Abstract
Multivalent dendrimeric conjugates of GPCR ligands may have increased potency or selectivity in comparison to monomeric ligands, a phenomenon that was tested in a model of cytoprotection in mouse HL-1 cardiomyocytes. Quantitative RT-PCR indicated high expression levels of endogenous A1 and A2A adenosine receptors (ARs), but not of A2B and A3ARs. Activation of the heterologously expressed human A3AR in HL-1 cells by AR agonists significantly attenuated cell damage following 4 h exposure to H2O2 (750 μM) but not in untransfected cells. The A3 agonist IB-MECA (EC50 3.8 μM) and the non-selective agonist NECA (EC50 3.9 μM) protected A3 AR-transfected cells against H2O2 in a concentration-dependent manner, as determined by lactate dehydrogenase release. A generation 5.5 PAMAM (polyamidoamine) dendrimeric conjugate of a N6-chain-functionalized adenosine agonist was synthesized and its mass indicated an average of 60 amide-linked nucleoside moieties out of 256 theoretical attachment sites. It nonselectively activated the A3AR to inhibit forskolin-stimulated cAMP formation (IC50 66 nM) and, similarly, protected A3–transfected HL-1 cells from apoptosis-inducing H2O2 with greater potency (IC50 35 nM) than monomeric nucleosides. Thus, a PAMAM conjugate retained AR binding affinity and displayed greatly enhanced cardioprotective potency.
Keywords: cardioprotection, nucleoside, G protein-coupled receptor, dendrimers, polymeric drugs, HL-1 cells
Introduction
Four subtypes of adenosine receptors (AR), which belong to the rhodopsin family of G protein-coupled receptors (GPCR), are activated by the endogenous ligand adenosine as well as by the non-selective agonist 5′-N-ethylcarboxamidoadenosine (NECA 1) [1]. There is increasing interest in the therapeutic potential of selective adenosine agonists for treating a wide range of diseases. For instance, the A3AR is known to be overexpressed in peripheral blood mononuclear cells and synoviocytes of rheumatoid arthritis patients [2], and receptor activation is known to reduce lung injury following reperfusion in cats [3]. Numerous AR ligands are already in or heading toward clinical trials as drug candidates. For example, selective agonists of the A3AR, N6-(3-iodobenzyl)-5′-N-methylcarboxamidoadenosine (IB-MECA 2) and 2-chloro-N6-(3-iodobenzyl)-5′-N-methylcarboxamidoadenosine (Cl-IB-MECA 3), are in trials for autoimmune inflammatory diseases [4,5] and hepatocarcinoma [6,7], respectively.
A3AR agonists have also displayed protective effects in ischemic models of the brain [8], nervous system [9], and skeletal muscle [10]. Activation of this receptor preconditions cardiomyocytes in culture [11,12], isolated hearts [13], and rabbit hearts in vivo to protect against the damaging effects of ischemia/hypoxia [14]. However, there is still uncertainty over the role of the A3AR in cardioprotection. While the expression of A1 and A2AARs in human (h) adult cardiomyocytes is known, direct evidence proving that the A2B and A3ARs are also expressed in these cells is lacking. In addition, the A1 and A2AARs are implicated in cardioprotection, although the signaling pathways have not yet been determined [15].
While activation of ARs has pronounced cardioprotective properties, more work is needed to further elucidate the effects of small molecular and multivalent agonists for the A3AR in models of cardioprotection. An immortalized atrial cardiomyocyte murine cell line, HL-1, is known to express all four ARs, although the levels of expression have not been reported [16]. These cells are able to continuously divide while maintaining a differentiated cardiac phenotype characterized by spontaneous action potentials and contractions [17]. HL-1 cells have been used to study pathophysiological conditions such as hypoxia and hyperglycemia [18], and can be transiently transfected using both transfection agents [19] and viral vectors [20]. A1 and A3ARs are involved in preconditioning HL-1 cells against damage from hypoxia and ischemic reperfusion [21,22]. Although cardioprotection induced by A3AR agonists has been well explored in a variety of systems and species, we adapted here the mouse HL-1 cell culture model for use as a model system in which the expression of AR subtypes could be measured and manipulated.
The structure activity relationship (SAR) of nucleoside derivatives as agonists at the ARs has been extensively studied [1]. Recently this analysis has been extended to multivalent nucleoside conjugates of polyamidoamine (PAMAM) dendrimers [23-27], which we are terming GPCR Ligand-Dendrimer (GLiDe) conjugates. PAMAM dendrimers are peptide-like in structure and as such generally biocompatible. Assuming proper functionalization of GPCR ligands for covalent conjugation [24], such multivalent dendrimeric conjugates of these ligands have displayed dramatically increased potency or selectivity in comparison to the monomeric, small molecular ligands. A multivalent agonist of the A2AAR effectively inhibited ADP-induced platelet aggregation [25]. A PAMAM dendrimeric conjugate (generation 2.5) 5 of a non-subtype selective adenosine agonist 4 (Figure 1A), which was chain-functionalized at the N6 position, displayed enhanced selectivity for the A3AR in both binding and functional assays [26]. Multivalent conjugates of the P2Y14 receptor agonist UDP-glucuronic acid activated that nucleotide receptor with up to 800-fold enhanced potency in comparison to the corresponding monomeric ligands [27].
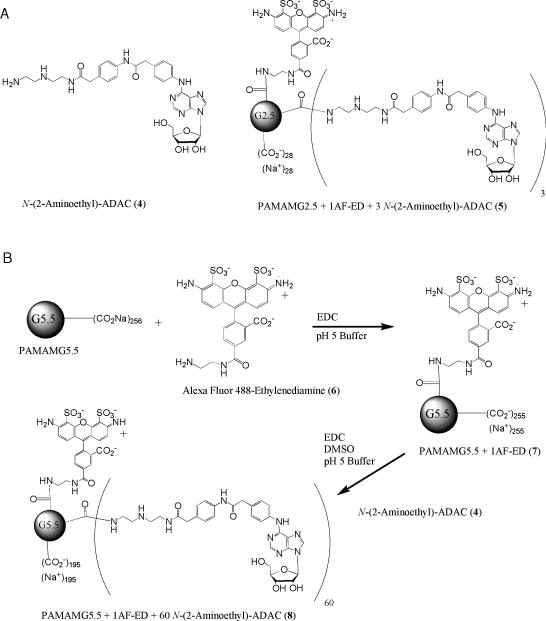
Figure 1.
(A) Structures of a non-selective amine-functionalized AR agonist (4) and an A3 selective dendrimeric conjugate (5) as reported [26]. (B) Synthesis of 8, a G5.5 PAMAM dendrimer with 1 AF488-ED and 60 N-(2-aminoethyl)-ADAC moieties. AF488-ED and N-(2-aminoethyl)-ADAC were conjugated to G5.5 PAMAM dendrimers using carbodiimide coupling.
The theoretical ability of such dendrimeric conjugates to bridge multiple protomers in a homodimeric AR structure was shown [28]. It is hoped that this design approach can be used to prepare pharmacological probes to act as selective agonists and antagonists at homomultimeric and heteromultimeric GPCRs.
The aim of our study was to investigate the cardioprotective effects of a novel, newly-synthesized high molecular weight (>88,000 D), multivalent AR agonist acting at the A3AR [18]. This agonist is similar to the A3AR-selective agonist 5 structurally and in the presence of terminal carboxylate groups (Figure 1A), but is derived from a higher generation (G5.5) PAMAM dendrimer and is more highly conjugated with a strategically functionalized adenosine derivative. In this study, cell death was induced using hydrogen peroxide (H2O2) in HL-1 cultured cardiomyocytes, in which the expression of the A3AR was controlled heterologously. A distinct protective effect of A3AR activation by either known monomeric A3AR agonists or a multivalent AR agonist was observed.
2. Materials and Methods
2.1. Materials
HL-1 mouse cardiomyocytes were a kind gift of Professor W.C. Claycomb, LSU Health Sciences Center, New Orleans, LA, USA [22]. Dulbecco's modified Eagle's medium (DMEM) and fetal bovine serum (FBS) were purchased from Life Technologies (Rockville, MD). Plastic cellware was purchased from Becton Dickinson (Bedford, MA). 2-Chloro-N6-cyclopentyladenosine (CCPA), and 2-[p-(2-carboxyethyl)phenylethyl-amino]- 5′-N-ethylcarboxamidoadenosine (CGS21680), and 3-iodobenzyl-5'-N-methylcarboxamidoadenosine (IB-MECA) were obtained from Tocris (Ellisville, MO). ADAC (N6-[4-[[[4-[[[(2-aminoethyl)amino]carbonyl]methyl]-anilino]carbonyl]methyl]phenyl]adenosine), PAMAM dendrimers (ethylenediamine core, generation 5.5 as 10 wt. % solution in methanol), N-(3-dimethylaminopropyl)-N'-ethylcarbodiimide (EDC), 2-(N-morpholino)ethanesulfonic acid (MES), magnesium chloride, methanol, triethylamine, methyl sulfoxide-d6 (DMSO-d6), N,N-dimethylformamide (DMF), Claycomb Media, fibronectin, ascorbic acid, norepinephrine, H2O2, Triton-X, rolipram, and gelatin were purchased from Sigma (St. Louis, MO). Bio-Beads® SX-1 beads were purchased from Bio-Rad (Hercules, CA). Alexa-Fluor® 488 carboxylic acid, 2,3,5,6-tetrafluorophenyl ester, 5-isomer (AF488-TFP) was purchased from Invitrogen (Carlsbad, CA). [125I]AB-MECA ([125I]4-amino-3-iodobenzyl-5'-N-methylcarboxamidoadenosine, 2200 Ci/mmol), [3H]CCPA (42.6 Ci/mmol), and [3H]CGS21680 (40.5 Ci/mmol) were purchased from PerkinElmer (Waltham, MA). 4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer, DMEM/F12 medium, and 1 M Tris-HCl (pH 7.5) were purchased from Mediatech, Inc. (Manassas, VA).
2.2. Chromatographic separation and spectroscopy
The column for size exclusion chromatography (SEC) was prepared by suspending 100 g of Bio-Beads® SX-1 beads in 1 L of DMF. After 24 h to allow for equilibration and expansion, the beads were added to the column as described previously [25]. High Performance Liquid Chromatography (HPLC) purification was performed using an 1100 Series HPLC (Agilent, Santa Clara, CA) equipped with a Luna 5μ C18(2) 100A analytical column (250 x 10 mm; Phenomenex, Torrance, CA). Peaks were detected by UV absorption using a diode array detector. Proton nuclear magnetic resonance spectra (NMR) were recorded on a Bruker DRX-600 spectrometer after being optimized for each sample using DMSO-d6 as a solvent unless otherwise noted. Electrospray ionization mass spectra (ESI MS) were taken using a LCT Premier mass spectrometer (Waters Corp., Waltham, MA). Matrix Assisted Laser Desorption/Ionization Time-of-Flight (MALDITOF) spectra were obtained with a Waters Micro mass spectrometer using Waters MassPREP Direct Ionization on Silica Desorption/ionization (DIOS) target plates.
The ESI MS data for the dendrimer complexes were obtained using a Waters LCT Premier TOF mass spectrometer. The mass spectrometer was operated in negative ion W mode with a resolution of 10,000 measured at half peak height. The capillary voltage was 2500 volts, the cone voltage was 40 volts, and the desolvation gas was dried nitrogen at 250 °C and a flow of 300 l/h. The sample was dissolved in a 1:1 solution of water:acetonitrile containing 0.2% formic acid and injected directly into the eluting stream flowing at 200 μl/min and consisting of 20:80 water:acetonitrile and 0.2% formic acid. The relevant spectra were summed using the MassLynx software, and the summed spectra were deconvoluted with the MaxEntI program (Waters).
2.3. Synthesis of dendrimeric derivatives PAMAMG5.5-1(AF488-ED) (7) and PAMAMG5.5-1(AF488-ED)-60(N-(2-aminoethyl)-ADAC) (8)
This procedure was adapted from a similar procedure to synthesize PAMAMG2.5-1AF488-ED [26]. 1.07 μmol of G5.5 PAMAM stock solution (0.93 mM in methanol, 56.9 mg) was added to a flask, and the methanol was evaporated. The remaining residue containing the polymer and the Alexa-Fluor 488-ethylenediamine (ED) derivative 6 (procedure in Supporting Information, 1.4 mg, 2.5 μmol) were dissolved in 1.0 ml of 0.1 M MES buffer, pH 5. EDC (18 mg, 94 μmol) dissolved in 1.0 ml of 0.1 M MES buffer, pH 5, was added, and the reaction was stirred for 60 h. After exhaustive dialysis with water, the mixture was lyophilized to give 33.1 mg of product (0.69 μmol, 65% yield) and redissolved in D2O for NMR measurements and further biological assays. The NMR spectrum was consistent with the assigned structure, but the signals resulting from AF488-ED could not be properly integrated due to the large G5.5 PAMAM peaks. Therefore, based on the MS results, it was assumed that each dendrimer 7 contained on average one moiety of 6. m/z (ESI- MS) calc: 53,435; found: 53,971.
Synthesis of PAMAMG5.5-1AF-ED-60(N-(2-aminoethyl)-ADAC) 8 30.9 mg of 7 (0.65 μmol) was dissolved in 2.0 ml of 0.1 M MES, pH 5, and placed under a nitrogen atmosphere. N-(2-Aminoethyl)-ADAC 4 (procedure in Supporting Information, 28.7 mg, 46.4 μmol) was dissolved in 3.0 ml of DMSO and was added to the solution of 7. Finally, 89 mg of EDC (146 μmol) was dissolved in 1 ml of 0.1 M MES, pH 5 and added to the mixture. After approximately 48 h, small molecule impurities were removed by filtration and extensive dialysis in water. After lyophilization, 26.8 mg (1.14 μmol, 46% yield) of product remained. The product was analyzed by MS, which indicated an average of 60 N-(2-aminoethyl)-ADAC moieties per dendrimer (of a possible 256 moieties). The NMR spectrum was consistent with the assigned structure, but the signals resulting from 7 could not be properly integrated due to the large G5.5 PAMAM peaks. m/z (ESI- MS) calc: 88,771; found: 88,595.
2.4. Cell culture and membrane preparation
HL-1 cells (murine cardiomyocytes) were grown in Claycomb Media supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, 2 μmol/ml glutamine, and 0.1 mM norepinephrine previously dissolved in 0.3 mM ascorbic acid [17,18]. All cell culture plates for the HL-1 cells were coated with a 25 μg/ml fibronectin solution prepared in a 0.02% gelatin. The murine cardiomyocyte cells were transiently transfected with the hA3AR gene expressed in the pcDNA5/FRT plasmid (Invitrogen) using GenJet (SignaGen Laboratories, Ijamsville, MD) as the transfection agent.
Chinese hamster ovary (CHO) cells stably expressing the recombinant hARs (except A2AAR, which was expressed in HEK293 cells) were cultured in Dulbecco's modified Eagle medium (DMEM) and F12 (1:1) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 μmol/ml glutamine [26,29]. After harvesting, cells were centrifuged at 500g for 10 min, and the pellet was resuspended in 50 mM Tris-HCl buffer (pH 7.5) containing 10 mM MgCl2. The suspension was homogenized and recentrifuged at 20,000g for 20 min at 4 °C. The resultant pellets were resuspended in Tris-HCl buffer and incubated with adenosine deaminase for 30 min at 37 °C. The suspension was stored at −80 °C until the binding experiments. The protein concentration was measured using the BCA Protein Assay Kit from Thermo Fisher Scientific Inc. (Waltham, MA).
2.5. Radioligand membrane binding studies
Radioligand binding assays at A1, A2A, and A3ARs were performed according to the procedures described previously [29]. Each tube in the binding assay contained 100 μL of membrane suspension (20 μg of protein), 50 μL of agonist radioligand, and 50 μL of increasing concentrations of the test ligands in Tris-HCl buffer (50 mM, pH 7.5) containing 10 mM MgCl2. The concentrations of the dendrimer-ligand complexes are measured by the concentration of the dendrimer, not the ligand. Therefore, all binding Ki values of dendrimers are expressed as Ki app (apparent inhibition constant). Nonspecific binding was determined using a final concentration of 10 μM NECA, a non-specific agonist, diluted with the buffer. The mixtures were incubated at 25 °C for 60 min. Binding reactions were terminated by filtration through Whatman GF/B filters under a reduced pressure using a MT-24 cell harvester (Brandell, Gaithersburg, MD). Filters were washed three times with 5 ml of 50 mM ice-cold Tris-HCl buffer (pH 7.5). The radioactive agonists [3H]CCPA and [3H]CGS21680 were used for the A1 and A2A assays, respectively, while [125I]AB-MECA was used for the A3 assays. All of the filters were washed 3 times with Tris-HCl, pH 7.5. Filters for A1 and A2AAR binding were placed in scintillation vials containing 5 ml of Hydrofluor scintillation buffer and counted using a PerkinElmer Tricarb 2810TR Liquid Scintillation Analyzer. Filters for A3AR binding were counted using a PerkinElmer Cobra II γ-counter. The Ki values were determined using Prism software (version 4.0, GraphPAD, San Diego, CA) for all assays.
2.6. cAMP assays
CHO cells expressing the A1, A2A, or A3AR were seeded in 24 well plates and incubated at 37 °C overnight. The following day the medium was removed and replaced with DMEM containing 50 mM HEPES, 10 μM rolipram, 3 U/ml adenosine deaminase and increasing concentrations of the test compound. After an incubation of 30 min at 37 °C, 10 μM of forskolin was added to stimulate cAMP levels in the A1 or A3 assays, and the cells were incubated at 37 °C for an additional 15 min. The A2A assay plates remained in the incubator for 45 min. Next, the medium was removed, and the cells were lysed with 200 μl of 0.1 M HCl. 100 μl of the HCl solution was used in the Sigma Direct cAMP Enzyme Immunoassay following the instructions provided with the kit. The results were calculated using an ELx808 Ultra Microplate reader (BioTek, Winooski, VT) at 405 nm and analyzed using Prism software.
2.7. Transfection of the A3AR in HL-1 cells
150 μl of GenJet and 25 μg of pcDNA5 plasmid encoding the cDNA of the hA3AR were each mixed in 750 μl of HL-1 media without serum or antibiotics. The solutions were combined together and added to 90% confluent HL-1 cells in a 75 cm2 flask. GenJet solution with no plasmid was added to a second flask of HL-1 cells as a control. After 5 h, the medium was removed and replaced with HL-1 media containing serum and antibiotics. After 24 h, the cells were trypsinized and split to 24 and 6 well plates for qRT-PCR or assays of lactate dehydrogenase (LDH) and apoptosis.
2.8. Quantitative RT-PCR of ARs in HL-1 cells
Non-transfected and hA3AR transfected HL-1 cells were grown in 6 well plates coated with fibronectin overnight. RNA from the cells was purified following the protocol of the RNeasy Kit (Qiagen Inc., Valencia, CA) with DNase I (Qiagen). Reverse transcription was completed using Superscript III First Strand Synthesis Supermix kit (Invitrogen). cDNA from ARs was quantified on a 7900HT Fast Real-Time PCR Machine (Applied Biosystems, Foster City, CA) instrument using SybrGreen PCR MasterMix (Sigma), 150 nM primers, and 50 ng/μl DNA (total volume is 20 μl). The following AR and β-actin primers were used: mA1-F: 5’-TGT GCC CGG AAA TGT ACT GG-3’, mA1-R: 5’-TCT GTG GCC CAA TGT TGA TAA-3’; mA2A-F: 5’-TGC CTC TTC TTC GCC TGC TTT-3’, mA2A-R: 5’-AAT CGC AAT GAT GCC CTT GCG C-3’; mA2B-F: 5’-GCG AGA GGG ATC ATT GCT GCT-3’, mA2B-R: 5’-CCC CCA GTT CTG TGC AGT TG-3’; mA3-F: 5’-CAC CCA TGC TTC CAT CAT GTC-3’, mA3-R: 5’-AGC CCC ACC AGA AAG GAA AC-3’; hA3-F: 5’-GGC CAA TGT TAC CTA CAT CAC C-3’, hA3-R: 5’-CCA GGG CTA GAG AGA CAA TGA A-3’. Differential expression of the nontransfected and transiently transfected cell lines was compared using the ΔΔCt method [30].
2.9. LDH assay for quantification of cytotoxicity
Transiently transfected or non-transfected HL-1 cells were added to 24 well plates coated with fibronectin and left at 37 °C overnight. Increasing concentrations of each agonist, diluted with PBS with calcium and magnesium, were incubated with the cells at 37 °C for 1 h. If an antagonist was used, it was incubated with the cells 1 h prior to the addition of the agonist. Without removing the agonist, H2O2 in PBS was added to the cells at a final concentration of 750 μM. The cells were then kept at 37 °C for 4 h. In all cases, cells that did not receive H2O2 served as a negative control for 0% cytotoxicity, and cells that received 0.02% Triton X served as a positive control for 100% cytotoxicity. 100 μl of each cell supernatant was added to a 96 well plate in triplicate, and the LDH assay (Roche Applied Sciences, Indianapolis, IN) was run following the manufacturer's instructions. The results were determined using an ELx808 Ultra Microplate reader at 490 nm and 650 nm and analyzed using Prism software.
2.10. Luminescent caspase assay for quantification of apoptosis
A3AR-transfected HL-1 cells were seeded in a 96-well opaque white bottom plate (30,000 cells/well) and incubated overnight at 37 °C. At the end of the incubation period, the media was replaced with DMEM with calcium and magnesium, which was used for the remainder of the experiment. The dendrimer compounds (10 μM) were added to the cells one hour prior to the addition of H2O2 and were left in the cell medium until the end of the experiment. Apoptosis was induced in the cells by the addition of 400 μM of H2O2, and cells were incubated for 3 h at 37 °C. The apoptosis induced by H2O2 was determined with the Caspase – Glo 3/7 assay kit (Promega Corporation, Madison, WI) which is based on the cleavage of the luminogenic caspase substrate. Equal volume of Caspase-Glo 3/7 reagent was added to the apoptosis induced cells, and the cells were incubated at room temperature for 90 min. After 90 min, caspase activity was quantitated using a 1420 Luminescence counter (PerkinElmer).
2.11. Statistical analysis of in vitro data
Pharmacological parameters were analyzed with Prism software. Data were expressed as mean ± standard error (n = 3). Statistical significance was calculated using the Student's t-test. There were 3 degrees of freedom (df) for the LDH assay, and 4 degrees of freedom for the caspase assay. P values less than 0.05 (P<0.05) were considered to be statistically significant. For the caspase assay, statistical significance between the results was analyzed by ANOVA followed by the Tukey-Kramer multiple comparison test.
3. Results
3.1. Synthesis of a G5.5 PAMAM dendrimer-nucleoside conjugate for AR activation
In order to introduce a fluorescent moiety on the parent dendrimer, we used compound 6, an amine-functionalized derivative of Alexa Fluor 488 (AF488) that was synthesized in our previous study and which contained a terminal primary amine located on an ED moeity [26]. Alexa Fluor dyes have previously been shown to have a more stable fluorescent signal than fluorescein [31]. Thus, G5.5-PAMAM-AF488 (7) was synthesized by water-soluble carbodiimide coupling (EDC, in 0.1 M MES, pH 5) to attach compound 6 to the carboxylic-functionalized G5.5 dendrimer, as shown in Figure 1B.The unreacted EDC and urea byproduct were removed by dialysis. Next, the terminal amino group of AR agonist 4 was amide conjugated to the fluorescent-labeled G5.5 dendrimer 7 also using a carbodiimide coupling.
The dendrimer conjugates were purified using SEC and characterized using NMR and electrospray ionization (ESI) mass spectrometry (MS). The parent G5.5 dendrimer had a measured molecular weight of 53,813, which was ~1 % higher than the theoretical mass of 52,900, as shown in Figure S1 (Supporting Information). The extra molecular weight and the significant fragmenting of the peaks were probably caused by the excess sodium ions in the sample. NOESY NMR showed no significant backfolding of arms attached to the G5.5 dendrimer. The fluorescent conjugate increased in molecular weight by 158 D for a total molecular weight of 53,917, close to the theoretical weight of 53,435, as shown in Figure S2. NMR of 7 showed that AF488-ED was attached, but the peaks were too small in comparison to the parent dendrimer to be properly integrated [32].
The nucleoside conjugate 8 was also analyzed by NMR and ESI MS. After removal of the monomers by dialysis, MS showed that, on average, approximately 60 moieties of 4 were attached per dendrimer, as shown in Figure S3. The NMR analysis was noisy due to the small sample size, so the peaks could not be properly integrated. However, peaks corresponding to 4 were seen in the NMR spectrum.
3.2. Pharmacological characterization of a G5.5 PAMAM dendrimer-nucleoside conjugate in AR binding and cAMP assays
Standard radioligand binding assays were used to measure the affinity of the dendrimer conjugate at three of the subtypes of ARs [29]. The affinity of similar derivatives at the A2BAR is very low [33]; thus, this subtype was not included in the assay. In radiologand saturation studies (data not shown), CHO cells stably transfected with the hA1 or the hA3AR had Bmax values of 530 ± 210 fmol/mg protein or 253 ± 19 fmol/mg protein, respectively, showing that there is similar receptor expression in both stably transfected cell lines. HEK cells stably transfected with the hA2AAR expressed 5000 ± 350 fmol/mg protein.
The hAR binding affinity of the functionalized congener 4 was prior to attachment to the dendrimers was previously reported [26]. Compound 4 displayed Ki values at the hA1, A2A and A3ARs of 43 ± 5 nM, 300 ± 20 nM, and 9.5 ± 2.0 nM, respectively. In an assay measuring the accummulation of cAMP (Table 1), compound 4 was also a potent full agonist at the A1 and A3ARs (inhibition) and the A2AAR (stimulation).
Table 1.
Ki or Ki apparent values for binding of nucleoside monomers and dendrimer conjugates and functional effects on cAMP at hA1, A2A, and A3ARs.a
| Binding, Ki app (nM) or % inhibition at 10 μM | Effects on cAMP, EC50 (nM) | |||||
|---|---|---|---|---|---|---|
| Compound | A1 | A2 | A3 | A1 | A2 | A3 |
| N-(2-Aminoethyl)-ADAC (4) | 43 ± 5 | 300 ± 20 | 9.5 ± 2.0 | 89 ± 17 | 36 ± 13 | 35 ± 12 |
| G5.5 | 25 ± 10% | 23 ± 2% | 26 ± 9% | NAb | NAb | NAb |
| G5.5 - 1 AF-ED (7) | 24 ± 1% | 37 ± 10% | 55 ± 10% | NAb | NAb | NAb |
| G5.5 - 1 AF-ED – 60 N-(2-Aminoethyl)-ADAC (8)c | 140 ± 65 | 80 ± 17 | 15 ± 4 | 100 ± 40 | 270 ± 90 | 66 ± 25 |
Binding experiments were completed in stably transfected CHO cells (A1, A3) or HEK cells (A2A). Cyclase experiments were completed in stably transfected CHO cells for all receptor types. Binding assays and functional assays using a cAMP kit were carried out as described in methods. Binding affinities are expressed as apparent inhibition constants (Ki app) and functional potencies as EC50 values (mean ± SEM, n = 3). As in previous studies, the results for the dendrimer derivatives are reported in dendrimer concentrations, rather than tethered monomer concentrations [18-20].
NA, not active – 10 μM of compound gives less than 20% activation at receptor compared to NECA.
MRS5212.
Although compound 5 displayed >100-fold selectivity for the A3AR in comparison to the A1 and A2AARs in both binding and functional cAMP assays [26], compound 8 was only slightly selective for the A3AR in binding and nonselective in an assay of adenylate cylase inhibition (Table 1). The binding Ki app values of compound 8 at the A1, A2A and A3 ARs were 140, 80, and 15 nM, respectively. The control dendrimer 7 at a 10 μM concentration was inactive in an assay of A3AR-mediated cAMP inhibtion and only weakly displaced radioligand at each of the three AR subtypes.
3.3. Quantification by qRT-PCR of the AR subtype gene expression in HL-1 cardiomyocytes
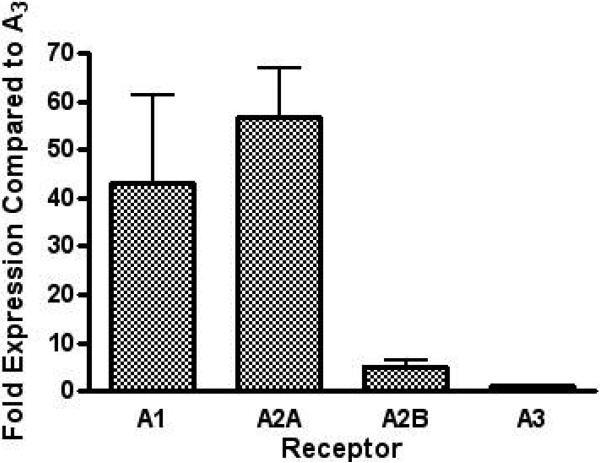
The levels of gene expression of all four mouse ARs in control HL-1 cells were measured using the ΔΔCT method of qRT-PCR using β-actin as an internal control. The endogenous level of the A3AR gene expression was the lowest among the four ARs. qRT-PCR indicated that A1 and A2AARs were expressed at 43 ± 18-fold and 56 ± 10-fold higher levels of expression than A3AR, respectively, but the A2BAR was only expressed at a 5-fold higher level than the A3AR (Figure 2).
Figure 2.
Gene expression levels of the A1, A2A, and A2B in HL-1 cells compared to A3 ARs measured using qRT-PCR. In three separate experiments, the fold expression of each AR is measured and normalized to the A3AR expression level, which is set to 1, using the ΔΔCT method.
3.4. Cytoprotection by AR agonists in an in vitro model of mouse cardiomyocte cell death
We used the HL-1 mouse cardiomyocyte model, in which cell damage was induced using H2O2, to test the cytoprotective ability of AR agonists. The degree of death in nontransfected HL-1 cells was shown using an LDH assay to be dependent on the concentration of H2O2 (Figure S4). The half-maximal increase in cell death occurred at ~1 mM, and it reached a plateau thereafter. 750 μM H2O2 produced between 35 – 45% cell death following a 4 h incubation, and this concentration was selected for further protection experiments.
In order to test the effect of greatly increasing the level of expression of the A3AR in the HL-1 cells on AR agonist-induced protection, we transfected the cells with cDNA coding for the receptor in the pcDNA5 plasmid. We measured the expression levels of the A3AR in the HL-1 cells following transfection using qRT-PCR (data not shown). Although the transfection level significantly varied between multiple transfections, in each case there was at least a 500-fold increase in the expression level after transfection compared to the endogenous level of A3AR. Therefore, all other ARs were expressed at a minimum of a 10-fold lower expression level than the A3AR.
H2O2 has previously been shown to induce death in a primary neuronal cell culture after an incubation of 3 h [34]. In the non-transfected HL-1 cells, only the nonselective agonist NECA and the A2A agonist CGS21680 protected against the H2O2–induced (750 μM) cytotoxicity as measured in an LDH assay, which reflects loss of cell membrane integrity. Interestingly, the nucleoside dendrimer 8 that bound to and activated the A2AAR, although less potently than A1 and A3ARs, did not show cytoprotection in nontransfected cells. In various models, A1, A2A, and A3ARs have all been shown to have cardioprotective properties.
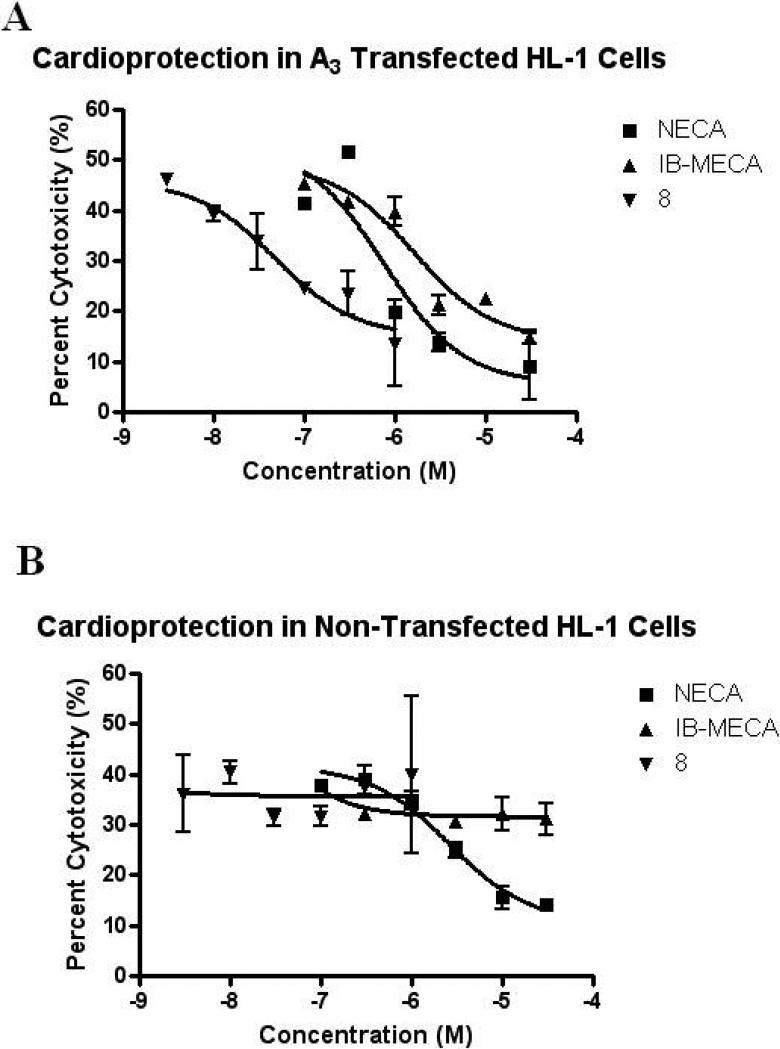
A comparison of NECA, IB-MECA, or 8 in A3AR-transfected HL-1 cells (Figure 3A) indicated that the dendrimeric derivative 8 was highly potent in cell protection. The results are tabulated as IC50 values in Table 2. The IC50 value of 8 was 100- to 200-fold less than those of the monomeric A3AR agonists IB-MECA and Cl-IB-MECA. The IC50 value for NECA did not significantly change between transfected and non-transfected cells, while neither IB-MECA nor Cl-IB-MECA gave protection in non-transfected cells (Figure 3B).
Figure 3.
Concentration-dependent protection against H2O2-induced cytotoxicity by A3 selective and non-selective AR agonists (3 nM – 30 μM) in A3AR-transfected (A) and non-transfected (B) HL-1 cells. HL-1 cells were pretreated with NECA (100 nM - 30 μM), IB-MECA (100 nM - 30 μM), or 8 (3 nM - 1 μM) for 1 h prior to the addition of H2O2 (final conc. 750 μM). After 4 h, 100 μl of media was added to 100 μl of the LDH measuring solution provided with the kit and incubated for 10 min. The results were analyzed with a microplate reader. Data shown are mean ± SD from three independent experiments in triplicate.
Table 2.
Protection from cell death induced by H2O2 in HL-1 cells using an LDH quantification assay.a
| Compound | Untransfected cells, IC50 (nM)b | Transfected cells, IC50 (nM)b |
|---|---|---|
| NECA (1) | 3700 ± 1000 | 3900 ± 1800 |
| CCPA | No protection | ND |
| CGS21680 | 7100 ± 2400 | ND |
| IB-MECA (2) | No protection | 3800 ± 1400 |
| Cl-IB-MECA (3) | No protection | 7900 ± 2300 |
| G5.5 – 1 AF-ED (7) | No protection | No protection |
| G5.5 – 1 AF-ED – 60(N-(2-Aminoethyl)-ADAC) (8) | No protection | 35 ± 8 |
Nucleoside derivative was administered 1 h prior to exposure to H2O2 and remained in the medium during the entire 4 h incubation in the presence of H2O2.
Either control cells (not transfected to express the A3AR) or cells transfected with cDNA for the hA3AR. No protection indicates lack of significant inhibition of cell death by the compound at a conc. up to 30 μM, except for 7 which was tested up to 1 μM. As in previous studies, the results for the dendrimer derivatives are reported in dendrimer concentrations, rather than tethered monomer concentrations [18-20].
ND not determined.
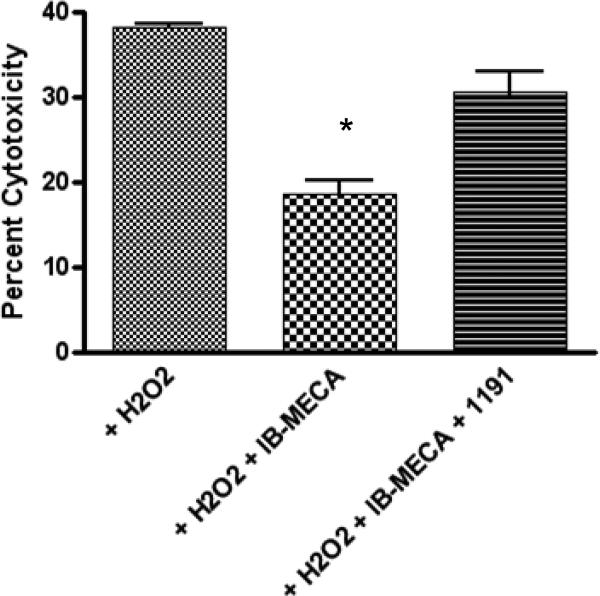
In order to determine if IB-MECA was acting at the A3AR, a selective A3 antagonist, 1,4-dihydropyridine derivative MRS1191 (10 μM) [35], was added prior to treatment with IB-MECA (30 μM). The antagonist prevented the protection afforded by IB-MECA from the H2O2-induced cell death in the transfected HL-1 cells (Figure 4). The difference between the cytotoxicity induced by H2O2 alone and the cytotoxicity induced with coadminstration of MRS1191 and IB-MECA was not statistically different using a student t-test (P = 0.05, df = 3).
Figure 4.
Effect of the A3 receptor antagonist MRS1191 on the protection by IB-MECA against H2O2-induced cell damage. HL-1 cells were pretreated with MRS1191 (10 μM), an antagonist of the A3 receptor, 1 h before treatment with IB-MECA (30 μM). One h after the addition of the agonist, H2O2 (750 μM) was incubated with the cells for 4 h. 100 μl of media was then added to 100 μl of the LDH measuring solution provided with the kit and incubated for 10 min. The results were analyzed with a microplate reader. Data shown are mean ± SD from three independent experiments in triplicate. Groups labeled * are significantly different from H2O2 control (P < 0.05).
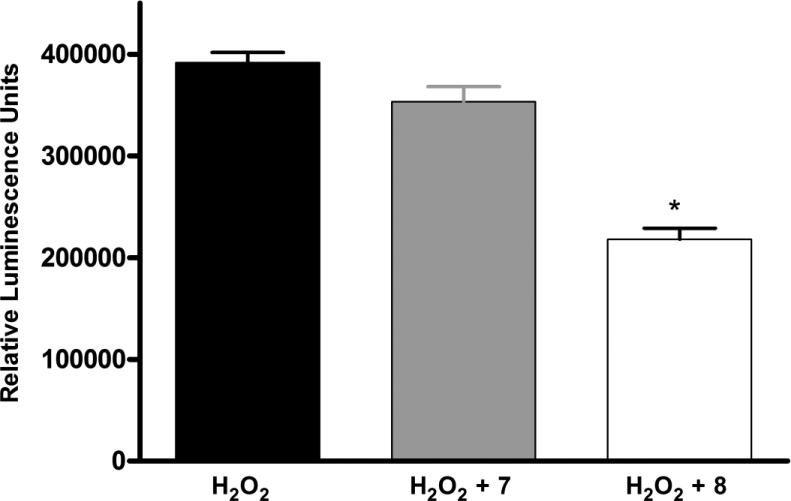
Finally, in order to determine if the cell death induced by the H2O2 was the result of apoptosis, an apoptosis-specific caspase 3/7 luminescent assay was used. There was an increase in the apoptosis signal in the control HL-1 cells when H2O2 was added (Figure 5). For the apoptosis assay, a lower concentration of H2O2 (400 μM) than in the LDH assay was sufficient. Apoptosis significantly decreased when 8, the dendrimer-A3 agonist conjugate was added prior to the H2O2, but did not significantly decrease when the cells were was incubated with 7, the control dendrimer (P = 0.05, df = 3).
Figure 5.
Protection afforded HL-1 cells by 8, a dendrimer-nucleoside conjugate, but not by 7, a control dendrimer, against H2O2-induced apoptosis. Dendrimer compounds (10 μM) were added 1 h prior to the addition of H2O2 (400 μM). The luminescent caspase 3/7 reagent was added 3 h after the addition of H2O2. After 1.5 h, apoptosis was quantified using a luminometer. Data shown are mean ± SD from three independent experiments in triplicate. Groups labeled * are significantly different from control treated with H2O2 alone (P < 0.05). Control cells in the absence of H2O2 gave 182,000 ± 20,000 relative luminescence units.
4. Discussion
This study investigated the ability of A3AR agonists, both multivalent GLiDe conjuates and monomers, to protect HL-1 cells against H2O2-induced cytotoxicity in HL-1 cells. While it was previously known that all ARs are expressed by HL-1 cells, the levels of expression were not known. Interestingly, the A2B and A3ARs are expressed at an almost 50-fold lower level than the A1 and A2AARs. A2B and A3ARs have not been definitively found in adult cardiac myocytes [15]. However, the HL-1 cells are believed to be a hybrid of adult and embryonic cardiomyocytes [17]. Although the A3AR may or may not be expressed in adult cardiac myocytes, protective effects in myocardial ischemia have been attributed to activation of the A3AR [15].
Interestingly, the selective A1AR agonist CCPA did not protect against H2O2-induced cell death in control HL-1 cells, while the A2AAR agonist CGS21680 weakly protected with an IC50 value of 7.1 μM. Although this concentration is high with respect to the Ki value of CGS21680 in activation of cAMP accumulation through the mouse A2AAR (7 nM) [36], the inactivity of A1 and A3AR agonists and the activity of NECA allow the conclusion that the A2AAR is likely responsible for the protection in control HL-1 cells. The observed protection by A2A but not A1AR activation is in contrast to some previous reports. For instance, Germack et al. found that CGS21680 does not protect neonatal rat cardiomyocytes against ischemia/reperfusion, while CPA, an A1AR agonist, does have protective effects [37]. However, H2O2-induced cell death may involve different signaling pathways than the damage resulting from ischemia/reperfusion. In fact, other studies have found that both the A1 and A2AARs are believed to be involved in cardioprotection. There is also evidence of cross-talk between ARs and that multiple ARs may be activated by endogenous adenosine in order to achieve cardioprotection [15].
In order to determine if the A3AR plays a role in cardioprotection in this cell culture model, we transfected the HL-1 cells with a plasmid coding for the hA3AR to greatly increase the level of A3AR expression. Although the transfection procedures remained constant, the levels of A3AR expression measured by qRT-PCR varied greatly between transfections. However, the RNA level always increased at least 500-fold over the endogenous level of A3AR RNA.There was not a significant difference in IC50 values of the A3AR agonists in the LDH assay based on the expression level of the A3AR.There are several explanations for why the IC50 values were not affected by the transfection level. For instance, the qRT-PCR measured the level of hA3 mRNA, not the level of receptor expression, which may have been much lower in all of the experiments. Attempts to measure the protein expression level using an A3-specific antibody were not successful due to non-specific binding by the available antibodies.
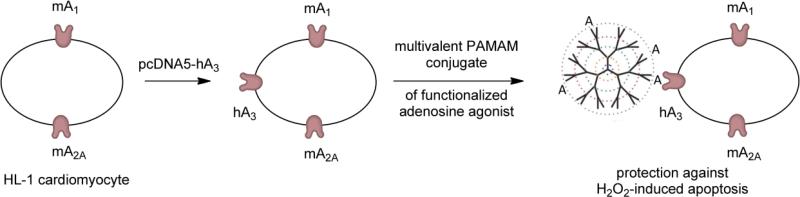
In the untransfected control cells, only NECA and CGS21680 were protective, likely through activation of the A2AAR. Increasing the expression level of the A3AR significantly revealed protection afforded by the A3 agonists IB-MECA and Cl-IBMECA, although the protective effects of the nonselective agonist NECA did not change. In the transfected cells, NECA may be protective through activation of the A2AAR, A3AR, or some combination of ARs, while protection against H2O2-induced apoptosis by the A3AR-selective agonists IB-MECA and Cl-IB-MECA is dependent on the expression of that subtype. This conclusion was further supported in the case of protection from cell death by the dendrimeric conjugate 8, which was blocked by an A3AR selective antagonist. Figure 6 shows a schematic representation of the basis for protection afforded to the HL-1 cells by compound 8. After mouse HL-1 cells were transfected with the hA3AR, the receptor protected the cells against H2O2-induced apoptosis when the protein was activated by 8. The A3AR has significant and unique cardioprotective properties [11,12]. While most previous work used whole animals or cultured cardiomyocytes from various species [11,13], the HL-1 cell line can now be used to study the signaling pathways involved in cardioprotection caused by AR activation.
Figure 6.
Schematic description of the mechanism of protection afforded HL-1 cells by multivalent dendrimeric AR agonist 8. The PAMAM dendrimer derivative is shown as a tree-like polymer with covalently attached nucleoside moieties “A”. Mouse HL-1 cells, which express a high basal level of mA1 and mA2AARs, were transfected with hA3AR mRNA in order to increase the expression levels of this subtype. Following transfection, activation of the hA3AR by conjugate 8 afforded significant protection to the HL-1 cells against H2O2-induced apoptosis.
Dendrimeric conjugation of GPCR ligands is a means of modulating their pharmacokinetic and pharmacodynamic characteristics [25,26], assuming that the linking chemistry is done in a way that preserves or enhances the pharmacological properties of the ligand [24]. In general, the chemical and biological properties of multivalent drugs bound to nanocarriers may differ greatly from those of the corresponding monomeric agents [38]. This approach provides an opportunity to tune the pharmacokinetics and pharmacodynamics in an otherwise unattainable manner and to introduce reporter or targeting moieties. It is also conceivable that the nucleoside bound to a polymeric carrier would have a reduced rate of metabolism in vivo [39], while preserving or enhancing the potency and selectivity.
In the present study, we synthesized a G5.5 PAMAM dendrimeric conjugate 8 containing an N6-chain-functionalized adenosine agonist amide-linked at approximately one quarter of the available carboxylic acid sites. The remaining terminal carboxylate groups afford it a negative charge. It was of much higher molecular weight than a previously synthesized G2.5 PAMAM conjugate of similar linkage chemistry, which was less densely substituted with ligand (~10% of the available sites) and selective in binding to and activating the A3AR [18]. The larger conjugate was designed because of the expected longer half-life of higher molecular weight PAMAM dendrimers in vivo [40]. Conjugate 8 activated ARs nonselectively and, similarly to the monomeric agonists of the A3AR, protected A3 transfected HL-1 cells using both an LDH assay and an apoptosis assay. However, the dendrimer conjugate had significantly greater potency (IC50 35 nM) than the corresponding monomeric nucleosides, which protected in the μmolar range. Thus, a multivalent conjugate retained binding affinity at the ARs and displayed greatly enhanced functional potency in an in vitro model of cardioprotection. GPCRs frequently exist as oligomeric complexes rather than single receptors [41], and the dendrimeric conjugate 8 could theoretically activate multiple receptor molecules in a dimer or higher order oligomer. While more research is necessary to determine the precise mechanism of the A3AR–induced protection and if simultaneous binding of multiple receptors is the basis for the increased potency, this research provides the groundwork for the use of multivalent drugs in treating cardiac diseases.
Supplementary Material
Acknowledgements
This research was supported by the Intramural Research Program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases. We thank Lena Yoo (NIDDK) for proofreading this manuscript.
Abbreviations
- ADAC
N6-[4-[[[4-[[[(2-aminoethyl)amino]carbonyl]methyl]-anilino]carbonyl]methyl]phenyl]adenosine
- AF488
Alexa-Fluor® 488
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate hydrate
- CHO
Chinese hamster ovary
- Cl-IB-MECA
2-chloro-N6-(3-iodobenzyl)-5′-N-methylcarboxamidoadenosine
- DMEM
Dulbecco's Modified Eagle Medium
- DMF
N,N-dimethylformamide
- DMSO
dimethyl sulfoxide
- ED
ethylenediamine
- EDC
N-(3-dimethylaminopropyl)-N'-ethylcarbodiimide
- EDTA
ethylenediaminetetraacetic acid
- FBS
fetal bovine serum
- [3H]CGS21680
2-[p-(2-carboxyethyl)phenylethylamino]-5′-N-ethylcarboxamido-adenosine
- GPCR
G protein-coupled receptor
- HEK
human embryonic kidney
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- [125I]AB-MECA
[125I]-4-aminobenzyl-5′-N-methylcarboxamidoadenosine
- IB-MECA
N6-(3-iodobenzyl)-5′-N-methylcarboxamidoadenosine
- MALDI-TOF
matrix assisted laser desorption/ionization time-of-flight
- MES
2-(N-morpholino)ethanesulfonic acid
- MS
mass spectrometry
- NMR
nuclear magnetic resonance
- PAMAM
polyamidoamine
- qRT-PCR
quantitative real-time polymerase chain reaction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Current address, Center for Drug Evaluation and Research, CDER-FDA, Silver Spring, MD 20993 USA.
Supporting information available: Synthetic procedures for compounds 4 and 6 and mass spectra of the dendrimer derivatives, concentration dependence of H2O2-induced cytotoxicity in HL-1 cells.
References
- 1.Jacobson KA, Gao ZG. Adenosine receptors as therapeutic targets. Nat Rev Drug Discov. 2006;5:247–64. doi: 10.1038/nrd1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Madi L, Cohen S, Ochayin A, Bar-Yehuda S, Barer F, Fishman P. Overexpression of A3 adenosine receptor in peripheral blood mononuclear cells in rheumatoid arthritis: involvement of nuclear factor-kappaB in mediating receptor level. J Rheumatol. 2007;34:20–6. [PubMed] [Google Scholar]
- 3.Matot I, Einav S, Weiniger CF, Pearl RG, Abramovitch R, Joshi BV, et al. Lung injury after in vivo reperfusion: outcome at 27 hours after reperfusion. Anesthesiology. 2008;109:269–78. doi: 10.1097/ALN.0b013e31817f5b90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bar-Yehuda S, Silverman MH, Kerns WD, Ochaion A, Cohen S, Fishman P. The anti-inflammatory effect of A3 adenosine receptor agonists: a novel targeted therapy for rheumatoid arthritis. Expert Opin Investig Drugs. 2007;16:1601–13. doi: 10.1517/13543784.16.10.1601. [DOI] [PubMed] [Google Scholar]
- 5.Silverman MH, Strand V, Markovits D, Nahir M, Reitblat T, Molad Y, et al. Clinical evidence for utilization of the A3 adenosine receptor as a target to treat rheumatoid arthritis: data from a phase II clinical trial. J Rheumatol. 2008;35:41–8. [PubMed] [Google Scholar]
- 6.Bar-Yehuda S, Stemmer SM, Madi L, Castel D, Ochaion A, Cohen S, et al. The A3 adenosine receptor agonist CF102 induces apoptosis of hepatocellular carcinoma via deregulation of the Wnt and NF-kappaB signal transduction pathways. Int J Oncol. 2008;33:287–95. [PubMed] [Google Scholar]
- 7.Jacobson KA, Klutz AM, Tosh DK, Ivanov AA, Preti D, Baraldi PG. Medicinal chemistry of the A3 adenosine receptor: agonists, antagonists, and receptor engineering. Handb Exp Pharmacol. 2009;(193):123–59. doi: 10.1007/978-3-540-89615-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen GJ, Harvey BK, Shen H, Chou J, Victor A, Wang Y. Activation of adenosine A3 receptors reduces ischemic brain injury in rodents. J Neurosci Res. 2006;84:1848–55. doi: 10.1002/jnr.21071. [DOI] [PubMed] [Google Scholar]
- 9.Schepp CP, Reutershan J. Bench-to-bedside review: adenosine receptors--promising targets in acute lung injury? Crit Care. 2008;12:226. doi: 10.1186/cc6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng J, Wang R, Zambraski E, Wu D, Jacobson KA, Liang BT. A novel protective action of adenosine A3 receptors: Attenuation of skeletal muscle ischemia and reperfusion injury. Am J Physiol. 2007;293:3685–91. doi: 10.1152/ajpheart.00819.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parsons M, Young L, Lee JE, Jacobson KA, Liang BT. Distinct cardioprotective effects of adenosine mediated by differential coupling of receptor subtypes to phospholipases C and D. FASEB J. 2000;14:1423–31. doi: 10.1096/fj.14.10.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stambaugh K, Jacobson KA, Jiang JL, Liang BT. A novel cardioprotective function of adenosine A1 and A3 receptors during prolonged simulated ischemia. Am J Physiol. 1997;273:H501–5. doi: 10.1152/ajpheart.1997.273.1.H501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Z, Jang Y, Mueller RA, Norfleet EA. IB-MECA and cardioprotection. Cardiovasc Drug Rev. 2006;24:227–38. doi: 10.1111/j.1527-3466.2006.00227.x. [DOI] [PubMed] [Google Scholar]
- 14.Takano H, Bolli R, Black RG, Jr, Kodani E, Tang XL, Yang Z, et al. A1 or A3 adenosine receptors induce late preconditioning against infarction in conscious rabbits by different mechanisms. Circ Res. 2001;88:520–8. doi: 10.1161/01.res.88.5.520. [DOI] [PubMed] [Google Scholar]
- 15.Peart JN, Headrick JP. Adenosinergic cardioprotection: multiple receptors, multiple pathways. Pharmacol Ther. 2007;114:208–21. doi: 10.1016/j.pharmthera.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 16.Chaudary N, Shuralyova I, Liron T, Sweeney G, Coe IR. Transport characteristics of HL-1 cells: a new model for the study of adenosine physiology in cardiomyocytes. Biochem Cell Biol. 2002;80:655–65. doi: 10.1139/o02-143. [DOI] [PubMed] [Google Scholar]
- 17.Claycomb WC, Lanson NA, Jr, Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, et al. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci USA. 1998;95:2979–84. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White SM, Constantin PE, Claycomb WC. Cardiac physiology at the cellular level: use of cultured HL-1 cardiomyocytes for studies of cardiac muscle cell structure and function. Am J Physiol Heart Circ Physiol. 2004;286:H823–9. doi: 10.1152/ajpheart.00986.2003. [DOI] [PubMed] [Google Scholar]
- 19.Brundel BJ, Henning RH, Ke L, van Gelder IC, Crijns HJ, Kampinga HH. Heat shock protein upregulation protects against pacing-induced myolysis in HL-1 atrial myocytes and in human atrial fibrillation. J Mol Cell Cardiol. 2006;41:555–62. doi: 10.1016/j.yjmcc.2006.06.068. [DOI] [PubMed] [Google Scholar]
- 20.Dandapat A, Hu CP, Li D, Liu Y, Chen H, Hermonat PL, et al. Overexpression of TGFbeta1 by adeno-associated virus type-2 vector protects myocardium from ischemia-reperfusion injury. Gene Ther. 2008;15:415–23. doi: 10.1038/sj.gt.3303071. [DOI] [PubMed] [Google Scholar]
- 21.Chaudary N, Naydenova Z, Shuralyova I, Coe IR. The adenosine transporter, mENT1, is a target for adenosine receptor signaling and protein kinase Cepsilon in hypoxic and pharmacological preconditioning in the mouse cardiomyocyte cell line, HL-1. J Pharmacol Exp Ther. 2004;310:1190–8. doi: 10.1124/jpet.104.067157. [DOI] [PubMed] [Google Scholar]
- 22.Yitzhaki S, Huang C, Liu W, Lee Y, Gustafsson AB, Mentzer RM, Jr, et al. Autophagy is required for preconditioning by the adenosine A1 receptor-selective agonist CCPA. Basic Res Cardiol. 2009;104:157–67. doi: 10.1007/s00395-009-0006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim Y, Klutz AM, Hechler B, Gao ZG, Gachet C, Jacobson KA. Application of the functionalized congener approach to dendrimer-based signaling agents acting through A2A adenosine receptors. Purinergic Signal. 2009;5:39–50. doi: 10.1007/s11302-008-9113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobson KA. Functionalized Congener Approach to the Design of Ligands for G Protein-Coupled Receptors (GPCRs). Bioconjug Chem. 2009;20:1816–35. doi: 10.1021/bc9000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim Y, Hechler B, Klutz AM, Gachet C, Jacobson KA. Toward Multivalent Signaling across G Protein-Coupled Receptors from Poly(amidoamine) Dendrimers. Bioconjug Chem. 2008;19:406–11. doi: 10.1021/bc700327u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klutz AM, Gao ZG, Lloyd J, Shainberg A, Jacobson KA. Enhanced A3 adenosine receptor selectivity of multivalent nucleoside-dendrimer conjugates. J Nanobiotechnology. 2008;6:12. doi: 10.1186/1477-3155-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das A, Zhou Y, Ivanov AA, Carter RL, Harden TK, Jacobson KA. Enhanced potency of nucleotide-dendrimer conjugates as agonists of the P2Y14 receptor: Multivalent effect in G protein-coupled receptor recognition. Bioconjug Chem. 2009;20:1650–9. doi: 10.1021/bc900206g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivanov AA, Jacobson KA. Molecular modeling of a PAMAM- CGS21680 dendrimer bound to an A2A adenosine receptor homodimer. Bioorg Med Chem Lett. 2008;18:4312–5. doi: 10.1016/j.bmcl.2008.06.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perreira M, Jiang JK, Klutz AM, Gao ZG, Shainberg A, Lu C, et al. “Reversine” and its 2-substituted adenine derivatives as potent and selective A3 adenosine receptor antagonists. J Med Chem. 2005;48:4910–8. doi: 10.1021/jm050221l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bookout AL, Mangelsdorf DJ. Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl Recept Signal. 2003;1:e012. doi: 10.1621/nrs.01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panchuk-Voloshina N, Haugland RP, Bishop-Stewart J, Bhalgat MK, Millard PJ, Mao F, et al. Alexa dyes, a series of new fluorescent dyes that yield exceptionally bright, photostable conjugates. J Histochem Cytochem. 1999;47:1179–88. doi: 10.1177/002215549904700910. [DOI] [PubMed] [Google Scholar]
- 32.Kim Y, Klutz AM, Jacobson KA. Systematic investigation of polyamidoamine dendrimers surface-modified with poly(ethylene glycol) for drug delivery applications: synthesis, characterization, and evaluation of cytotoxicity. Bioconjug Chem. 2008;19:1660–72. doi: 10.1021/bc700483s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Zwart M, Link R, von Frijtag Drabbe Künzel JK, Cristalli G, Jacobson KA, Townsend-Nicholson A, IJzerman AP. A screening of adenosine analogues on the human adenosine A2B receptor as part of a search for potent and selective agonists. Nucleos Nucleotid. 1998;17:969–86. doi: 10.1080/07328319808004215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whittemore ER, Loo DT, Watt JA, Cotman CW. A detailed analysis of hydrogen peroxide-induced cell death in primary neuronal culture. Neuroscience. 1995;67:921–32. doi: 10.1016/0306-4522(95)00108-u. [DOI] [PubMed] [Google Scholar]
- 35.Jacobson KA, Park KS, Jiang J-l, Kim YC, Olah ME, Stiles GL, Ji Xd. Pharmacological characterization of novel A3 adenosine receptor-selective antagonists. Neuropharmacology. 1997;36:1157–65. doi: 10.1016/s0028-3908(97)00104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ge ZD, Peart JN, Kreckler LM, Wan TC, Jacobson MA, Gross GJ, et al. Cl-IB-MECA [2-chloro-N6-(3-iodobenzyl)adenosine-5'-N-methylcarboxamide] reduces ischemia/reperfusion injury in mice by activating the A3 adenosine receptor. J Pharmacol Exp Ther. 2006;319:1200–10. doi: 10.1124/jpet.106.111351. [DOI] [PubMed] [Google Scholar]
- 37.Germack R, Dickenson JM. Adenosine triggers preconditioning through MEK/ERK1/2 signalling pathway during hypoxia/reoxygenation in neonatal rat cardiomyocytes. J Mol Cell Cardiol. 2005;39:429–42. doi: 10.1016/j.yjmcc.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 38.Tomalia DA. In quest of a systematic framework for unifying and defining nanoscience. J Nanoparticle Res. 2009;11:1251–310. doi: 10.1007/s11051-009-9632-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee CC, MacKay JA, Frechet JM, Szoka FC. Designing dendrimers for biological applications. Nat Biotechnol. 2005;23:1517–26. doi: 10.1038/nbt1171. [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi H, Brechbiel MW. Nano-sized MRI contrast agents with dendrimer cores. Adv Drug Deliv Rev. 2005;57:2271–86. doi: 10.1016/j.addr.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 41.Gandia J, Galino J, Amaral OB, Soriano A, Lluis C, Franco R, et al. Detection of higher-order G protein-coupled receptor oligomers by a combined BRET-BiFC technique. FEBS Lett. 2008;582:2979–84. doi: 10.1016/j.febslet.2008.07.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.