Abstract
Analysis of count data from clinical trials using mixed effect analysis has recently become widely used. However, algorithms available for the parameter estimation, including LAPLACE and Gaussian quadrature (GQ), are associated with certain limitations, including bias in parameter estimates and the long analysis runtime. The stochastic approximation expectation maximization (SAEM) algorithm has proven to be a very efficient and powerful tool in the analysis of continuous data. The aim of this study was to implement and investigate the performance of a new SAEM algorithm for application to count data. A new SAEM algorithm was implemented in MATLAB for estimation of both, parameters and the Fisher information matrix. Stochastic Monte Carlo simulations followed by re-estimation were performed according to scenarios used in previous studies (part I) to investigate properties of alternative algorithms (1). A single scenario was used to explore six probability distribution models. For parameter estimation, the relative bias was less than 0.92% and 4.13 % for fixed and random effects, for all models studied including ones accounting for over- or under-dispersion. Empirical and estimated relative standard errors were similar, with distance between them being <1.7 % for all explored scenarios. The longest CPU time was 95s for parameter estimation and 56s for SE estimation. The SAEM algorithm was extended for analysis of count data. It provides accurate estimates of both, parameters and standard errors. The estimation is significantly faster compared to LAPLACE and GQ. The algorithm is implemented in Monolix 3.1, (beta-version available in July 2009).
Keywords: Algorithms; Binomial Distribution; Biometry; Humans; Likelihood Functions; Markov Chains; Models, Statistical; Monte Carlo Method; Nonlinear Dynamics; Normal Distribution; Numerical Analysis, Computer-Assisted; Poisson Distribution
Keywords: Nonlinear Mixed Effects Model, Count data, Maximum Likelihood Estimation, Fischer information matrix, SAEM algorithm, MONOLIX
Introduction
The analysis of the count data with mixed linear models has been frequently reported in recent history (2, 3). This approach offers several advantages, as it allows for estimation of the central tendency of population parameters as well as quantification of the inter-individual variability (4, 5). These parameter estimates are further utilized in many different aspects, e.g. for simulation of novel scenarios with respect to new dosing schedules or new patient populations. Therefore it is important that these parameter estimates are unbiased and reliable.
Using maximum likelihood methods to estimate parameters for count data models requires approximation of the true likelihood since the integral of the likelihood cannot be explicitly solved(6).
For the discrete data, these approximations include LAPLACE and Gaussian quadrature methods (7). In mixed effect analysis, model parameters enter non-linearly into the model function form, which impose certain difficulties to approximate the likelihood integral precisely. Consequences of different integral approximation include difficulties in the estimation procedure, bias in parameter estimates, and prolonged runtime for the analysis (1, 8–10).
In recent years, there have been several approaches/algorithms developed for the analysis of continuous data which provide a solution to the likelihood function without approximation of the model(11). A stochastic approximation version of EM algorithm (SAEM) linked to a Markov Chain Monte Carlo procedure has been suggested for maximum likelihood estimation within the non-linear mixed effects framework. This procedure has been demonstrated to possess excellent statistical convergence properties as well as the ability to provide an estimate close to the maximum likelihood estimate in only few iterations (12). In addition to the maximization of the likelihood of the observations, the SAEM algorithm also provides the user with the Fisher Information Matrix, which is further used to assess parameter estimate uncertainty. However, to our knowledge, there have not been studies reported with respect to application of a stochastic algorithm to the analysis of discrete data.
The aims of this study were (i) to extend the SAEM algorithm for estimation of parameters in count data models, (ii) to evaluate its performance via stochastic Monte Carlo simulations followed by the re-estimation procedure, (iii) to implement estimation of the Fisher information matrix for this case, and (iv) to evaluate precision of the standard error estimates using the proposed implementation.
Methods
Notation: the mixed count data models
A parametric count data model is defined by the probabilities PΨ(Y = k), for k=0,1,…. Here, Ψ is a vector of parameters. Consider a Poisson model for example. Then, Ψ=λ and PΨ(Y = k) = e−λλk/k!.
In the population approach, each subject i has their own vector of parameters ψi=(ψil). We assume here that ψi =h(φi) where φι is a Gaussian vector with mean μ and variance-covariance matrix Ω (extension to the case where the mean of φι depends on some covariates is straightforward). We can use different transformations h, according to the constraints on ψi. For example, we can set ψil=exp(φil) to constrain the I’th parameter ψil to be non negative, or φil =logit(ψil) to constrain ψil to take its values between 0 and 1.
Implementation of the SAEM algorithm for count data models
We propose to extend the SAEM algorithm described in (12) for continuous data models to count data models. SAEM is an iterative procedure where, at iteration k, a new set of individual parameters is drawn with the conditional distribution p(ϕ|y; μ(k), Ω(k)). Then, the new population parameters (μ(k+1), Ω(k+1)) are obtained by maximizing Qk+1(μ, Ω) defined as follows:
| (1) |
where l(y, ϕ(k);μ, Ω) is the complete log-likelihood
| (2) |
Here, p(μ,Ω) is the probability density function of the normal distribution with mean μ and variance-covariance Ω.
The simulation step is identical to the simulation step of SAEM for continuous data. The maximization step is slightly different, since there is no error model to estimate. The only parameters to estimate are the fixed effects and the variance-covariance matrix of the random effects.
For the numerical experiments presented below, we used γk=1 during the first 200 iterations of SAEM and γk=1/(k−200) during the next 100 iterations.
An MCMC algorithm was used for the simulation step (see (12, 13) for more details).
Estimation of the Fisher information matrix
Let θ= (μ, Ω) be the set of population parameters to be estimated, and let θ̂ be the maximum likelihood estimate of θ computed with SAEM. The Fisher Information matrix is defined as where l(y; θ̂) is the log-likelihood of the observations, computed with θ=θ̂.
For continuous data models, it is now widely acknowledged that techniques based on the linearization of the model (first order and first order conditional estimation, FO and FOCE) present severe drawbacks for the estimation of the population parameters (14). In contrast, several numerical experiments have shown that linearization of the model for estimating the Fisher information matrix (as implemented in MONOLIX 2.4) is satisfactory.
However, this approach is not applicable for discrete data models. As alternative we propose to compute a stochastic approximation of the Fisher Information matrix using the Louis formula (see(12) for more details):
| (3) |
The procedure consists in computing first θ̂ with SAEM then applying the Louis formula with θ=θ̂ which requires the computation of the conditional expectation and conditional variance. These quantities are estimated by Monte-Carlo simulation: we performed 300 iterations of MCMC for the numerical experiments – these will provide 300 simulated values used to compute empirical means and variances as defined in the equation 3. For discrete data models, this method is a default method for computation of Fisher information matrix in MONOLIX.
Evaluation of the SAEM algorithm
The performance of the SAEM algorithm was evaluated via Monte Carlo simulation-estimation exercises. The settings were identical to ones presented previously in the part I, where a single scenario was studied to evaluate performance of the algorithm with six different count data models, including Poisson (PS), Zero-inflated Poisson (ZIP), Generalized Poisson (GP), Poisson with Markovian Features (PMAK), Poisson with a mixture distribution for individual observations (PMIX) and Negative binomial (NB) model. As this communication is focused on application of SAEM to count data, the reader is kindly asked to refer to Part I of this report (for detailed description of the study design, model parameters and specific notation (1)). For each scenario, one hundred datasets were simulated in MATLAB and parameters were estimated using the SAEM algorithm implemented in MATLAB. All procedures were performed on laptop DELL D830 2.4 GHz
To assess statistical properties of the method, relative bias (RB), relative estimation error (REE) and root mean square errors (RMSE) were computed for each scenario using equations 4, 5 and 6, were θ̂k is the kth estimated parameter, θ* is the true parameter and θ̂med is a median of n estimated parameters, where n=100. CPU runtime was also measured to assess the efficiency of the algorithm and the runtime for the analysis.
| (4) |
| (5) |
| (6) |
Evaluation of the standard error estimates
The Fisher information matrix was estimated for each data set, and its inverse was used to assess the standard error estimates. These were further used to assess the relative standard errors, as a ratio of the standard error and parameter estimate, expressed as a percentage (%), denoted as ( ). Uncertainty of parameter estimates θ̂k around model parameter θ* was assessed by computing the empirical relative standard errors(sd*(θ)) using equation 7.
Comparison between estimated relative standard errors and empirical relative standard errors was computed by determining the relative distance between those, as shown in equation 8, expressed as a percentage (%). This quantity we denote as absolute error estimate (AEE).
| (7) |
| (8) |
Outcomes of all Monte-Carlo simulation studies exploring both, the parameter estimation process and estimation of Fischer information matrix, were presented as box-plots where bias and imprecision of the method can easily be visualized.
Visual evaluation
Visual evaluation was performed by generating visual predictive check (VPC) and normalized prediction distribution errors (NPDEs)(15, 16). For visual predictive check, 1000 new datasets were simulated and in order to perform statistical calculations, a randomly chosen simulated dataset was treated as the observed data. The visual predictive check was derived on the quantity such as individual variances versus individual means of counts. The median (50th percentile), quartiles (25th and 75th percentiles) as well as 10th and 90th percentiles were calculated. These visual evaluation techniques were generated by using the true parameter values, which were quite similar to the estimated parameters across all 100 datasets. In order to show the strength of these visual simulation-based diagnostics for detecting the impact of the model misspecification by means of biased parameter estimates (variances), we also generated VPCs and NPDEs using the biased variances (1.5 and 2 times of the original value).
Results
Overall, the estimation procedure with the SAEM algorithm in a non-linear mixed effect modelling framework for count data models, showed satisfactory performance with low bias and high precision. For parameter estimation, the absolute value of relative bias was less than 0.92% and 4.13 % for fixed and random effects and RMSE was less than 12.34 % and 13.13% for fixed and random effects, across all tested models. For standard error estimation, the absolute value of relative bias was less than 1.7 and 1.6 % for fixed and random effects, and RMSE was less than 1 and 1.54% for fixed and random effects. The variances of over-dispersion parameters, shown to be biased when estimated with LAPLACE, were precisely estimated with SAEM, exhibiting relative bias of 1.62%, 1.26% and 2.38% for p0, δ and OVDP. Detailed results are listed below. The distribution of REE and AEE for all models and all parameters is shown in Figure 1a–f, while the numerical results are represented in Table I The summary for imprecision estimate (RMSE) for all parameters and their standard error estimates across all models using SAEM is shown in Table II.
Figure 1.
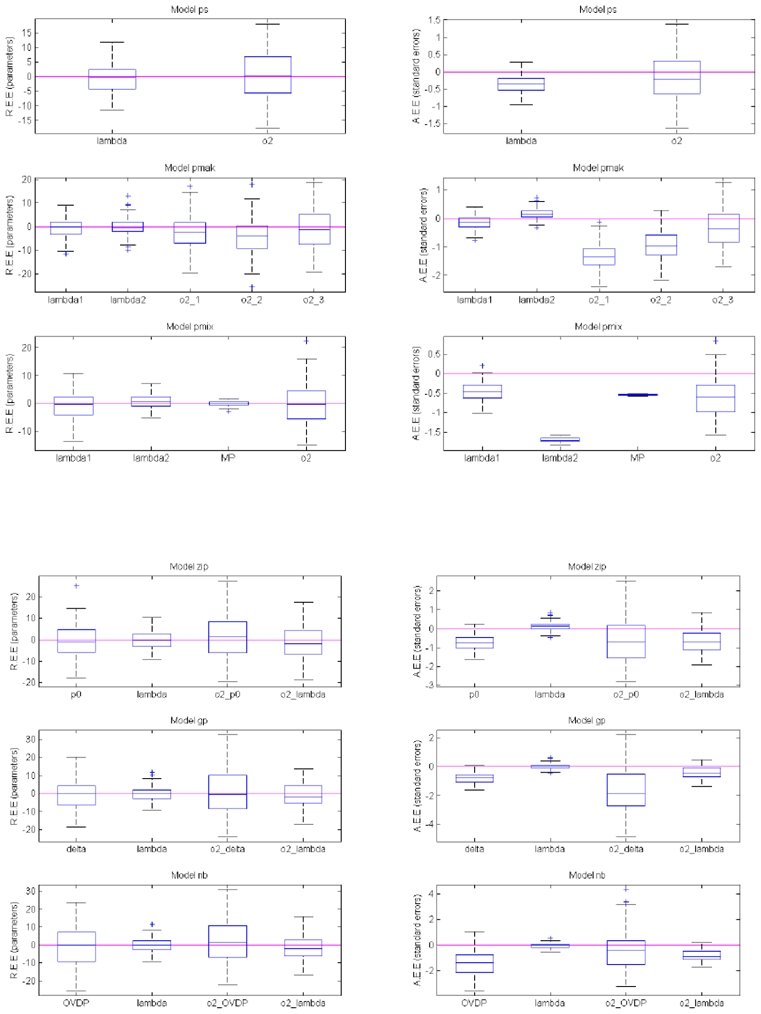
Distribution of relative estimation error (REE) for all parameters (left panel) and standard errors (right panel) across all the models. The errors (y axes) are given as percentages (%). (Abbreviations: PS = Poisson, PMAK = Poisson with Markovian Features, PMIX = Poisson with a mixture distribution for individual observations, ZIP = Zero-inflated Poisson, GP = Generalized Poisson, and NB = Negative binomial model)
Table 1.
Summary of relative bias (RB) for parameter estimates and absolute estimation error (AEE) for standard error estimates for all parameters and all models.
| Poisson model | |||||
| λ | Ω(λ) | ||||
| RB (parameter) (%) | −0.6 | 0.75 | |||
| AEE (standard error) (%) | −0.37 | −0.13 | |||
| Poisson model with Markov element | |||||
| λ1 | λ2 | Ω(λ1) | Ω(λ2) | Cov(λ1–λ2) | |
| RB (parameter) (%) | −0.41 | −0.07 | −2.39 | −4.13 | −1.36 |
| AEE (standard error) (%) | −0.13 | 0.17 | −1.33 | −0.93 | −0.33 |
| Poisson model with mixtures in individual observations | |||||
| λ1 | λ2 | MP | Ω(λ1) | ||
| RB (parameter) (%) | −0.4 | 0.54 | −0.05 | −0.13 | |
| AEE (standard error) (%) | −0.47 | −1.70 | −0.55 | −0.61 | |
| Zero inflated Poisson model | |||||
| λ1 | P0 | Ω(λ1) | Ω(P0) | ||
| RB (parameter) (%) | −04.05 | −0.75 | −1.08 | 1.62 | |
| AEE (standard error) (%) | 0.14 | −0.74 | −0.64 | −0.59 | |
| Generalized Poisson model | |||||
| λ1 | δ | Ω(λ1) | Ω(δ) | ||
| RB (parameter) (%) | −0.19 | −0.21 | −1.03 | 1.26 | |
| AEE (standard error) (%) | 0.02 | −0.79 | −0.41 | −1.61 | |
| Negative binomial model | |||||
| λ1 | Ovdp | Ω(λ1) | Ω(Ovdp) | ||
| RB (parameter) (%) | −0.02 | −0.92 | −1.42 | 2.38 | |
| AEE (standard error) (%) | −0.01 | −1.36 | −0.79 | −0.33 | |
Table 2.
Summary of RMSE for parameter estimates and standard error estimates for all parameters and all models.
| Poisson model | |||||
| λ | Ω(λ) | ||||
| RMSE(parameter) | 5.23 | 8.56 | |||
| RMSE (standard error) | 0.26 | 0.66 | |||
| Poisson model with Markov element | |||||
| λ1 | λ2 | Ω(λ1) | Ω(λ2) | Cov(λ1–λ2) | |
| RMSE(parameter) | 4.23 | 3.91 | 7.60 | 8.68 | 7.91 |
| RMSE (standard error) | 0.23 | 0.20 | 0.46 | 0.52 | 0.64 |
| Poisson model with mixtures in individual observations | |||||
| λ1 | λ2 | MP | Ω(λ1) | ||
| RMSE(parameter) | 4.97 | 2.47 | 0.79 | 7.14 | |
| RMSE (standard error) | 0.24 | 0.05 | 0.02 | 0.47 | |
| Zero inflated Poisson model | |||||
| λ1 | P0 | Ω(λ1) | Ω(P0) | ||
| RMSE(parameter) | 4.35 | 7.85 | 8.07 | 10.81 | |
| RMSE (standard error) | 0.23 | 0.36 | 0.62 | 1.22 | |
| Generalized Poisson model | |||||
| λ1 | δ | Ω(λ1) | Ω(δ) | ||
| RMSE(parameter) | 3.98 | 8.05 | 6.87 | 13.13 | |
| RMSE (standard error) | 0.20 | 0.36 | 0.44 | 1.54 | |
| Negative binomial model | |||||
| λ1 | Ovdp | Ω(λ1) | Ω(Ovdp) | ||
| RMSE(parameter) | 4.24 | 12.34 | 7.29 | 11.28 | |
| RMSE (standard error) | 0.22 | 1.00 | 0.47 | 1.43 | |
Poisson model
Parameters of this model were precisely estimated with relative biases of −0.6 % and 0.75 % for fixed and random effect. Relative bias for standard errors estimate was also very low: −0.37 % and −0.13 % for fixed and random effect, indicating good accuracy for uncertainty estimation.
Poisson model with Markov element
Fixed effects of this model along with their uncertainty were accurately estimated with relative bias less than 0.41 %, for both, parameter estimates and standard error estimates. The relative bias of random effects estimate ranged from −1.36 % to −4.13 %, indicating satisfactory accuracy. Also uncertainty of random effects was accurately assessed, with relative bias <1.33 %.
Poisson model with a mixture distribution for individual observations
Both, fixed and random effects of this model were accurately estimated with relative bias values ranging from −0.4 % to 0.54 %. The same was true for estimates of uncertainty for these parameters, with relative bias ranging from −0.47 % to 1.7 %.
Zero inflated Poisson model
Fixed effects of this model along with their uncertainty were accurately estimated with relative bias less than −0.75 %, for both, parameter estimates and standard error estimates. The same holds for random effect estimates and their uncertainty with relative bias < 1.62 %.
Generalized Poisson model
Similar to all other models, parameter estimates, including fixed and random effects and their uncertainty, were accurately estimated with relative bias ranging between −1.61 % and 1.26 %.
Negative binomial model
Relative bias in fixed effects and their uncertainty was less than −1.36 % for this model. Random effects and their uncertainty were also accurately estimated with relative bias < 2.38 %.
Overall convergence was 100% for both parameter and standard error estimation. The average CPU time per run was 48s for parameter estimation and 34s for standard error estimation, when the algorithm was implemented in Matlab. Test runs were performed with all models implemented in C++, and this sped up the estimation process by approximately 1/3 for 5 out of 6 models. Comparison of CPU times between SAEM and other algorithms, including Gaussian quadrature with one quadrature point (equivalent to LAPLACE) and Gaussian quadrature with nine quadrature points, is shown in Table III.
Table 3.
Overview of the CPU runtimes for different models with different algorithms
| Model | SAS GQ (1 point) | SAS GQ (9 points) | SAEM (Matlab) | SAEM (C++) |
|---|---|---|---|---|
| PS | 0.04s | NA | 18s + 19s (s.e.) | 13s + 13s (s.e.) |
| PMAK | 58s + 39s (s.e.) | 30s + 20s (s.e.) | ||
| PMIX | 95s + 56s (s.e.) | 112s + 63s (s.e.) | ||
| ZIP | 60s | 12min 51s | 32s + 31s (s.e.) | 20s + 19s (s.e.) |
| GP | 59s | 13min 37s | 30s + 29s (s.e.) | 24s + 21s (s.e.) |
| NB | 4min 27s | 60min | 52s + 30s (s.e.) | 27s + 25s (s.e.) |
Visual predictive check revealed good accordance between observed and simulated data no matter which variances were used for simulations, indicating the weak power of this visual technique to detect model misspecification (Figure 2a–c). Numerical predictive check showed somewhat more sensitivity, indicating higher disagreement between expected and observed number of data points outside prediction intervals for mis-specified models (Table IV). However NPDEs showed to be most sensitive towards indicating model misspecification as illustrated in Figure 3a–c. Visual findings of normalized prediction distribution errors (NPDEs) were confirmed by statistical tests, including Kolmogorov-Smirnov and chi-squared tests: hypotheses that NPDEs are normally distributed with zero mean and variance of 1 was rejected for two mis-specified models (p<10−32).
Figure 2.
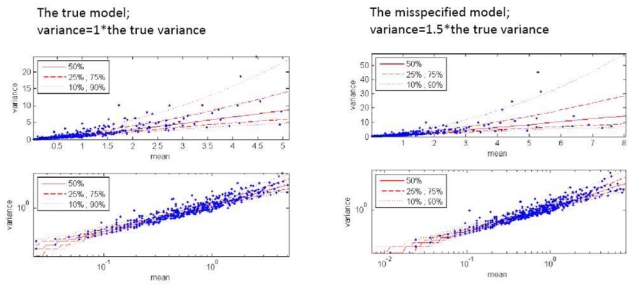
Visual predictive check of variance versus mean on normal (upper panel) and log (lower panel) scale. Observed data are given as scatter points, while prediction intervals are represented as red lines. VPC is shown for the true (left panel) and for a mis-specified zero-inflated Poisson models, when variance used for simulations is equal to 1.5 the true variance (right panel).
Table 4.
Numerical predictive check shown as empirical cumulative distribution function when the true and two misspecified zero-inflated Poisson models were used for simulations.
| Empirical cumulative distribution function | |||||
|---|---|---|---|---|---|
| Expected | 10 | 25 | 50 | 75 | 90 |
| Variance = 1*the true | 9.3 | 25.2 | 51.7 | 75.7 | 90.5 |
| Variance = 1.5*the true | 7.7 | 23 | 51 | 78.1 | 94.1 |
| Variance = 2*the true | 16.6 | 37 | 67.6 | 88 | 97.4 |
Figure 3.
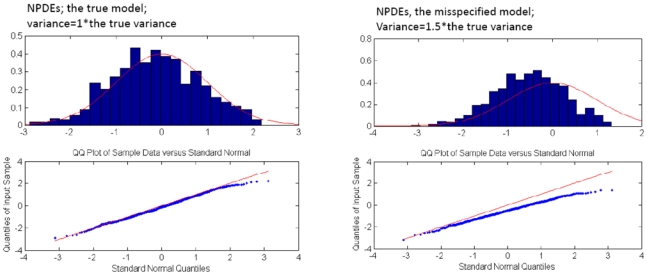
Comparison of NPDEs and normal distribution with zero mean and variance of one, represented as histogram (upper panel) and q-q plot (lower panel). NPDEs are shown for the true (left panel) and for a mis-specified zero-inflated Poisson models, when variance used for simulations is equal to 1.5 times the true variance (right panel).
Discussion
A new SAEM algorithm has been developed, implemented and evaluated for application to count data models in the non-linear mixed effects framework. Six relevant models for analyzing this type of data were evaluated, including those that can deal with overdispersion, when individual variances exceed the individual means. The algorithm was also implemented for computation of Fischer Information matrix in order to assess the uncertainty estimate.
The SAEM algorithm performed well under all tested model scenarios resulting in accurate and precise estimation of all parameters. Even, variances of over-dispersion parameters were accurately and precisely estimated, which was not the case reported previously in part I of this report in analysis with LAPLACE method (1). The explanation for previously observed biases was presumed related to the poor approximation of the likelihood integral, when models are highly non-linear, which is the case for models handling over- and under-dispersion cases. In addition, random effects often enter models in a non-linear fashion; therefore these are most likely to suffer from the poor integral approximation. Observed poor approximation of the likelihood integral by LAPLACE was explained by the asymmetric shrinkage of Empirical Bayes Estimates (EBEs); indeed the LAPLACE method is conditioned on point estimate of EBEs, however reliability of this estimate is not taken into account. Poor performance of other estimation methods due to EBE shrinkage has also been previously demonstrated (17) When better approximation of the likelihood integral was used in previous work involving nine quadrature points of the Gaussian quadrature method, the bias disappeared (1). Similar observations were made when analyzing ordinal type of data (9). The SAEM algorithm does not involve any approximation of the model in computation of the likelihood integral; it provides estimation of both the likelihood and Fischer information matrix, without linearization of the model. Moreover, SAEM does not condition the likelihood computation on the EBE estimate and therefore it is not affected by poor estimates of the likelihood particularly in the case of shrinkage. Clearly, this is a favorable property of the algorithm, which leads to accurate and unbiased parameter and standard error estimates.
Pharmacokinetic and pharmacodynamic (PKPD) models are becoming more widely used as decision support tools in drug development, particularly towards the goal of individualized therapy. Therefore, having both an appropriate model as well as precisely and accurately estimated parameters, is critical for the forward looking utility of these models. Correct approximation of the likelihood integral plays an important role, as it allows for both, unbiased parameter estimation, but also for performing a reliable likelihood ratio test in the model building procedure. It is well known that employing different types of model linearization leads to poor properties of the likelihood ratio test, which is widely used as a tool for model discrimination (18). Having a reliable likelihood ratio test is important for utilizing other techniques in the model building procedure, such as log-likelihood profiling used for assessment of parameter uncertainty.
The importance of unbiased parameters has been discussed previously (9, 14); however the importance of unbiased standard error estimates has seldom been the topic of discussion. Unbiased SE estimates are an important aspect of prospective simulations and exploration of competing study design scenarios. The SAEM algorithm appeared to satisfy the goal toward required precision and unbiased estimates of parameter uncertainty.
The results of the part I showed that when employing Gaussian quadrature method with nine quadrature points, precision and accuracy appeared to be good. However, the consequences of the implementation of this method results in a lower convergence rate (ranging from 77% – 100%) and increased runtimes (up to 1h) All these potential limitations are circumvented with the new SAEM algorithm. All studied models (100%) converged successfully, for both parameter estimation and standard error estimation. This is a convenient property and strength of the algorithm, as it almost always guarantees the user with successful runs and usable results. With respect to CPU runtime, the SAEM algorithm appeared to be fast. The longest time for parameter estimation was still in range of seconds (93s, when ran on laptop DELL D830 2.4 GHz)), implying that, for example for estimation of parameters of the negative binomial model, it performs 90 times faster than Gaussian quadrature method. The runtime can be improved even further by compiling SAEM via in C++ instead of MATLAB. The SAEM algorithm, which forms the core of MONOLIX is a freeware available at http://www.monolix.org and is based on the evaluated and documented thorough statistical theory and it is an ongoing project implementing new statistical developments in a dynamic environment (12, 19). The new version of Monolix program will include the extension of the algorithm for the analysis of count and ordered categorical data.
In conclusion, the SAEM algorithm was extended for analysis of count data. It provides accurate estimates of both, parameters and standard errors with convergence rate of 100%. The estimation is significantly faster compared to other algorithms. The algorithm will be implemented in Monolix 3.1.
Acknowledgments
Radojka Savic was financially supported by a Postdoc grant from the Swedish Academy of Pharmaceutical Sciences (Apotekarsocieteten). We thank Dr Shasha Jumbe for valuable comments on the manuscript.
Contributor Information
Radojka Savic, Modèles et méthodes de l'évaluation thérapeutique des maladies chroniques INSERM : U738, Université Paris-Diderot - Paris VII, FR.
Marc Lavielle, INRIA Saclay - Ile de France, SELECT INRIA, Université Paris Sud - Paris XI, CNRS : UMR, FR.
References
- 1.Plan EL, Maloney A, Troconiz IF, Karlsson MO. Maximum Likelihood Approximations: Performance in Population Models for Count Data. 2008. p. 17. Abstr 1372 http://www.page-meeting.org/?abstract=1372. [DOI] [PubMed]
- 2.Frame B, Miller R, Lalonde RL. Evaluation of mixture modeling with count data using NONMEM. J Pharmacokinet Pharmacodyn. 2003;30:167–183. doi: 10.1023/a:1025564409649. [DOI] [PubMed] [Google Scholar]
- 3.Troconiz IF, Plan EL, Miller R, Karlsson MO. Modelling Overdispersion and Markovian Features in Count Data. American Conference on Pharmacometrics; Tucson, Arizona. 2008. [DOI] [PubMed] [Google Scholar]
- 4.Beal SL, Sheiner LB, Boeckmann AJ. NONMEM Users Guides. Icon Development Solutions; Ellicott City, Maryland, USA: 1989–2006. [Google Scholar]
- 5.Ette EI, Williams PJ. Pharmacometrics: The Science of Quantitative Pharmacology. John Wiley & Sons, Inc; 2007. [Google Scholar]
- 6.Wang Y. Derivation of various NONMEM estimation methods. J Pharmacokinet Pharmacodyn. 2007;34:575–593. doi: 10.1007/s10928-007-9060-6. [DOI] [PubMed] [Google Scholar]
- 7.SAS Institute Inc SAS Online Doc. SAS/STAT User’s Guide, The NLMIXED Procedure. [Google Scholar]
- 8.Clausen WHO, Ronn BB, Skoovgard IM. Maximum likelihood estimation in nonlinear mixed effect models: Adaptive Gaussian Quadrature by sparse grid sampling. 2009. p. 18. Abstr 1584 http://www.page-meeting.org/?abstract=1584.
- 9.Jonsson S, Kjellsson MC, Karlsson MO. Estimating bias in population parameters for some models for repeated measures ordinal data using NONMEM and NLMIXED. J Pharmacokinet Pharmacodyn. 2004;31:299–320. doi: 10.1023/b:jopa.0000042738.06821.61. [DOI] [PubMed] [Google Scholar]
- 10.Verbeke G. Mixed models for the analysis of categorical repeated measures. 2006. p. 15. Abstr 930 http://www.page-meeting.org/?abstract=930.
- 11.Bauer RJ, Guzy S, Ng C. A survey of population analysis methods and software for complex pharmacokinetic and pharmacodynamic models with examples. AAPS J. 2007;9:E60–83. doi: 10.1208/aapsj0901007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuhn E, Lavielle M. Maximum likelihood estimation in nonlinear mixed effects models. Computational Statistics and Data Analysis. 2005;49:1020–1038. [Google Scholar]
- 13.MONOLIX 2.4 User Guide. http://software.monolix.org.
- 14.Girard P, Mentre F. Pamplona, Spain: 2005. 2005. A comparison of estimation methods in nonlinear mixed effects models using a blind analysis; p. 14. Abstr 834 http://www.page-meeting.org/?abstract=834. [Google Scholar]
- 15.Brendel K, Comets E, Laffont C, Laveille C, Mentre F. Metrics for external model evaluation with an application to the population pharmacokinetics of gliclazide. Pharm Res. 2006;23:2036–2049. doi: 10.1007/s11095-006-9067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karlsson MO, Holford N. Marseille: 2008. A Tutorial on Visual Predictive Checks; p. 17. Abstr 1434 http://www.page-meeting.org/?abstract=1434. [Google Scholar]
- 17.Savic RM, KMO . Copenhagen: 2007. Importance of Shrinkage in Empirical Bayes Estimates for Diagnostics and Estimation: Problems and Solutions; p. 16. Abstr 1087 http://www.page-meeting.org/?abstract=1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wahlby U, Jonsson EN, Karlsson MO. Assessment of actual significance levels for covariate effects in NONMEM. J Pharmacokinet Pharmacodyn. 2001;28:231–252. doi: 10.1023/a:1011527125570. [DOI] [PubMed] [Google Scholar]
- 19.Lavielle M, Mentre F. Estimation of population pharmacokinetic parameters of saquinavir in HIV patients with the MONOLIX software. J Pharmacokinet Pharmacodyn. 2007;34:229–249. doi: 10.1007/s10928-006-9043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]


