR experiments performed on RNA samples harvested from new cultures (Table 2 and Table 4).
Table 3. VjbR and BabR Shared Targets: ORFs Identified by the Proteomic and Transcriptomic Analyses and Regulated by Both LuxR Type Regulators.
| target | identity/similarity/function | ratio ΔvjbR/wt | ratio ΔbabR/wt | |
|---|---|---|---|---|
| Co-regulated targets | BMEI0056 | LSU Ribosomal Protein L28P | 2.27 | 1.34 |
| BMEI0195 | ATP-Dependent Clp Protease, ATP-Binding Subunit ClpB | 1.26 | 1.57 | |
| BMEI0223 | Membrane Bound Lytic Murein Transglycolase | 2.56 | 1.38 | |
| BMEI0742 | Protein Translation Elongation Factor Tu (EF-Tu) | 1.81 | 1.30 | |
| BMEI0753 | SSU Ribosomal Protein S7P | 1.37 | 1.26 | |
| BMEI0754 | Protein Translation Elongation Factor G (EF-G) | 1.71 | 1.28 | |
| BMEI0874 | ATP-Dependent Clp Protease Proteolytic Subunit | 1.66 | 1.49 | |
| BMEI1480 | SSU Ribosomal Protein S6P | 2.09 | 1.35 | |
| BMEI1481 | SSU Ribosomal Protein S18P | 1.76 | 1.27 | |
| BMEI1747 | Aldehyde Dehydrogenase | 2.37 | 1.98 | |
| BMEI1915 | SSU Ribosomal Protein S1P | 1.26 | 1.46 | |
| BMEII0593 | Glucose ABC Transporter ATPase | 2.92 | 1.93 | |
| Differentially regulated targets | BMEI0219 | Malonate-Semialdehyde Dehydrogenase (Acylating)/Methylmalonate-Semialdehyde Dehydrogenase (Acylating) | 3.27 | 0.67 |
| BMEI0469 | Purine Nucleoside Permease | 0.45 | 1.35 | |
| BMEI0668 | Calcium Binding Protein | 5.77 | 0.59 | |
| BMEI0727 | d-Alanine-d-Alanine Ligase A | 0.60 | 1.28 | |
| BMEI0851 | Enolase | 0.56 | 1.34 | |
| BMEII0025 | Attachment Mediating Protein VirB1 Homologue | 0.15 | 1.43 | |
| BMEII0026 | Attachment Mediating Protein VirB2 Homologue | 0.11 | 1.57 | |
| BMEII0027 | Channel Protein VirB3 Homologue | 0.15 | 1.56 | |
| BMEII0028 | ATPase VirB4 Homologue | 0.33 | 1.56 | |
| BMEII0029 | Attachment Mediating Protein VirB5 Homologue | 0.26 | 1.37 | |
| BMEII0030 | Channel Protein VirB6 Homologue | 0.61 | 1.26 | |
| BMEII0032 | Channel Protein VirB8 Homologue | 0.50 | 1.55 | |
| BMEII0033 | Channel Protein VirB9 Homologue | 0.72 | 1.58 | |
| BMEII1047 | 10 kDa Chaperonin GroES | 0.49 | 2.95 | |
| BMEII1048 | 60 kDa Chaperonin GroEL | 0.35 | 3.19 |
Table 4. Validation of Some Targets by qRT-PCR and Analysis of C12−HSL Effecta.
| babR | vjbR | dnaK | virB2 | groEL | groES | BMEI0433 | BMEI0668 | BMEII0625 | |
|---|---|---|---|---|---|---|---|---|---|
| wt + ACN | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| wt + C12−HSL | 2.6 | 0.5 | 1.6 | 0.2 | 2.0 | 2.2 | 2.1 | 4.5 | 1.8 |
| ΔbabR + ACN | 0 | 1.7 | 1.9 | 1.9 | 3.1 | 2.7 | 1.1 | 0.6 | 0.8 |
| ΔbabR + C12−HSL | 0 | 0.3 | 1.8 | 0.1 | 4.0 | 3.3 | 0.8 | 3.5 | 2.2 |
| ΔvjbR + ACN | 0.7 | 0 | 0.9 | 0.1 | 0.1 | 0.1 | 2.9 | 4.5 | 3.8 |
| ΔvjbR + C12−HSL | 1.5 | 0 | 1.2 | 0.1 | 0.2 | 0.2 | 4.4 | 9.7 | 4.9 |
Comparison of fold change ratios for mRNA from wt, ΔbabR and ΔvjbR strains with or without C12−HSL. RNA was extracted at an equivalent OD600 for the transcriptomic and the qRT-PCR experiments. ACN: Acetonitrile: C12−HSL solvent. C12−HSL: dodecanoyl-L-homoserine lactone (added to the culture media at a final concentration of 5 mM). We considered that gene expression is different between wt and mutant strain when the ratio is >1.3 or <0.7.
A recent study by Rambow-Larsen and collaborators identified 36 BabR (that they called BlxR) target genes based on a microarray analysis restricted to 289 genes selected for their potential involvement in virulence.(30) Among these 36 targets, only 8 were common to our analysis (8 genes encoding VirB proteins). Strikingly, whereas these genes appeared to be activated by BabR in the study of Rambow-Larsen, they appeared to be repressed in our analysis. These discrepancies could be in part explained by the differences in the experimental design of these two experiments (growth phase, culture medium, and microarray design).
Impact of C12−HSL on Selected QS Targets
To assess the effect of C12−HSL on selected target genes, we performed qRT-PCR on total RNA extracted from B. melitensis 16 M and isogenic ΔvjbR and ΔbabR mutants grown with or without C12−HSL to an OD600 nm of 0.7. Results are presented in Table 4; wt strain cultivated without addition of C12−HSL was used as a benchmark. Regarding the expression of the genes encoding the two LuxR regulators in the parental strain, vjbR expression is repressed when exogenous C12−HSL is added whereas babR expression is activated. The fact that the C12−HSL effect on vjbR expression was observed in both the B. melitensis 16 M and the babR mutant suggests that VjbR regulates its own negative feedback loop. A similar proposal could be also suggested for BabR, but with a positive feedback loop.
Table 4 shows that, except for virB2 and vjbR, C12−HSL activates the expression of target genes in B. melitensis 16 M. Interestingly, depending on the target gene, the C12−HSL activating effect seems to be mostly dependent either on BabR (e.g., BMEI0433), VjbR (e.g. dnaK) or both regulators (e.g., BMEI0668 and BMEII0625). Because the effect of C12−HSL on some targets (e.g., BMEI0433) is still observed in the ΔvjbR strain (but not in the ΔbabR strain) and VjbR and BabR are the only predicted proteins possessing a predicted AHL-binding domain in B. melitensis 16 M, this result is the first evidence suggesting that BabR can respond to C12−HSL. The fact that two regulators react to the same signal molecule is quite unusual. One possibility could be that the two regulators have a different affinity for the C12−HSL. For example VjbR may respond to a lower level of AHLs once inside the cell and when a higher AHL concentration is reached, BabR may be activated. This will be an interesting hypothesis to test since we propose that BabR can modulate VjbR activity. Nevertheless, we cannot exclude the possibility that other unidentified AHLs may act preferentially on one or the other LuxR-type regulator.
Global Impact of QS on Brucella melitensis 16 M
Cell Wall/Envelope Biogenesis and Transport/Secretion Proteins
As shown in Table 2A, VjbR and BabR affect many genes involved in cell envelope biogenesis and membrane transport. These genes constitute the largest class identified in the B. melitensis 16 M QS regulon. As expected from previous work in our laboratory,19,28 the involvement of VjbR in the regulation of genes encoding components of the type four secretion system (T4SS) and outer membrane proteins (OMP) is observed. The identification of numerous membrane proteins whose genes are regulated by VjbR in this analysis further emphasized the role of VjbR in the control of membrane components. Interestingly, in addition to genes encoding OMPs, several genes predicted to be involved in murein and polysaccharide synthesis and LPS biogenesis are also regulated by VjbR.
Regarding the T4SS, a major component in Brucella virulence, we note a clear and inverse regulatory effect between the two LuxR regulators. VjbR activates the transcription of the virB operon (as previously described19,28), while BabR had a repressing effect on these genes. This observation was confirmed by two independent qRT-PCR experiments (Tables 2 and 4).
Numerous genes predicted to be involved in amino acid, oligopeptide and sugar transport were found to be QS targets in B. melitensis 16 M (Table 2A) and many of these genes appear to be regulated by VjbR. The fact that a lot of genes putatively involved in amino acid and sugar transport are part of the QS regulon suggests that a metabolic switch could be initiated by QS.
Metabolism Pathways
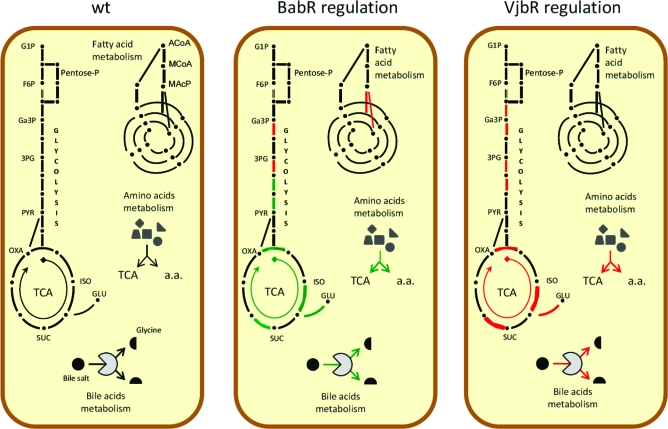
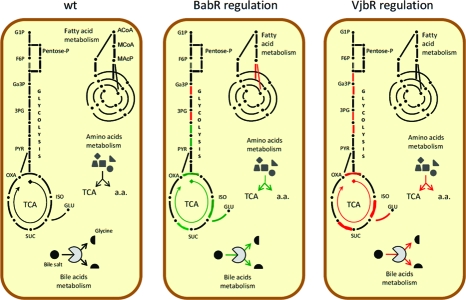
As can be seen in Table 2B, our analyses of vjbR and babR mutants revealed that numerous genes and/or proteins involved in metabolic pathways are regulated by QS in the parental 16 M strain. Figure 1 presents a schematic view of the main central metabolic pathways in B. melitensis 16 M, and the effects of vjbR and babR mutations on these pathways. Transcriptomic analysis revealed that VjbR exerts a repressive effect on numerous genes encoding enzymes involved in TCA cycle and glycolysis. As for BabR, proteomic analysis showed an activation effect on these two pathways. Interestingly, this same group of targets, constituted by BMEI0851 (enolase), BMEI0836 (citrate synthase), BMEI0791 (isocitrate dehydrogenase), BMEI0161 and BMEI0162 (succinate dehydrogenases) and BMEI0231 (NAD specific glutamate dehydrogenase) was regulated differentially depending upon the LuxR regulator, suggesting that QS could have a global reorganization effect on central metabolic processes. BabR also exerts a repressive effect on fatty acid metabolism genes in the parental strain.
Figure 1.
Diagram representing the main metabolic pathways in the wt strain and the regulation effect of VjbR and BabR. Pentose-P, pentose phosphate pathway; TCA, tricarboxylic acid cycle; G1P, glycerol-1-P; F6P, fructose-6-P; Ga3P, glyceraldehyde-3-P; 3PG, 3-P-glycerate; PYR, pyruvate; OXA, oxaloacetate; ISO, isocitrate; SUC; succinate; GLU, glutamate; Bile salt, glycocholate or taurocholate. Red lines/arrows represent repressed pathways while green lines/arrows represent activated pathways by the regulator. a.a., amino acid.
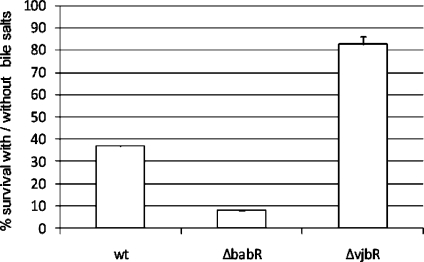
Both LuxR regulators also have a strong regulatory effect on BMEI0543 (a gene coding for a choloylglycine hydrolase). VjbR repressed the transcription of cgh (transcriptional fold change = 1.58) while BabR strongly activated the production of CGH (proteomic fold change = 0.11). A recent study in B. abortus has demonstrated the involvement of cgh in successful infection of mice through the oral route.(48) Interestingly CGH is found in Brucella culture supernatants and its secretion seems to be VirB-dependent as demonstrated by the analysis of B. abortus wt and virB mutant strains.(49)Brucella QS regulators could thus be involved not only in the regulation of the genes encoding the VirB machinery but also in the regulation of the genes encoding the effectors it secretes. Consequently, we tested the resistance of QS mutants to bile salts. As shown Figure 2, the ΔbabR strain was significantly more sensitive to bile salts than the B. melitensis16 M. In contrast, the ΔvjbR strain displayed an enhanced resistance to bile salts, supplying a biological validation of our proteomic/transcriptomic analysis.
Figure 2.
wt, ΔvjbR and ΔbabR resistance to bile salts. Strains were growth in 2YT with bile salts and CFU were compared with cultures in 2YT (100% of survival). Error bars represent standard deviation from three independent experiments. CFU, colony forming unit.
Despite the fact that VjbR and BabR regulate in an opposite way the same group of genes encoding central metabolic enzymes, we never observed a growth delay for the vjbR and babR mutant strains in liquid or solid culture in rich media (see for example Figure 1A). However, using the Biotype 100 system (Biomerieux), we noted some differences in carbon substrate assimilation between the parental strain and the vjbR and babR mutants (data not shown). So the role of the corresponding LuxR regulators in regulating metabolic pathways is worthy of further investigation.
Protein Synthesis and Respiration
Numerous genes coding for ribosomal proteins (LSU and SSU ribosomal proteins) and translation factors (EF-Tu, EF-G) are repressed by VjbR and to a lesser extent by BabR suggesting that these regulators depress protein synthesis (Table 2C). As can be seen in Table 2D, VjbR modulates the expression of genes encoding the terminal oxidases of the respiratory chain (activating the ubiquinol oxidase gene (cyo) and repressing the cytochrome C oxidase genes coxA (BMEI1465), coxB (BMEI1466) ccoN (BMEI1565) and ccoO (BMEI1564). BabR does not appear to control the expression of these cytochrome genes.
Stress Responses
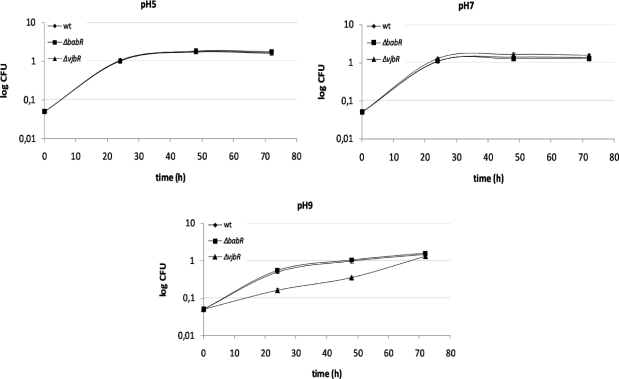
Our study suggests that a fraction of the QS targets in B. melitensis 16 M may be involved in stress responses (Table 2E). VjbR targets belonging to this category are essentially involved in protein folding (groES and groEL are activated by VjbR and repressed by BabR) and thiol-disulfide exchange (BMEI1129 and BMEI2022 encoding respectively a glutaredoxin and a thioredoxin are repressed by VjbR). BabR repressed many genes belonging to this functional group. These include clpP, clpA, and genes coding for the chaperones GroES, GroEL and DnaK, a chaperone identified as necessary for B. suis survival in macrophages.(50) To further examine the role of QS in stress responses in B. melitensis 16 M, we tested the resistance of both ΔvjbR and ΔbabR mutants to several kinds of stresses. The two QS mutants behave as the parental strain during the growth at pH 5, pH 7 (figure 3) and at pH 4, pH 6 and pH 8 (data not shown). In contrast, the vjbR mutant seems to be delayed its the adaptation to alkaline pH (pH 9). The response of Brucella strains to alkaline stress has not been described, but Appelbe et al. have shown that dnaK and groEL are induced during alkaline stress in Enterococcus faecalis.(51) While numerous genes encoding stress response proteins involved in adaptation to oxidative stress (hfq, clpA, clpB, sodC...) are regulated trough VjbR and BabR in B. melitensis 16 M, neither of the QS mutants displayed a higher sensitivity to H2O2 than the parent strain (data not shown). Likewise, the vjbR and ΔbabR mutants were also insensitive to cold or heat shock (data not shown).
Figure 3.
B. melitensis wt, ΔvjbR and ΔbabR response to acid and alkaline stresses. Strains were growth in 2YT pH 5, 7, or 9 and CFU were compared with cultures in 2YT (100% of survival). Error bars represent standard deviation from three independent experiments. CFU, colony forming unit.
Regulation, DNA Replication and Transcription
In addition to the cross talk between the two QS regulators described above, other regulators are part of the QS regulon (Table 2F): PdhS, a histidine kinase involved in cell cycle control,(52) and four putative transcriptional regulators of the families ArsR, GntR, IclR and MerR.
VjbR represses the transcription of hfq, a RNA chaperone that binds small regulatory RNA (sRNAs) and mRNAs to facilitate translational regulation in response to envelope stress, environmental stress and changes in metabolite concentrations and that was described as being crucial both for stationary phase survival and infection in murine model.(53) An additional interesting observation resulting from the analysis of the VjbR transcriptomic data is the presence of Bru-RS1 sequences near several VjbR targets. Bru-RS1 are conserved palindromic DNA sequences of 103 bp.(54) Their function is still unknown, but 30% of the 41 full length Bru-RS1 detected in the B. melitensis 16 M genome are located upstream (2/12 Bru-RS1) or downstream (10/12 Bru-RS1) of VjbR targets, and all of these genes are repressed by this regulator. We propose that these Bru-RS, when transcribed, could act as regulatory RNAs in conjunction with the Hfq protein, whose gene is also repressed by VjbR. The involvement of sRNA in QS regulatory systems is widespread55,56 and often allows a supplementary level of control on QS targets in response to environmental conditions.57,58
In general, genes involved in DNA replication and transcription appeared to be activated by BabR and repressed by VjbR in B. melitensis 16 M.
Identification of Direct VjbR Targets Using Chromatin Immunoprecipitation
The proteomic and transcriptomic approaches used in the study are complementary, but lead to the identification of both direct and indirect targets. The identification of the DNA binding sites recognized by VjbR and BabR would allow the subsequent identification of the whole direct regulon of Brucella QS regulators. Given the involvement of VjbR in B. melitensis virulence, we focused on the identification of direct targets of VjbR using a chromatin immunoprecipitation assay (ChIP), a technique allowing the detection of protein−DNA interactions in vivo. In order to be able to detect a direct binding between VjbR and a target promoter, we used a strain expressing a constitutive VjbR regulator (unresponsive to AHLs). Specifically, the ΔvjbR/pSB502 strain expresses the vjbRHTH-FLAG allele coding for the helix turn helix domain of VjbR fused with a C-terminal FLAG tag,(28) under the control of the E. coli lac promoter (Plac) in a vjbR deficient background. As VjbR is essential for the expression of the virB operon, we verified that the ΔvjbR/pSB502 strain produces the VirB8 protein, indicating the functionality of the VjbRHTH-FLAG regulator (data not shown). The immunoprecipitation experiments were performed in parallel with the ΔvjbR/pSB502 (ΔvjbR, Plac-vjbRHTH-FLAG) and the ΔvjbR strain harboring the empty plasmid (pBBR1-MCS5) as a negative control. Real-Time PCR was then used to quantify upstream regions of the targets displaying the highest ratios observed by the transcriptomic analysis and we performed RT-PCR to quantify the immunoprecipitated upstream regions.
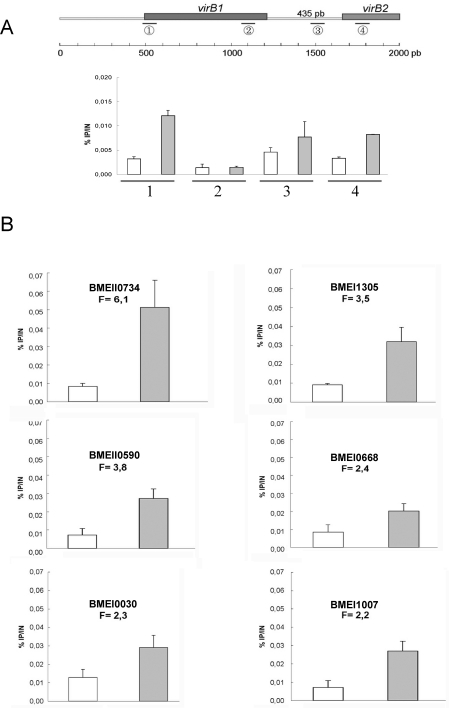
Figure 4 illustrates ChIP analysis showing an enrichment of target genes in the VjbRHTH-FLAG immunoprecipitation compared to the control immunoprecipitation (nontagged strain). Given that the DNA was sonicated to obtain fragments with an average size of 500 bp, these results suggest that VjbR is able to bind to the promoter region of virB operon, omp25b (BMEI1007), omp36 (BMEI1305), BMEI0668 coding for a putative calcium binding protein, BMEI0030 coding for a hypothetical protein conserved in P. aeruginosa (36% of identity, 60% of similarity), and BMEII0590 and BMEII0734 both encoding for components of ABC transporters (specific for sugars and oligopeptides, respectively). Interestingly, three regions of the virB1-virB2 locus seem to be bound by VjbR. The first one (locus 1 on the top of figure 2) corresponds to the previously defined PvirB promoter.(59) The other two correspond to the virB1-virB2 intergenic region (435 bp). The group of Sieira et al. has demonstrated by primer extension that virB transcription starts at a unique site, however the virB1-virB2 intergenic region also seems to include regulatory site(s).(59) This proposition has been recently confirmed by the study of de Jong and collaborators(36) where the authors demonstrated by EMSA that VjbR is able to bind both the B. abortus PvirB and virB1-virB2 intergenic regions. Our ChIP experiment demonstrated that in B. melitensis, VjbR was also able to bind these regions in vivo.
Figure 4.
ChIP experiments showing direct binding of VjbR on several promoter regions. (A) Detection of several virB1-virB2 regions. (B) Detection of BMEII0734, BMEII0590, BMEI0030, BMEI1305, BMEI0668 and BMEI1007 promoter regions. All ChIP were performed with a C-terminal Flag-tagged VjbR-HTH protein expressed from a high copy plasmid (pSB502). The y-axis represents the ratio of immunoprecipited product (IP) versus input (IN) (%IP/IN). White columns represent IP from control strain (ΔvjbR, empty plasmid), gray columns represent IP from (ΔvjbR, pSB502). Error bars represent standard deviation from three independent experiments.
In an attempt to find the DNA motif recognized by VjbR, we analyzed the upstream regions of genes directly bound by VjbR using the RSAT web resource(60) and the MEME motif discovery tool,(61) without success.
Among the 144 genes predicted to be under the control of a consensus virB promoter box in B. abortus,(36) we found only 10 of their B. melitensis homologues in our screens (Table 2). However, the consensus virB promoter VjbR box defined in the study of de Jong(36) was found in only 3 promoter sequences of the 8 VjbR targets found in our ChIP analysis (BMEI1007, BMEII0025 and BMEII0026). This observation suggests that the VjbR binding site is not well-conserved or is not present in all promoters that are direct targets of VjbR.
Conclusions
Our study is the first report of the impact of QS at the genome scale in an intracellular pathogen. Brucella QS was initially discovered through its impact on virulence both in cellular and mouse models,(19) and the present study confirms that QS regulates numerous genes previously identified as being essential for the full virulence of Brucella (Supplementary Table 2, Supporting Information). Nevertheless, the main conclusion of this paper is that QS should not be considered anymore only as a virulence regulatory system, but should also be viewed as a major global coordinator of crucial cellular and metabolic processes related to the adaptation of Brucella to its intracellular niche.
Indeed, the proteomic and transcriptomic analyses of the B. melitensis QS mutants showed that genes whose products are predicted to perform the following function are regulated by QS: (i) response to oxidative stress (sodC, hfq...), (ii) general stress response and protein folding (groES, groEL...), (iii) respiration under aerobic conditions (coxA, coxB, coxC), (iv) response to varied nutrients availability (sugar and amino acid transporters...), (v) enzymes of the glycolytic and TCA pathway and (vi) numerous ribosomal proteins. These observations are in agreement with previous claims that Brucella strains meet nutritionally poor and microaerobic environments during their infectious cycle50,62 and that they engage an adaptive response by quantitative reduction of cellular processes participating in energy, protein, and nucleic acid metabolism.(63)
We propose that VjbR is required early in host cell infection not only to activate the genes encoding the T4SS (necessary to reach the permissive replicative-compartment)(35) but also for the early adaptation of B. melitensis 16 M to the stressful conditions encountered in the vacuole and in the slowdown of this strain’s basic metabolism. This would prevent multiplication until the replicative compartment is reached. This proposal is well supported by the recent kinetic analysis of the B. abortus proteome during macrophage infection.(17) After an initial shut down of the intracellular Brucellae’s basic cellular processes in the early steps of macrophage infection, a majority of these proteins return to their initial level later during the infection. We propose that BabR could be a player in this latter step since it acts in an opposite way compared to VjbR on several key QS targets including the genes encoding the VirB proteins, GroESL and key central metabolic enzymes.
Particularly striking also is the parallelism that can be drawn between the targets identified as part of the QS regulon in this study and the direct or indirect targets of another major regulator of Brucella virulence: the two component system (TCS) BvrS/BvrR. The results presented here, along with those from a previous study(28) strongly support the involvement of VjbR in the control of envelope properties in Brucella strains, and this appears also to be the case for BvrS/BvrR.64−66 More importantly, a recent proteomic analysis of outer membrane fragments released by B. abortus bvrR/bvrS mutants(67) pointed out an important increase of periplasmic proteins, ABC transporters and chaperones in these mutants compared to the parent strain. The expression of genes encoding products belonging to these same functional categories are also clearly increased in our vjbR mutant (see Table 2A, E). In both the bvrS/R mutants and in the vjbR mutant, these kinds of changes seem to mimic nutrient starvation. Consequently, BvrS/BvrR was suggested to be directly or indirectly involved in adjusting the metabolism of Brucella(67) and, considering the impact of the QS system on central metabolism (see Table 2B), a similar proposition can be clearly put forth for this latter system. However, neither the analysis of Lamontagne(67) nor our analyses have demonstrated a link between the BvrS/BvrR system and VjbR. Nevertheless, these analyses have been performed under very dissimilar conditions and we cannot exclude the possibility that these two regulatory pathways could be connected (directly or indirectly through other global starvation sensing mechanisms like the stringent response(68) and/or the PTS system(18)). Altogether, these systems should contribute to the adaptation of the metabolic network during the nutrient shift faced by Brucella all along its intracellular trafficking.
In summary, our results demonstrate that B. melitensis 16 M possesses a nonclassical QS regulatory system since: (i) despite the lack of a classical AHL-synthase in this pathogen, QS regulates a large fraction of its genome under the conditions tested, (ii) BabR can behave as a modulator of VjbR activity, (iii) C12−HSL have an effect both on BabR and VjbR, and (iv) QS is involved in the intracellular survival of B. melitensis through VjbR.
The use of a QS system in the individual vacuole surrounding the Brucellae in the host cell represents a good example of “efficiency sensing”, in agreement with the definition of Hense and co-workers,(25) since the diffusion of AHLs in these compartments should be delayed compared to the environments encountered by these bacteria before their entry into host cells. This proposal and the biosynthetic pathway responsible of the production of low amounts of C12−HSL(32) should be further investigated to get further insights on QS in Brucella strains.
Abbrevations:
3PG, 3-P-glycerate; AA, amino acid; AcoA, acetyl coenzyme A; AHL, acyl-homoserine lactone; BCV, Brucella containing vacuole; CDS, coding sequence; CFU, colony forming unit; ChIP, chromatin immunoprecipitation; DNA, DNA; EPS, exopolysaccharide; ER, endoplasmic reticulum; F6P, fructose-6-P; FC, fold change; Ga3P, glyceraldehyde-3-P; G1P, glycerol-1-P; GLU, glutamate; Gnt, gentamycin; HTH, helix-turn-helix; ISO, isocitrate; LPS, lipopolysaccharide; MacP, malonyl acyl carrier protein; McoA, malonyl coenzyme A; Nal, nalidixic acid; OMP, outer membrane protein; OXA, oxaloacetate; Pentose-P, pentose phosphate pathway; PYR, pyruvate; QS, Quorum Sensing; RNA, ribonucleic acid; SUC, succinate; T4SS, type four secretion system; TCS, two component system; TCA, tricarboxylic acid cycle; wt, wild type.
Acknowledgments
We thank past and present members of the Brucella team of the URBM for fruitful discussions and R. Martin Roop, II for critical reading of the manuscript and for improving the English. This work was supported by the Commission of the European Communities (contract no. QLK2-CT-1999-00014), the FRFC (Fonds de la Recherche Fondamentale Collective, conventions 2.4521.04 and 2.4521.08), and the ARC fellowship (Actions de Recherches Concertées, conventions 04/09-325 and 08/13-015). S.U. and J.L. hold a specialization grant from the Fonds pour la Formation à la Recherche dans l’Industrie et l’Agriculture (FRIA).
Supporting Information Available
Supplementary Tables 1−3. This material is available free of charge via the Internet at http://pubs.acs.org.
Supplementary Material
References
- Pappas G.; Papadimitriou P.; Akritidis N.; Christou L.; Tsianos E. V. The new global map of human brucellosis. Lancet Infect. Dis. 2006, 6 (2), 91–9. [DOI] [PubMed] [Google Scholar]
- Smith L. D.; Ficht T. A. Pathogenesis of Brucella. Crit. Rev. Microbiol. 1990, 17 (3), 209–30. [DOI] [PubMed] [Google Scholar]
- Corbel M. J. Brucellosis: an overview. Emerg Infect. Dis. 1997, 3 (2), 213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center for Disease Control and Prevention: Select agent program. http://www.cdc.gov/od/sap.
- Celli J. Surviving inside a macrophage: the many ways of. Res. Microbiol. 2006, 157 (2), 93–8. [DOI] [PubMed] [Google Scholar]
- Detilleux P. G.; Deyoe B. L.; Cheville N. F. Penetration and intracellular growth of Brucella abortus in nonphagocytic cells in vitro. Infect. Immun. 1990, 58 (7), 2320–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficht T. A. Intracellular survival of Brucella: defining the link with persistence. Vet. Microbiol. 2003, 92 (3), 213–23. [DOI] [PubMed] [Google Scholar]
- Pizarro-Cerda J.; Meresse S.; Parton R. G.; van der Goot G.; Sola-Landa A.; Lopez-Goni I.; Moreno E.; Gorvel J. P. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect. Immun. 1998, 66 (12), 5711–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barquero-Calvo E.; Chaves-Olarte E.; Weiss D. S.; Guzman-Verri C.; Chacon-Diaz C.; Rucavado A.; Moriyon I.; Moreno E. Brucella abortus uses a stealthy strategy to avoid activation of the innate immune system during the onset of infection. PLoS ONE 2007, 2, e631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.; Ng T. W.; Wehrly T. D.; Knodler L. A.; Celli J. Brucella intracellular replication requires trafficking through the late endosomal/lysosomal compartment. Traffic 2008, 9 (5), 678–94. [DOI] [PubMed] [Google Scholar]
- Moreno E.; Moriyon I.. The genus Brucella. In The prokaryotes. Electronic version.; Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E., Eds.; Springer: New York, 2001. [Google Scholar]
- Kohler S.; Foulongne V.; Ouahrani-Bettache S.; Bourg G.; Teyssier J.; Ramuz M.; Liautard J. P. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc. Natl. Acad. Sci. U.S.A. 2002, 99 (24), 15711–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roop R. M. 2nd; Bellaire B. H.; Valderas M. W.; Cardelli J. A. Adaptation of the Brucellae to their intracellular niche. Mol. Microbiol. 2004, 52 (3), 621–30. [DOI] [PubMed] [Google Scholar]
- Porte F.; Liautard J. P.; Kohler S. Early acidification of phagosomes containing Brucella suis is essential for intracellular survival in murine macrophages. Infect. Immun. 1999, 67 (8), 4041–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler S.; Michaux-Charachon S.; Porte F.; Ramuz M.; Liautard J. P. What is the nature of the replicative niche of a stealthy bug named Brucella. Trends Microbiol 2003, 11 (5), 215–9. [DOI] [PubMed] [Google Scholar]
- Kohler S.; Porte F.; Jubier-Maurin V.; Ouahrani-Bettache S.; Teyssier J.; Liautard J. P. The intramacrophagic environment of Brucella suis and bacterial response. Vet. Microbiol. 2002, 90 (1−4), 299–309. [DOI] [PubMed] [Google Scholar]
- Lamontagne J.; Forest A.; Marazzo E.; Denis F.; Butler H.; Michaud J. F.; Boucher L.; Pedro I.; Villeneuve A.; Sitnikov D.; Trudel K.; Nassif N.; Boudjelti D.; Tomaki F.; Chaves-Olarte E.; Guzman-Verri C.; Brunet S.; Cote-Martin A.; Hunter J.; Moreno E.; Paramithiotis E. Intracellular Adaptation of Brucella abortus. J. Proteome Res. 2009, 8 (3), 1594–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letesson J. J.a. D. B., X., Brucella - Molecular and Cellular Biology; Horizon Bioscience: Norfolk, 2004; pp 117−58. [Google Scholar]
- Delrue R. M.; Deschamps C.; Leonard S.; Nijskens C.; Danese I.; Schaus J. M.; Bonnot S.; Ferooz J.; Tibor A.; De Bolle X.; Letesson J. J. A quorum-sensing regulator controls expression of both the type IV secretion system and the flagellar apparatus of Brucella melitensis. Cell Microbiol. 2005, 7 (8), 1151–61. [DOI] [PubMed] [Google Scholar]
- Sjoblom S.; Brader G.; Koch G.; Palva E. T. Cooperation of two distinct ExpR regulators controls quorum sensing specificity and virulence in the plant pathogen Erwinia carotovora. Mol. Microbiol. 2006, 60 (6), 1474–89. [DOI] [PubMed] [Google Scholar]
- Smith R. S.; Iglewski B. H. P. aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 2003, 6 (1), 56–60. [DOI] [PubMed] [Google Scholar]
- Waters C. M.; Bassler B. L. Quorum Sensing: Cell-to-Cell Communication in Bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–46. [DOI] [PubMed] [Google Scholar]
- Hastings J. W.; Nealson K. H. Bacterial bioluminescence. Annu. Rev. Microbiol. 1977, 31, 549–95. [DOI] [PubMed] [Google Scholar]
- Fuqua W. C.; Winans S. C.; Greenberg E. P. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 1994, 176 (2), 269–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hense B. A.; Kuttler C.; Muller J.; Rothballer M.; Hartmann A.; Kreft J. U. Does efficiency sensing unify diffusion and quorum sensing. Nat. Rev. Microbiol. 2007, 5 (3), 230–9. [DOI] [PubMed] [Google Scholar]
- Redfield R. J. Is quorum sensing a side effect of diffusion sensing. Trends Microbiol. 2002, 10 (8), 365–70. [DOI] [PubMed] [Google Scholar]
- DelVecchio V. G.; Kapatral V.; Redkar R. J.; Patra G.; Mujer C.; Los T.; Ivanova N.; Anderson I.; Bhattacharyya A.; Lykidis A.; Reznik G.; Jablonski L.; Larsen N.; D’Souza M.; Bernal A.; Mazur M.; Goltsman E.; Selkov E.; Elzer P. H.; Hagius S.; O’Callaghan D.; Letesson J. J.; Haselkorn R.; Kyrpides N.; Overbeek R. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. U.S.A. 2002, 99 (1), 443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzureau S.; Godefroid M.; Deschamps C.; Lemaire J.; De Bolle X.; Letesson J. J. Mutations of the Quorum Sensing-dependent regulator VjbR lead to drastic surface modifications in Brucella melitensis. J. Bacteriol. 2007, 189, 6035–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letesson J. J.; Lestrate P.; Delrue R. M.; Danese I.; Bellefontaine F.; Fretin D.; Taminiau B.; Tibor A.; Dricot A.; Deschamps C.; Haine V.; Leonard S.; Laurent T.; Mertens P.; Vandenhaute J.; De Bolle X. Fun stories about Brucella: the “furtive nasty bug”. Vet. Microbiol. 2002, 90 (1−4), 317–28. [DOI] [PubMed] [Google Scholar]
- Rambow-Larsen A. A.; Rajashekara G.; Petersen E.; Splitter G. Putative quorum-sensing regulator BlxR of Brucella melitensis regulates virulence factors including the type IV secretion system and flagella. J. Bacteriol. 2008, 190 (9), 3274–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taminiau B., Etude du Quorum Sensing chez Brucella melitensis 16 M. Thesis, ISBN: 2-87037-399-6, 2003.
- Taminiau B.; Daykin M.; Swift S.; Boschiroli M. L.; Tibor A.; Lestrate P.; De Bolle X.; O’Callaghan D.; Williams P.; Letesson J. J. Identification of a quorum-sensing signal molecule in the facultative intracellular pathogen Brucella melitensis. Infect. Immun. 2002, 70 (6), 3004–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delrue R., Contribution à l’analyse des mécanismes moléculaires impliqués dans le traffic intracellulaire de Brucella melitensis 16 M. Thesis, ISBN: 2−87037−372−4 , 2002.
- Fretin D.; Fauconnier A.; Kohler S.; Halling S.; Leonard S.; Nijskens C.; Ferooz J.; Lestrate P.; Delrue R. M.; Danese I.; Vandenhaute J.; Tibor A.; DeBolle X.; Letesson J. J. The sheathed flagellum of Brucella melitensis is involved in persistence in a murine model of infection. Cell Microbiol. 2005, 7 (5), 687–98. [DOI] [PubMed] [Google Scholar]
- Comerci D. J.; Martinez-Lorenzo M. J.; Sieira R.; Gorvel J. P.; Ugalde R. A. Essential role of the VirB machinery in the maturation of the Brucella abortus-containing vacuole. Cell Microbiol. 2001, 3 (3), 159–68. [DOI] [PubMed] [Google Scholar]
- de Jong M. F.; Sun Y. H.; den Hartigh A. B.; van Dijl J. M.; Tsolis R. M. Identification of VceA and VceC, two members of the VjbR regulon that are translocated into macrophages by the Brucella type IV secretion system. Mol. Microbiol. 2008, 70 (6), 1378–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach M. E.; Elzer P. H.; Hill D. S.; Robertson G. T.; Farris M. A.; Roop R. M.; Peterson K. M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995, 166 (1), 175–6. [DOI] [PubMed] [Google Scholar]
- Dricot A.; Rual J. F.; Lamesch P.; Bertin N.; Dupuy D.; Hao T.; Lambert C.; Hallez R.; Delroisse J. M.; Vandenhaute J.; Lopez-Goni I.; Moriyon I.; Garcia-Lobo J. M.; Sangari F. J.; Macmillan A. P.; Cutler S. J.; Whatmore A. M.; Bozak S.; Sequerra R.; Doucette-Stamm L.; Vidal M.; Hill D. E.; Letesson J. J.; De Bolle X. Generation of the Brucella melitensis ORFeome version 1.1. Genome Res. 2004, 14 (10B), 2201–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F. D.; H B.; Pierre M.; Gaigneaux A.; Depiereux E The “Window t-test”: a simple and powerful approach to detect differentially expressed genes in microarray datasets. Cent. Eur. J. Biol. 2008, 3 (3), 327–344. [Google Scholar]
- Irizarry R. A.; Hobbs B.; Collin F.; Beazer-Barclay Y. D.; Antonellis K. J.; Scherf U.; Speed T. P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003, 4 (2), 249–64. [DOI] [PubMed] [Google Scholar]
- Lamanda A.; Zahn A.; Roder D.; Langen H. Improved Ruthenium II tris (bathophenantroline disulfonate) staining and destaining protocol for a better signal-to-background ratio and improved baseline resolution. Proteomics 2004, 4 (3), 599–608. [DOI] [PubMed] [Google Scholar]
- Shevchenko A.; Wilm M.; Vorm O.; Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996, 68 (5), 850–8. [DOI] [PubMed] [Google Scholar]
- Kuras L.; Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 1999, 399 (6736), 609–13. [DOI] [PubMed] [Google Scholar]
- DeLisa M. P.; Wu C. F.; Wang L.; Valdes J. J.; Bentley W. E. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 2001, 183 (18), 5239–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M.; Lostroh C. P.; Ogi T.; Greenberg E. P. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 2003, 185 (7), 2066–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner V. E.; Bushnell D.; Passador L.; Brooks A. I.; Iglewski B. H. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 2003, 185 (7), 2080–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner V. E.; Gillis R. J.; Iglewski B. H. Transcriptome analysis of quorum-sensing regulation and virulence factor expression in Pseudomonas aeruginosa. Vaccine 2004, 22 (Suppl 1), S15–20. [DOI] [PubMed] [Google Scholar]
- Delpino M. V.; Marchesini M. I.; Estein S. M.; Comerci D. J.; Cassataro J.; Fossati C. A.; Baldi P. C. A bile salt hydrolase of Brucella abortus contributes to the establishment of a successful infection through the oral route in mice. Infect. Immun. 2007, 75 (1), 299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpino M. V.; Comerci D. J.; Wagner M. A.; Eschenbrenner M.; Mujer C. V.; Ugalde R. A.; Fossati C. A.; Baldi P. C.; Delvecchio V. G. Differential composition of culture supernatants from wild-type Brucella abortus and its isogenic virB mutants. Arch. Microbiol. 2009, 191 (7), 571–81. [DOI] [PubMed] [Google Scholar]
- Kohler S.; Ekaza E.; Paquet J. Y.; Walravens K.; Teyssier J.; Godfroid J.; Liautard J. P. Induction of dnaK through its native heat shock promoter is necessary for intramacrophagic replication of Brucella suis. Infect. Immun. 2002, 70 (3), 1631–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelbe O. K.; Sedgley C. M. Effects of prolonged exposure to alkaline pH on Enterococcus faecalis survival and specific gene transcripts. Oral Microbiol. Immunol. 2007, 22 (3), 169–74. [DOI] [PubMed] [Google Scholar]
- Hallez R.; Mignolet J.; Van Mullem V.; Wery M.; Vandenhaute J.; Letesson J. J.; Jacobs-Wagner C.; De Bolle X. The asymmetric distribution of the essential histidine kinase PdhS indicates a differentiation event in Brucella abortus. Embo J. 2007, 26 (5), 1444–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G. T.; Roop R. M. Jr. The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol. Microbiol. 1999, 34 (4), 690–700. [DOI] [PubMed] [Google Scholar]
- Halling S. M.; Bricker B. J. Characterization and occurrence of two repeated palindromic DNA elements of Brucella spp.: Bru-RS1 and Bru-RS2. Mol. Microbiol. 1994, 14 (4), 681–9. [DOI] [PubMed] [Google Scholar]
- Hammer B. K.; Bassler B. L. Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 2007, 104 (27), 11145–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P.; Ross H. F.; Projan S. J.; Kornblum J.; Kreiswirth B.; Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. Embo J. 1993, 12 (10), 3967–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano-Sagie M.; Xavier K. B. The role of small RNAs in quorum sensing. Curr. Opin. Microbiol. 2007, 10 (2), 189–98. [DOI] [PubMed] [Google Scholar]
- Toledo-Arana A.; Repoila F.; Cossart P. Small noncoding RNAs controlling pathogenesis. Curr. Opin. Microbiol. 2007, 10 (2), 182–8. [DOI] [PubMed] [Google Scholar]
- Sieira R.; Comerci D. J.; Pietrasanta L. I.; Ugalde R. A. Integration host factor is involved in transcriptional regulation of the Brucella abortus virB operon. Mol. Microbiol. 2004, 54 (3), 808–22. [DOI] [PubMed] [Google Scholar]
- van Helden J. Regulatory sequence analysis tools. Nucleic Acids Res. 2003, 31 (13), 3593–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L.; Williams N.; Misleh C.; Li W. W. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34 (Web Server issue), W369–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Dahouk S.; Loisel-Meyer S.; Scholz H. C.; Tomaso H.; Kersten M.; Harder A.; Neubauer H.; Kohler S.; Jubier-Maurin V. Proteomic analysis of Brucella suis under oxygen deficiency reveals flexibility in adaptive expression of various pathways. Proteomics 2009, 9 (11), 3011–21. [DOI] [PubMed] [Google Scholar]
- Al Dahouk S.; Jubier-Maurin V.; Scholz H. C.; Tomaso H.; Karges W.; Neubauer H.; Kohler S. Quantitative analysis of the intramacrophagic Brucella suis proteome reveals metabolic adaptation to late stage of cellular infection. Proteomics 2008, 8 (18), 3862–70. [DOI] [PubMed] [Google Scholar]
- Guzman-Verri C.; Manterola L.; Sola-Landa A.; Parra A.; Cloeckaert A.; Garin J.; Gorvel J. P.; Moriyon I.; Moreno E.; Lopez-Goni I. The two-component system BvrR/BvrS essential for Brucella abortus virulence regulates the expression of outer membrane proteins with counterparts in members of the Rhizobiaceae. Proc. Natl. Acad. Sci. U.S.A. 2002, 99 (19), 12375–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manterola L.; Moriyon I.; Moreno E.; Sola-Landa A.; Weiss D. S.; Koch M. H.; Howe J.; Brandenburg K.; Lopez-Goni I. The lipopolysaccharide of Brucella abortus BvrS/BvrR mutants contains lipid A modifications and has higher affinity for bactericidal cationic peptides. J. Bacteriol. 2005, 187 (16), 5631–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola-Landa A.; Pizarro-Cerda J.; Grillo M. J.; Moreno E.; Moriyon I.; Blasco J. M.; Gorvel J. P.; Lopez-Goni I. A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol. Microbiol. 1998, 29 (1), 125–38. [DOI] [PubMed] [Google Scholar]
- Lamontagne J.; Butler H.; Chaves-Olarte E.; Hunter J.; Schirm M.; Paquet C.; Tian M.; Kearney P.; Hamaidi L.; Chelsky D.; Moriyon I.; Moreno E.; Paramithiotis E. Extensive cell envelope modulation is associated with virulence in Brucella abortus. J. Proteome Res. 2007, 6 (4), 1519–29. [DOI] [PubMed] [Google Scholar]
- Dozot M.; Boigegrain R. A.; Delrue R. M.; Hallez R.; Ouahrani-Bettache S.; Danese I.; Letesson J. J.; De Bolle X.; Kohler S. The stringent response mediator Rsh is required for Brucella melitensis and Brucella suis virulence, and for expression of the type IV secretion system virB. Cell Microbiol. 20068, (11), 1791–802. [DOI] [PubMed] [Google Scholar]
- Kohler S.; Ouahrani-Bettache S.; Layssac M.; Teyssier J.; Liautard J. P. Constitutive and inducible expression of green fluorescent protein in Brucella suis. Infect. Immun. 1999, 67 (12), 6695–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulongne V.; Bourg G.; Cazevieille C.; Michaux-Charachon S.; O’Callaghan D. Identification of Brucella suis genes affecting intracellular survival in an in vitro human macrophage infection model by signature-tagged transposon mutagenesis. Infect. Immun. 2000, 68 (3), 1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Watarai M.; Kondo Y.; Erdenebaatar J.; Makino S.; Shirahata T. Isolation and characterization of mini-Tn5Km2 insertion mutants of Brucella abortus deficient in internalization and intracellular growth in HeLa cells. Infect. Immun. 2003, 71 (6), 3020–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong P. C.; Tsolis R. M.; Ficht T. A. Identification of genes required for chronic persistence of Brucella abortus in mice. Infect. Immun. 2000, 68 (7), 4102–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen C. A.; Adams L. G.; Ficht T. A. Transposon-derived Brucella abortus rough mutants are attenuated and exhibit reduced intracellular survival. Infect. Immun. 1998, 66 (3), 1008–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler S.; Teyssier J.; Cloeckaert A.; Rouot B.; Liautard J. P. Participation of the molecular chaperone DnaK in intracellular growth of Brucella suis within U937-derived phagocytes. Mol. Microbiol. 1996, 20 (4), 701–12. [DOI] [PubMed] [Google Scholar]
- Tibor A.; Wansard V.; Bielartz V.; Delrue R. M.; Danese I.; Michel P.; Walravens K.; Godfroid J.; Letesson J. J. Effect of omp10 or omp19 deletion on Brucella abortus outer membrane properties and virulence in mice. Infect. Immun. 2002, 70 (10), 5540–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delrue R. M.; Martinez-Lorenzo M.; Lestrate P.; Danese I.; Bielarz V.; Mertens P.; De Bolle X.; Tibor A.; Gorvel J. P.; Letesson J. J. Identification of Brucella spp. genes involved in intracellular trafficking. Cell Microbiol. 2001, 3 (7), 487–97. [DOI] [PubMed] [Google Scholar]
- O’Callaghan D.; Cazevieille C.; Allardet-Servent A.; Boschiroli M. L.; Bourg G.; Foulongne V.; Frutos P.; Kulakov Y.; Ramuz M. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 1999, 33 (6), 1210–20. [DOI] [PubMed] [Google Scholar]
- Sieira R.; Comerci D. J.; Sanchez D. O.; Ugalde R. A. A homologue of an operon required for DNA transfer in Agrobacterium is required in Brucella abortus for virulence and intracellular multiplication. J. Bacteriol. 2000, 182 (17), 4849–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latimer E.; Simmers J.; Sriranganathan N.; Roop R. M. 2nd; Schurig G. G.; Boyle S. M. Brucella abortus deficient in copper/zinc superoxide dismutase is virulent in BALB/c mice. Microb. Pathog. 1992, 12 (2), 105–13. [DOI] [PubMed] [Google Scholar]
- Lestrate P.; Dricot A.; Delrue R. M.; Lambert C.; Martinelli V.; De Bolle X.; Letesson J. J.; Tibor A. Attenuated signature-tagged mutagenesis mutants of Brucella melitensis identified during the acute phase of infection in mice. Infect. Immun. 2003, 71 (12), 7053–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.