Abstract
Background
Testicular germ cell tumors (TGCT) are the most frequent cancers among young men. There is a clear familial component to TGCT etiology, but no high-penetrance susceptibility gene has been identified. Epigenetic aberrations of the genome represent an alternative mechanism for cancer susceptibility; and, studies suggest that epigenetic changes that influence cancer risk can be inherited through the germline. Global DNA hypomethylation has been associated with the risk of cancers of the bladder and head/neck.
Methods
We performed a pilot study of global methylation at long interspersed nuclear elements-1 (LINE-1) in peripheral blood DNA isolated from 466 family members of 101 multiple-case testicular cancer families.
Results
Investigating the correlation of LINE-1 methylation levels among parent-child pairs independent of affection status (n = 355) revealed a strong positive association only between mother-daughter (r = 0.48, P = <0.001) and father-daughter pairs (r = 0.31, P = 0.02), suggesting gender-specific inheritance of methylation. Incorporating cancer status, we observed a strong correlation in LINE-1 methylation levels only among affected father-affected son pairs (r = 0.49, P = 0.03). There was a marginally significant inverse association between lower LINE-1 methylation levels and increased TGCT risk, compared with healthy male relatives (P = 0.049).
Conclusions
Our data suggest that heritability of LINE-1 methylation may be gender-specific. Further, the strong correlation between LINE-1 methylation levels among affected father-affected son pairs suggests that transgenerational inheritance of an epigenetic event may be associated with disease risk. Larger studies are needed to clarify these preliminary observations.
Background
Testicular germ cell tumors (TGCT) are the most commonly diagnosed cancer among young American men aged 20-35 years, and their incidence has doubled over the last 40 years [1]. Family history of TGCT is one of the strongest and most consistent risk factors for this tumor. Brothers of affected cases have an 8-to 10-fold increased relative risk compared with the general population and fathers/sons a 4-and 6-fold higher risk [2,3]. These high familial risks suggest that inherited susceptibility and/or environmental factors that cluster in families may account for a significant portion of TGCT cases. Testicular microlithiasis (TM), a condition characterized by the presence of calcium deposits within the seminiferous tubules, aggregates in families and has been associated with testicular malignancy [4,5]. Linkage and candidate gene studies [6-10] have identified several genomic regions of interest, including Xq27, 2p23, 3p12, 3q26, 12p13-q21 and 18q21-q23. Candidate gene studies have identified two loci of interest, i.e., the Y-chromosome gr/gr deletion [9] and the PDE11A gene [8], while recent GWAS analyses identified KIT-ligand, SPRY4 and BAK1) [6,7]. However, a specific high-penetrance susceptibility gene has yet to be proven, suggesting that the combined contribution of multiple common genetic variants of lower penetrance may account for the inherited component of TGCT susceptibility [10].
Epigenetic changes in the genome, such as aberrant DNA methylation, are an increasingly recognized contributor to cancer development. TGCTs have distinct DNA methylation profiles, and parallels have been observed between the epigenetic properties of TGCTs and embryogenesis (e.g., DNA methylation increases with differentiation) [11]. The seminoma genome is essentially devoid of DNA methylation, while the nonseminoma genome is less extensively hypomethylated and has variable CpG island hypermethylation levels [12,13]. A few tumor suppressor genes are inactivated by DNA promoter hypermethylation in a small proportion of TGCTs, while DNA hypomethylation of testis-and cancer-associated genes and unmethylated XIST are frequently observed in TGCTs (Reviewed in [11]). Imprinting defects have also been observed in TGCT somatic tissues, suggesting that epigenetic defects may be present. In the aggregate, the data suggest that methylation may provide an alternate genetic mechanism for TGCT susceptibility.
Transgenerational epigenetic inheritance has been well studied and documented in many eukaryotic organisms (e.g., maize, yeast, Drosophila, mice) [14-16], and recent human studies suggest that epigenetic aberrations that influence cancer risk can be inherited through the germline from parent to child [17-20]. Epigenetic patterns have been observed to segregate in both Mendelian and non-Mendelian patterns, as well as in a pattern consistent with environmental exposure. Non-Mendelian and environmentally-induced transgenerational inheritance of epimutations is supported by model organism data [21-27]. There are several potential modes of transgenerational epigenetic inheritance, including paramutations, aberrant gene imprinting, and dsRNA processes (Reviewed in [28]). Retrotransposons are thought to be resistant to epigenetic reprogramming during embryogenesis in mice [29], and thus may play a role in epigenetic heritability (e.g., if inserted in or near the affected gene). A recent mouse model study found that transgenerational epigenetic inheritance through the germline controls susceptibility to TGCTs [21].
Global DNA hypomethylation, characterized by a global loss of 5-methylcytosine (5-mC), contributes to malignant transformation by activating oncogenes or latent retrotransposons, such as LINE-1, and by loss of imprinting [30]. LINE-1 retrotransposons are hypomethylated in many cancers and their lack of repression may contribute to genome disorganization, expression changes, and chromosomal instability [31]. The nucleotide patterns of LINE-1 repeats have been shown to both segregate in human pedigrees, and to provide an individual-specific fingerprint [32]. LINE-1 insertion dimorphisms may reflect DNA mutations or methylation changes [32]. Methylation levels at LINE-1 loci have been shown to differ among different loci in normal tissues, suggesting that cis-elements near each LINE-1 may influence their epigenetics [33]. In addition, locus-specific LINE-1 methylation has been shown to be differentially influenced by carcinogenic processes, depending on where the LINE-1s are located in the genome [33]. LINE-1 retrotransposons are strongly expressed in TGCTs [31,34,35], and it was hypothesized that active LINE-1 retrotransposon events are involved in primordial germ cell differentiation, and the consequential genome instability contributes to malignant transformation [35].
In recent case-control studies, hypomethylation of peripheral blood DNA was found to be associated with increased risks of bladder and head/neck cancers, suggesting that global demethylation in genomic DNA is a potential biomarker of cancer susceptibility [36,37]. We performed a pilot analysis of global DNA methylation levels in peripheral blood among family members from 101 multiple-case familiar testicular cancer families. Methylation at LINE-1 regions using pyrosequencing was used as a proxy for measuring global methylation levels. We examined the heritability of the global genomic methylation phenotype in our families, as well as the association between global (LINE-1) methylation levels and testicular cancer.
Results
DNA from 152 patients with TGCT and 314 unaffected family members from 101 multiple-case testicular cancer families were available for LINE-1 methylation analysis. Table 1 shows the characteristics of the 466 study participants. Males had significantly higher levels of LINE-1 methylation than females (P = 0.002). There were no significant effects of age, smoking, or alcohol consumption on LINE-1 methylation levels (Additional file 1). There was no significant difference in mean LINE-1 methylation between men with and without microlithiasis (P = 0.608).
Table 1.
Global (LINE-1) methylation levels and characteristics of all subjects.
| Variables | No. (%) | Mean level* | 95% CI | P |
|---|---|---|---|---|
| Family, healthy | 255 (54.7) | 79.31 | 79.06-79.56 | |
| Family, other cancer | 39 (8.4) | 79.11 | 78.47-79.74 | |
| Family, other condition§ | 20 (4.3) | 79.71 | 78.84-80.58 | |
| TGCT patients | 152 (32.6) | 79.13 | 78.78-79.45 | 0.377¶ |
| Multi-case families | 118 (77.6) | 79.50 | 79.12-79.88 | |
| Sporadic bilateral TGCT | 34 (22.4) | 78.91 | 78.21-79.60 | 0.142¥ |
| Gender‡ | ||||
| Male | 114 (44.7) | 79.60 | 79.26-79.95 | |
| Female | 141 (55.3) | 78.87 | 78.55-79.18 | 0.002† |
| Patient TGCT type | ||||
| NOS | 4 (2.6) | 77.74 | 75.68-79.81 | |
| Seminoma | 73 (48) | 79.19 | 78.70-79.68 | |
| Non-seminoma | 75 (49.3) | 79.59 | 79.11-80.08 | 0.267a |
| Choriocarcinoma | 2 (2.7) | 79.53 | 76.86-82.20 | |
| Embryonal | 15 (20) | 79.88 | 78.90-80.86 | |
| Germinoma | 2 (2.7) | 78.58 | 75.91-81.26 | |
| Mixed germ cell | 34 (45.3) | 79.19 | 78.52-79.86 | |
| Teratocarcinoma | 20 (26.7) | 80.78 | 79.84-81.73 | |
| Yolk sac tumor | 2 (2.7) | 76.06 | 74.27-79.85 | |
| Total participants | 466 | 79.25 | 79.08-79.43 | |
* Adjusted for age;
§ Other testicular conditions (e.g., hernia or cryptorchidism);
‡ Healthy family only;
¶ For methylation level difference between TGCT patients and healthy family, adjusted for age and sex;
¥ For methylation level difference between patients from multiple case families vs. sporadic bilateral TGCT patients, adjusted for age;
† For methylation level difference between males and females, adjusted for age;
a For methylation level difference between TGCT patients with seminoma vs. non-seminoma, adjusted for age;
NOS = not otherwise specified.
Inheritance of global (LINE-1) methylation
We assessed whether LINE-1 methylation levels in offspring were correlated with either paternal or maternal levels (n = 355 pairs), irrespective of patient status (Table 2). These analyses revealed a high correlation in LINE-1 methylation between mother-daughter (r = 0.48, P = 0.0002; Figure 1) and father-daughter pairs (r = 0.31, P = 0.021), but only a weak correlation between mother-son (r = 0.18; P = 0.047) and father-son pairs (r = 0.16; P = 0.114).
Table 2.
Intra-familial correlations adjusted for age.
| Relationship | No. of pairs | r† | P value |
|---|---|---|---|
| Parent-children | 355 | 0.228 | <0.0001 |
| Mother-children | 190 | 0.271 | 0.0002 |
| Mother-daughter | 59 | 0.476 | 0.0002 |
| Mother-son | 131 | 0.176 | 0.047 |
| Father-children | 165 | 0.187 | 0.018 |
| Father-daughter | 58 | 0.308 | 0.022 |
| Father-son | 107 | 0.157 | 0.114 |
†Spearman rank correlation coefficient adjusted for parental age, offspring age, and disease status of the males.
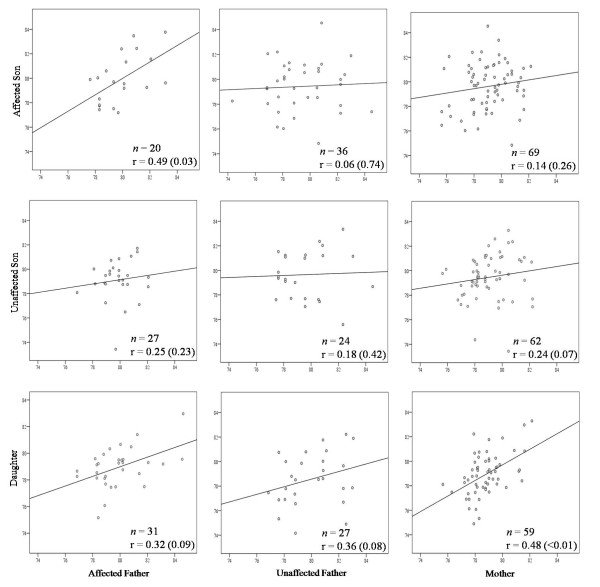
Figure 1.
Familial relations of global (LINE-1) methylation levels by patient status. Each plot shows the number of pairs (n), correlation coefficient (r) and corresponding P value. Affected refers to patients with cancer, and unaffecteds are cancer-free.
Figure 1 illustrates LINE-1 methylation level correlations between parent-child pairs after stratifying by testicular cancer status. There was a statistically significant correlation in LINE-1 methylation levels between affected father-affected son pairs (r = 0.49, P = 0.034). Affected father methylation levels were not significantly correlated with unaffected son levels (P = 0.228). Furthermore, when comparing unaffected father-and mother-affected son pairs, no significant associations were demonstrated (r = 0.06, P = 0.738, and r = 0.14, P = 0.257, respectively). There were no significant differences between the correlations among unaffected sons and daughters with affected and unaffected fathers (Figure 1).
Patients with testicular germ cell tumors and global (LINE-1) methylation
As previously shown, younger age (26-50 years of age) and the presence of testicular microlithiasis were each significantly associated with testicular cancer risk (independent of methylation levels; Table 3). There was no statistically significant difference between LINE-1 methylation levels in TGCT patients and healthy male family members (patient mean: 79.1 [95% CI 78.8-79.5] versus healthy male mean: = 79.6 [95% CI 79.3-79.9]; P = 0.297). A comparison of LINE-1 methylation as a continuous variable yielded a marginally statistically significant association between decreasing LINE-1 methylation level and testicular cancer (OR per unit change = 1.15, 95% CI 1.00-1.32; P = 0.049). Males subjects were categorized into tertiles of LINE-1 methylation level based on the methylation distribution among healthy male family members. An upward trend, not statistically significant, was observed when comparing the lowest and middle tertiles to the highest (OR = 1.41 [95% CI 0.71-2.80] and 1.83 [95% CI 0.91-3.66], respectively; Ptrend = 0.089) (Table 4). Similarly, with LINE-1 methylation dichotomized at the median in healthy male controls, lower methylation was associated with a non-significant increase in testicular cancer risk (OR = 1.48, 95% CI 0.85-2.55).
Table 3.
Association of individual variables with patient status.
| Variables | Healthy Males | TGCT Cases | OR† (95% CI) | ||
|---|---|---|---|---|---|
| No. | (%) | No. | (%) | ||
| Age: | |||||
| 0-25 | 27 | (23.68) | 11 | (7.28) | 1.00 (ref) |
| 26-50 | 34 | (29.82) | 108 | (71.52) | 8.30 (3.62, 19.05) |
| 51-75 | 47 | (41.23) | 31 | (20.53) | 1.72 (0.72, 4.10) |
| 76-100 | 6 | (5.26) | 1 | (0.66) | 0.52 (0.05, 4.92) |
| Alcohol use: | |||||
| Never | 25 | (30.86) | 39 | (26.90) | 1.00 (ref) |
| Former | 14 | (17.28) | 33 | (22.76) | 1.12 (0.45, 2.77) |
| Current | 42 | (51.85) | 73 | (50.34) | 0.78 (0.38, 1.64) |
| Smoking status: | |||||
| Never | 41 | (50.00) | 97 | (66.44) | 1.00 (ref) |
| Former | 28 | (34.15) | 38 | (26.03) | 0.67 (0.33, 1.35) |
| Current | 13 | (15.85) | 11 | (7.53) | 0.42 (0.16, 1.14) |
| Microlithiasis | |||||
| No | 23 | (88.46) | 32 | (59.26) | 1.00 (ref) |
| Yes | 3 | (11.54) | 22 | (40.74) | 5.69 (1.41, 23.04) |
† Odds ratios (95% confidence intervals) adjusted for age and/or microlithiasis status.
Table 4.
Association of TGCT with blood-derived DNA global (LINE-1) methylation levels.
| Relative methylation level in tertiles | Healthy Males§ | TGCT Cases* | OR† (95% CI) | P value |
|---|---|---|---|---|
| No. (%) | No. (%) | |||
| All patients | ||||
| Low (≤78.8) | 38 (33.3) | 57 (37.8) | 1.83 (0.91, 3.66) | 0.089 |
| Middle (78.9-80.6) | 39 (34.2) | 52 (34.4) | 1.41 (0.71, 2.80) | |
| High (>80.6) | 37 (32.5) | 42 (27.8) | 1.00 (ref) | |
| Patients with seminoma | ||||
| Low | 38 (33.3) | 29 (39.7) | 2.21 (0.90, 5.43) | 0.085 |
| Middle | 39 (34.2) | 27 (37.0) | 1.76 (0.72, 4.30) | |
| High | 37 (32.5) | 17 (23.3) | 1.00 (ref) | |
| Patients with non-seminoma | ||||
| Low | 38 (33.3) | 26 (35.1) | 1.56 (0.67, 3.66) | 0.306 |
| Middle | 39 (34.2) | 23 (31.5) | 1.18 (0.50, 2.78) | |
| High | 37 (32.5) | 25 (33.8) | 1.00 (ref) | |
| Multi-case families‡ | ||||
| Low | 38 (33.3) | 41 (35.3) | 1.68 (0.80, 3.49) | 0.174 |
| Middle | 39 (34.2) | 41 (35.3) | 1.51 (0.74, 3.10) | |
| High | 37 (32.5) | 34 (29.3) | 1.00 (ref) | |
| Bilateral TGCT families¶ | ||||
| Low | 38 (33.3) | 15 (44.1) | 2.12 (0.75, 6.01) | 0.218 |
| Middle | 39 (34.2) | 11 (32.4) | 1.23 (0.43, 3.46) | |
| High | 37 (32.5) | 8 (23.4) | 1.00 (ref) | |
† Odds ratios (95% confidence intervals) adjusted for age and microlithiasis status;
§ Males without cancer or other testicular abnormalities;
* Only 151 total TGCT cases, one subject with teratocarcinoma failed bisulfite conversion;
‡ Families with two or more cases of TGCT;
¶ A single family member had bilateral testicular cancer.
Associations between lower LINE-1 methylation and TGCT risk were stronger in patients with seminoma versus non-seminomatous tumors, and in bilateral TGCT patients versus patients from multi-case families (Table 4). We also examined the association between TGCT and LINE-1 methylation after stratification by age (data not shown). For those < 50 years, the OR for the lowest LINE-1 methylation level (1st tertile) was 1.61 (95% CI 0.57-2.59), versus 1.48 (95% CI 0.48-4.60) for those ≥50.
Discussion
We found that males had significantly higher global (LINE-1) methylation levels than females, consistent with prior reports [37-39]. This difference has been variably attributed to gender-related dietary differences, reduced circulating levels of folate due to female menstruation, and/or the presence of an additional X chromosome in female cells [37,39]. We found no association between LINE-1 methylation levels and age [36,37,40], smoking [36,38], or alcohol usage [36,37], also consistent with prior studies.
LINE-1 methylation levels of offspring were found to be significantly positively correlated with parental levels, particularly between mother-daughter, father-daughter, and affected father-affected son pairs. There are limited data investigating epigenetic inheritance in humans, although it has been suggested that epimutations may be maintained more frequently during oogenesis [41,42] than during spermatogenesis [43]. Differences in epigenetic reprogramming between maternal and paternal genomes have been observed in mouse and drosophila studies [22,23,25-27]. In mice, epigenetic inheritance at a locus involved in fur color demonstrated that there was incomplete erasure of an epigenetic modification at a retrotransposon when passed through the female germ line [23].
However, others have shown that exposure to an endocrine disruptor during embryonic gonadal sex determination in rats induced permanent DNA methylation imprinting of the male germline (i.e., sperm) that was transmitted to subsequent generations [25-27]. This epigenetic alteration was associated with transgenerational disease states (approximately 85% frequency), including abnormalities in testis function, male infertility, and tumor development. This observation illustrates how an environmental exposure can epigenetically reprogram the germline and promote adult-onset defects in the germline, providing one possible mechanism by which environmental exposures may promote TGCTs [44].
Transgenerational epigenetic interactions have been shown to control susceptibility to TGCTs in mice with Dead-end homologue 1 (Dnd1) mutations [21]. It was suggested that these epigenetic modifiers may mark the germline, and offspring that also carry a genetic susceptibility factor will have an increased risk of TGCTs; or, environmental factors may influence genes to act as mediators and to monitor cellular conditions that then induce epigenetic alterations [28]. Lam et al. [21] suggested that, in humans, transgenerational epigenetics may explain the 2-3 fold risk difference in brothers and sons of TGCT affected individuals. The strong correlation we observed between LINE-1 methylation levels among affected father-affected son pairs may reflect an inherited or germline LINE-1 hypomethylation abnormality. Such an inherited epigenetic event could be associated either with an environmental factor or genetic susceptibility.
We observed an association between TGCT and global hypomethylation that was marginally significant when considering LINE-1 methylation as a continuous variable after controlling for potential confounders and testicular cancer risk factors (age and microlithiasis). Our study had ≥80% statistical power to detect ORs of 1.6 and 2.4 with LINE-1 methylation in the second and first tertiles, respectively, assuming a type I error of 0.05. Other studies have found that global hypomethylation in DNA from blood was associated with bladder and head/neck cancers [36,37]. Genomic global (LINE-1) methylation may be an independent risk factor or a phenotypic marker of cancer risk associated with genetic instability, aberrant epigenetic regulation, or other factors.
Our small sample size with attendant reduced statistical power to detect differences in LINE-1 methylation levels is a limitation of this study, although ours is the largest cohort of multiple-case families available for analyses of this kind. The family-based design of this study also limits our exploring the association between TGCT and global hypomethylation levels, since we have observed that this may be an heritable phenotype; a non-family based matched control group would be a useful follow-up. However, the unique multiple-case family design of this study is a strength for investigating inheritance, which allowed us to study multiple individuals per family with and without cancer, and to eliminate potential confounding due to ethnic background or population stratification.
LINE-1 retrotransposons are highly-activated in TGCTs and may lead to insertional mutations, transcriptional deregulation, DNA breaks, and an increased frequency of recombinational events [31,34,35]. Little is known regarding the mechanisms underlying hypomethylation changes of LINE-1 sequences; however, available data suggest that LINE-1 retrotransposon events may contribute to genome instability and malignant transformation. This is the first study to show that global LINE-1 methylation levels may be inherited from parent to child in humans, and that hypomethylation is associated with testicular cancer risk. More research is needed to help explain differences in maternal/paternal effects and mechanisms of transgenerational epigenetic inheritance.
Conclusions
The results of this study suggest that LINE-1 methylation is heritable in humans, and that transgenerational inheritance of LINE-1 methylation levels may be associated with testicular cancer risk. The incomplete penetrance and non-Mendelian inheritance of epigenetic variation are consistent with complex disease phenotypes, which familial testicular cancer displays, and thus it may provide some insight into the basis of this complex disease. It has proved difficult to identify high-penetrance TGCT susceptibility genes in humans, despite a high familial risk and likely inherited susceptibility. Transgenerational epigenetic mechanisms could help explain the difficulty of finding classical TGCT susceptibility genes in humans. Larger studies are warranted to further explore the association between genomic global hypomethylation and TGCT risk, and to investigate the hypothesis that LINE-1 epigenetic inheritance is associated with TGCT susceptibility.
Methods
Study population
Families with two or more cases of TGCT or a single family member with bilateral testicular cancer were ascertained as part of the NCI Clinical Genetics Branch Familial TGCT study, details of which are described elsewhere [4]. Men with sporadic bilateral TGCT were included because of the known excess risk of bilateral affection in familial TGCT [45]. In brief, "cases" were men with histologically-confirmed cancer of one or both testes. Family members were invited to participate in the study if they were first-degree relatives of a case (aged ≥12yrs), spouses of a case who had participating children, non-first-degree blood relatives who provided a genetic link between cases, or blood relatives with cancer. All participants completed detailed questionnaires of medical and family history and risk factors, provided blood samples, and a subset of participants underwent a detailed medical evaluation at the NIH Clinical Center. Blood was collected between 2003 and 2006 and stored at the NCI/CGB Biorepository. The majority of patients underwent blood collection 5 or more years after they were diagnosed with TGCT (mean time between diagnosis and blood draw was 9 yrs; median time = 5 yrs). We observed no significant correlation between number of years from cancer diagnosis/treatment to study blood draw and methylation level (P = 0.492; data not shown). The parent study has been reviewed and approved by the NCI Institutional Review Board (NCI Protocol 02-C-0178; NCT00039598), and all participants provided written informed consent.
DNA extraction and bisulfite treatment
Genomic DNA was extracted from fresh whole blood by standard methods. For bisulfite conversion, DNA was treated using EZ DNA Methylation-Gold™ Kit (Zymo Research Corp., CA, USA) according to the manufacturer's recommendations. The final elution was performed with 30 μl of M-Elution Buffer.
LINE-1 PCR and pyrosequencing
DNA methylation (%5-mC) of LINE-1 was quantified using PCR-pyrosequencing of the bisulfite-treated DNA by EpigenDx Laboratory Service (Worcester, MA), as previously described [46]. In brief, the bisulfite-treated DNA was amplified by PCR using primers designed toward a consensus LINE-1 sequence. A 50-μL PCR was carried out in 25-μL GoTaq Green Master mix (Promega, Madison, WI, USA), 1 pmol of the forward primer (TTTTGAGTTAGGTGTGGGATATA), 1 pmol of the biotinylated reverse primer (biotin-AAAATCAAAAAATTCCCTTTC), 50 ng of bisulfite-treated genomic DNA, and water. Biotin-labeled primers were used to purify the final PCR product using Sepharose beads. The PCR product was bound to Streptavidin Sepharose HP (Amersham Biosciences, Uppsala, Sweden) and the Sepharose beads containing the immobilized PCR product were purified, washed, and denatured using a 0.2 M NaOH, and washed again using the Pyrosequencing Vacuum Prep Tool (Pyrosequencing, Inc., Westborough, MA, USA), as recommended by the manufacturer. Then, 0.3 μM of the pyrosequencing primer was annealed to the purified single-stranded PCR product and pyrosequencing was performed using the PSQ-HS 96 Pyrosequencing System (Pyrosequencing, Inc.). The relative 5-mC content was expressed as percentage of methylated cytosines divided by the sum of methylated and unmethylated cytosines (5-mC/[5-mC + unmethylated cytosine] = %5-mC). Built-in controls were used to verify bisulphite conversion efficiency. To increase precision, each sample was tested four times for LINE-1 methylation, and the 4-test mean was used in statistical analyses. The coefficient of variation (CV) among 48 blinded replicate samples was 5.7%, and the inter-plate CV was 2.9%.
Statistical analysis
We tested the intra-familial (parent-offspring pairs) correlation of LINE-1 methylation using Spearman rank correlations. Scatterplots were also used to explore the relationship between LINE-1 methylation levels and parent-child pairs. In this analysis, male participants were stratified according to their testicular cancer status. These correlations were adjusted for parental and offspring age, and the un-stratified analyses were additionally adjusted for case status. Spearman rank correlations and general linear models were used to investigate the strength of the associations between %5-mC and age, smoking status (never, current, former), and alcohol consumption (never, current, former). We adjusted for age in all analyses, since age has been associated with global hypomethylation [47,48]. For microlithiasis, only men who have had a testicular ultrasound were included in the analysis as lacking microlithiasis. We used linear regression models to evaluate potential predictors of LINE-1 methylation levels, as a continuous variable. To evaluate the association between TGCT and LINE-1 methylation, we used unconditional logistic regression models to calculate the odds ratio (OR) and 95% confidence intervals (CI) adjusting for potential confounders (e.g., age). LINE-1 methylation levels were categorized in tertiles and dichotomized at the median, based on the distribution in the healthy male family controls; females were excluded from these analyses. We computed the variance of the OR estimates using a robust variance estimator [49] to adjust for the correlations between participants from the same family. Covariate data were taken from study questionnaires, physician history and physical exams. All tests were two-sided. Statistical significance refers to a P ≤ 0.05 or an OR 95% CI that excludes 1.0. All analyses were carried out using SAS software version 9.1 (SAS Institute, Cary, NC).
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
LM designed the study, performed the methylation and statistical analyses, and drafted the manuscript. SAS helped design the study, participated in statistical analyses and manuscript preparation. LK performed medical evaluation of patients, case management and patient/family data collection. SMG helped with the statistical analysis accounting for relatedness. MHG designed the Familial Testicular Cancer study, and provided critical input to the current analysis strategy, access to resources, and manuscript editing. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Global (LINE-1) methylation levels among strata by patient status. The effects of age, smoking, and alcohol consumption on LINE-1 methylation levels are shown.
Contributor Information
Lisa Mirabello, Email: mirabellol@mail.nih.gov.
Sharon A Savage, Email: savagesh@mail.nih.gov.
Larissa Korde, Email: lkorde@seattlecca.org.
Shahinaz M Gadalla, Email: gadallas@mail.nih.gov.
Mark H Greene, Email: greenem@mail.nih.gov.
Acknowledgements
We thank Drs Marston Linehan and Jennifer Loud, Susan Pfeiffer, and June Peters for their contributions to the CGB Familial Testicular Cancer study. We thank Dr. Nicolas Wentzensen for helpful manuscript comments. We are grateful to the patients and families who participated in this study. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Division of Cancer Epidemiology and Genetics, and by support services contracts NO2-CP-11019-50 and NO2-CP-65504-50 with Westat Inc.
References
- Huyghe E, Matsuda T, Thonneau P. Increasing incidence of testicular cancer worldwide: a review. J Urol. 2003;170:5–11. doi: 10.1097/01.ju.0000053866.68623.da. [DOI] [PubMed] [Google Scholar]
- Hemminki K, Li X. Familial risk in testicular cancer as a clue to a heritable and environmental aetiology. Br J Cancer. 2004;90(9):1765–1770. doi: 10.1038/sj.bjc.6601714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Hemminki K. Modification of cancer risks in offspring by sibling and parental cancers from 2,112,616 nuclear families. Int J Cancer. 2001;92(1):144–150. doi: 10.1002/1097-0215(200102)9999:9999<::AID-IJC1147>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Korde LA, Premkumar A, Mueller C, Rosenberg P, Soho C, Bratslavsky G, Greene MH. Increased prevalence of testicular microlithiasis in men with familial testicular cancer and their relatives. Br J Cancer. 2008;99(10):1748–1753. doi: 10.1038/sj.bjc.6604704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid H, Cos L, Weinberg E, Messing E. Testicular microlithiasis: a review and its association with testicular cancer. Urol Oncol. 2004;22(4):285–289. doi: 10.1016/S1078-1439(03)00177-7. [DOI] [PubMed] [Google Scholar]
- Rapley EA, Turnbull C, Al Olama AA, Dermitzakis ET, Linger R, Huddart RA, Renwick A, Hughes D, Hines S, Seal S. A genome-wide association study of testicular germ cell tumor. Nat Genet. 2009;41(7):807–810. doi: 10.1038/ng.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanetsky PA, Mitra N, Vardhanabhuti S, Li M, Vaughn DJ, Letrero R, Ciosek SL, Doody DR, Smith LM, Weaver J. Common variation in KITLG and at 5q31.3 predisposes to testicular germ cell cancer. Nat Genet. 2009;41(7):811–815. doi: 10.1038/ng.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath A, Korde L, Greene MH, Libe R, Osorio P, Faucz FR, Raffin-Sanson ML, Tsang KM, Drori-Herishanu L, Patronas Y. Functional phosphodiesterase 11A mutations may modify the risk of familial and bilateral testicular germ cell tumors. Cancer Res. 2009;69(13):5301–5306. doi: 10.1158/0008-5472.CAN-09-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson KL, Kanetsky PA, Hawes R, Vaughn DJ, Letrero R, Tucker K, Friedlander M, Phillips K-A, Hogg D, Jewett MAS. The Y deletion gr/gr and susceptibility to testicular germ cell tumor. Am J Hum Genet. 2005;77(6):1034–1043. doi: 10.1086/498455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crockford G, Linger R, Hockley S, Dudakia D, Johnson L, Huddart R, Tucker K, Friedlander M, Phillips K, Hogg D. Genome-wide linkage screen for testicular germ cell tumour susceptibility loci. Hum Mol Genet. 2006;15(3):443–451. doi: 10.1093/hmg/ddi459. [DOI] [PubMed] [Google Scholar]
- Lind G, Skotheim R, Lothe R. The epigenome of testicular germ cell tumors. APMIS. 2007;115(10):1147–1160. doi: 10.1111/j.1600-0463.2007.apm_660.xml.x. [DOI] [PubMed] [Google Scholar]
- Smiraglia D, Szymanska J, Kraggerud S, Lothe R, Peltomäki P, Plass C. Distinct epigenetic phenotypes in seminomatous and nonseminomatous testicular germ cell tumors. Oncogene. 2002;21(24):3909–3916. doi: 10.1038/sj.onc.1205488. [DOI] [PubMed] [Google Scholar]
- Netto G, Nakai Y, Nakayama M, Jadallah S, Toubaji A, Nonomura N, Albadine R, Hicks J, Epstein J, Yegnasubramanian S. Global DNA hypomethylation in intratubular germ cell neoplasia and seminoma, but not in nonseminomatous male germ cell tumors. Mod Pathol. 2008;21(11):1337–1344. doi: 10.1038/modpathol.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I, Jacobsen S. Epigenetic inheritance in plants. Nature. 2007;447(7143):418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- Chong S, Whitelaw E. Epigenetic germline inheritance. Curr Opin Genet Dev. 2004;14(6):692–696. doi: 10.1016/j.gde.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Rakyan V, Beck S. Epigenetic variation and inheritance in mammals. Curr Opin Genet Dev. 2006;16(6):573–577. doi: 10.1016/j.gde.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Frazier M, Xi L, Zong J, Viscofsky N, Rashid A, Wu E, Lynch P, Amos CI, Issa J. Association of the CpG Island Methylator Phenotype with Family History of Cancer in Patients with Colorectal Cancer. Cancer Res. 2003;63(16):4805–4808. [PubMed] [Google Scholar]
- Chan T, Yuen S, Kong C, Chan Y, Chan A, Ng W, Tsui W, Lo M, Tam W, Li V. Heritable germline epimutation of MSH2in a family with hereditary nonpolyposis colorectal cancer. Nat Genet. 2006;38:1178–1183. doi: 10.1038/ng1866. [DOI] [PubMed] [Google Scholar]
- Raval A, Tanner SM, Byrd JC, Angerman EB, Perko JD, Chen S-S, Hackanson B, Grever MR, Lucas DM, Matkovic JJ. Downregulation of Death-Associated Protein Kinase 1 (DAPK1) in Chronic Lymphocytic Leukemia. Cell. 2007;129(5):879–890. doi: 10.1016/j.cell.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchins MP, Wong JJL, Suthers G, Suter CM, Martin DIK, Hawkins NJ, Ward RL. Inheritance of a Cancer-Associated MLH1 Germ-Line Epimutation. N Engl J Med. 2007;356(7):697–705. doi: 10.1056/NEJMoa064522. [DOI] [PubMed] [Google Scholar]
- Lam M-YJ, Heaney JD, Youngren KK, Kawasoe JH, Nadeau JH. Trans-generational epistasis between Dnd1Ter and other modifier genes controls susceptibility to testicular germ cell tumors. Hum Mol Genet. 2007;16(18):2233–2240. doi: 10.1093/hmg/ddm175. [DOI] [PubMed] [Google Scholar]
- Xing Y, Shi S, Le L, Lee CA, Silver-Morse L, Li WX. Evidence for Transgenerational Transmission of Epigenetic Tumor Susceptibility in Drosophila. PLoS Genet. 2007;3(9):e151. doi: 10.1371/journal.pgen.0030151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan H, Sutherland H, Martin D, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- Rakyan V, Chong S, Champ M, Cuthbert P, Morgan H, Luu K, Whitelaw E. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc Natl Acad Sci USA. 2003;100:2538–2543. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, Anway M, Rekow S, Skinner M. Transgenerational Epigenetic Imprinting of the Male Germline by Endocrine Disruptor Exposure during Gonadal Sex Determination. Endocrinology. 2006;147(12):5524–5541. doi: 10.1210/en.2006-0987. [DOI] [PubMed] [Google Scholar]
- Anway M, Memon M, Uzumcu M, Skinner M. Transgenerational effect of the endocrine disruptor vinclozolin on male spermatogenesis. Journal of Andrology. 2006;27:868–879. doi: 10.2164/jandrol.106.000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway M, Cupp A, Uzumcu M, Skinner M. Epigenetic Transgenerational Actions of Endocrine Disruptors and Male Fertility. Science. 2005;308(5727):1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming JL, Huang THM, Toland AE. The role of parental and grandparental epigenetic alterations in familial cancer risk. Cancer Res. 2008;68(22):9116–9121. doi: 10.1158/0008-5472.CAN-08-2184. [DOI] [PubMed] [Google Scholar]
- Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- Dobrovic A, Kristensen L. DNA methylation, epimutations and cancer predisposition. Int J Biochem Cell Biol. 2009;41(1):34–39. doi: 10.1016/j.biocel.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Schulz W. L1 retrotransposons in human cancers. J Biomed Biotechnol. 2006;2006:1–12. doi: 10.1155/JBB/2006/83672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badge RM, Alisch RS, Moran JV. ATLAS: A system to selectively identify human-specific L1 insertions. American Journal of Human Genetics. 2003;72:823–838. doi: 10.1086/373939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phokaew C, Kowudtitham S, Subbalekha K, Shuangshoti S, Mutirangura A. LINE-1 methylation patterns of different loci in normal and cancerous cells. Nucleic acids research. 2008;36:5704–5712. doi: 10.1093/nar/gkn571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratthauer G, Fanning T. Active LINE-1 retrotransposons in human testicular cancer. Oncogene. 1992;7(3):507–510. [PubMed] [Google Scholar]
- Su Y, Davies S, Davis M, Lu H, Giller R, Krailo M, Cai Q, Robison L, Shu X. Expression of LINE-1 p40 protein in pediatric malignant germ cell tumors and its association with clinicopathological parameters: a report from the Children's Oncology Group. Cancer Lett. 2007;247(2):204–212. doi: 10.1016/j.canlet.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Moore L, Pfeiffer R, Poscablo C, Real F, Kogevinas M, Silverman D, Garcia-Closas R, Chanock S, Tardon A, Serra C. Genomic DNA hypomethylation as a biomarker for bladder cancer susceptibility in the Spanish Bladder Cancer Study: a case-control study. Lancet Oncol. 2008;9(4):359–366. doi: 10.1016/S1470-2045(08)70038-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiung D, Marsit C, Houseman E, Eddy K, Furniss C, McClean M, Kelsey K. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16:108–114. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- Rusiecki J, Baccarelli A, Bollati V, Tarantini L, Moore L, Bonefeld-Jorgensen E. Global DNA Hypomethylation Is Associated with High Serum-Persistent Organic Pollutants in Greenlandic Inuit. Environ Health Perspect. 2008;116:1547–1552. doi: 10.1289/ehp.11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Maarri O, Becker T, Junen J, Manzoor S, Diaz-Lacava A, Schwaab R, Wienker T, Oldenburg J. Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Hum Genet. 2007;122:505–514. doi: 10.1007/s00439-007-0430-3. [DOI] [PubMed] [Google Scholar]
- Chalitchagorn K, Shuangshoti S, Hourpai N, Kongruttanachok N, Tangkijvanich P, Thong-ngam D, Voravud N, Sriuranpong V, Mutirangura A. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23(54):8841–8846. doi: 10.1038/sj.onc.1208137. [DOI] [PubMed] [Google Scholar]
- Hitchins M, Wong J, Suthers G, Suter C, Martin D, Hawkins N. Inheritance of a cancer-associated MLH1 germ-line epimutation. N Engl J Med. 2007;356:697–705. doi: 10.1056/NEJMoa064522. [DOI] [PubMed] [Google Scholar]
- Morak M, Schackert H, Rahner N, Betz B, Ebert M, Walldorf C, Royer-Pokora B, Schulmann K, von Knebel-Doeberitz M, Dietmaier W. Further evidence for heritability of an epimutation in one of 12 cases with MLH1 promoter methylation in blood cells clinically displaying HNPCC. Eur J Hum Genet. 2008;16(7):804–811. doi: 10.1038/ejhg.2008.25. [DOI] [PubMed] [Google Scholar]
- Hitchins M, Ward R. Erasure of MLH1 methylation in spermatozoa-implications for epigenetic inheritance. Nat Genet. 2007;39:1289. doi: 10.1038/ng1107-1289. [DOI] [PubMed] [Google Scholar]
- Skinner M. Epigenetic transgenerational toxicology and germ cell disease. Int J Androl. 2007;30:393–396. doi: 10.1111/j.1365-2605.2007.00796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai PL, Friedlander M, Tucker K, Phillips K-A, Hogg D, Jewett MAS, Lohynska R, Daugaard G, Richard S, Bonaïti-Pellié C. The International Testicular Cancer Linkage Consortium: A clinicopathologic descriptive analysis of 461 familial malignant testicular germ cell tumor kindred. Urol Oncol. 2009. in press . [DOI] [PMC free article] [PubMed]
- Bollati V, Baccarelli A, Hou L, Nonzini M, Fustioni S, Cavallo D, Byun H, Jiang J, Marinelli B, Pesatori A. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- Fraga M, Agrelo R, Esteller M. Cross-talk between aging and cancer: the epigenetic language. Ann NY Acad Sci. 2007;1100:60–74. doi: 10.1196/annals.1395.005. [DOI] [PubMed] [Google Scholar]
- Liu L, Wylie R, Andrews L, Tollefsbol T. Aging, cancer and nutrition: the DNA methylation connection. Mech Ageing Dev. 2003;124:989–998. doi: 10.1016/j.mad.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Zeger S, Liang K. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42:121–130. doi: 10.2307/2531248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Global (LINE-1) methylation levels among strata by patient status. The effects of age, smoking, and alcohol consumption on LINE-1 methylation levels are shown.