Abstract
Whole-genome sequencing (WGS) of organisms displaying a specific mutant phenotype is a powerful approach to identify the genetic determinants of a plethora of biological processes. We have previously validated the feasibility of this approach by identifying a point-mutated locus responsible for a specific phenotype, observed in an ethyl methanesulfonate (EMS)-mutagenized Caenorhabditis elegans strain. Here we describe the genome-wide mutational profile of 17 EMS-mutagenized genomes as assessed with a bioinformatic pipeline, called MAQGene. Surprisingly, we find that while outcrossing mutagenized strains does reduce the total number of mutations, a striking mutational load is still observed even in outcrossed strains. Such genetic complexity has to be taken into account when establishing a causative relationship between genotype and phenotype. Even though unintentional, the 17 sequenced strains described here provide a resource of allelic variants in almost 1000 genes, including 62 premature stop codons, which represent candidate knockout alleles that will be of further use for the C. elegans community to study gene function.
INDUCING molecular lesions in a genome is an effective approach to interrogate the genome for its functional elements. Molecular lesions can be induced using a variety of methods. Because of their efficiency and their ability to generate alleles with various different alterations in gene activity (e.g., amorphic, antimorphic, hypomorphic, and hypermorphic), chemical mutagens, such as ethyl methanesulfonate (EMS), are frequently used in genetic mutant screens (Anderson 1995). However, due to mutagen efficiency, a mutant animal selected for a single-locus phenotype invariably contains EMS-induced “background mutations” in its genome. Experimenters try to minimize the potential impact of background mutations through outcrossing to animals with a wild-type genome. Yet no full snapshots of genome sequences right after EMS mutagenesis and after outcrossing have so far been provided to illustrate the extent of background mutations and the extent to which they can indeed be eliminated.
Another caveat of using base-changing chemical mutagens is the relative difficulty associated with identifying the phenotype-causing molecular lesion. In multicellular genetic model organisms, mutant identification involves time-consuming positional cloning approaches, usually involving breeding with genetically marked strains that allow pinpointing of the location of a molecular lesion. Even with rapid, SNP-based mapping approaches in animals with short generation times, such as Caenorhabditis elegans, substantial time hurdles, particularly in the final, fine-mapping stages, still exist. Conceptually similar problems in defining the location of a molecular lesion are encountered by human geneticists who attempt to identify disease-causing genetic lesions.
Whole-genome sequencing (WGS) is beginning to emerge as an efficient and cost-effective tool to shortcut time-consuming mapping and positional cloning efforts (Hobert 2010). The sequencing of an entire genome and its ensuing comparison to a wild-type reference genome can potentially directly pinpoint the molecular lesion that results in the mutant phenotype the animal has been selected for. Proof-of-concept studies in bacteria, yeast, plants, worms, and flies have validated the applicability of this approach (Sarin et al. 2008; Smith et al. 2008; Srivatsan et al. 2008; Blumenstiel et al. 2009; Irvine et al. 2009; Flowers et al. 2010).
Present-day deep sequencing platforms used for WGS generate relatively short sequence reads, thereby posing the bioinformatic challenge to align those reads to a reference genome. We previously described a software pipeline, MAQGene, which is based on the standard alignment program MAQ (Li et al. 2008) and facilitates this bioinformatic step by providing the end user with an extensively curated list of sequence variants from a WGS run of a mutated genome compared to a reference genome (Bigelow et al. 2009). This pipeline can be used for well-annotated, assembled genomes, such as C. elegans or Drosophila. In this article, we describe that this pipeline can identify not only point mutations but also deletions. We then use this pipeline to analyze a total of 17 EMS-mutagenized genomes. We find that EMS-mutagenized genomes carry a significant mutational load including presumptive loss-of-function alleles in several protein-coding genes that can lead to synthetic genetic interactions, one of which we describe here in more detail. We show that outcrossing to wild-type animals can lighten the mutational load; however, a substantial number of sequence variants are also introduced during outcrossing. Even though background mutations uncovered by WGS may complicate the interpretation of mutant phenotypes, they do provide a potentially useful source for functional studies of the affected genes.
MATERIALS AND METHODS
Strains used in this study:
Several strains whose sequence we analyzed on a genome-wide level were previously described: lsy-12(ot177), lsy-5(ot240), and lsy-22(ot114) (Sarin et al. 2008; Flowers et al. 2010). Other strains were isolated by screens for neuronal specification defects in ASE, dopaminergic neurons, AIY, or OLL. With the exception of ttx-3(ot358), we do not describe here the variant that causes the respective mutant phenotype that we selected for. All strains were mutagenized with 50 mm EMS, as described (Brenner 1974). F2 animals (from the mutagenized P0) displaying the mutant phenotype of interest were singled out from the population.
Genetic screen for ot358:
The genetic screen for AIY cell fate mutants was done as first described in (Bertrand and Hobert 2009). Briefly, animals containing the chromosomally integrated ttx-3prom∷gfp reporter mgIs18 or otIs173 (an AIY cell fate marker) were mutagenized with EMS and the F2 generation was screened for defects in gfp expression under a standard dissecting microscope or using a worm sorting machine. We isolated 19 mutant alleles from screening ∼200,000 haploid genomes. One allele was the previously described ref-2 allele (Bertrand and Hobert 2009) and 6 alleles failed to complement ttx-3. One of them was ot358, described here as an enhancer deletion allele, and 5 others were missense, nonsense, or splice site mutations in the ttx-3 coding region (Figure 1C). Other alleles isolated from this screen will be described elsewhere.
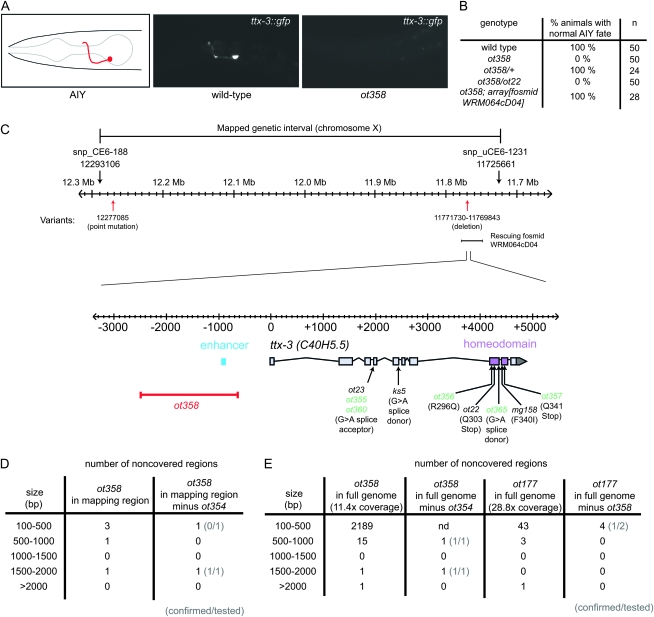
Figure 1.—
WGS identifies a cis-regulatory deletion in ot358 animals. (A) Expression of a GFP-based AIY cell fate marker (otIs173) in wild-type or ot358 adults (red: AIY interneuron). (B) Percentage of adults showing expression of the AIY GFP reporter otIs173 (array[fosmid WRM064cD04] is a chromosomally integrated array containing a ttx-3 rescuing genomic fragment in the context of the fosmid shown in C, kindly provided by V. Reinke). (C) ttx-3 genomic locus. The ot358 deletion removes a cis-regulatory element regulating ttx-3 expression (enhancer in blue). Our screen and previous screens have also identified strains bearing mutations in splice sites or coding regions of the ttx-3 locus (in green are mutations reported here for the first time). (D) Number of noncovered regions in ot358 in the 0.6-cM mapping region. Left, noncovered in ot358; right, noncovered in ot358 but covered in ot354. Tested and confirmed deletions are denoted in parentheses. (E) Number of noncovered regions in ot358 or ot177 in the whole genome. ot358 left, noncovered in ot358; ot358 right, noncovered in ot358 but covered in ot354. ot177 left, noncovered in ot177; ot177 right, noncovered in ot177 but covered in ot358. Tested and confirmed deletions are denoted in parentheses.
WGS procedure:
All runs were done in-house on an Illumina Genome Analyzer II platform, with the exception of ot177, ot114, and ot340, which were done by Illumina's sequence service. For genomic DNA preparation, four 15-cm plates of a confluent population of worms were washed two times in M9 and then incubated in M9 for 30 min at room temperature to purge the worms' intestines. The worms were then washed twice with M9. One of two methods was used to isolate the genomic DNA, both of which worked equally well. The first requires the Qiagen Gentra Puregene Kit (cat. nos. 158622, 158667, and 158689) and instructions therein. The second method uses the following steps: wash with 1× NTE, and lyse worms for 1 hr at 65° with 2 mL proteinase K solution. Perform three phenol/chloroform extractions, always keeping the aqueous phase. Add 1/10 vol 3 m NaOAC, pH 5.2, and 2.5 vol 100% EtOH and spool DNA with a glass pipette. Dissolve in 400 ul TE, add RNAse, and then perform another phenol/chloroform extraction. Precipitate DNA with 1/10 vol 3 m NaOAC, pH 5.2, and 2.5 vol −20° EtOH. Pellet DNA by centrifugation at full speed for 15 min. Redissolve in 200 μl TE.
Sequencing outputs (qseq format) were converted to fastq and directly put into the MAQGene pipeline (Bigelow et al. 2009).
Validation of variants:
We validated variants for two purposes: first, to gauge the reliability of the WGS bioinformatic pipeline (see below) and second, to provide the C. elegans community with validated putative loss-of-function alleles for further functional analysis. We validated variants called by different filtering criteria, a low filter and a stringent filter. The low filter included variants supported by ≥2 reads on either strand, but <60 reads (to exclude highly repetitive regions that are prone to mismapping). The stringent filter included variants supported by ≥3 reads on either strand, but <60 reads. Both filtering criteria used a consensus score >2, loci multiplicity score <2, and variant read:total read ratio >0.86. The consensus score is the probability of the consensus base being correct but expressed in the Phred form (Li et al. 2008; Bigelow et al. 2009). Validation involved creating 200- to 500-bp amplicons surrounding the variant and then Sanger sequencing. We found the highly stringent criteria to be a good measure for accurately called variants (those that could be confirmed by manual resequencing). Sanger sequencing confirmed 83/90 variants called with the stringent filter while 0/23 variants called with the low filter were confirmed.
Finding variants in conserved coding sequences:
We previously described the incorporation into MAQGene (Bigelow et al. 2009) of the six-way PhastCons score that identifies conserved elements between multiply aligned species. For nematodes, Phastcons gives a conservation score for each nucleotide, a numerical value (0–1) based on sequence similarities of sliding 20-bp windows between C. elegans and five related nematode species. The score is weighted on the basis of best-fit models to phylogenetic data (Siepel et al. 2005). As the MAQGene output includes conservation score, we assessed the ability of this score to predict conservation between C. elegans and human sequences. We used the UCSC genome browser and manually inputted each position with conservation score >0 in the C. elegans genome bearing a missense variant, searching for a homologous human gene in which the affected amino acid was also conserved between the two species. Seventeen of 400 of the variant positions unique to one data set tested met these criteria (Table S6). We found that the conservation score did not predict homology between C. elegans and human coding sequences. Specifically, nucleotides with low conservation scores, i.e., not well conserved between all nematode species, may indeed be conserved in the corresponding human homologs. Similarly, high conservation scores (>0.9) did not ensure sequence similarity between worms and humans.
MAQGene:
Update:
We implemented a series of improvements to the previously published MAQGene platform (available at http://maqweb.sourceforge.net). As described in the original article (Bigelow et al. 2009), the core MAQGene pipeline runs MAQ and a long set of SQL commands to associate, sort, and filter the set of found variants with annotation information. Originally implemented as a series of shell commands, if one failed for any reason, there was no facility in place to gracefully abort with meaningful error messages. It would continue, producing empty results files and misleading log messages. Now, the pipeline dependencies are expressed as GNU make rules.
The old pipeline exposed the host machine to the risk of becoming overloaded if users submitted more than one run at a time or if other unrelated processes were running. The new pipeline automatically detects total CPU load and keeps any submitted runs on hold until the machine's CPU usage falls to the requested level.
The old pipeline implemented parallelism in a fragile way involving monitoring of shell processes. This design was liable to be misled by any other unrelated processes running MAQ. The new pipeline automatically detects any processes that can be run in parallel and uses as many CPUs as the user specifies.
Because each link in the dependency graph is expressed as a standalone rule, new links may be added, rearranged, and connected without damaging the existing structure. This will greatly facilitate incorporation of new components to MAQGene, for example, the addition of a new aligner or a new annotation category. Robustness to change is of great importance for software in general, since it is time consuming for authors and users to find bugs that may creep in. Incidentally, the Illumina pipeline is also based on GNU make.
Besides these code design changes, a few bugs have been found and repaired. First, a bug in the supporting CisOrtho software (Bigelow et al. 2004) prevented the reporting of variants occurring in the first exon of any gene. Second, the gene relations “5′”, “genic”, “3′”, have been replaced by “upstream”, “into”, and “downstream”, respectively. The offset field now complies with convention, such that it is the negative of the distance from gene start for “upstream”, the positive of the distance from gene start for “into”, and the positive of the distance from transcription stop for “downstream”.
The installation procedure now automates a few previously required manual steps. The required versions of MAQ, CisOrtho, and samtools binaries are now automatically retrieved, compiled, and tested. While the mysql server must still be configured by hand, the MAQGene installation script now tests to make sure it is correctly configured before proceeding or alerts the user in case requirements are not met. This greatly reduces confusion for users who have not yet met the requirements. For users wanting to update their existing copy, note that the installation script preserves existing results files, although it may overwrite locally configured settings. Once the MAQGene code is updated, build_annotation_tables.sh must also be run for each species previously installed for MAQGene. Typically, this is just the C. elegans genome, but this may change in the future.
As a postinstall test, MAQGene now comes with a set of real reads from the ot340 mutant previously known to localize to chromosome I at 11–12 M. Once installation and the building of annotation tables is complete, a folder called ot340 will appear on the web page and the user may immediately test the installation by selecting these and launching a run. Previously, a much smaller set of reads came with the distribution, which was insufficient to do a real test, but provided only a placeholder for the location where new reads data should be placed. This led to confusion as new users attempted to perform actual runs with this prepackaged data.
We added to the MAQGene distribution a new tool for comparing pileup lines for loci of interest across multiple MAQGene runs. The script, pileup_loci_comparison, takes a list of pileup files (each generated by a separate MAQGene run on a different mutant) and a list of loci as <chromosome> <position>, one entry per line, and then produces a list of the pileup lines at these specific loci sorted by chromosome, locus, and mutant. This facilitates at-a-glance detection of common background mutations.
Now, in addition to exonic and intronic annotations, MAQGene associates sequenced variants with 5′- and 3′-UTRs, known SNPs, and the noncoding RNA types miRNA, rRNA, scRNA, snoRNA, snRNA, and tRNA. This extra annotation is available for C. elegans only, but may be augmented to support other species if the demand is sufficient and time allows.
Finally, we have taken initial steps to port MAQGene to MAC OS X; however, this is still pending. Updates will be available at http://maqweb.sourceforge.net.
All of the above improvements and bug fixes have been submitted to the MAQGene sourceforge code repository and are the default current version for new users. Users who have the original version are urged to update. In either case, a fresh install is recommended. Note that existing MAQGene output data or input reads will not be touched by the installation process. To install, run the following commands (assuming a prompt of ‘$’):
—This will download the MAQGene source into a directory called “MAQGene” (or whatever word you choose at the end of this command)
—The destination directory, MAQGene, in this case, should not exist. If it remains from a previous download, rename it or choose a different name.
$ svn checkout https://maqweb.svn.sourceforge.net/svnroot/maqweb/trunkMAQGene
$ cd MAQGene
Next edit the settings in scr/config.sh:
$ su root
$ ./install.sh
After the prompt, cut-and-paste lines into the Apache httpd.conf file.
If the identical cut-and-paste lines already existed in your httpd.conf file, there is no need to restart httpd. Otherwise, type the following:
$ /sbin/service httpd restart
Also, the install.sh script will note the following:
$ cd /usr/bin; ./build_annotation_tables.sh
Note that it cannot be run like this:
$ /usr/bin/build_annotation_tables.sh
During the running of build_annotation_tables.sh, in C. elegans, choose “ce8” for genome and “wormbase” for data source.
How to find deletions with MAQGene:
To identify deletions in WGS data sets we used the “uncovered.txt” output file. This file reports any noncovered region larger than a chosen threshold. Noncovered regions can reflect true deletions, regions difficult to sequence/map (like repetitive sequences), or missampling due to low coverage. Regions noncovered due to difficulty in sequencing/mapping are removed by comparing with another independent WGS data set. The ‘uncovered.txt’ file in previous versions of MAQGene is now integrated in the final output as class “uncovered” and description “codons FIRST to LAST of TOTAL (LENGTH%)” for affected protein-coding genes. As usual, the parent_feature column will contain the name of the gene affected. For example, “codons 5 to 27 of 130 (17%)” means that codons 5 through 27 were in an uncovered region of a 130 codon gene, comprising 17% of the total. Uncovered regions entirely within introns of genes are also reported. The original uncovered.txt file is still available for referencing non-genic uncovered regions.”
RESULTS
Using WGS data to identify deletions:
WGS using second-generation deep sequencing technology, such as that utilized by the Illumina Genome Analyzer or ABISolid platform, provides a large amount of short DNA sequence reads (Shendure and Ji 2008). These reads require extensive bioinformatic processing and analysis to identify “variants,” i.e., sequence alterations relative to a reference genome. We previously described a software pipeline, MAQGene, that “wraps” the standard alignment program MAQ (Li et al. 2008) into a user-friendly platform and provides simple customized mining of WGS data (Bigelow et al. 2009). One feature of MAQ—and WGS in general—that we aimed to subject to “real life,” proof-of-principle validation is the ability to reliably identify larger deletions, introduced by various mutagens, including EMS or irradiation. In principle, such deletions should appear as “lack of coverage” for genomic intervals; i.e., there should be regions in the genome to which no sequencer-generated reads align. We tested the ability of the WGS approach to reliably identify deletions through sequencing the entire genome of a strain, OH9331, which carries the ot358 allele. This allele was identified in a screen for EMS-induced mutants in which the fate of the AIY interneuron pair is not appropriately executed, as assessed by the expression of a gfp-tagged AIY cell fate marker (see materials and methods) (Bertrand and Hobert 2009) (Figure 1, A and B). The ot358 allele mapped onto the X chromosome within a 0.6-map-unit interval. WGS (single 75 nucleotide reads) of ot358 animals revealed only one, noncoding point variant in this interval (Figure 1C). As EMS can also generate deletions of up to many kilobases, albeit at a lower frequency than base transitions (Anderson 1995), we used an output file of MAQGene, which lists regions that are not covered by the short sequencing reads produced by the sequencer (see materials and methods). We found five noncovered regions of >100 bp in the interval to which we had mapped ot358 (Figure 1D). Three of them are also present in a different mutant strain independently isolated in the same screen (ot354) (Figure 1D); they likely reflect either regions difficult to sequence/map (e.g., repetitive sequences) or deletions initially present in the starting strain for the screen. We analyzed the remaining two ot358-specific noncovered regions by PCR and Sanger sequencing. While the smaller one (103 bp) could not be confirmed, the larger one corresponds to a real deletion of 1888 bp located just a few hundred base pairs upstream of the ttx-3 gene, a homeobox gene known to be involved in AIY development (Hobert et al. 1997). This deletion removes a cis-regulatory element that we recently identified as regulating ttx-3 expression in the AIY lineage (Bertrand and Hobert 2009) (blue box in Figure 1C). Consistent with the possibility that this variant is indeed phenotype causing and affecting the ttx-3 locus, ot358 fails to complement the canonical ttx-3 allele ot22 and was rescued by a ∼40-kb piece of genomic DNA (fosmid) containing a wild-type copy of ttx-3 (Figure 1, B and C).
To further investigate the reliability with which deletions can be accurately discovered by WGS, we examined the entire ot358 mutant genome for deletions. The ot358 WGS data set contains a high number of noncovered regions of <500 bp (Figure 1E) likely reflecting missampling due to the low coverage [11.4×; note that in theory such fold coverage is sufficient to reliably identify missense variants (Shen et al. 2008)]. Consistent with this hypothesis, we find that a previously published WGS data set (ot177 allele) (Sarin et al. 2008), which has a much higher coverage (28.8×), displays far fewer noncovered regions (Figure 1E). Apart from these small noncovered regions, the ot358 WGS data set contains 17 noncovered regions of >500 bp. Fifteen of them are also present in the WGS data set from another mutant, ot354, which was retrieved by the same screen (and therefore has the same genetic starting background). Lack of coverage in these 15 common regions reflects either difficult regions to sequence or deletions initially present in the screening strain (Figure 1E). The ot358 WGS data set therefore contains only 2 unique noncovered regions of >500 bp, one of them corresponding to the 1888-bp deletion responsible for the phenotype and the other to a true deletion of 475 bp located on another chromosome (true deletion sizes are usually smaller than the “uncovered regions” output of MAQGene because of the difficulty to obtain coverage in the immediate flanking sequences of a deletion). This illustrates that this approach can be efficiently used at the whole-genome scale to identify deletions of >500 bp for a genome with coverage of ∼10×, even if one only employs single read WGS.
Can one reliably call deletions of <500 bp if the sequence coverage is higher? To answer this question, we again turned to the ot177 WGS data set (28.8× coverage). This data set contains four noncovered regions of 100–500 bp (Figure 1E). We tested two of them by Sanger sequencing and found that one corresponds to a true deletion of 418 bp while the other reflects missampling (the last two regions are too repetitive to be amplified by PCR). This suggests that at ∼30× coverage, the background is low enough to allow the use of this simple approach to identify deletions of as little as 100 bp at the whole-genome scale level.
EMS-mutagenized strains display a high mutational load, even after outcrossing:
Together with the ttx-3(ot358) case described above, we have now reported on a total of 4 cases [ttx-3 (this article), lsy-12 (Sarin et al. 2008), and lsy-5 and lsy-22 (Flowers et al. 2010)] in which WGS has successfully identified a lesion in a single genetic locus that is responsible for a specific mutant phenotype. Having these genome data sets at hand, we took a much broader view of the overall, genome-wide landscape of EMS-induced effects. Apart from the 4 above-mentioned WGS data sets, we also consider an additional 13 WGS data sets that we generated by WGS of EMS-mutagenized strains, which we isolated on the basis of specific phenotypic features (Table 1) (materials and methods). Some genomes had been outcrossed several times before being subjected to WGS; others were not outcrossed at all. The parameters for all 17 genome sequence runs (read length, coverage, and number of lanes used on a flow cell) are shown in supporting information, Table S1. After MAQGene identified sequence variants relative to the reference genome using standard filtering criteria (Bigelow et al. 2009), we validated a subset of the variants by manual Sanger resequencing. We focused on 113 detected variants from all data sets that resulted in premature stops or splice site mutations. Seventy-nine of 113 variants were confirmed to be “true” variants by Sanger sequencing. All confirmed variants were supported by three or more reads. This validation therefore allowed us to further sharpen our criteria for variant calling with MAQGene by raising the minimum number of reads required to call a variant from two to three (see materials and methods for the recommended complete parameter set). In other words, this new parameter set reliably identifies all true, spot-checked 79 variants and was then used to assess genome-wide mutation signatures (see below).
TABLE 1.
Mutational profile of strains analyzed by WGS
| Variants |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain name | Phenotype selecteda | Allele selected and LG | Transgenic reporter background | Number of times outcrossed | Strain against which it was outcrossed | Nonsense | Splice site | Missense conserved in human homologb | Missensetotal | Total variants | Variants per covered basec (×10−6) | Origin |
| OH9482 | Lol | ot567 X | otIs138 | 10 | N2 | 2 | 0 | 2 | 50 | 479 | 5.42 | Background |
| 0 | 0 | 1 | 68 | 669 | 7.57 | EMS induced | ||||||
| 2 | 0 | 3 | 118 | 1,148 | 13.00 | Total | ||||||
| OH8001 | Lsy | ot177 V | otIs114 | 5 | N2 (3) and otIs114 (2) | 2 | 0 | 2 | 63 | 519 | 5.23 | Background |
| 3 | 1 | 3 | 89 | 771 | 7.77 | EMS induced | ||||||
| 5 | 1 | 5 | 152 | 1290 | 13.00 | Total | ||||||
| OH6087 | Dopy | ot340 III | vtIs1 vsIs33 | 4 | N2 | 0 | 0 | 0 | 6 | 39 | 3.39 | Background |
| 0 | 0 | 0 | 42 | 285 | 24.78 | EMS induced | ||||||
| 0 | 0 | 0 | 48 | 324 | 28.17 | Total | ||||||
| OH7116 | Lsy | ot114 I | otIs3 | 4 | N2 | 0 | 1 | 2 | 49 | 425 | 4.35 | Background |
| 1 | 0 | 1 | 39 | 530 | 5.42 | EMS induced | ||||||
| 1 | 1 | 3 | 88 | 955 | 9.77 | Total | ||||||
| OH7317 | Lsy | ot86 II | ntIs1 | 4 | N2 | 1 | 0 | 1 | 37 | 217 | 5.39 | Background |
| 1 | 0 | 2 | 88 | 744 | 18.50 | EMS induced | ||||||
| 2 | 0 | 3 | 125 | 961 | 23.89 | Total | ||||||
| OH7677 | Lsy | ot104 I | ntIs1; otIs220 | 3 | N2 | 2 | 0 | 2 | 58 | 465 | 5.17 | Background |
| 2 | 1 | 1 | 68 | 745 | 8.29 | EMS induced | ||||||
| 4 | 1 | 3 | 126 | 1210 | 13.46 | Total | ||||||
| OH8421 | Lsy | ot219 V | otIs114 | 3 | N2 (2) and otIs114 (1) | 3 | 1 | 1 | 57 | 452 | 4.93 | Background |
| 3 | 1 | 0 | 66 | 758 | 8.27 | EMS induced | ||||||
| 6 | 2 | 1 | 123 | 1210 | 13.21 | Total | ||||||
| OH4303 | Dopy | ot280 I | vtIs1 | 2 | N2 | 3 | 0 | 2 | 68 | 409 | 7.16 | Background |
| 1 | 0 | 1 | 50 | 473 | 8.29 | EMS induced | ||||||
| 4 | 0 | 3 | 118 | 882 | 15.45 | Total | ||||||
| OH8545 | Dopy | ot477 I | vtIs1 vsIs33 | 2 | N2 | 2 | 0 | 2 | 72 | 467 | 6.38 | Background |
| 0 | 1 | 0 | 39 | 477 | 6.52 | EMS induced | ||||||
| 2 | 1 | 2 | 111 | 944 | 12.90 | Total | ||||||
| OH8547 | Dopy | ot479 III | vtIs1 vsIs33 | 2 | N2 | 2 | 0 | 2 | 64 | 372 | 6.27 | Background |
| 0 | 0 | 1 | 68 | 557 | 9.38 | EMS induced | ||||||
| 2 | 0 | 3 | 132 | 929 | 15.65 | Total | ||||||
| OH9330 | Loss of AIY fate | ot354 II | otIs173 | 1 | N2 | 1 | 1 | 2 | 65 | 418 | 4.62 | Background |
| 3 | 1 | 2 | 77 | 720 | 7.95 | EMS induced | ||||||
| 4 | 2 | 4 | 142 | 1138 | 12.57 | Total | ||||||
| OH9331 | Loss of AIY fate | ot358 X | none | 1 | N2 | 1 | 1 | 2 | 54 | 321 | 3.59 | Background |
| 0 | 1 | 0 | 23 | 247 | 2.76 | EMS induced | ||||||
| 1 | 2 | 2 | 77 | 568 | 6.35 | Total | ||||||
| OH9305 | Lsy | ot240 I | otIs114 | 1 | HAe | 1 | 1 | n/a | 60 | 509 | 5.27 | Background |
| 1 | 0 | n/a | 327 | 3238 | 33.55 | EMS induced | ||||||
| 2 | 1 | n/a | 387 | 3747 | 38.83 | Total | ||||||
| OH6071 | Dopy | ot337 I | vtIs1 vsIs33 | 0 | NA | 4 | 0 | 2 | 63 | 375 | 7.00 | Background |
| 3 | 0 | 1 | 78 | 629 | 11.74 | EMS induced | ||||||
| 7 | 0 | 3 | 141 | 1004 | 18.74 | Total | ||||||
| OH4240 | Dopy | ot260 I | vtIs1 | 0 | NA | 6 | 1 | 1 | 106 | 633 | 6.85 | Background |
| 5 | 0 | 1 | 76 | 753 | 8.14 | EMS induced | ||||||
| 11 | 1 | 2 | 182 | 1382 | 14.95 | Total | ||||||
| OH4247 | Dopy | ot263 X | vtIs1 | 0 | NA | 5 | 2 | 2 | 107 | 604 | 6.85 | Background |
| 2 | 1 | 3 | 87 | 740 | 8.40 | EMS induced | ||||||
| 7 | 3 | 5 | 194 | 1344 | 15.25 | Total | ||||||
| OH2042 | Ssy | ot83 V | otIs3 dpy20 lin-49(ot78) | NAd | N2 | 0 | 1 | 2 | 61 | 392 | 4.12 | Background |
| 2 | 1 | 0 | 41 | 677 | 7.11 | EMS induced | ||||||
| 2 | 2 | 2 | 102 | 1069 | 11.23 | Total | ||||||
| Totals | 35 | 9 | 27 | 1,040 | 7,096 | Background | ||||||
| 27 | 8 | 17 | 1,326 | 13,013 | EMS induced | |||||||
| 62 | 17 | 44 | 2,366 | 20,105 | Total | |||||||
| Average per genomef | 2.1 | .5 | 1.6 | 61.3 | 411.7 | 5.42 | Background | |||||
| 1.6 | .5 | 1.0 | 62.4 | 610.9 | 9.43 | EMS induced | ||||||
| 3.7 | 1.0 | 2.6 | 123.7 | 1022.4 | 14.84 | Total | ||||||
Deletions are separately shown in Table S5. The nature and location of all the nonsense, conserved missense, and splice site mutations are shown in Table S2, Table S3, and Table S6.
Lsy, laterally symmetric ASE fate; Dopy, dopaminergic fate atypical; Ssy, suppressor of ASE symmetry; Lol, loss of OLL fate.
Homology determined by sequence BLAT against the human genome as described by the UC Santa Cruz genome browser. Only those mutations that change an amino acid conserved between C. elegans and Homo sapiens were included as those have the highest likelihood to disrupt protein function when mutated. See Table S6 for identities of 17 EMS-induced conserved missense mutations.
See Table S1 for WGS run statistics, including average sequencing depth, for each strain.
Strain was isolated as a suppressor of lin-49(ot78)'s effect on ASE fate specification, a mutant that was isolated itself from an independent mutagenesis and backcrossed an undetermined number of times.
HA = Hawaiian C. elegans isolate. Annotated HA SNPs are removed during variant analysis. The large number of variants uncovered in this strain is likely due to unannotated HA SNPs resulting from the outcrossing scheme; those could not be eliminated since this is the only HA strain that we sequenced.
Averages exclude OH9305, see (e).
Variants relative to the wild-type reference genome can be attributed to three different sources: (1) variants that already existed in our N2 wild-type strain or are sequencing errors in the reference genome; (2) variants that existed in our transgenic reporter strain, which was generated through chromosomal integration into our N2 wild-type strain (the radiation-mediated “chromosomal integration” is itself mutagenic); and (3) variants caused by the EMS mutagenesis that produced the specific mutant strain. To distinguish between these possibilities, we compared variants between individual data sets. Variants found in all data sets were considered to be present in our N2 starting strain (totaling >600 variants); variants found in a subset (particularly those found in strains with the same transgenic marker) were also considered to be unrelated to the EMS mutagenesis, even though we cannot completely rule out the possibility that they may be independent EMS hits.
After filtering out the variants that were found in multiple data sets (“background” rows in Table 1), we examined the molecular identity of the remaining individual variants, present uniquely in only 1 strain. We found that each sequenced genome displayed not only a large number of noncoding sequence variants, but also a large number of predicted protein-coding single-nucleotide variants, between 24 and 96 per genome (Table 1). This substantial number of protein-affecting background mutations was surprising because some of the sequenced genomes were outcrossed several times (Table 1), but they nevertheless maintained a high mutational load. Intriguingly, the number of outcrosses did not correlate significantly with the number of variants (Figure 2; see legend for how we dealt with issues such as linkage). For example, a 5-fold outcrossed strain, OH8001, selected for a single-neuron-specific differentiation defect, still contains a mutational load of >90 protein-coding allelic variants and 682 non-protein-coding variants (Table 1). Similarly, the 10-fold outcrossed strain OH9482, selected for another neuron specification defect, also still contains 68 protein-coding allelic variants and 601 non-protein-coding variants (Table 1). Several possibilities may explain this lack of correlation. Below, we provide evidence for two specific scenarios that may contribute to the retention of a high mutational load. These are (1) linkage disequilibrium between unlinked chromosomes that may select for viability (or other phenotypes) and (2), surprisingly, the introduction of novel variants through outcrossing, rather than a retention of the initial, EMS-induced mutational load.
Figure 2.—
Lack of correlation between fold outcross and variant number. (A) y-axis represents unique variant numbers divided by total number of sequenced bases (those with at least 3× and at most 60× sequencing depth) for each data set. This accounts for differences in coverage between different data sets. Unique variants exclude those shared between two or more data sets. (B) This graph presents unique variants per analyzed chromosome, excluding those linked to the mutation or transgene reporter. To deal with linkage, we did not include variants on chromosomes that contained (a) the phenotype-causing mutation isolated for, (b) the integrated fluorescent transgene used to score the mutant phenotype, and (c) the X chromosome, which is subject to fewer recombination events due to method of outcrossing: F1 males from the first outcross (which are XO for this chromosome and therefore do not undergo meiotic recombination) are mated with the wild-type strain. F2 progeny from this second outcross containing the mutant phenotype are then isolated. This analysis accounts for mutation hitchhiking effects.
Balancing selection may retain variants
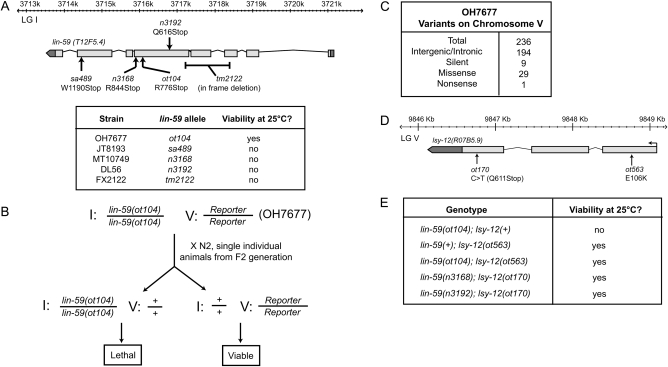
The EMS-mutagenized strain OH7677 was isolated for a specification defect of gustatory neurons (Lsy phenotype) (Sarin et al. 2007), subjected to WGS, and found to contain a premature stop codon in an essential gene, lin-59 (Figure 3). This was curious as the OH7677 strain is viable and fertile (note that the lin-59 allele, ot104, resides in a position between two previously characterized, lethality-causing nonsense alleles of lin-59; Figure 3). Upon outcrossing OH7677 animals, we noted that even though ot104 resides on chromosome I, we could not unlink it as a viable allele from a gfp reporter transgene integrated on chromosome V. We therefore surmised that chromosome V may contain a locus, linked to the reporter gene, that when mutated allows ot104 homozygous animals to live. We examined sequence variants on chromosome V in the OH7677 genome sequence data set and noted the presence of a missense mutation (E106K) in the lsy-12/R07B5.9 locus. To test whether lsy-12 is indeed causative for the lin-59(ot104) lethality suppression, we combined other lin-59 nonsense alleles, n3168 and n3192, with another lsy-12 allele, ot170, and found that ot170 also suppressed the lethality exhibited by the other lin-59 alleles (Figure 3). We have therefore uncovered through WGS a genetic interaction that would have been much harder to molecularly elucidate through conventional mapping schemes. Importantly, this example makes a case for a scenario in which a mutational load is retained through balancing selection. During outcrossing, which selected for the lin-59(ot104) Lsy phenotype, inadvertently, chromosome V was “carried along” as it contained the lsy-12 suppressor of the ot104 lethality. Variants linked to lsy-12 on chromosome V are therefore more difficult to outcross and a larger mutational load remains. However, this can still not fully explain why a 3× outcrossed lin-59(ot104) strain holds a substantial mutational load that is equivalent to other strains backcrossed fewer times (Figure 2). Furthermore, a high mutational load is still present after disregarding variants on chromosome V (data not shown).
Figure 3.—
lin-59 lethality is suppressed by lsy-12. (A) lin-59 gene structure. The table at the bottom displays viability of each lin-59(−) strain. (B) Outcrossing of OH7677 reveals a modifier locus on chromosome V that suppresses the observed lethality likely due to lin-59(ot104). (C) Variants discovered on chromosome V in OH7677. Variants were processed and filtered with MAQGene (Bigelow et al. 2009). One of the missense variants resides within the lsy-12 locus. (D) lsy-12 gene structure. This schematic shows an incomplete version of the lsy-12 locus per the last release of WormBase. Transcript mapping shows that the lsy-12 locus extends in the more upstream located mys-3 gene, a histone acetyltransferase (M. M. O'Meara and O. Hobert, unpublished results). (E) A second independently isolated allele of lsy-12 (Sarin et al. 2007) suppresses two other independently isolated lin-59 alleles (Potts et al. 2009) that display lethality at 25°.
Outcrossing EMS-mutagenized strains relieves initial mutational load but introduces new variants:
To pursue the matter of mutational load further, we sequenced two strains carrying the ot354 allele that causes a differentiation defect of the AIY interneuron similar to the ot358 allele shown in Figure 1. One ot354-containing strain was backcrossed once against N2 wild type, and the other was backcrossed an additional six times to a total of seven times. We found that the 1× outcrossed ot354 strain contained a total of 730 unique variants (Figure 4). We analyzed each variant position in the 7× outcrossed ot354 genome sequence and found that most of them (549/730) had been replaced, through outcrossing, by the wild-type, reference genome base. One hundred thirty-four existed in a heterozygous state while 30 remained homozygous variants, i.e., were maintained after 6 outcrosses (Figure 4) (the remaining 17 were in areas of low coverage and therefore a reliable consensus base could not be obtained). As expected, most of these retained homozygous variants, 24/30, were linked to the chromosomes harboring either the phenotype-causing mutant allele (II:ot354) or the integrated gfp reporter used to assess this phenotype (III:otIs173). Thus, considering only unlinked chromosomes, 6 outcrosses reduced the observed variant number from 428 to 6. After the first outcross, the number of variants that are predicted to exist in the homozygous state should be 25% of the original load (note that variants are not detected by our filtering criteria in their heterozygous state), and then each subsequent outcross would reduce this number by 50%. Our observed number of variants unlinked to the phenotype-causing mutation or reporter (6) is comparable to the predicted number (3). Thus, outcrossing to a wild-type strain indeed has the intended effect of removing background variants.
Figure 4.—
Outcrossing reduces total variant number but introduces new variants. Variant numbers that are unique to the ot354 data set, i.e., not found in any other of the 16 sequenced mutant genomes, are considered. ot354; otIs173 was outcrossed against the laboratory wild-type N2 Bristol C. elegans strain and F2 homozygous mutant; reporter animals were singled out after each outcross. x/x are numbers that indicate the state of each unique variant position (wild type, heterozygous, etc.) in the other strain's genome, revealing both the loss and the gain of variants after six additional outcrossing events. Homozygous mutations were defined by those positions covered by more than ten reads holding a variant:total read ratio x > 0.86. Heterozygous mutations were defined by those positions covered by more than two reads holding a variant:total read ratio 0.4 > x ≥ 0.86. See materials and methods for the value of the other parameters.
However, we noted that outcrossing against our wild-type N2 strain also had unintended effects: although a total of 30 variants were common between the 1× and 7× outcrossed ot354-strains, we were surprised to note that ot354 (7× outcrossed) carried 202 unique variants of its own, i.e., those not found in any other strain (several of which were outcrossed with our N2 strain, too) (Figure 4). One hundred twelve of 202 of these variants were sufficiently covered in the 1× outcrossed data set for further analysis: a closer inspection revealed that 69 of these variant positions were in fact wild type in the 1× outcrossed strains, implying that new variants had been introduced by outcrossing. These new variants are likely the result of previously observed, substantial genetic drift in the N2 population (Denver et al. 2009; Flibotte et al. 2010, accompanying article in this issue) to which the mutagenized strain was outcrossed or in the ot354 population that was grown for several months during the outcrossing. Such drift could, at least in part, explain the surprising lack of correlation observed between fold outcross and variant number (Figure 2) (see discussion).
Identifying putative loss of protein function alleles:
Summing up all 17 strains whose genome we sequenced, our analysis identified unique allelic variants in >1000 genes. With the exception of lsy-22, all sequenced strains are completely viable even though a substantial number of the many mutations in the sequenced strains are supposed to have deleterious effects on protein function (Table 1). Such a deleterious effect is assumed if one of four criteria is fulfilled:
Premature stop codons, which truncate a protein: We surmise likely loss (either complete or partial) of protein function if the premature stop codon occurs before a conserved protein domain or in an exon before the last exon, which presumably subjects the message to nonsense-mediated mRNA decay. All allelic variants of this type are shown in Table S2.
Splice site mutations: Even though their effect on protein function is more difficult to predict as alternate, cryptic splice sites may be used, many cases in the literature show highly deleterious effects of splice site mutations, usually because aberrant splicing generates reading frame changes. All allelic variants of this type are shown in Table S3.
Deletions usually have the most obvious effects on protein function. As described above, extracting deletions from WGS data requires an analysis of the genomic regions with lack of coverage. For reasons described above in the section on the ttx-3(ot358) deletion allele, we consider only such regions that are >500 bp and excluded those regions that are similarly noncovered between data sets sharing the same transgenic background. All allelic variants of this type are shown in Table S4 and Table S5. We manually confirmed the boundaries of these deletions (Table S5).
Finally, we considered missense mutations, whose impact on protein function is difficult to assess. We used the criteria of phylogenetic conservation as an indicator of potential impact of an allele on protein function. We first used the six-way PhastCons score integrated into the MAQGene platform, which compares successive 20-bp C. elegans sequence windows to five closely related species and weights homology using phylogenetic criteria (Siepel et al. 2005;Bigelow et al. 2009). Amino acids found to be conserved by these criteria were then assessed for whether they are conserved in human homologs as well (see materials and methods). All allelic variants of this type are shown in Table S6.
The total number of alleles from all 17 strains falling into these four categories is 57. Sixty-two percent of the genes had no allelic variant described before. The number of protein-altering allelic variants doubles to >120 if one takes into account variants that were already present in the starting strain, before mutagenesis (Table 1). In sum, each mutant strain that we genome sequenced contains, on average, four premature stop codons and one splice site mutation.
As mentioned above, premature stop codons and deletions are excellent candidates for genetic loss-of-function alleles. The strains that contain these alleles are therefore a valuable resource that complements the efforts of the C. elegans knockout consortia (Moerman and Barstead 2008; Mitani 2009). Because of this value, we confirmed all premature stop codon variants shown in Table 1 by Sanger resequencing. All strains with these alleles have been submitted to the Caenorhabditis Genetics Center (CGC) and the sequence information has been submitted to WormBase (www.wormbase.org) for annotation.
WGS reveals bias in variant distribution within C. elegans autosomes:
Examination of the genome-wide distribution of variants uncovered by WGS reveals that the distribution of these variants is not completely uniform. Variants appear slightly more concentrated on the arms of each chromosome (Figure S1 A), away from gene-rich regions (Barnes et al. 1995). Quantification of variant rates inside and outside such regions revealed that indeed variants were more likely to be retrieved on the arms of the chromosomes by an average factor of 1.5× among all five autosomes (Figure S1 B). This trend remained even when variants on chromosomes bearing the phenotype-causing mutations and transgenes were removed from this analysis (Figure S1 C). Such an analysis could not be conducted on the X chromosome as it does not hold a similar gene-rich region (Barnes et al. 1995). The fact that the strains analyzed by WGS display fewer variants in gene-rich regions than in gene-poor ones may result from the negative selection of lethal or sterile mutations. A mutation in a gene-rich region is more likely to affect an essential gene and result in the death or sterility of the animal, which introduces a bias against its isolation. In support of this idea, synonymous changes, which should not be under negative selection, were more randomly distributed across the autosomes (Figure S1 D).
Specificity and efficiency of the EMS mutagen:
We also considered the efficiency and specificity with which EMS induces mutations. A previous study analyzed 245 mutations in coding regions and reported a frequency of 92% G/C > A/T transitions (Anderson 1995). Through sequencing five EMS-mutagenized strains, a similar mutational spectrum of EMS is also reported by Flibotte et al. (2010). In our three unbackcrossed strains we find a broader range of specificity as 66% are G/C > A/T transitions (considering 649 supported by more than five reads). The rest are G/C > T/A (5%), G/C > C/G (2%), T/A > G/C (9%), T/A > C/G (12%), or T/A > A/T (5%) mutations. Not all these variants are necessarily EMS induced, considering the genetic drift that we discussed above.
To assess EMS efficiency, we considered three nonoutcrossed strains that we sequenced. At the doses that we use (50 mm), we find that EMS induces roughly one change per 100,000 nucleotides (Table S7). This number refers only to variants in a homozygous state. To get a rough idea for the number of heterozygous variants in the background, we needed to take into consideration the reliability with which MAQGene can assess a “mixed” position. To optimize the chances of correctly calling such a mixed state, we considered only those positions that are covered >10× and for which ∼40–60% of the bases are called by the software as a mutant. Such positions may be either true heterozygotes or sequencing errors resulting from base calling and/or mapping of highly repetitive regions differing at only a few positions. Three of 10 were manually validated from the original genomic sample used for genome resequencing. For one such strain, OH9482, we find 2067 variants that fit these criteria among 65,165,624 bases covered by at least 10 reads. If 0.3 of these are true heterozygotes, this means 1 heterozygous mutation per 94,581 bases. Previous predictions of the frequency of G/C to A/T transitions induced by standard concentrations of EMS (50 mm) are 7 × 10−6 per mutagenized G/C base (Anderson 1995). We find the frequency to be quite close, 1 × 10−5 per mutagenized G/C base. Given that ∼26% of the genome is protein coding (Spieth and Lawson, 2006), this number is also not far off from Brenner's original estimate of the forward mutation rate of 5 × 10−4 per gene (Brenner 1974).
These numbers have to be taken with caution due to founder effects and the drive for homozygosity of selfing strains. Invariably, upon picking individuals during strain maintenance one generates a founder effect, with a 25% probability for each variant initially induced by EMS to have been lost.
DISCUSSION
Our key results can be summarized as follows:
Through deep sequencing of multiple genomes, followed by validation of selected variants, we have further optimized our parameter set and bioinformatic pipeline (MAQGene) for application of WGS for mutation identification, an approach that we expect to become routine in the model system community (Hobert 2010).
We provide two additional examples for the use of WGS to identify phenotype-causing mutations in an organism; one particular example shows that WGS can be used to detect deletions, while another example demonstrates the use of WGS to identify a genetic interaction between two chromatin-modifying loci (lin-59; lsy-12 interaction).
We show that even though outcrossing removes many EMS-induced variants, mutagenized genomes are still littered with protein-coding sequence variants. The variant landscape that we describe here also provide a first pass estimate for how many variants to expect in a genetic interval in which a genetic trait has been mapped to [Table 1; (1.6 + 0.5 + 62.4)/6 which equals about 11 protein-altering variants per chromosome].
The fortuitously identified allelic variants described here are potentially useful tools for further studies on the respective genes. On the basis of this observation, it can be anticipated that more and more allelic variants of all sorts of genetic loci will be identified in future WGS data sets of mutagenized genomes.
Our most unanticipated finding is the mutational load (“dirtiness”) even of backcrossed strains. This is a somewhat troubling finding as a basic premise of genetic mutant analysis is that one compares a strain that differs in its genetic makeup from the wild-type control strain only in the single locus under investigation. Our analysis shows that this is very far from being the case; in reality, even backcrossed strains contain multiple, likely protein-changing sequence variants. We do not believe that this is due to some unusual laboratory-specific strain problem with our N2 wild-type strain since some strains were also backcrossed with nonmutagenized transgenic starter strains, which are different for different WGS data sets (Table 1). We also examined WGS data sets that we produced for several other laboratories, which provided us with strains of varying degrees of outcrossing. We made the same observations in those data sets, i.e., a high mutational load observed even after outcrossing.
One explanation for this mutational load is genetic drift. WGS studies on mutation-accumulation lines indicate that independently isolated animals from a parental wild-type N2 strain acquire, on average, 40 unique spontaneous mutations over 200 generations (Denver et al. 2009). The consequence of genetic drift may be that the wild-type N2 strain, considered to be isogenic, is in fact quite heterogeneous, containing perhaps many heterozygous mutations that became fixed in unique combinations after outcrossing and selection for the mutant phenotype. In support of this idea is the observation that many variants are present in only a subset of our 17 sequenced strains (despite the variant position being covered by sufficient reads in all). As this subset does not correlate with strain history (i.e., one vs. another transgenic starter strain), this suggests that they may have been present in a fraction of our N2 worms and became homozygous in a random subset of manipulated populations after outcrossing. Furthermore, a 1.4-kb deletion was found to be common between only two strains, OH7116 and OH9330, which do not share the same original transgenic background (Table S5). No other sequenced strain carried this deletion, which was likely present in our wild-type N2 strain as a heterozygote, introduced by outcrossing, and randomly driven to homozygosity in only these two strains. Similarly, genetic drift likely also acts on the mutant strain itself, introducing spontaneous variants that are unique to its genome.
Some of the mutational load may also be due to instances of linkage disequilibrium between alleles. Extensive linkage disequilibrium, extending to unlinked chromosomes, has been observed in recombinant inbred C. elegans wild isolates (Rockman and Kruglyak 2009). Our reported lin-59; lsy-12 interaction provides an extreme example of linkage, or balanced selection, of two physically unlinked variants. The presence of lin-59 fixes lsy-12 (and variants physically linked to lsy-12) in the population. Another potential reason for the lack of correlation that we observe between outcross number and variant number may be variable efficiency of the EMS mutagenesis procedure that may result in variable initial mutational loads between strains before outcrossing.
In effect, the standard comparison found in most C. elegans research reports between a premutagenesis and a postmutagenesis mutant strain, selected for a specific, single-locus phenotype, is therefore not a comparison between two strains that differ only in one locus, but rather a comparison between strains that bear many hundreds of sequence variants, including a double-digit number of protein-coding variants that likely severely affect protein function. It therefore is extremely important to make sure that one can indeed ascribe a mutant phenotype to the loss of one specific locus alteration, rather than the phenotype being a composite one. Traditionally, the four most commonly used criteria to assign gene function to a single locus are (1) mapping of a mutation to a specific interval, (2) rescue of the mutant phenotype, (3) observation of the same phenotype with multiple, independently isolated alleles that fail to complement one another, and (4) RNAi phenocopy of the mutant phenotype [all those four criteria were applied to pinpoint the molecular identity of lsy-12(ot177) in our initial WGS study (Sarin et al. 2008)]. Even though quite good, none of these approaches alone deals perfectly well with the possibility of modifier mutations in the background. Mapping may just identify one component of the trait, and a modifier locus contained in the background may easily go undetected; this holds particularly for the commonly used rapid SNP mapping approaches in which mapping to a specific interval is not necessarily absolute. Transformation rescue also does not exclude a modifier effect for the simple technical reason that copy number issues of transgenic arrays do not allow an experimenter to interpret the extent of rescue. The availability of multiple, independently isolated alleles is also not a sure way to exclude the possibility that due to selective pressure, the retrieval of multigenic loci may be favored. Even the very simple approach of checking whether a specific, recessive trait segregates in a 1/4 Mendelian ratio can be misleading as there may be selective pressure on maintaining the modifier mutation in the background. Finally, phenocopy by RNAi against a candidate gene also leaves open the possibility of modifier effects. The bottom line is that ideally the experimenter wants to have not one, but several pieces of independent lines of evidence that one deals indeed with a single-locus effect.
Background variants have the following practical impact on cloning a gene by WGS. In principle, sequencing a single mutant genome can identify the phenotype-causing mutation of interest [as shown in our proof-of-principle study (Sarin et al. 2008)]. However, due to the amount of background variants, some rough mapping is recommended to hone in on a smaller number of candidate variants that may be responsible for the mutant phenotype (Sarin et al. 2008). Preferably, one has sequenced multiple, independently isolated mutant strains that originated from the same starting strain, which allows one to eliminate all strain background variants (this study and Flowers et al. 2010). Such data set comparisons are more cost effective than sequencing the starting strain, which will yield no other information than background variants, while sequencing another nonallelic mutant strain may also lead to the identification of another phenotype-causing mutation. Most preferably, one undertakes WGS of two alleles of the same locus. Comparing these data sets will eliminate all EMS/outcrossing-induced random variants that will affect distinct sets of genes; only those variants that affect the same locus are candidates for the phenotype-causing mutation. In such a case, at least in theory, minimal to no mapping is required before undertaking WGS. Yet, even though several scenarios can be envisioned in which WGS does not require any previous, classic mapping, we still recommend coarse (and therefore fast) classic mapping through simple linkage analysis. Such analysis not only helps in analyzing WGS data, but also implicitly validates genetic complementation data (i.e., alleles that fail to complement should map to the same interval) and assesses whether a trait is mono- or polygenic.
Acknowledgments
We thank Stephen Nurrish and Iva Greenwald for extensive discussions on the subject and Qi Chen for expert DNA injection. We thank Iva Greenwald, Eric Alani, Don Moerman and members of the Hobert lab for comments on the manuscript. We acknowledge funding by the National Institutes of Health to O.H. (R01NS039996-05 and R01NS050266-03) and to S.S. (NS054540-01) V.B. was funded by postdoctoral fellowships by the European Molecular Biology Organization and Human Frontier Science Program Organization. O.H. is an Investigator of the Howard Hughes Medical Institute.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.116319/DC1.
References
- Anderson, P., 1995. Mutagenesis, pp. 31–58 in Caenorhabditis elegans—Modern Biological Analysis of an Organism, edited by H. F. Epstein and D. Shakes. Academic Press, New York/London/San Diego.
- Barnes, T. M., Y. Kohara, A. Coulson and S. Hekimi, 1995. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics 141 159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand, V., and O. Hobert, 2009. Linking asymmetric cell division to the terminal differentiation program of postmitotic neurons in C. elegans. Dev. Cell 16 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigelow, H., M. Doitsidou, S. Sarin and O. Hobert, 2009. MAQGene: software to facilitate C. elegans mutant genome sequence analysis. Nat. Methods 6 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigelow, H. R., A. S. Wenick, A. Wong and O. Hobert, 2004. CisOrtho: a program pipeline for genome-wide identification of transcription factor target genes using phylogenetic footprinting. BMC Bioinformatics 5 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenstiel, J. P., A. C. Noll, J. A. Griffiths, A. G. Perera, K. N. Walton et al., 2009. Identification of EMS-induced mutations in Drosophila melanogaster by whole-genome sequencing. Genetics 182 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver, D. R., P. C. Dolan, L. J. Wilhelm, W. Sung, J. I. Lucas-Lledo et al., 2009. A genome-wide view of Caenorhabditis elegans base-substitution mutation processes. Proc. Natl. Acad. Sci. USA 106 16310–16314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flibotte, S., M. L. Edgley, I. Chaudhry, J. Taylor, S. E. Neil et al., 2010. Whole-genome profiling of mutagenesis in Caenorhabditis elegans. Genetics 185 431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers, E. B., R. J. Poole, B. Tursun, E. Bashllari, I. Pe'er et al., 2010. The Groucho ortholog UNC-37 interacts with the short Groucho-like protein LSY-22 to control developmental decisions in C. elegans. Development 137 1799–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert, O., 2010. The impact of whole genome sequencing on model system genetics: get ready for the ride. Genetics 184 317–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert, O., I. Mori, Y. Yamashita, H. Honda, Y. Ohshima et al., 1997. Regulation of interneuron function in the C. elegans thermoregulatory pathway by the ttx-3 LIM homeobox gene. Neuron 19 345–357. [DOI] [PubMed] [Google Scholar]
- Irvine, D. V., D. B. Goto, M. W. Vaughn, Y. Nakaseko, W. R. McCombie et al., 2009. Mapping epigenetic mutations in fission yeast using whole-genome next-generation sequencing. Genome Res. 19 1077–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., J. Ruan and R. Durbin, 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18 1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani, S., 2009. Nematode, an experimental animal in the national BioResource project. Exp. Anim. 58 351–356. [DOI] [PubMed] [Google Scholar]
- Moerman, D. G., and R. J. Barstead, 2008. Towards a mutation in every gene in Caenorhabditis elegans. Brief. Funct. Genomic. Proteomic. 7 195–204. [DOI] [PubMed] [Google Scholar]
- Potts, M. B., D. P. Wang and S. Cameron, 2009. Trithorax, Hox, and TALE-class homeodomain proteins ensure cell survival through repression of the BH3-only gene egl-1. Dev. Biol. 329 374–385. [DOI] [PubMed] [Google Scholar]
- Rockman, M. V., and L. Kruglyak, 2009. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 5 e1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin, S., M. O'Meara, E. B. Flowers, C. Antonio, R. J. Poole et al., 2007. Genetic screens for Caenorhabditis elegans mutants defective in left/right asymmetric neuronal fate specification. Genetics 176 2109–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin, S., S. Prabhu, M. M. O'Meara, I. Pe'er and O. Hobert, 2008. Caenorhabditis elegans mutant allele identification by whole-genome sequencing. Nat. Methods 5 865–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Y., S. Sarin, Y. Liu, O. Hobert and I. Pe'er, 2008. Comparing platforms for C. elegans mutant identification using high-throughput whole-genome sequencing. PLoS ONE 3 e4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure, J., and H. Ji, 2008. Next-generation DNA sequencing. Nat. Biotechnol. 26 1135–1145. [DOI] [PubMed] [Google Scholar]
- Siepel, A., G. Bejerano, J. S. Pedersen, A. S. Hinrichs, M. Hou et al., 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D. R., A. R. Quinlan, H. E. Peckham, K. Makowsky, W. Tao et al., 2008. Rapid whole-genome mutational profiling using next-generation sequencing technologies. Genome Res. 18 1638–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieth, J., and D. Lawson, 2006. Overview of gene structure (January 18, 2006), WormBook, edited by The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.65.1, http://www.wormbook.org.
- Srivatsan, A., Y. Han, J. Peng, A. K. Tehranchi, R. Gibbs et al., 2008. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet. 4 e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]