Abstract
The slicer activity of the RNA-induced silencing complex resides within its Argonaute (Ago) component, whose PIWI domain provides the catalytic residues governing guide-strand mediated site-specific cleavage of target RNA. We report on structures of ternary complexes of T. thermophilus Ago catalytic mutants with 5′-phosphorylated 21-nt guide DNA and complementary target RNAs of length 12-, 15- and 19-nt, which define the molecular basis for Mg2+-facilitated site-specific cleavage of the target. We observe pivot-like domain movements within the Ago scaffold on proceeding from nucleation to propagation steps of guide-target duplex formation, with duplex zippering beyond one turn of helix requiring release of the 3′-end of the guide from the PAZ pocket. Cleavage assays on targets of various lengths supported this model, and sugar-phosphate backbone modified target strands revealed the importance of structural and catalytic divalent metal ions observed in the crystal structures.
An extensive body of research has demonstrated that Argonaute (Ago) is the key component of the RNA-induced silencing complex (RISC) and plays an essential role in guide-strand mediated target RNA recognition, cleavage and product release1–8. Ago adopts a bilobal architecture, composed of N-terminal PAZ-containing (N and PAZ domains) and C-terminal PIWI-containing (Mid and PIWI) lobes. Structural studies have established that the PIWI domain of Ago adopts an RNase H fold9–11, whose catalytic Asp-Asp-Asp/His residues contribute to slicer activity11–13, while its Mid domain sequesters the 5′-phosphate of the guide strand14,15, and its PAZ domain recognizes the 2-nt overhang at the 3′-end of the guide strand16,17. Ago-mediated target RNA cleavage requires Watson-Crick pairing between guide and target spanning both the seed segment (positions 2 to 8) and the cleavage site (10–11 step) as counted from the 5′-end of the guide strand3,4. Endonucleolytic cleavage is mediated by Mg2+ cations18,19 and generates fragments containing a 3′-OH for the 5′-segment and a 5′-phosphate for the 3′-segment20. Molecular insights into target RNA recognition and cleavage have emerged from chemical21,22, biophysical23, and structural5,24–27 studies, with potential application of RNA interference-based approaches as a therapeutic modality against a range of human diseases28,29.
We have previously reported on crystal structures of T. thermophilus Ago bound to 5′-phosphorylated 21-nt guide DNA (binary complex)30, and with added 20-nt target RNA (ternary complex)31 (see Supplementary materials for a summary of these results). A major limitation of the earlier structural study of the ternary complex31 was that the bases of the target RNA could not be monitored due to disordered electron density at the 10–11 cleavage site as a result of mismatch incorporation at these steps so as to inactivate cleavage activity. The catalytic activity of the RNase H fold of the PIWI domain of T. thermophilus Ago originates in Asp residues D478, D546 and D660, and hence, in the present study, single Asp (D) to Asn (N), Glu (E) or Ala (A) mutants were incorporated at these positions to inactivate the cleavage activity. The ternary complexes of these catalytic mutants were then generated with 5′-phosphorylated 21-nt guide DNA and fully-complementary target RNAs of varying length (12-, 15- and 19-nt), conditions under which both the seed segment and the cleavage site could be potentially monitored, thereby providing insights into cleavage mechanism. Structural studies were then undertaken on those ternary complexes of Ago mutants with bound guide DNA and varying target RNA lengths that yielded diffraction quality crystals.
Cleavage site in Ago ternary complexes
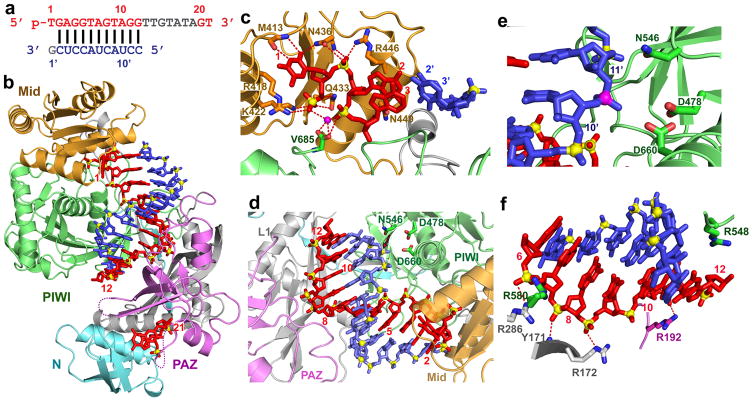
We have solved the 2.6 Å crystal structure of the N546 catalytic mutant of T. thermophilus Ago bound to 5′-phosphorylated 21-nt guide DNA and a 12-nt target RNA that is fully complementary along the length of the duplex (Fig. 1a). This is our highest resolution structure of a ternary complex to date (Fig. 1b; stereo view in a different perspective31 in Supplementary Fig. 1a; x-ray statistics are listed in Supplementary Table 1) and has provided detailed insights into the alignment of the guide and target strands that span both the seed segment and the cleavage site. The guide DNA strand in red can be monitored from positions 1–12 spanning the 5′-half and for positions 20–21 at the 3′-end, while the target RNA strand in blue can be monitored for positions 2′–12′ (Fig. 1b). Both ends of the guide strand are anchored in their respective binding pockets despite formation of an 11-bp DNA-RNA duplex. Intermolecular contacts within the target 12-mer ternary complex are highlighted in Supplementary Fig. 2. Bases 1 and 2 are splayed apart, with thymine at position 1 stacked over the side chain of R418, and its N3 nitrogen and O4 oxygen hydrogen-bonded to the backbone (M413) and side chain (N436) of the Ago scaffold (Fig. 1c). Base 1 is the only residue on the guide strand that makes base-specific contacts with the Ago scaffold and this observation is consistent with the reported sorting of small RNAs in Arabidopsis Ago complexes by the 5′-terminal nucleotide32,33.
Figure 1. Crystal structure of T. thermophilus Ago containing N546 catalytic mutant bound to 5′-phosphorylated 21-nt guide DNA and 12-nt target RNA.
a, Sequence of the guide DNA-target RNA duplex. The traceable segments of the bases of the guide DNA and target RNA in the structure of the ternary complex are shown in red and blue, respectively. Disordered segments of the bases on both strands that cannot be traced are shown in grey. b, A view of the 2.6 Å crystal structure of the Ago ternary complex. The Ago protein is color-coded by domains (N in cyan, PAZ in magenta, Mid in orange and PIWI in green) and linkers (L1 and L2 in grey). The bound 21-nt guide DNA is in red and traced for bases 1–12 and 20–21, whereas the bound 12-nt target RNA is in blue and traced for bases 2′–12′. Backbone phosphorus atoms are in yellow. Both ends of the bound guide DNA are anchored in this ternary complex. c, An expanded view of the ternary complex highlighting the alignment of guide DNA (1–3) and target RNA (2′–3′), where the bases of the 1–2 step of the guide strand are splayed apart. Note the intermolecular hydrogen-bonding of the Watson-crick edge of T1 with the backbone amide carbonyl of M413 and side chain of N436, as well as the positioning of phosphate 1 of the guide strand in the Mid binding pocket. A Mg2+ cation (purple) coordinates to phosphates 1 and 3 of the guide strand, as well as to an inserted carboxylate of V685 from the C-terminus of the protein. d, An expanded view of the ternary complex highlighting the guide DNA (1–12) - target RNA (2′–12′) duplex, together with the catalytic residues (D478, D660 and N546 mutant) of the RNase H fold of the PIWI domain. The scissile phosphate group at the 10–11 step of the target RNA is indicated by a red arrow. e, An expanded view highlighting the positioning of the backbone phosphate linking the 10–11 step (phosphorus colored in magenta) of the target RNA relative to the catalytic residues (D478, D660 and N546 mutant) in the ternary complex. f, Positioning of the side chain of R548 relative to the guide DNA (6 to 12) - target RNA (6′ to 12′) duplex. Note the intermolecular contacts between the sugar-phosphate backbone of the guide strand and side chains of the protein in the ternary complex.
The guide DNA-target RNA duplex spanning positions 2 to 12 (Fig. 1d) superpositions better with an A-form helix compared to its B-form counterpart (Supplementary Figs. 3a and 3b, respectively), with the scissile phosphate (10–11 step) on the target strand positioned opposite the catalytic residues (D478, D660 and N546 mutant) of the RNase H fold of the PIWI domain (Figs. 1d and 1e). Bases 10 and 11 of the target strand stack on each other in a catalytically competent helical conformation in the ternary Ago complex (Fig. 1f), in contrast to the orthogonal arrangements of these bases due to insertion of R548 between them in the binary Ago complex30 (compare Supplementary Fig. 4a [binary] with 4b [ternary]). Conformational changes in both the guide strand (Supplementary Fig. 5a) and Ago (Supplementary Fig. 5b) accompany the transition from binary to ternary complex formation (Supplementary Fig. 6 and movie 1 provided in the Supplementary information).
Release of guide 3′-end on propagation of guide-target duplex
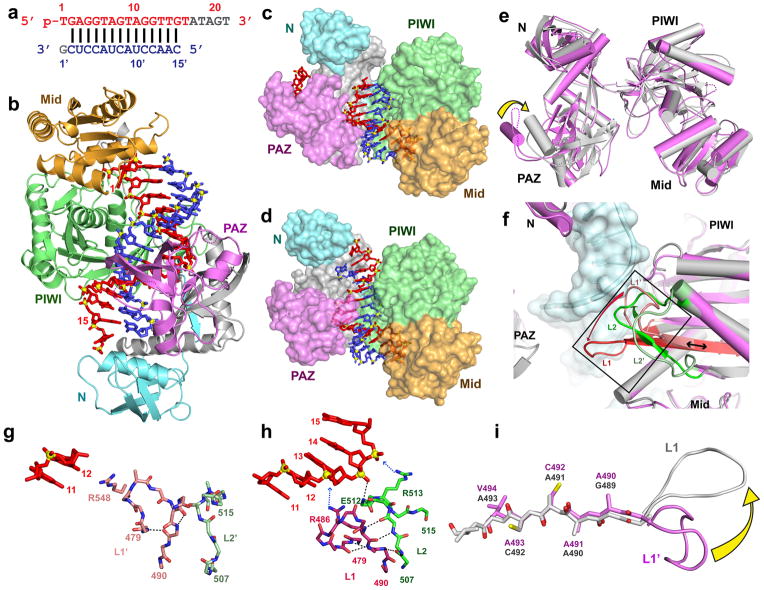
We next solved the 3.05 Å crystal structure of the E546 catalytic mutant of T. thermophilus Ago bound to 5′-phosphorylated 21-nt guide DNA and a 15-nt target RNA that is fully complementary along the length of the duplex (Fig. 2a; stereo view in Supplementary Fig. 1b; x-ray statistics are listed in Supplementary Table 1). The guide DNA strand can be monitored from positions 1–16, while the target RNA strand can be monitored from positions 2′–15′ (Fig. 2b). The 5′-phosphate of the guide strand is still anchored in the Mid pocket but the 3′-end (positions 17–21 are disordered and cannot be traced) is released from the PAZ pocket on formation of the 14-bp duplex spanning positions 2–15 of the guide strand. The molecular basis for the release of the 3′-end of the guide strand is that the helical conformation for nucleotides 12–15 disallows the 3′-end from reaching the binding pocket in the PAZ domain.
Figure 2. Crystal structure of T. thermophilus Ago containing E546 catalytic mutant bound to 5′-phosphorylated 21-nt guide DNA and 15-nt target RNA.
a, Sequence of the guide DNA-target RNA duplex, with traceable segments color-coded as in Fig. 1a. b, A view of the 3.05 Å crystal structure of the Ago ternary complex, color-coded as outlined in Fig. 1b. The bound 21-nt guide DNA is in red and traced for bases 1–16, whereas the bound 15-nt target RNA is in blue and traced for bases 2′–15′. Only the 5′-end of the guide DNA is anchored in this ternary complex. Comparison of crystal structures of c, Ago N546 mutant - 12-mer target and d, Ago E546 mutant - 15-nt target ternary complexes. The Ago protein is shown in a surface representation with labeled domains and linkers color-coded as in Fig. 1b. The guide DNA (red) and target RNA (blue) are shown in stick representation with backbone phosphorus atoms in yellow. e, View of alignment of Ago N546 mutant - 12-mer target complex (magenta) and Ago E546 mutant - 15-nt target complex (silver), after superpositioning of their PIWI-containing (Mid and PIWI domains) modules. The yellow arrow indicates the magnitude of the conformational change on proceeding from the 12-mer target to 15-mer target ternary complexes. f, Conformational changes in loop 1 (residue 479-488, red arrow) and loop 2 (residues 505-516, green arrow) of the PIWI domain on proceeding from the 12-nt target ternary complex (magenta) to the 15-nt target ternary complex (silver). Only the DNA-RNA duplex for the 15-nt target ternary complex is shown in cyan in a surface representation. Loops 1 and 2 are colored in light red (labeled L1′) and light green (labeled L2′) in the 12-nt target ternary complex, while they are colored in dark red (labeled L1) and dark green (labeled L2) in the 15-nt target ternary complex. The β-strand involved in sliding is highlighted by black double-edged arrow. g, The ternary complex containing 12-nt target RNA. Residues 11 and 12 of the guide strand are in red and loops L1′ and L2′ are in light red and light green, respectively. h, The ternary complex containing 15-nt target RNA. Residues 11 and 15 of the guide strand are in red and loops L1 and L2 are in dark red and dark green, respectively. Loop L1 switches to a β-turn aligned through hydrogen bonding both within the turn and also with loop L2, thereby stabilizing this turn conformation. The main-chain of Glu512 forms a hydrogen bond with phosphate group of residue 14 of the guide DNA. The positively-charged side chains of Arg513 and Arg486 interact with the backbone of the DNA guide strand, as indicated by blue arrows. i, A ribbon representation of the sliding of the β-strand (Gly489 to Val494) by one residue and conformational transition in adjacent L1 loop on proceeding from the 12-mer RNA target ternary complex (magenta) to 15-mer target RNA ternary complex (silver).
We observe conformational changes on proceeding from the ternary Ago complex with bound 12-mer target (Fig. 2c) to its counterpart with bound 15-mer target (Fig. 2d), and these changes can be visualized following superpositioning of the PIWI-containing (Mid and PIWI) lobe as shown by the yellow arrow in Fig. 2e (also, see movie 2 provided in the Supplementary information). These changes involve a pivotal rotation of the PAZ domain (compare PAZ domain alignments in Supplementary Figs. 7a and 7b), as well as movement of loops L1 and L2 located on the nucleic acid-interfacing surface of the PIWI domain (Fig. 2f).
Details of intermolecular contacts between loop L1 and the guide DNA 11–12 segment in the 12-mer target RNA ternary complex is shown in Fig. 2g, while intermolecular contacts between loops L1 and L2 and the guide DNA 11–15 segment in the 15-mer target RNA ternary complex is shown in Fig. 2h. Interestingly, L1 changes from a loop (Fig. 2g) to a β-turn (Fig. 2h) on proceeding from the 12-mer to the 15-mer target RNA ternary complexes, resulting in several additional hydrogen-bonds within this β-turn and with loop 2, thereby stabilizing this new conformation. The conformational transitions in loops L1 and L2 are mandated so as to avoid steric clashes with the DNA guide strand (Supplementary Fig. 8) on addition of three more base pairs on proceeding from the 12-mer to 15-mer target RNA ternary complexes. Unexpectedly, changes in loop L1 conformation forces the attached β-strand encompassing residues 489–493, as part of a multi-stranded β-sheet, to slide by a single residue with the accompanying flip of the entire β-strand and its side chains, on proceeding from the 12-mer to the 15-mer ternary complex (Fig. 2i, identified by a black double-edged arrow in Fig. 2f and Supplementary Fig. 9).
In mechanistic terms, we favor the view that the conformational transitions in loops L1 and L2 and associated sliding and flipping of the β-strand is triggered by widening of the substrate binding channel between the PIWI and N domains so as to accommodate a lengthening of the A-form duplex from 11-bp in the 12-mer target RNA complex to 14-bp in the 15-mer target RNA complex. Such changes not only push the PAZ domain away but also release the 3′ end of guide strand from the PAZ-binding pocket (Figs. 1b and 2b and Supplementary Fig. 7). Moreover, we note that the sliding and flipping of the β-strand occurs with minimal perturbation of β-sheet formation (schematic in Supplementary Fig. 9), and the flipping of the entire β-strand did not disrupt specific side chain interactions.
We have compared the structures of Ago mutant ternary complexes with 12-mer (Fig. 1b) and 15-mer (Fig. 2b) target RNAs reported in this study with the structure of the ternary complex of wild-type Ago with 20-mer target RNA containing a pair of mismatches at the cleavage site reported previously31. The previously reported structure of the ternary complex (two molecules in the asymmetric unit) 31 and one solved recently in a different crystal form (one molecule in the asymmetric unit; x-ray crystallographic statistics in Supplementary Table 2) in which segment 2–9 is fully paired and both ends of the guide strand are anchored is most similar to the ternary complex with 12-mer target RNA in the present study in which segment 2–12 is fully-paired and both ends of the guide strand are also anchored (comparison outlined in Supplementary Figs. 10a, b).
Our structural studies resolve a mechanistic issue related to guide strand-mediated recognition and cleavage of target RNA within Ago complexes. Several groups have proposed a ‘two-state’ model wherein the guide strand is anchored at both its ends during the nucleation step of target recognition, but its 3′-end is released from the PAZ pocket due to topological constraints, following propagation of the duplex towards the 3′-end of the guide strand4,11,34. An alternate ‘fixed-end’ model was also considered where both ends of the guide strand remain anchored during both the nucleation and propagation steps of RNA recognition34. Our results support a ‘two-state’ mechanism for the system under study, given that our structural studies demonstrate that both ends of the guide strand are anchored in a ternary complex containing one turn of A-form helix (12-mer target RNA) spanning the seed segment and cleavage site (Fig. 1b), but the 3′-end is released from the PAZ pocket on extending this duplex by three more base pairs (15-mer target RNA) towards the 3′-end of the guide strand (Fig. 2b).
N domain blocks guide-target pairing beyond position 16
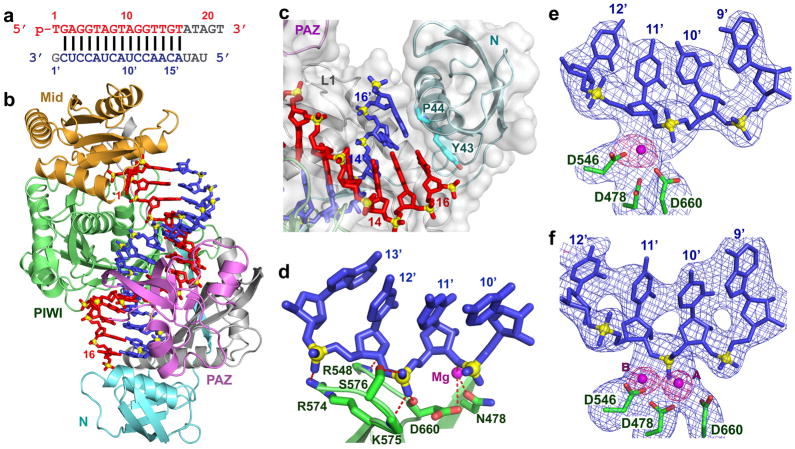
The 2.8 Å crystal structure of the N478 catalytic mutant of T. thermophilus Ago bound to 5′-phosphorylated 21-nt guide DNA and a 19-nt target RNA (sequence in Fig. 3a; structure in Fig. 3b; stereo view in Supplementary Fig. 1c; x-ray statistics are listed in Supplementary Table 1) is similar to the Ago E546 catalytic mutant ternary complex with 15-nt target RNA (Fig. 2b), except that one additional base pair can be traced, allowing monitoring of 15-bp of guide-target duplex spanning positions 2 to 16 of the guide strand (electron density maps of the guide and target strands are shown in stereo in Supplementary Fig. 11). Intermolecular contacts within the 19-mer target ternary complex are highlighted in Supplementary Fig. 12). In addition, the sugar-phosphate backbone of the target strand is intact at the 10–11 step, and on either side of it, for both 15-mer and 19-mer target ternary complexes (see Fo-Fc omit maps contoured at 3.7σ in Supplementary Figs. 13a and 13b, respectively).
Figure 3. Crystal structure of T. thermophilus Ago containing N478 catalytic mutant bound to 5′-phosphorylated 21-nt guide DNA and 19-nt target RNA and identification of Mg2+ binding sites within the catalytic pocket of the wild-type Ago complex.
a, Sequence of the guide DNA-target RNA duplex, with traceable segments color-coded as in Fig. 1a. b, A view of the 2.8 Å crystal structure of the ternary complex, color-coded as outlined in Fig. 1b. The bound 21-nt guide DNA is in red and traced for bases 1–16, whereas the bound 19-nt RNA is in blue and traced for bases 2′–16′. Only the 5′-end of the guide strand is anchored in this ternary complex. c, An expanded view of the 19-mer target ternary complex highlighting blocking of propagation of the guide DNA - target RNA duplex beyond pair 16 by the N domain. Base 16 of the guide strand stacks over the aromatic ring of Tyr43, while base 16′ stacks over Pro44. d, Intermolecular hydrogen-bonding contacts between the sugar-phosphate backbone of the 10′ to 13′ target RNA segment and backbone and side chains of the PIWI domain in the 19-mer target ternary complex. e, f, Fo-Fc omit maps (blue color, contoured at 3.5σ) of the 9′–12′ segment of bound RNA and catalytic D478, D546 and D660 residues in the 3.3 Å structures of the ternary complexes in 50 mM Mg2+ (space group P43212, one molecule in the asymmetric unit, panel e) and in 80 mM Mg2+ (space group P212121, two molecules in asymmetric unit, panel f). Bound Mg2+ cation(s) were identified in omit maps contoured in purple at 6.0σ as outlined in (e) and (f), based on coordination to several oxygen atoms in an approximate octahedral geometry. One bound Mg2+ cation can be assigned in the ternary complex in 50 mM Mg2+ in (e) and two bound Mg2+ cations can be assigned in the ternary complex in 80 mM Mg2+ in (f).
An unexpected mechanistic insight to emerge from our structural studies of the three ternary Ago complexes outlined above is that the guide DNA-target RNA duplex retains the A-form duplex architecture spanning the seed segment, the cleavage site and observable elements towards the 3′-end of the guide strand (up to position 16), and it is solely the Ago scaffold that adjusts through pivot-like domain movements, to relieve the topological stress associated with zippering up the RNA target through pairing with its guide strand template. A second unanticipated observation is that the N domain blocks propagation of the guide DNA - target RNA duplex beyond position 16 in the 19-mer target ternary complex (Fig. 3c), with the base at position 16 of the guide strand stacking on the aromatic ring of Tyr43 and the base at 16′ of the target strand stacked over the Pro44 ring. Thus, base pairing is disrupted for steps 17, 18 and 19, with anticipated trajectories for the separated guide and target strands schematized in Supplementary Fig. 14.
The sugar-phosphate backbone spanning the seed segment of the guide but not target strand is hydrogen-bonded to the protein, (see movie 3 provided in the Supplementary information). We also note that the sugar-phosphate backbone of the target RNA spanning the 10′ to 13′ segment forms intermolecular hydrogen bonds with the Ago scaffold in the 12-mer (Supplementary Fig. 15a), 15-mer (Supplementary Fig. 15b) and 19-mer (Fig. 3d) target ternary complexes, establishing the potential for photochemically-facilitated cross links between this segment of the target RNA and its spatially identified proximal sites on the protein35,36
A pair of Mg2+ cations mediate cleavage chemistry
The PIWI domain of Ago adopts an RNase H fold9–11,30,31, with catalytic D478, D546 and D660 residues lining the active site of the T. thermophilus enzyme. Two Mg2+ cations have been shown to facilitate RNA hydrolysis during catalytic cleavage by RNase H-containing nucleases, with cation A assisting nucleophilic attack by positioning and activating a water molecule and cation B stabilizing the transition state and leaving group37,38. Since catalytic mutations could induce distortions of the optimal geometry for coordination to divalent cations, we attempted to identify bound Mg2+ cation(s) in the catalytic pocket of the ternary complex of wild-type T. thermophilus Ago with 19-nt target RNA, that is fully complementary to positions 2–19 of the guide strand (Fig. 3a).
Crystals of the Ago ternary complex were grown as a function of Mg2+ concentration, with 3.3 Å data sets collected for crystals in 50 mM Mg2+ (space group P43212, one molecule in the asymmetric unit) and 80 mM Mg2+ (space group P212121, two molecules in asymmetric unit) solution (x-ray statistics listed in Supplementary Table 3). Gel electrophoresis of the crystals established that the target RNA was not cleaved in either ternary complex, presumably because T. thermophilus Ago-mediated cleavage is optimal at higher temperatures, and with a marked preference for Mn2+ over Mg2+ as the divalent cation11. The Fo-Fc omit maps (blue color, contoured at 3.5σ) of the target strand residues 9′–12′ and catalytic Asp residues for the Ago ternary structures in 50 mM Mg2+ and 80 mM Mg2+ are shown in Figs. 3e and 3f, respectively. A single bound Mg2+, positioned towards the leaving group side of the scissile phosphate (cation B), can be identified in the structure in 50 mM Mg2+ (Fig. 3e, omit map contoured in purple at 6.0σ), with an intact target RNA readily traceable for the 9′–12′ segment. A pair of Mg2+ cations separated by 3.9 Å, that coordinate the hydrolysis of the scissile phosphate, were identified in the structure in 80 mM Mg2+ (Fig. 3f). The assignment of the extra density to Mg2+ site(s) at 3.3 Å resolution is based on coordination of the divalent cation(s) to several oxygen atoms in an approximate octahedral geometry (stereo views in Supplementary Figs. 16a and 16b). Of the three catalytic Asp residues lining the catalytic pocket, D478 is the only one that coordinates to both Mg2+ cations (Fig. 3f and Supplementary Fig. 16b). The structures of the catalytic residues, Mg2+ sites and RNA backbone for B. halodurans RNase H (1.85 Å) and T. thermophilus Ago (3.3 Å) complexes are superpositioned in stereo for comparative purposes in Supplementary Fig. 17. Given that the crystals of the ternary complexes grown from both 50 mM and 80 mM Mg2+ diffract to 3.3 Å resolution, it is currently not possible to identify the position of the water molecule that would participate and be positioned for in-line attack on the scissile phosphate.
We observe detectable conformational changes following superpositioning of the single and pair of Mg2+-bound ternary complex structures through their PIWI-containing lobes. The observed changes are restricted to the PAZ domain (Supplementary Fig. 18a) and the target RNA strand (Supplementary Fig. 18b). The catalytic residues are optimally positioned for cleavage in the structure of the ternary complex with a pair of Mg2+ cations.
Thus, the Ago protein, capitalizing on the RNase H fold of its PIWI domain9–11, uses three catalytic Asp residues and two Mg2+ cations to facilitate site-specific cleavage of RNA targets, thereby yielding products containing 5′-phosphate and 3′-OH ends20, a feature in common with members of the retroviral integrase superfamily38.
Biochemical analysis of the catalytic activity of T. thermophilus Ago
Target RNA cleaving bacterial complexes are most effectively reconstituted using single-stranded guide DNA rather than RNA11,15,30,31. In order to explore whether DNA might also function as a target, we subjected chemically synthesized DNA and RNA targets (Supplementary Table 4) to DNA-guided Ago cleavage reactions. DNA is resistant against hydrolysis by divalent metal ions and high temperature incubation, thereby yielding a clearer picture of target cleavage. T. thermophilus Ago loaded with guide DNA derived from luciferase sequence studied previously30,31 cleaved DNA as well as RNA targets; however, several unexpected minor cleavage products were also observed (Supplementary Fig. 19). These side products resulted from partial self-complementarity of the guide DNA, leading to cleavage of guide DNA during the Ago loading process and acceptance of the shorter cleavage products as guide DNAs. We therefore tested new guide and target sequence pairs, identical to the let-7 sequence selected for crystallography. The let-7 guide and target molecules yielded a single cleavage band, with DNA being a better substrate than RNA (Supplementary Fig. 20a). Target DNA cleavage occurred in the presence of Mg2+ or Mn2+, but not Ca2+ (Supplementary Fig. 20b), supporting single and multiple turnover (Supplementary Fig. 20c). Cleavage products started to accumulate after a short (about 2 min) lag phase, at an approximate rate constant of 0.1 min−1 under single turnover (0.5 μM target, Supplementary Fig. 20c) and 0.2 to 0.4 min−1 under multiple turnover conditions (5 μM target, Supplementary Fig. 20c). These rate constants indicate that our cleavage conditions are approaching substrate saturation and that product release is not rate limiting. We also included cleavage experiments using mutant Ago proteins that were utilized for the crystal structures (Figs. 1–3) and tested for DNA-guided RNA (Supplementary Fig. 21a) or DNA (Supplementary Fig. 21b) target cleavage. Of the mutant Agos, only the N546 mutant showed some residual activity, and product formation was reduced >500-fold.
Minimal target DNA requirements
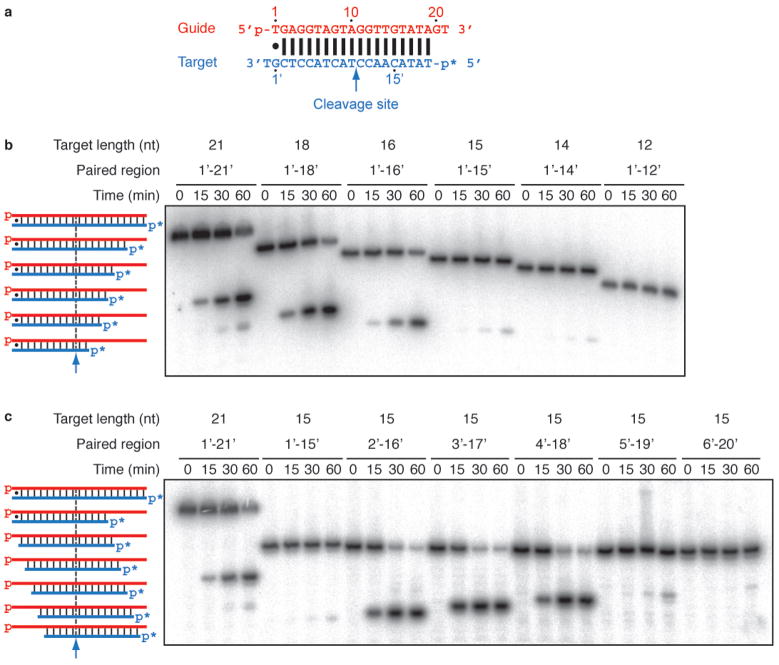
Previously, we showed that luc guide DNA strands as short as 9-nt promoted target RNA cleavage but the minimal target length was not addressed31. We first shortened the let-7 DNA target (Fig. 4a) from its 5′ end (Fig. 4b). Truncation of the target to 16-nt did not alter cleavage activity, but 15- and 14-nt targets showed 120- and 400-fold reduced cleavage rates, respectively, and a 12-nt target was not cleaved. This indicates that residues 17′ and higher do not contribute to cleavage, and further supported by our finding that 21-nt or 24-nt DNA targets, in which regions 17′–21′, or 17′–24′ were unpaired with same size guides, showed similar activity compared to their fully-paired versions (Supplementary Fig. 22).
Figure 4. Effect of complementarity and length on target DNA cleavage by T. thermophilus Ago.
0.5 μM 5′-phosphorylated guide oligodeoxynucleotides were incubated with 0.5 μM T. thermophilus Ago at 55 °C for 30 min in the presence of 5 mM Mg2+ followed by the addition of 0.5 μM 5′-32P-radiolabeled (*) DNA target and further incubation at 75 °C for the indicated time. Cleavage products were resolved on denaturing polyacrylamide gels and visualised by phosphoimaging; for DNA sequences, see Supplementary Table 4. a, Schematic of the DNA duplex that was manipulated in this experiment; the cleavage site is indicated by an arrow, the position of the 32P label by an asterix. b, Shortening of the target DNA from its 5′ end. Alterations of the target DNA and corresponding paired structure are illustrated to the left. Target DNA cleavage was performed at 65 °C rather than 75 °C to facilitate hybridization of shortened targets. c, Positional variation of 15-nt target DNAs. For labelling and reaction conditions, see panel b.
To examine the importance of the 3′ end of the target, we tested 15-nt DNA target strands displaced in 1-nt steps relative to the let-7 target (Fig. 4c). DNA targets covering 2′–16′, 3′–17′, and 4′–18′ showed cleavage activity similar or better than 21-nt long targets, but 100- and 500-fold reduced rates were obtained for targets covering 5′–19′ and 6′–20′. These experiments indicate that positions 1′ to 3′ were dispensable for target cleavage.
In summary, positions 4′ to 16′ need to be paired to facilitate efficient target DNA cleavage when presented to T. thermophilus Ago loaded with 21-nt guide DNA. On the other hand, guide DNA as short as 9-nt promoted T. thermophilus Ago cleavage of target RNA, indicating that base-pairing involving residues 10′ to 16′ per se is not essential. Short guides, in contrast to 21-nt guides, are unable to occupy the PAZ domain with their 3′ ends. Therefore, we speculate that transitioning of the Ago ternary complex into a cleavage-active conformation requires either the release of the guide 3′-end PAZ interaction or its initial absence as seen for short guide strands. Release of the PAZ guide 3′-end interactions is driven by base-pairing including position 16′ of a target.
In the light of this discussion, it may appear surprising that the Ago conformation of the 15- and 19-nt target-RNA-containing structures were similar. However, the thermodynamic stability of DNA-RNA duplexes is different from DNA/DNA duplexes39 and fewer but more stable base-pairs may facilitate the switch to the active conformation. In support of this view, we observed that the cleavage activity for the 15-nt (position 1′–15′) and a 16-nt (position 1′–16′) target RNAs (Supplementary Fig. 23) only differed 1.4-fold and was comparable to that of the longer target RNA (Supplementary Fig. 21).
Our crystal structures also indicated that base pairs involving positions 17′ or higher could not form due to steric clashes with the N-terminal domain. To test, if the propagation of the duplex beyond position 16′ could contribute to catalysis, we tested Ago deletion mutants Del 1–106 and Del 1–177, but found that they lost all activity (Supplementary Figs. 21a and 21b). This suggests that the N domain also plays an important role for transitioning or stabilizing the active conformation of the ternary complex, and could possibly even affect other steps including loading of the guide DNA, which were not tested.
Role of target DNA sugar-phosphate backbone during cleavage
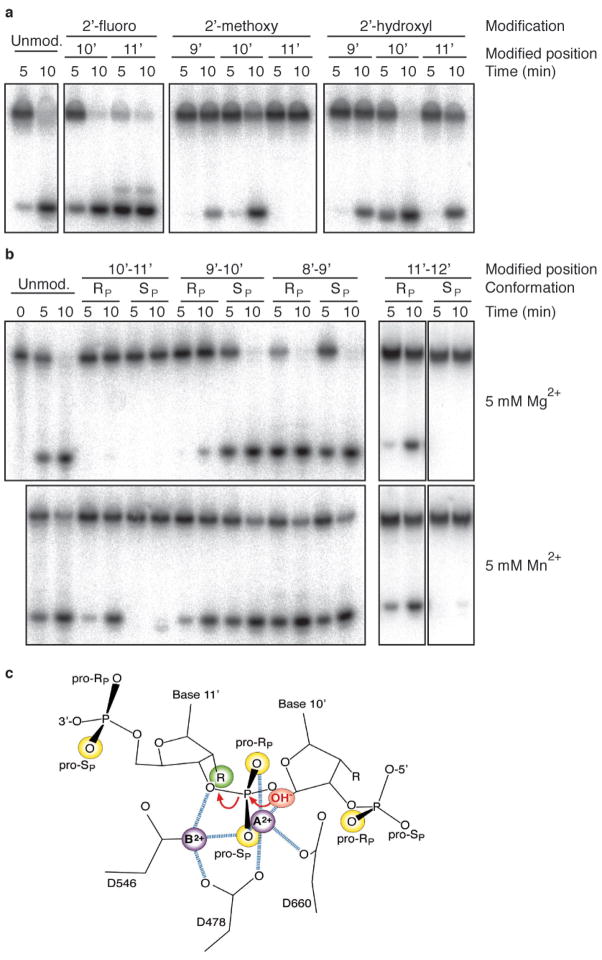
To assess the contribution of sugar and phosphate residues during target DNA recognition and cleavage, we introduced 2′-hydroxyl (OH) and 2′-methoxy (Ome) modifications at positions 9′, 10′, or 11′, as well as 2′-fluoro (F) at positions 10′ or 11′ (Fig. 5a, Supplementary Fig. 24, Supplementary Table 4). OH, Ome, and F 2′ modified ribonucleosides favour the A-helical C3′-endo ribose conformation, whereas deoxynucleotides are preferable in the B-helical C2′-endo conformation40, and therefore stabilize double-helical structures. 2′-Substitutions at residue 11′, which are immediately adjacent to the cleaved phosphodiester bond, showed the most profound effects on cleavage. The 2′-F substitution enhanced the single (Fig. 5a) and multiple (Supplementary Fig. 24) turnover cleavage rate approximately 4-fold to 6-fold, respectively, compared to 2′-H, presumably because the electronegative 2′-F group is able to stabilize the developing negative charge of the 3′ oxygen leaving group during the transition state. 2′-OH at residue 11′ reduced the cleavage rate 2-fold compared to 2′-H, and 2′-Ome completely abrogated cleavage, presumably by affecting the hydration pattern optimal for stabilization of the transition state. Also, there is no evidence for hydrogen bonding of the 2′ residue to neighbouring nucleotides or amino acid side chains. Taken together, the drastic effects on reaction rates by 2′ modifications at the 11′ position cannot be rationalized by simple differences in sugar conformation, but by a combination of electronic and steric effects differentially affecting the transition state. Modifications of the 2′ position one nucleotide removed form the cleavage site showed less or no effect; by contrast, position 9′ showed an unanticipated 3-fold reduction in rate for 2′-OH and 2′-Ome (Fig. 5a).
Figure 5. Effect of sugar-phosphate backbone modifications on target DNA cleavage by T. thermophilus Ago.
Cleavage experiments were performed as described in Fig. 4. a, 2′-Fluoro-, 2′-methoxy-, and 2′-hydroxyl-substitutions of single 2′-deoxyribose residues of the target DNA strand at and nearby the cleavage site. The control target (unmod.) was the unmodified oligodeoxynucleotide. b, Phosphorothioate modification of the target DNA. The phosphate configuration (RP or SP) of the phosphorothioate diastereomers is indicated. Cleavage assays were performed in the presence of either Mg2+ or Mn2+ cations. Note that the experiment for the 11′–12′ isomers was a different experiment, where overall reaction rates were slower. For the complete experiment see Supplementary Fig. 25. For sequences of oligonucleotides see Supplementary Table 4. c, Structure of the cleavage site modelling the attack of the hydroxyl nucleophile. Phosphate oxygen and active site carboxylate oxygens coordinated to metal ions A and B (purple spheres), with distances less than 2.5 Å are shown as blue dashed lines. The coordination of the carboxylate oxygen from D546 to metal ion B is hidden in the projection. The phosphate oxygens and 2′ residues sensitive to modification are shown in yellow and green spheres, respectively; R = 2′-H, OH, F, or Ome. The red arrows indicate the attack of the hydroxyl nucleophile modelled to be directly coordinated by metal ion A, and the stabilization of the developing negative charge of the 3′ oxyanion leaving group by metal ion B.
To probe the role of phosphate oxygens, which can coordinate structurally or catalytically important divalent metal ions41, we synthesized the mixed phosphorothioate diastereomers located between residues 8′–9′, 9′–10′, 10–11′, or 11′–12′, and purified by reverse phase HPLC the SP form to >85%, and the RP form to >97% purity. Cleavage reactions were performed in the presence of either 5 mM Mg2+, which preferably coordinates to oxygen, or 5 mM Mn2+, which preferably coordinates to sulphur. Phosphorothioate substitution at the cleavage site, position 10′–11′, showed the most profound effects (Fig. 5b). In Mg2+-containing buffer, the SP form was inactive and the RP form reduced 200-fold in single-turnover cleavage rates. The loss in activity of the RP form was rescued by Mn2+ yielding a less than 2-fold reduction compared to 2′-H; however, the SP form remained inactive. Phosphorothioate substitutions more distant to the cleavage site either had no effect (RP and SP at position 8′–9′, SP at position 9′–10′ and RP at position 11′–12′) or were reduced by 15-fold and greater than 80-fold for RP at position 9–10′ and SP at position 11′–12′, respectively, and rescued by Mn2+ to less than 2-fold and greater than 20-fold, respectively. Non-bridging phosphate oxygens sensitive to sulphur substitution and responsive to Mn2+ rescue are believed to directly coordinate to Mg2+, and the interaction stabilizes ground and transition states of the cleavage reaction to a similar degree. A phosphate oxygen sensitive to phosphorothioate substitution, but without metal ion rescue feature, such as the pro-SP oxygen at the cleavage site, is likely to differentially stabilize the transition state versus the ground state. Substituting the 10′–11′ pro-SP oxygen by sulphur increases the bond length by about 0.6 Å, a distance sufficient to perturb the complex network of interactions coordinated at this phosphate oxygen (Fig. 5c). The pro-SP oxygen is coordinated to metal ions A and B, with A positioning the attaching hydroxyl ion nucleophile and B stabilizing the leaving 3′ oxyanion. The importance of stabilizing the leaving group was also documented above by the effects of modifications at the adjacent 2′ position. In contrast, the pro-RP oxygen at the cleavage site is only coordinated to metal ion A, and the sulphur substitution was rescued with Mn2+, indicating more flexibility for positioning the nucleophile by metal ion A.
Structural overview and functional implications
In summary, our current structures of ternary complexes with catalytic mutants of T. thermophilus Ago have defined the positioning of the guide DNA-target RNA A-form duplex relative to the catalytic Asp residues of the RNase H fold of the PIWI domain, thereby establishing the molecular basis for site-specific cleavage at the phosphate bridging the 10–11 step of the target strand. Additional structural studies of ternary complexes with wild-type Ago have identified two Mg2+ cations within the catalytic pocket, located on either side of the cleavable phosphate, thereby positioned to mediate the cleavage chemistry. Both ends of the guide strand are anchored in the ternary complex composed of one turn of DNA-RNA duplex spanning the seed segment and cleavage site, but consistent with a ‘two-state’ model, the 3′-end is released from the PAZ pocket following propagation of the guide-target duplex by three additional base pairs. Strikingly, the guide DNA and target RNA form a regular A-form helix spanning a maximum of 15 base pairs (positions 2–16), with the Ago scaffold undergoing pivot-like domain movements as the target RNA zippers up through pairing with its guide template.
The kinetic effect of target site phosphorothioate substitution and 2′ modification during Ago-mediated DNA cleavage are rationalized by the crystal structure, and consistent with the mechanism of RNase H cleavage studied in other systems38. The absence of amino acid side chains able to interrogate whether the target presented at the active site is RNA or DNA might suggest that DNA could be a more likely target of this bacterial Ago protein, as seen for other members of the retroviral integrase superfamily into which Ago proteins belong38.
METHODS SUMMARY
Wild-type and mutant T. thermophilus Ago proteins were over-expressed from E. coli and purified by chromatography as described previously30. Crystals were obtained by the hanging-drop or sitting-drop vapor diffusion. The ternary Ago complex was generated in a stepwise manner by initially mixing the protein with 21-mer 5′-phosphorylated guide DNA, followed by addition of different length target RNAs. All wild-type and mutant Ago complex structures were determined by molecular replacement using the domains of the binary Ago complex structure (Protein Data Bank accession code 3DLH)30 as search models. Cleavage assays were undertaken with let-7 guide and target oligodeoxynucleotides. Details of all crystallographic and biochemical procedures are listed in Methods.
Supplementary Material
Acknowledgments
The research was supported by funds from the NIH and the Starr Foundation to DJP and NIH and from HHMI to TT. We would like to thank the staff of NE-CAT beam lines at the Advanced Photon Source (APS), Argonne National Laboratory, and the X-29 beamline at the Brookhaven National Laboratory, supported by the US Department of Energy, for assistance with data collection. We thank Dr. Zhanxin Wang for assistance with x-ray data collection at APS.
Footnotes
Full Methods and associated references are available in the on line version of the paper at www.nture.com/nature.
Supplementary Information is linked to the on line version of the paper at www.nature.com/nature.
Conflict of interest
TT is a cofounder and scientific advisor to Alnylam Pharmaceuticals and an advisor to Regulus Therapeutics.
Author Contributions YW and GS expressed and purified wild-type T. thermophilus Ago and its catalytic mutants, as well as grew crystals of the various ternary complexes. HL collected x-ray diffraction data on the various NE-CAT beam lines, and YW solved the structures of these ternary complexes. The structural studies were undertaken under the supervision of DJP. SJ was responsible for the cleavage assays on Ago with modified DNA and RNA target strands, and GSW purified the phosphorothioate diastereomers and quality controlled oligonucleotides, under the supervision of TT. DJP and TT were primarily responsible for writing the structural and biochemical contents of the paper, respectively and all authors read and approved the submitted manuscript.
Author Information The structures of ternary complexes of T. thermophilus Ago have been deposited to the Protein Data Bank. The accession codes are
N546 mutant Ago - 12-nt target RNA: 3HO1
E546 mutant Ago - 15-nt target RNA: 3HJF
N478 Ago mutant - 19-nt target RNA: 3HK2
Wild-type Ago - 19-mer target RNA, 50 mM Mg: 3HM9
Wild-type Ago - 19-mer target RNA, 80 mM Mg: 3HVR
Second crystal form of wild-type Ago - 20-mer target RNA containing two mismatches31: 3HXM
References
- 1.Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- 2.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 3.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 4.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 5.Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol. 2005;15:331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 7.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 8.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 10.Parker JS, Roe S, Barford D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 2004;23:4727–4737. doi: 10.1038/sj.emboj.7600488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan YR, et al. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, et al. Argonaute2 is the catalytic engine of RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 13.Rivas FV, et al. Purified Ago2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 14.Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma JB, et al. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma J, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the Paz domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3′-end recognition by the Argonaute2 Paz domain. Nature Struct Mol Biol. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- 18.Martinez J, Tuschl T. RISC is a 5′-phosphomonoester-producing RNA endonuclease. Genes Dev. 2004;18:975–980. doi: 10.1101/gad.1187904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwartz DS, Tomari Y, Zamore PD. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr Biol. 2004;14:787–791. doi: 10.1016/j.cub.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 20.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21-and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ameres LA, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 22.Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8:23–26. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 23.Parker JS, et al. Enhancement of the seed-target recognition step in RNA silencing by a PIWI/MID domain protein. Mol Cell. 2009;33:204–214. doi: 10.1016/j.molcel.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker JS, Barford D. Argonaute: a scaffold for the function of short regulatory RNAs. Trends in Biochem Scis. 2006;31:622–630. doi: 10.1016/j.tibs.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 25.Patel DJ, et al. Structural biology of RNA silencing and its functional implications. Cold Spring Harbor Symposia on Quantitative Biology. 2006;71:81–93. doi: 10.1101/sqb.2006.71.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tolia NH, Joshua-Tor L. Slicer and the argonautes. Nature Chem Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 27.Jinek M, Doudna J. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 28.De Fougerolles A, Vornlocher HP, Maraganore L, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nature Rev Drug Discovery. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, et al. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, et al. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mi S, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′-terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montgomery TA, et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 34.Filipowicz W. The nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 35.Chu SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PURE-CLIP. Cell. 2009 doi: 10.1016/j.cell.2010.03.009. under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: Substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 38.Nowotny M. Retroviral integrase superfamily: the structural perspective. EMBO reports. 2009;10:144–151. doi: 10.1038/embor.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner DH. Thermodynamics of base pairing. Curr Opin Strut Biol. 1996;6:299–304. doi: 10.1016/s0959-440x(96)80047-9. [DOI] [PubMed] [Google Scholar]
- 40.Freier SM, Altmann KH. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1997;25:4429–4443. doi: 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verma S, Eckstein F. Modified oligonucleotides: synthesis and strategy for users. Annu Rev Biochem. 1998;67:99–134. doi: 10.1146/annurev.biochem.67.1.99. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.