Abstract
With the most recent releases of the Drosophila melanogaster genome sequences, much of the previously absent heterochromatic sequences have now been annotated. We undertook an extensive genetic analysis of existing lethal mutations, as well as molecular mapping and sequence analysis (using a candidate gene approach) to identify as many essential genes as possible in the centromeric heterochromatin on the right arm of the second chromosome (2Rh) of D. melanogaster. We also utilized available RNA interference lines to knock down the expression of genes in 2Rh as another approach to identifying essential genes. In total, we verified the existence of eight novel essential loci in 2Rh: CG17665, CG17683, CG17684, CG17883, CG40127, CG41265, CG42595, and Atf6. Two of these essential loci, CG41265 and CG42595, are synonymous with the previously characterized loci l(2)41Ab and unextended, respectively. The genetic and molecular analysis of the previously reported locus, l(2)41Ae, revealed that this is not a single locus, but rather it is a large region of 2Rh that extends from unextended (CG42595) to CG17665 and includes four of the novel loci uncovered here.
THE term “heterochromatin” was introduced by Heitz (1928) to describe regions of mitotic chromosomes that remain condensed throughout the cell cycle, in contrast to regions of euchromatin, which condense only during cell division. Heterochromatin was later divided into two classes: constitutive and facultative heterochromatin (Brown 1966). Constitutive heterochromatin is found in large blocks near centromeres and telomeres, while facultative heterochromatin can be described as silenced euchromatin that undergoes heterochromatization at specific developmental stages. Other properties of constitutive heterochromatin include late replication in S phase, low gene density, strikingly reduced level of meiotic recombination, enrichment in transposable element sequences and highly repetitive satellite DNA sequences, and the ability to silence euchromatic gene expression in a phenomenon called position effect variegation.
Approximately 30% of the Drosophila melanogaster genome consists of constitutive heterochromatin (Gatti and Pimpinelli 1992). Centromeric heterochromatin in D. melanogaster is composed of mainly middle-repeat satellite DNA sequences and clusters of transposable element sequences (Lohe et al. 1993; Pimpinelli et al. 1995). Genes that reside in the heterochromatin are scattered like islands between the satellites and clusters of transposable elements. On average, heterochromatic genes are larger than euchromatic genes, primarily due to the prevalent accumulation of transposable element sequences in their introns (Devlin et al. 1990; Biggs et al. 1994; Dimitri et al. 2003a,b; Hoskins et al. 2007). Heterochromatic genes also tend to be AT-rich compared to their euchromatic counterparts; there is some evidence suggesting that the coding sequences of heterochromatic genes evolve toward AT richness in response to being located in heterochromatin (Yasuhara et al. 2005; Díaz-Castillo and Golic 2007).
Drosophila heterochromatin is vastly under-replicated in polytene chromosomes, so heterochromatic genes cannot easily be mapped through polytene analysis. However, by using Hoechst 33258 and N-chromosome banding techniques, Dimitri (1991) was successful in dividing heterochromatin in mitotic chromosomes into distinct cytological bands; this was an important step in mapping the precise location of heterochromatic genes because before this time heterochromatic genes could be mapped only relative to one another. Here we focus on further refining the previous mapping work on essential genes in the proximal heterochromatin of the right arm of the second chromosome (2Rh) in cytological region h41–h46 of D. melanogaster (Hilliker 1976; Hilliker et al. 1980; Coulthard et al. 2003; Myster et al. 2004).
Early mapping studies in D. melanogaster putatively placed the light (lt) and rolled (rl) genes in, or near, chromosome 2 heterochromatin (Schultz 1936; Hannah 1951; Hessler 1958). The first large-scale mutagenesis specifically directed at finding vital loci in second chromosome heterochromatin was conducted by Hilliker (1976). Using heterochromatic deletions created by Hilliker and Holm (1975), Hilliker (1976) set out to map vital loci using the mutagen ethyl methanesulfonate (EMS). He identified seven individual lethal complementation groups in 2Rh that were interpreted as representing seven vital loci. One of these heterochromatic loci was identified as the previously described rl gene. Two of the remaining vital loci have since been identified: Nipped-A is synonymous with the l(2) 41Ah complementation group (Rollins et al. 1999) and RpL38 is synonymous with Minute(2)41A and Hilliker's (1976) l(2)41Af complementation group (Marygold et al. 2005; also referred to as l(2)Ag in FlyBase). In addition, Rollins et al. (1999) found the Nipped-B gene to be located in 2Rh, but how this locus fit into the data from Hilliker (1976) was unclear.
With the limited release of some of the more distal heterochromatic sequences (Hoskins et al. 2002), a more recent mutagenesis screen focusing on distal 2Rh was conducted by Myster et al. (2004). In the region defined by the overlap between Df(2R)41A8 and Df(2R)41A10 (the latter was previously shown to be deficient for most of 2Rh; Hilliker and Holm 1975), Myster et al. (2004) reported the existence of 15 vital loci, considerably more than the 4 essential loci predicted by Hilliker (1976). The discrepancy between these two studies was the catalyst for this current work. Each group used the same mutagen, EMS, yet each group came up with very different interpretations of the number of vital loci.
Hilliker's interpretation relied on earlier evidence that EMS preferentially produced point mutations and not large-scale aberrations (Lim and Snyder 1974). Assuming that the mutants isolated in his study were point mutations, or small aberrations limited to one locus, Hilliker found that some of the loci that he identified exhibited complex interallelic complementation; the most complex complementation pattern was observed with locus l(2)41Ae. On the other hand, the interpretation of Myster et al. (2004) was that heterochromatin was more sensitive to EMS and that EMS could produce large heterochromatic deletions; they proposed that the complex interallelic complementation in l(2)41Ae was due to the presence of deletions and that l(2)41Ae represented a region of 2Rh containing many genes, rather than being a single locus.
To resolve these different interpretations of the genomic segment containing l(2)41Ae (i.e., is it a single locus or a region of 2Rh), we set out to map l(2)41Ae and the region surrounding the presumed location of l(2)41Ae (as in Myster et al. 2004) by performing a large-scale inter se complementation analysis between all available mutant lines that were previously mapped to l(2)41Ae (including Nipped-B). In addition, we undertook a molecular mapping and sequence analysis, using a candidate gene approach with the most recent annotation of 2Rh (Hoskins et al. 2007), to characterize the region and identify as many essential genes as possible. We also used these approaches to map l(2)41Ab and unextended [two of the more proximal complementation groups identified by Hilliker (1976)]. Finally, we also utilized available RNA interference (RNAi) lines to knock down the expression of 12 genes in 2Rh in an attempt to identify essential genes.
MATERIALS AND METHODS
Fly strains:
A total of 69 mutant lines that had been previously mapped to, or near, the presumed location of l(2)41Ae were obtained (Table 1). The majority of these lines were EMS-induced homozygous lethals. Of the 49 EMS lines obtained, 12 were created by Hilliker (1976), while the 28 EMS lines, named NC##, were created by Myster et al. (2004). The remaining 9 EMS lines were created by Eberl (1990). In addition to the EMS-induced lines, we had access to 10 transposable element insertion lines, 7 known heterochromatic deletions, and 2 γ ray-induced lines. A total of 5 EMS mutant lines (Table 1) were used to map loci l(2)41Ab and unextended.
TABLE 1.
Mutant lines obtained for this study
| Line obtained | Known synonyms | Mutagen or method of creation | Reference |
|---|---|---|---|
| NC1 | l(2)IR3[NC1] | EMS | Myster et al. (2004) |
| NC6 | Nipped-BNC6 | EMS | Gause et al.(2008) |
| NC7 | EMS | Myster et al. (2004) | |
| NC9 | EMS | Myster et al. (2004) | |
| NC19 | l(2)NC19NC19 | EMS | Myster et al. (2004) |
| NC21 | RpL38NC21 | EMS | Myster et al. (2004); Marygold et al. (2005) |
| NC23 | Nipped-BNC23 | EMS | Gause et al. (2008) |
| NC28 | l(2)NC28NC28 | EMS | Myster et al.(2004) |
| NC30 | l(2)NC30NC30 | EMS | Myster et al. (2004) |
| NC37 | l(2)NC37NC37 | EMS | Myster et al. (2004) |
| NC38 | l(2)NC38NC38 | EMS | Myster et al. (2004) |
| NC39 | Nipped-BNC39 | EMS | Gause et al. (2008) |
| NC41 | Nipped-BNC41 | EMS | Gause et al. (2008) |
| NC59 | Nipped-BNC59 | EMS | Gause et al. (2008) |
| NC60 | EMS | Myster et al. (2004) | |
| NC70 | l(2)NC70NC70 | EMS | Myster et al. (2004) |
| NC71 | Nipped-BNC71 | EMS | Gause et al. (2008) |
| NC77 | Nipped-BNC77 | EMS | Gause et al. (2008) |
| NC78 | Nipped-BNC78 | EMS | Gause et al. (2008) |
| NC83 | l(2)NC83NC83 | EMS | Myster et al. (2004) |
| NC89 | Df(2R)NC89 | EMS | Myster et al. (2004) |
| NC91 | EMS | Myster et al. (2004) | |
| NC109 | Df(2R)NC109 | EMS | Myster et al. (2004) |
| NC110 | l(2)NC110NC110 | EMS | Myster et al. (2004) |
| NC133 | l(2)NC133NC133 | EMS | Myster et al. (2004) |
| NC138 | Df(2R)NC138 | EMS | Myster et al. (2004) |
| NC173 | EMS | Myster et al. (2004) | |
| NC195 | EMS | Myster et al. (2004) | |
| 34-2 | EMS34-2 | EMS | Hilliker (1976) |
| 34-3 | EMS34-3 | EMS | Hilliker (1976) |
| 34-13 | EMS34-13 | EMS | Hilliker (1976) |
| 34-14 | EMS34-14 | EMS | Hilliker (1976) |
| 34-25 | EMS34-25 | EMS | Hilliker (1976) |
| 45-20 | EMS 45-20 | EMS | Hilliker (1976) |
| 45-23 | EMS45-23 | EMS | Hilliker (1976) |
| 45-33 | EMS45-33 | EMS | Hilliker (1976) |
| 45-34 | EMS45-34 | EMS | Hilliker (1976) |
| 45-53 | EMS45-53 | EMS | Hilliker (1976) |
| 45-61 | EMS45-61 | EMS | Hilliker (1976) |
| 45-71 | EMS45-71 | EMS | Hilliker (1976) |
| L10 | EMS | Eberl (1990) | |
| L2 | EMS | Eberl (1990) | |
| L5 | EMS | Eberl (1990) | |
| L7 | EMS | Eberl (1990) | |
| La4 | EMS | Eberl (1990) | |
| La6 | EMS | Eberl (1990) | |
| Lc | EMS | Eberl (1990) | |
| Lf | EMS | Eberl (1990) | |
| LL | EMS | Eberl (1990) | |
| 407 | Nipped-B407 | γ rays | Rollins et al. (1999) |
| 292.1 | Nipped-B292.1 | γ rays | Rollins et al. (1999) |
| IR13 | l(2)41AeIR13 | IR-hybrid dysgenesis | Dimitri et al. (1997) |
| IR23 | IR-hybrid dysgenesis | Dimitri et al. (1997) | |
| IR3 | IR-hybrid dysgenesis | Dimitri et al. (1997) | |
| IR4 | IR-hybrid dysgenesis | Dimitri et al. (1997) | |
| k02601 | Nipped-Bk02601a | P-element insertion | Corradini et al. (2003) |
| EY09264 | P-element insertion | Bellen et al. (2004) | |
| 2047 | Nipped-B02047 | P-element insertion | Rollins et al. (1999) |
| Lp5 | P-element insertion | P. Dimitri (Unpublished data) | |
| 309 | l(2)3091 | P-element insertion | Myster et al. (2004) |
| CH(2)54 | Nipped-Bch(2)54 | P-element insertion | Zhang and Spradling (1994) |
| Df(2R)41A2 | Df(2R)M41A2 | X ray | |
| Df(2R)247 | Df(2R)247 | Imprecise P-element excision | Myster et al. (2004) |
| Df(2R)244 | Imprecise P-element excision | Myster et al. (2004) | |
| Df(2R)Dark2 | Imprecise P-element excision | Myster et al. (2004) | |
| Df(2R)Nipped-D 263.3 | Rollins et al. (1999) | ||
| Df(2R)Nipped-D 341.1 | Rollins et al. (1999) | ||
| Df(2R)Nipped-E43 | Rollins et al. (1999) | ||
| 739b | Unknown | Unknown | |
| EMS 45-84 | 45-10 | EMS | Hilliker (1976) |
| EMS 45-87 | 45-10 | EMS | Hilliker (1976) |
| EMS 45-10 | 45-10 | EMS | Hilliker (1976) |
| EMS 34-7 | uex2 | EMS | Hilliker (1976) |
| EMS 45-17 | uex4 | EMS | Hilliker (1976) |
The second chromosome balancer line w ; KrIf-1/CyO, P[GAL4-twi.G], P[UAS-2xEGFP] was used to rebalance all mutant lines in preparation for sequencing or deletion mapping (see below). The heterochromatic deletion line, Df(2R)M41A10/CyO, S cn bw, was used to test any lethal line to ensure that its lethality mapped to 2Rh. Df(2R)M41A10 lacks most of the heterochromatin on the right arm of the second chromosome (Hilliker and Holm 1975; Dimitri 1991).
Lethal complementation analysis:
Complementation analysis was performed by crossing three to five males from one mutant line to three to five females of a second line in a glass vial containing standard Drosophila medium. All crosses were maintained at 25° with a 12-hr light/day cycle. Progeny from each cross were scored for the presence or absence of the balancer chromosome, using both the dominant Curly and Star markers on the balancer chromosome. Each crossing vial was scored until exhaustion (i.e., all animals had either emerged as adults or died in earlier stages).
Detecting lethal combinations of mutant chromosomes:
Chi square analysis was performed to test the ratio of observed progeny from each cross against the 2:1 expected ratio for a viable combination of chromosomes. When observed ratios were significantly different from the expected ratios, due to a small number of unbalanced progeny, the chromosome combination was considered to have some degree of lethality. Typically, if the unbalanced progeny comprised <5% of the total progeny, the cross was considered lethal; if the unbalanced progeny comprised >5% of the total progeny, and observed ratios were significantly different from the expected ratios, the cross was considered semi-lethal.
Interallelic complementation maps:
Interallelic complementation maps were drawn in the same manner as Hilliker (1976). In brief, each mutant strain was represented with a line. If two mutant strains were lethal in combination, then the lines representing them were drawn on the map such that they were parallel and could be joined by a perpendicular transversal.
Using recombination to resolve potential second-site lethal interactions:
Any two mutant chromosomes that were believed to be lethal due to a shared second-site lethal were allowed to undergo recombination with a wild-type chromosome to try and separate the second-site lethal from the 2Rh mutation under study. Independent putative recombinant lines were recovered as described below.
Males of the mutant line were crossed to virgin wild-type females, and the resulting unbalanced F1 female progeny (in which recombination could take place) were crossed to males of a second chromosome balancer line containing the CyO, S, cn, bw balancer chromosome. In the next generation, a single male offspring containing the CyO, S, cn, bw balancer chromosome and a putative recombinant chromosome was backcrossed to the second chromosome balancer line. The resulting balanced progeny were used to create a putative recombinant stock containing a single putative recombinant chromosome. A total of 10 single-male-derived putative recombinant lines were obtained for each mutant line.
Putative recombinant lines were then tested for (1) lethality with the parental line from which it was derived and (2) lethality with the chromosome believed to share a second-site lethal. Because recombination will not remove the 2Rh mutation from the original chromosome, viability between putative recombinant lines whose parental lines are lethal reveals the presence of a second-site lethal mutation.
Isolating genomic DNA from homozygous lethal mutant strains:
All lethal chromosomes of interest were first balanced over the CyO, P[GAL4-twi.G], P[UAS-2xEGFP] balancer chromosome by crossing mutant lines to w ; KrIf-1/EGFP[w+] CyO and selecting against the dominant KrIf-1 marker and selecting for CyO progeny. Embryo collection began by placing ∼50 males and 50 females from each line into individual collection cages consisting of an empty Drosophila mating bottle with small air holes poked in its bottom and one collection plate (a 35- × 100-mm culture dish lid filled with grape juice agar) used as the bottle's sealing cap. A small amount of active yeast was sprinkled on the surface of the collection plate to promote egg laying. Flies were allowed to lay eggs overnight.
The next day, a fresh collection plate (sprinkled with yeast) was added to each collection cage. Flies were allowed to lay eggs for 2 hr after which the adults were removed to a fresh collection cage so the process could be repeated. Embryos on collection plates were allowed to develop for 2 hr. Embryos were then dechorionated by gently rolling on Scotch tape and placed in 13 μl of embryo lysis buffer (10 mm Tris pH 8.2, 1 mm EDTA, 25 mm NaCl) and 2 μl of proteinase K (20 mg/ml). The proteinase K in the lysis solution was then activated by incubating the PCR tubes at 37° for 30 min. After the 30-min incubation, the proteinase K was inactivated by increasing the temperature to 95° for 10 min.
Enhanced green fluorescent protein diagnostic PCR:
Enhanced green fluorescent protein (EGFP) diagnostic PCR is a method used to detect balancer chromosome DNA in a single-fly-embryo genomic DNA preparation. This method relies on the fact that EGFP protein is not present in the Drosophila genome, so there are unique DNA sequences in the EGFP marker. Primers made to the EGFP sequence were used to detect the presence of the EGFP marker in genomic DNA isolated from balanced embryos and to reveal the absence of the marker in homozygous embryos. A control primer set was run simultaneously to ensure that the PCR reaction was successful in reactions lacking the EGFP PCR product.
Primers to the EGFP sequence are listed in Table 2 and produce a 683-bp PCR product when using a DNA template containing the EGFP-CyO balancer chromosome. The EGFP− diagnostic control primer pair produces a 350-bp PCR product corresponding to a segment of the Drosophila SOD1 gene. All primers were designed using the Primer 3 program available at http://frodo.wi.mit.edu/primer3/.
TABLE 2.
Primer pairs for all diagnostic PCR and sequencing analysis
| Locus/marker | Primer pair specifics | Forward primer | Reverse primer |
|---|---|---|---|
| EGFP | ctggtcgagctggacggcgacg | cacgaactccagcaggaccatg | |
| EGFP diagnostic control | SOD1 | gagcagcggtacgcccgtga | gcaccagcgttgcccgttga |
| Deletion mapping control | 3′ essential end of crumbs | ggcaactgcacggatcttat | gggcggtacgtatgtcatct |
| CG41265 (l(2)41Ab) | Exon 1 | tttcatttctggaggaattagcc | ggaaagaataaacgccgttgg |
| Exon 2–3 | ggtcctcaaaatgactattgaacg | ggacatgcacagatatgaaaacg | |
| Exon 4 | ccagcctctatcttggatagttcc | gctaatcgctttcataccagacg | |
| Exon 5 | tgaaattttgctttctcatttcg | aaaaactatgttatgcccaaaaagc | |
| Exon 6–7 | aagagtgatcagcaatcgtaaacc | tccagaattttccgttattttgg | |
| CG41592 | 3′ end | tcagtggcagaggaatcaga | tcatcattggtgcagttgct |
| CG40270 | 5′ end | attgccgttacgaaaacacc | tgccgctctccaaactaact |
| CG40733 | 5′ end | ctattctcatgctggggtttc | tcgcatcagaaagtgtttgg |
| CG41278 | 5′ end | caagctaacgcagttccaca | cggtgcactcatgatacctg |
| CG42595 (uex) | Exon 1 | tgcatgttagctctttattacaatcg | ggtgactgtatggatgtttctcagg |
| Exon 2 | tggttattcatcttgtttagactcacg | aaatataaggcaataggagttctacacc | |
| Exons 3–4 | tgatattggcaagaagaataaaatcc | caaattgacttattttcccgttagc | |
| Exons 5–6 | gaattttgctgtttctgtgtgtcg | ttttgacccataccctgtgaagg | |
| Exons 7–9 | atttctagttcgcgctttcactagc | atttcgcgtaagcattcacactagc | |
| Exon 10 | gcattcgtattttgtgtttttcaacc | ggaatataattaatatgggaaaacctatgg | |
| Exons 11–13 | taatgctctcatcttcaagctttgc | gcataagtgcgaatttaatagtgc | |
| Haspin | Exons 1–2 | aagcaaataaataaacaacgtcacc | ccattacacagatacagatccaagc |
| Exon 5 | tgcacacgaatttattgaaattaagc | gtaatcggaattcaaagaactcagc | |
| CG41323 | Exons 3–4 | atcttccaattcttctggtctgtcc | ataaccggaatctataaccttgtcg |
| CG40085 | atcttccaattcttctggtctgtcc | ataaccggaatctataaccttgtcg | |
| CG41363 | 5′ end | cctttgtaacatgccgtgtg | tccaacagcccacaacagta |
| CG17683 | 5′ end | gaggtttctgcggtttgtga | gcagcatcctccacacctat |
| Exons 1–3 | attacgccctcacaggtttg | tgccttgctgaaaaagtcct | |
| CG17683 | Exons 3–4 | ctaccatgtgaccgtgatgc | ttaaggagttttcgggttcg |
| 3′ end | cggaggagcacaaatacgtc | aacggccatgcattctacat | |
| CG40133 | 5′ end | cacagggaagagaatgacca | ccgacttttcgtcaaactgc |
| 3′ end | ccataacagcagtggtggaa | cacaaggtcgtgttggtgtc | |
| CG40127 | actaaaacggcccttgttcc | cactccgccccttaatacaa | |
| CG40131 | tcgttcgttcagtttggtga | gcatttgtgttacggattgg | |
| CG41449 | tggagtcattggtcagcaaa | gtgggcttaatctccactcg | |
| CG17665 | 5′ end | cggttgcattttattgtggtc | acgaattcccgtaacaacca |
| Exon 2 | aattcgagcgttcgtttgtc | ccggaacgcttaactactgc | |
| Exon 3 | tcccttgaaaagtgcaaatgt | agaaatccaacagcgtgctac | |
| Exons 3–5 | gatgggcaatcataggttgg | catcgtcgtcatccgtattg | |
| Exons 5–6 | ttttagcgacgaaactgctg | tcgaaacaaaccgtacaaagg | |
| Exons 6–7 | gggagactttgcctcaacaa | gacttccggcttccagttct | |
| Exons 6–9 | cggctctcgactatttcactg | gaacgtaaaggccagttcca | |
| Exons 8–11 | tgcttccctgcttacatcaa | tccttctcttagccgtttgc | |
| 3′ end | gcagcggaagaagataccac | aaccatcttgcgatcctgtc | |
| Gprk1 | 5′ end | aatacctcgggtcgggtaaa | gccatccacggtcacacta |
| 3′ end | ataaaagtctgcgggccaat | ttctgaggtatatggtaaagcgttaat | |
| CG17883 | 3′ end | cggaggagcacaaatacgtc | aacggccatgcattctacat |
| Nipped-B | 5′ end | gtttgtttggttgtagcgtaatg | atgttggtccggagagtctg |
| 3′ end | gaggagaaattggaccttgttg | gttgcttagcttcgacttcttc | |
| CG17706 | agcacttctcattgcacatcc | cacttccactgtttccagttc |
All primers were designed to have a melting temperature of ∼60°–64°.
PCR reaction products were run on 2% agarose gels to separate and visualize the two potential PCR products in the diagnostic reaction: the EGFP product and the SOD1 control product. PCR products that gave a strong SOD1 control product and no EGFP product revealed that the original embryo from which DNA was extracted was homozygous for the desired mutant second chromosome.
Mapping deletion breakpoints:
To map the breakpoints of homozygous lethal deletions, genomic DNA was isolated from homozygous embryos using the embryo collection protocols described above. Deletion breakpoints were mapped using diagnostic PCR with primer pairs designed against the annotated 2Rh heterochromatic map (Table 2). Deletion breakpoints were roughly mapped by determining which primer pair product could be amplified through PCR and which could not be amplified. A control primer set was run simultaneously to ensure that the DNA was intact and that the PCR reaction was successful in reactions lacking the other primer pair product.
Sequencing and polymorphisms:
Genomic DNA used in sequencing was obtained from homozygous embryos using the embryo collection protocols described above. To help identify mutations and polymorphisms, each sequence obtained from a mutant line was compared to three groups of sequences: (1) the published heterochromatin sequence, (2) other lines from the same mutagenesis screen, and (3) lines from other mutagenesis screens.
DNA sequences were compared using the ClustalW2 alignment software available at http://www.ebi.ac.uk/Tools/clustalw2/index.html. The ExPASy translation software (available at http://ca.expasy.org/tools/dna.html) was used to translate DNA sequences into amino acid sequences. ClustalW2 was also used to align amino acid sequences.
RNAi:
RNAi lines from the Fly Stocks of the National Institute of Genetics (NIG-fly; http://www.shigen.nig.ac.jp/fly/nigfly/index.jsp) and the Vienna Drosophila RNAi Center (VDRC; http://stockcenter.vdrc.at/control/main) were used to knock down the expression of 12 putative genes in 2Rh. The transgenic lines contained inverted repeat (IR) sequences, corresponding to the heterochromatic genes of interest and under the regulation of a UAS-promoter. UAS-IR males were crossed to UAS-dcr2; daughterless-GAL4 virgin females (created from VDRC line 60011), in which the DCR2 protein is overexpressed, inducing a highly efficient RNAi pathway. As a control for lethality due to the double inserts, the males were also crossed to UAS-dcr2; Ly/TM3. All crosses were brooded for 2–3 days, kept at 25°, and subsequently scored for lethality. Numbers of viable offspring, as well as those of dead larvae and pupae, were recorded. In some cases, ratios of balanced vs. nonbalanced offspring were tested with chi-square analysis, as described above.
Cytology:
Mitotic chromosome preparations and DAPI staining were as described by Dimitri (2004). Chromosome preparations were analyzed using a computer-controlled Leica fluorescence microscope equipped with a cooled CCD camera (Photometrics, Tucson, AZ). The DAPI staining was recorded by IP Spectrum Lab Software and edited with Adobe PhotoShop 8.
RESULTS
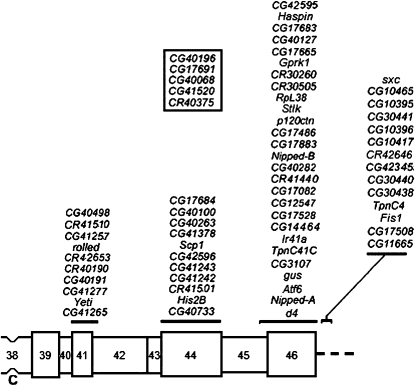
A schematic map of the heterochromatic segment of the right arm of mitotic chromosome 2 is shown in Figure 1 (modified from figure 1 of Dimitri et al. 2009). In the current work, we focus on the region of 2Rh extending from CG42595 to Nipped-B in cytological bands h45–h46. Our initial objective was to resolve the issue of the genetic identity of l(2)41Ae, which was mapped to h46 (Dimitri 1991; Coulthard et al. 2003; Dimitri et al. 2009). However, we also planned to characterize as many of the other essential loci in the region as possible.
Figure 1.—
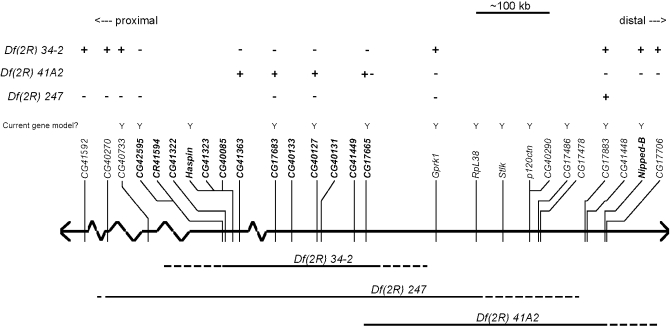
A schematic map of the heterochromatic segment of the right arm of mitotic chromosome 2. This map, modified from figure 1 of Dimitri et al. (2009), includes only current gene models (recently withdrawn models have been removed). The approximate positions of the current gene models located in this segment are shown above the chromosome. Cytological regions are labeled 38–46, as determined by Dimitri (1991), and the centromere is labeled “C.” The five gene models inside the box belong to contig CP000218; their exact cytological location within 2Rh is unknown.
Creating a new complementation map For 41Ae:
Sixty-eight mutant lines were tested inter se to create a large complementation map for l(2)41Ae (Table 1; Table S1). The first complementation map produced from this analysis showed a level of complexity similar to the original 41Ae complementation map produced by Hilliker (1976). This initial map needed to be refined due to the presence of shared second-site lethal mutations and complex rearrangements that unduly complicated the map (see Table S2).
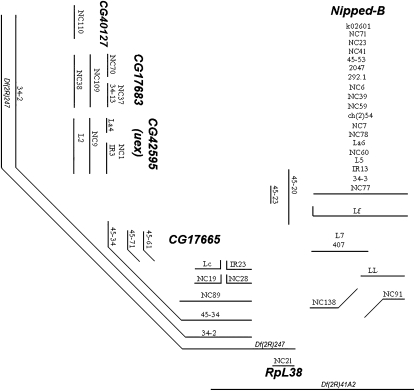
The refined complementation map is presented in Figure 2 and contains a total of 50 mutant lines. This map contains six complementation groups: Nipped-B, RpL38, CG17665, CG40127, CG17683, and CG42595 (uex). Four of these complementation groups were identified molecularly in this study (see below). The Nipped-B complementation group was identified by the eight Nipped-B alleles sequenced by Gause et al. (2008) (see Table 1). Similarly, Marygold et al. (2005) previously sequenced line NC21 showing that it is an allele of RpL38.
Figure 2.—
The simplified complementation map of l(2)41Ae was drawn, as in Hilliker (1976), with each mutant strain being represented with a line. If two mutant strains were lethal in combination, then the lines representing them were drawn on the map such that they were parallel and could be joined by a perpendicular transversal. To fully complete the map, some mutant strains had to be represented by lines that ran horizontal, angled, and vertical. The molecular identity of six complementation groups, some of which were determined here, is labeled on the map.
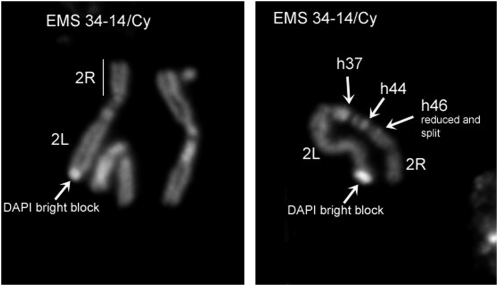
Of particular interest with respect to Figure 2 was the exclusion of line EMS 34-14 because EMS 34-14 has been the traditional tester allele for l(2)41Ae. Cytological analysis of EMS 34-14 (and EMS 34-25) mitotic chromosomes from larval brain squashes using DAPI staining yielded a surprising result. While the structure and heterochromatin pattern of EMS 34-25 chromosome is cytologically normal (data not shown), the EMS 34-14 chromosome clearly contains a complex rearrangement associated with the occurrence of multiple breaks (Figure 3). The EMS 34-14 chromosome has a submetacentric structure, shows a reduction of region h46 on 2Rh, and carries a large DAPI bright block of unknown origin on the 2L tip. The new submetacentric organization may be explained by assuming the presence of a pericentric inversion with a breakpoint in h46 of 2Rh and somewhere within 2L. The breakpoint in h46, possibly associated with a partial deletion of this region, may inactivate several vital genes and thus be responsible for the complex complementation pattern of EMS 34-14. Because a complex heterochromatic rearrangement such as EMS 34-14 can join non-adjacent regions of 2Rh, it could unduly complicate a lethal complementation map, and thus it was not included in Figure 2.
Figure 3.—
EMS 34-14 mitotic chromosomes from larval brain squashes using DAPI staining. The 34-14 chromosome is clearly a complex rearrangement due to the occurrence of multiple breaks. It has a submetacentric structure, shows the reduction of region h46 on 2Rh, and carries a large DAPI bright block of unknown origin on the 2L tip. The new submetacentric organization may be explained assuming the presence of a pericentric inversion with a breakpoint in h46 of 2Rh and somewhere within 2L. The breakpoint in h46, possibly associated with a partial deletion of this region, may inactivate several vital genes and thus be responsible for the complex complementation pattern of EMS 34-14.
EMS 34-2 is a heterochromatic deletion overlapping Df(2R)247 and Df(2R)41A2:
Figure 2 contains two known deletions: Df(2R)247 and Df(2R)41A2. To better align the complementation map to the published heterochromatin sequences, the endpoints of Df(2R)247 and Df(2R)41A2 were roughly mapped using EGFP diagnostic PCR. EMS 34-2 was included in the analysis because its complementation profile closely matched that of Df(2R)247, suggesting that it might also be a deletion.
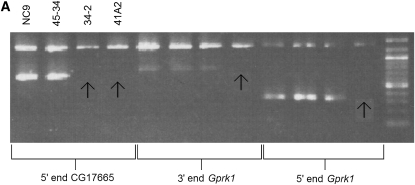
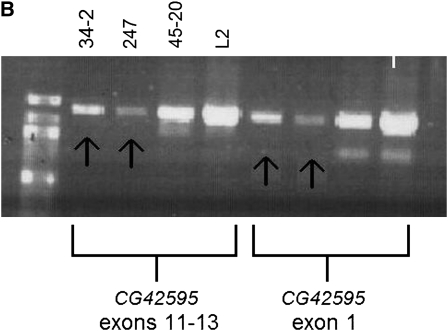
Representative molecular data for Df(2R)41A2 and EMS 34-2 are shown in Figure 4. Figure 4A shows that EMS 34-2 is deficient for the 5′ end of CG17665, while the gene Grpk1 (CG40129) is present in EMS 34-2. Figure 4B shows that CG42595 cannot be amplified in EMS 34-2 or Df(2R)247. Taken together with the results of other diagnostic tests (not shown), the data indicate that EMS 34-2 is a deletion that extends from the 5′ end, or upstream of, CG17665 to exon 11–12, or downstream of CG42595. Df(2R)41A2 is deficient for Gprk1 and the 5′ end of CG17665 (Figure 4A). Further analysis (data not shown) found that Df(2R)41A2 was not deficient for exon 2 of CG17665. Df(2R)41A2 and Df(2R)34-2 are lethal in combination, so these diagnostic tests reveal that Df(2R)41A2 and Df(2R)34-2 overlap at the 5′ end of CG17665. Diagnostic PCR tests on Df(2R)247 show that it extends from some point proximal to CG17883 to some point between CG41592 and CG40270 (data not shown), placing the proximal breakpoint of Df(2R)247 in contig CP000163 in cytological region h44. The mapping data are summarized in Figure 5.
Figure 4.—
(A) Df(2R)41A2 and Df(2R)34-2 overlap at the 5′ end of CG17665. (B) Gene CG42595 cannot be amplified in Df(2R)34-2 or Df(2R)247. The top band in each gel is the deletion mapping control band that should be present in all PCR reactions, and the second, shorter product is the test product amplified from the DNA template using primers designed for specific sequences in 2Rh (Table 2). The DNA templates are written above the gel, and the test primers are listed on the bottom. The order of the DNA templates is the same for each test group on the same gel. Arrows point to missing test products.
Figure 5.—
Mapping heterochromatic deletions in the vicinity of l(2)41Ae. The thick horizontal line represents the second chromosome, and the thin vertical lines represent the position of known or predicted genes. Solid horizontal lines below the chromosome show the areas of the chromosome that are missing in the mapped deficiencies. Dashed areas of the deletion lines are used to represent uncertainty about the exact endpoint of the deletions. The “+” and “−” signs above the genes represent diagnostic PCR deletion mapping results. A “+” indicates that a PCR product was obtained for the deletion indicated on the left, and a “−”indicates that no product was obtained for the deletion. Note that Df(2R)41A2 breaks in the first exon of CG17665, so the diagnostic results are represented by “+ −” to indicate that part of the gene is present in the deletion and part is absent. Current gene models are indicated by a “Y”; the other gene models have been withdrawn.
Molecular identification of four complementation groups:
After establishing that Df(2R)34-2 is a large heterochromatic deletion, a direct sequencing strategy was employed to identify the essential complementation groups within the boundaries of Df(2R)34-2. We began our sequencing with CG17665 because both Df(2R)41A2 and Df(2R)34-2 break in CG17665 and are lethal with the large EMS 45-34 complementation group (Figure 2).
CG17665 was first sequenced in EMS 45-34 and a G-to-A nucleic acid substitution was discovered (see Table 3 for all mutations discovered in this study); this substitution gives rise to a premature stop codon in exon 3. Two other stop codons in CG17665 were found in lines EMS 45-61 and NC19. Line NC28 was found to contain a T-to-A substitution in CG17665 that produces an L-to-P amino acid change, while line EMS 45-71 contains a G-to-A substitution that leads to a G-to-E amino acid change. Four polymorphisms were detected in CG17665, and these are listed in Table 4, together with other polymorphisms discovered in this study. These data confirm that the EMS45-34 complementation group represents CG17665. These data also confirm that the complex complementation pattern observed in the EMS45-34 complementation group (Figure 2) is due to interallelic complementation.
TABLE 3.
The nature and location of mutations found in this study
| Gene | Sequenced mutant line | Nucleotide changes | Amino acid changes | Location of mutation |
|---|---|---|---|---|
| CG41265 (l(2)41Ab) | EMS45-10 | G → A | (W → stop) | +93785 (exon 7) |
| CG42595 (uex) | L2 | C → T | Q → STOP | +10139 (exon 4) |
| NC1 | 13-bp deletion and 4-bp deletion | frameshift + possible loss of a splice junction motif | +9949 to +9961 (exon 11) and +9989 to +9991 (intron 11) | |
| uex34-7 | G → C | M → I | +3 (putative transcription start site) | |
| CG17683 | NC109 | T → A and T → G | V → D and S → A | +559 and +564 (exon 3) |
| NC38 | T → A | C → STOP | +602 (exon 3) | |
| EMS 34-13 | C → T | S → F | +658 (exon 3) | |
| CG40127 | NC110 | T → A | L → H | +375 (exon 3) |
| CG17665 | EMS 45-34 | G → A | W → STOP | +3917 (exon 3) |
| EMS 45-61 | C → T | R → STOP | +6088 (exon 7) | |
| EMS 45-71 | G → A | G → E | +4168 (exon 3) | |
| NC19 | G → A | W → STOP | +3736 (exon 3) | |
| NC28 | T → A | L → P | +3796 (exon 3) |
Genes are listed in the proximal to distal direction. The location of the mutation is written as the number of amino acids downstream (+) or upstream (−) from the predicted translation start site (+0).
TABLE 4.
Polymorphisms found in all sequences analyzed
| Gene model | Location of polymorphism | Polymorphism detected | Lines with polymorphism | Lines without polymorphism |
|---|---|---|---|---|
| CG41265 | +48 (exon 1) | T → A | 45-10, 45-84, 45-87, 34-7 | |
| +94167 (exon 7) | C → T (A → V) | 45-10, 45-84, 45-87, 34-7 | ||
| CG42595 | +22662 (intron 6) | T → C | L2, NC9, NC1, uex34-7 | |
| CG17683 | −142 to −177 | 36 missing nucleotides | NC9, NC109, NC70, NC1, NC38 | 34-13 |
| CG40133 | +1876 (3′ UTR) | G → A | 34-13, NC109 | |
| +8197 (exon 2) | G → T (S → I) | NC9, NC1, Lf | 45-20 | |
| +119 (intron 1) | G → T | 45-20, NC9, NC1, Lf, NC110 | 45-34 | |
| CG40127 | +289 to +293 (intron 2) | ATAC absent | 45-20, 45-34 | |
| +307 (intron 2) | A → C | 45-20, NC9, NC1, Lf, NC110 | ||
| +319 (intron 2) | T → C | 45-20, NC9, NC1, Lf, NC110 | ||
| CG40131 | +235 (exon 1) | G → C (G → R) | 45-20, NC9, NC1, Lf | |
| CG41449 | +186 (exon 1) | C → G (silent) | NC9, NC1, Lf | 45-20 |
| CG17665 | +49 (exon 1) | A → G (I → V) | 45-20, 45-34, 45-61, NC9, NC19, Lf | |
| +4358 (intron 3) | T → C | 45-20, NC9, NC28, NC19, Lf | 45-34, 45-61, 45-71, Lf | |
| +6176 (intron 7) | A → G | 45-20, 45-34, NC9, NC19, Lf | 45-61 | |
| +6460 (intron 8) | G → A | 45-20, 45-34, 45-61, NC9, NC19, Lf |
The location of the polymorphism is written as the number of amino acids downstream (+) or upstream (−) from the predicted translation start site (+0). All polymorphisms are relative to the published genomic sequence.
Table 3 shows the three mutations that were found in gene CG17683. All three mutations were in lines that compose the NC38 complementation group (Figure 2), showing that the NC38 group represents CG17683. Line NC38 contains a T-to-A nucleic acid substitution that results in a premature stop codon. Line EMS 34-13 contains a C-to-T substitution in exon 3 that leads to an S-to-F amino acid change. NC109 contains two closely linked amino acid substitutions in exon 3: a T-to-A and T-to-G that lead to a V-to-D and an S-to-A amino acid substitution, respectively. In addition, two polymorphisms were detected in, or near, CG17683 (Table 4).
The L2 line was found to contain a premature stop codon in exon 4 of CG42595 while line NC1 was found to have two small deletions: a frameshifting 13-bp deletion in exon 11 and a 4-bp deletion of the first four nucleotides of intron 11 that potentially removes a splice junction motif (Table 3). One polymorphism has been identified in CG42595 (Table 4).
Finally, the NC110 line was found to contain an L-to-H amino acid substitution in exon 3 of the small CG40127 gene. There were several intronic polymorphisms detected in CG40127 (Table 4).
The unextended gene [l(2)41Ad] is synonymous with CG42595:
Preliminary data (not shown) revealed that Df(2R)247 uncovers the unextended (uex) gene; thus, having already mapped the breakpoints of Df(2R)247, as described above (Figure 5), we conducted a genetic analysis of the uex34-7 and uex45-17 alleles, originally described by Hilliker (1976), to map uex.
Lines uex34-7 and uex45-17 were lethal in combination, with most trans-heterozygotes dying in the pupal stage or during eclosion (Table 5). In crosses between Df(2R)247 and either uex allele, the majority of uex/Df(2R)247 heterozygotes died in the pupal stage or during eclosion (Table 5). The rare adult escapers exhibited smaller body size, misshapen hind legs, and unextended wings. These phenotypes were not fully penetrant, with escapers showing any combination of the phenotypes, and sometimes these escapers appeared fully wild type (Figure 6). Specific hind-leg phenotypes observed included the following: shortened and bulbous leg segments, leg segments fused together, missing or extra leg segments (usually tarsal segments), and a tarsal “hook” where the last tarsal segment is held out at almost 90°.
TABLE 5.
Genetic analysis leading to the discovery of the molecular basis of uex
| Progeny count |
CyO[+] adults with |
||||||
|---|---|---|---|---|---|---|---|
| Line 1 | Line 2 | Cyo | CyO[+] | Dead pupae | Eclosion deaths | Misshapen hind leg | Unextended wings |
| uex34-7 | uex45-17 | 108 | 0 | 46 | 26 | ||
| uex34-7 | Df(2R)247 | 79 | 5 | 21 | 34 | 3 | 4 |
| uex45-17 | Df(2R)247 | 92 | 21 | 32 | 13 | 5 | 9 |
| uex34-7 | Df(2R)M41A10 | 90 | 0 | 53 | 4 | 0 | 0 |
| uex45-17 | Df(2R)M41A10 | 68 | 5 | 34 | 16 | 0 | 5 |
| uex34-7 | Df(2R)34-2 | 58 | 16 | 8 | 1 | 2 | 0 |
| uex45-17 | Df(2R)34-2 | 69 | 34 | 8 | 3 | 0 | 0 |
| uex34-7 | L2 | 32 | 31 | 6 | 0 | 9 | 0 |
| uex45-17 | L2 | 98 | 68 | 28 | 2 | 0 | 0 |
| uex34-7 | La4 | 82 | 42 | 13 | 2 | 1 | 1 |
| uex34-7 | NC9 | 43 | 28 | 15 | 1 | 0 | 0 |
| Df(2R)34-2 | La4 | 93 | 0 | 44 | 3 | 0 | 0 |
| Df(2R)34-2 | NC1 | 68 | 0 | 12 | 6 | 0 | 0 |
| Df(2R)34-2 | L2 | 82 | 0 | 4 | 0 | 0 | 0 |
| Df(2R)34-2 | NC9 | 108 | 0 | 1 | 0 | 0 | 0 |
| La4 | NC1 | 150 | 16 | 77 | 6 | 1 | 6 |
| NC1 | L2 | 75 | 0 | 3 | 1 | 0 | 0 |
| NC1 | NC9 | 67 | 0 | 22 | 0 | 0 | 0 |
| L2 | NC9 | 89 | 0 | 2 | 1 | 0 | 0 |
Figure 6.—
Three rare adult escapers of the genotype Df(2R)247/uex34-7 showing varying severities of uex phenotypes. (A) The uppermost fly appears to be wild type while the bottom two flies have unextended wings and smaller body size. (B) A closer look at the hind legs of the bottom two flies in A; one fly has legs with a wild-type appearance (C) while the other fly has a single misshapen hind leg (D).
Crosses between uex34-7 and Df(2R)34-2 revealed a semi-lethality with a few adults exhibiting misshapen hind legs (Table 5), so representative alleles of the complementation groups in Figure 2 that were lethal with Df(2R)34-2 were crossed to the uex alleles. Crosses between the CG42595 stop codon mutant L2 and uex34-7 produced hind-leg phenotypes similar to uex hind-leg phenotypes, while crosses between L2 and uex45-17 yielded a considerable amount of pupal lethality (Table 5). These data suggest that CG42595 might correspond to uex. Therefore, the genetic analysis was extended to the entire CG42595 complementation group. Crosses between uex alleles and CG42595 alleles produced characteristic uex phenotypes: pupal and eclosion deaths, misshapen hind legs, and unextended wings (Table 5).
CG42595 was sequenced using DNA from homozygous uex34-7 embryos, and a G-to-C nucleotide change was found in the third position of the predicted translation start site of CG42595. Together, the genetic and molecular data show that the CG42595 complementation group represents the uex gene.
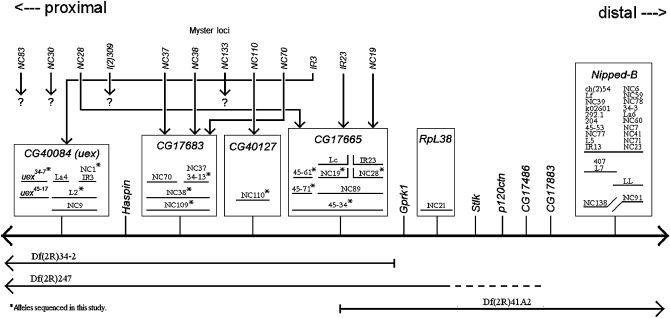
Combining all the deletion mapping, complementation data and sequencing gives the flat complementation map in Figure 7. EMS 45-20 was not included in Figure 7 because it was lethal with both the Nipped-B complementation group and the CG17683 complementation group from the map, yet diagnostic PCR and sequencing of EMS 45-20 found no detectable aberrations in EMS 45-20 in CG17665 or between CG17683 and CG17665 (data not shown). Thus, our genetic and molecular data show that, of the 12 current protein-coding gene models in the region spanning from Nipped-B to CG42595 (see Figure 5), at least 7 are essential. Although this is likely a minimal estimate (see below), it is considerably less than the 15 essential genetic loci previously reported for this segment by Myster et al. (2004).
Figure 7.—
Flat complementation map showing the identity of six complementation groups and how the 12 loci reported by Myster et al. (2004) align with our results. The thick horizontal line represents the second chromosome, and the thin vertical lines represent the positions of known genes or current gene models. Solid horizontal lines below the chromosome show the areas of the chromosome that are missing in the mapped deficiencies. Dashed areas of the deletion lines are used to represent uncertainty about the exact endpoint of the deletions. Boxes enclose the lethal complementation groups identified in this study. The 12 loci reported by Myster et al. that were mapped proximal to RpL38 are listed, in order, above the map. Arrows originating from the Myster loci point to their true identity, as determined in this study.
Molecular basis for l(2)41Ab uncovered:
Mapping studies originally placed l(2)41Ab proximal to the rl gene (for review see Coulthard et al. 2003). On the basis of the recent annotations of heterochromatin (Hoskins et al. 2007 and Tweedie et al. 2009), there are only two protein-coding gene models located proximal to rl (see Figure 1). Sequencing these candidate genes from DNA from EMS 45-10 embryos failed to identify l(2)41Ab. Because we thought it possible that the mapping of this locus was incorrect due to the existence of inversion polymorphisms, we turned next to the protein-coding gene models that are distal to rl. Thus, we sequenced these candidate genes from DNA from EMS 45-10 embryos and uncovered the molecular basis of l(2)41Ab (Table 3); i.e., EMS 45-10 contains a premature stop codon in exon 7 of CG41265.
Identification of vital loci in 2Rh through RNAi:
We wanted to investigate whether we could identify essential genes in 2Rh that had not yet been identified with conventional mutagenesis (EMS or transposable element mutagenesis). To this end, we carried out a pilot study using RNAi lines from VDRC and National Institute of Genetics to knock down the expression of 14 genes in the 2Rh area (see materials and methods; at the time this work was begun only these 2Rh lines were available). None of the lethal RNAi constructs had any off-target effects, meaning that the knock-down is specific to one mRNA [see Dietzl et al. (2007) for VDRC's methods of detecting off-target genes]. The UAS-RNAi transgenes were expressed under the control of the daughterless-GAL4 transgene because the da promoter drives GAL4 expression in a large proportion of tissues (S. Mukai and J. Hu, Hilliker laboratory, unpublished observations). The knock-down of 9 of the genes led to a lethal, or semi-lethal, phenotype, whereas no effect was observed for the other 3 (Table 6). Four of the genes that were identified as essential—Nipped-A, Nipped-B, gus, and RpL38—had been identified as essential in earlier studies and served as positive controls in our study (Rollins et al. 1999; Styhler et al. 2002; Marygold et al. 2005). Interestingly, the remaining 5 essential genes had not been identified as essential in earlier studies, but the semi-lethality of the RNAi knock-down lines for genes CG17665 and CG17683 is consistent with our genetic and molecular identification of these genes as essential, as described above.
TABLE 6.
Characteristics of genes in 2Rh knocked down by RNAi
| Gene | RNAi line | Source | Non-knockdown progeny classes | dcr2 RNAi da-GAL4 progeny | Expected progeny ratio | Lethality | Stage |
|---|---|---|---|---|---|---|---|
| Nipped-B | 17704R-2 | NIG | 86 | 0 | 1:1 | Lethal | L |
| Nipped-A | 2905R-7 | NIG | 79 | 0 | 1:1 | Lethal | E |
| gusa | 8688 | VDRC | 62 | 0 | 1:1 | Lethal | E |
| CG17684 | 46596 | VDRC | 76 | 0 | 1:1 | Lethal | E |
| CG17665b | 46743 | VDRC | 41 | 0 | 1:1 | Lethal | P |
| CG17683 | 19180 | VDRC | 53 | 0 | 1:1 | Lethal | L, P |
| CG17883c | 30277 | VDRC | 89 | 59 | 1:1 | Semi-lethal | E, P |
| 17883R-4 | NIG | 98 | 48 | 1:1 | Semi-lethal | E, P | |
| Atf6 | 36504 | VDRC | 53 | 7 | 1:1 | Semi-lethal | L |
| CG2981 | 2981R-3 | NIG | 43 | 33 | 1:1 | Viable | NA |
| CG17486 | 17486R-1 | NIG | 60 | 17 | 3:1 | Viable | NA |
All progeny counts are for dcr2-enhanced RNAi knockdown with a daughterless (da)-GAL4 driver. Expected progeny ratios differ depending on the insertion location and viability of the RNAi construct. NIG, National Institute of Genetics; VDRC, Vienna Drosophila RNAi Center.
RNAi knockdown with gus was not enhanced with dcr2. Used da-GAL4 driver only.
Semi-lethal without dcr2 enhancement.
Weak semi-lethality present only in dcr2-enhanced knockdown. Viable without dcr2 enhancement.
The existence of eight novel essential loci was verified through the combined genetic and molecular approaches. Table 7 lists the eight novel loci and their putative homologs.
TABLE 7.
A summary of the essential loci in this study, listed from proximal to distal
| Essential locus uncovered | Methods used to uncover locus | Homologs |
|---|---|---|
| CG41265 (l(2)41Ab) | Sequencing 1 allele | Similar to myosin heavy polypeptide 9, nonmuscle, (rat), 2e-47 |
| Ccdc40 mouse 2e-17 | ||
| Myosin heavy chain A (Schmidtea mediterranea), 3e-15 | ||
| Centromere protein E (CENP-E) (human), 9e-10 | ||
| CG17684 | RNAi | DPPY (DPP10, dipeptidyl peptidase) (human), 6e-74 |
| uex (CG42595) | Genetic analysis and sequencing three alleles | Cyclin M2, 3E-168 |
| CG17683 | Genetic analysis, RNAi, and sequencing three alleles | Nuclear prelamin A recognition factor-like (rat), 8.00E-147 |
| CG40127 | Genetic analysis and sequencing 1 allele | Salivary secreted ribonucleaes (Culex pipiens quinquefasciatus), 1e-27 |
| RnaseK (mouse), 3e-10 | ||
| CG17665 | Genetic analysis, RNAi and sequencing five alleles | Integrator complex subunit 3 (mouse), E = 0 |
| CG17883 | RNAi | TBC1 domain family member 20 (human), 4.00E-65 |
| Atf6 | RNAi | ATF6 (human), 2e-16 |
Six novel essential loci were uncovered while the molecular identity of two previously reported essential loci, l(2)41Ab and uex, was discovered. Putative homologs were determined using BLASTp available at http://blast.ncbi.nlm.nih.gov.
DISCUSSION
The heterochromatic locus l(2)41Ae was previously mapped to cytological band h44 in the centromeric heterochromatin on the right arm of the second chromosome (Dimitri 1991) and was originally believed to be a single locus exhibiting a complex interallelic complementation pattern (Hilliker 1976). A more recent study by Myster et al. (2004) concluded that l(2)41Ae represented a region of 2Rh and not a single locus. To resolve these opposing views for l(2)41Ae, we completed a large inter se complementation analysis with mutant lines that were known to map to, or near, l(2)41Ae. From the initial complementation map, we were able to resolve the map to six complementation groups (Figure 2), two of which, Nipped-B and RpL38, had already been defined molecularly. The molecular identities of the remaining four complementation groups were discovered here to be CG17665, CG40127, CG17683, and CG42595.
It is evident from the current work that l(2)41Ae is not a single locus, but rather a large region of 2Rh that we now refer to as the 41Ae region. The borders of the 41Ae region are defined by the breakpoints of Df(2R)34-2, which includes the five gene models from CG42595 (uex) to CG17665 (see Figures 1 and 7; Tweedie et al. 2009). The Nipped-B locus was represented in Hilliker's (1976) original complementation map of l(2)41Ae, but we do not include Nipped-B in the 41Ae region because Nipped-B does not fall within the breakpoints of Df(2R)34-2 and because locus Rpl38 (41Af), which has always been separated genetically from 41Ae (Hilliker 1976; Marygold et al. 2005), is located between Nipped-B and CG17665 (Figure 5). It is likely that complex EMS-induced rearrangements, such as EMS 34-14, caused Nipped-B to be included in Hilliker's (1976) original complementation map of 41Ae. We found that EMS 34-14 failed to complement alleles from CG17665 and Nipped-B, as well as failed to complement numerous other lethal mutant lines, including insertion lines with distant insertion sites. EMS 34-14 was often used as a representative allele for what was believed to be a single locus, l(2)41Ae. In hindsight, because of its complex genetic properties, it was not a good choice.
Myster et al. (2004) were correct in their conclusion that EMS can induce large chromosome aberrations and that 41Ae is a region, not a locus. However, they considerably overestimated the number of vital genetic loci in 2Rh. Of the 12 vital loci reported by Myster et al. (2004) that lie proximal to RpL38/41Af (Figure 7), 2 of them, NC19 and NC28, have been shown here, through sequencing, to be alleles of CG17665. Seven of the 12 loci reported by Myster et al. (2004) were shown to lie between NC19 and NC28; this cannot be the case as there are not 7 essential loci embedded within CG17665. The identification of these as 7 separate loci by Myster et al. (2004) was likely incorrect because they did not create as many heterochromatic deletions in the area as they believed (see Table 8). The single alleles of the last 3 of the 12 loci reported by Myster et al., 309, NC83, and NC30, were all viable with Df(2R)M41A10 when tested here, so their location in 2Rh could not be determined. Table 8 summarizes the evidence from this study that resolves the 12 loci proximal to 41Af reported by Myster et al. (2004) into the 4 essential loci identified here. Table 8 also lists four deletions reported by Myster et al. (2004) that do not appear to be deletions joining separate essential loci; if they are deletions, they are likely limited to one essential locus or, as in the case of Df(2R)NC109, have now been shown to be associated with point mutations in one essential locus.
TABLE 8.
Reconsideration of 12 essential loci and 4 deletions that were reported by Myster et al. (2004) as mapping proximal to l(2)41Af
| Groupings as reported by Mysteret al. | Mysteret al. locus/deletion | Nature of the locus/deletion | Evidence from this study |
|---|---|---|---|
| Most proximal complementation group. Loci were unordered. | NC83 | Unclear if line maps to 2Rh | Not lethal with Df(2R)M41A10. |
| NC30 | Unclear if lines maps to 2Rh | Not lethal with Df(2R)M41A10. | |
| NC28 | CG17665 allele | A sequenced allele in the CG17665 complementation group (Table 3; Figure 7). | |
| l(2)309 | Unclear if line maps to 2Rh | Not lethal with Df(2R)M41A10. | |
| Second most proximal complementation group. Loci were unordered. | NC37 | CG17683 allele | Fails to complement three sequenced alleles in the CG17683 complementation group (Table S1; Figure S1; Table 3). |
| NC38 | CG17683 allele | A sequenced allele (Table 3) in the CG17683 complementation group (Figure 7). | |
| NC133 | Unclear | NC133 was lethal only with line 45-20 and that lethality has been shown to be secondary (Table S2). | |
| NC110 | CG40127 allele | A sequence allele of CG40127 (Table 3). | |
| NC70 | CG17683 allele | Fails to complement two sequenced alleles in the CG17683 complementation group (Table S1; Figure S1; Table 3). | |
| IR3 | uex allele | Fails to complement three alleles in the uex complementation group (Table S1 and Figure S1), two of which have been sequenced (Table 3). | |
| IR23 | CG17665 allele | Fails to complement four alleles in the CG17665 complementation group (Table S1; Figure S1), two of which have been sequenced (Table 3). | |
| NC19 | CG17665 allele | A sequenced allele (Table 3) in the CG17665 complementation group (Figure 7). | |
| Df(2R)NC138 | Nipped-B allele. | Fails to complement 24 alleles in the Nipped-B complementation group (Table S1; Figure S1), 11 of which have been sequenced (Gause et al. 2008). | |
| Df(2R)NC109 | CG17683 allele | A sequenced allele (Table 3) in the CG17683 complementation group (Figure 7). | |
| Df(2R)NC89 | CG17665 allele | Fails to complement five alleles in the CG17665 complementation group (Table S1; Figure S1), three of which have been sequenced (Table 3). | |
| Df(2R)NC9 | uex alelle | Fails to complement three alleles in the uex complementation group (Table S1; Figure S1), two of which have been sequenced (Table 3). |
Loci in column 2 are listed in proximal to distal order as reported by Myster et al. Complementation groups, indicated in column 1, are unordered.
One puzzling example of interallelic complementation uncovered in this study involves line EMS 45-71, a missense allele in exon 3 of CG17665 (Table 3). This missense mutation fails to complement EMS 45-34, a stop codon mutant in exon 3 of CG17665, yet EMS 45-71 fully complements NC19, another stop codon muant of CG17665 that is upstream of the EMS 45-34 stop codon mutation. The opposite result would be more intuitive since NC19 produces the more truncated protein and may be missing a critical functional domain present in the longer EMS 45-34 protein. With only the genetic data available, a proper explanation of the EMS 45-71 complementation data is not possible.
The classic example of homodimers vs. heterodimers provides a simple mechanism of interallelic complementation that could explain the complementation data for EMS45-71. In this case, flies homozygous for EMS 45-34, EMS 45-71, or NC19 would produce a homodimer (or larger multimer) that does not function adequately, resulting in lethality. In the case of a NC19/EMS 45-71 trans-heterozygote genotype, nonfunctioning homodimers would be formed alongside heterodimers. The truncated protein encoded by the 45-34 contains ∼60 more amino acids than the NC19 protein. This stretch of 60 amino acids may not contain any vital functional domain, but rather may allow 45-71/45-34 heterodimers to establish a better and more functional conformation than 45-71/NC19 heterodimers. The differing functional abilities of each heterodimer could be the factor that allows one combination to complement while the other fails to complement.
In addition to characterizing the 41Ae region, we have discovered the molecular basis for the locus l(2)41Ab to be CG41625. It should be emphasized that early mapping studies placed l(2)41Ab proximal to the rl gene (for review see Coulthard et al. 2003), but based on the recent annotation of heterochromatin (Hoskins et al. 2007) and the more recent evidence from Dimitri et al. (2009), l(2)41Ab should be located distal to rl. Indeed, CG41265 is located distal to rl (see Figure 1). We believe it likely that there was an inversion in 2Rh in the genetic background of the original mapping strains produced by Hilliker and Holm (1975). This may also be true for the uex region, as indicated by the fact that uex (CG42595) was originally mapped to cytological region h44 (Dimitri 1991), when in fact the gene is located in the distal end of cytological region h45 (Rossi et al. 2007; see also Figure 1).
The apparent prevalence of polymorphic inversions in second-chromosome heterochromatin may explain other mapping difficulties for this segment. For example, Rollins et al. (1999) placed Nipped-B distal to Nipped-A (l(2)41Ah); however, the recent annotation of 2Rh (Hoskins et al. 2007), and our discovery that Nipped-B was part of the original l(2)41Ae complementation map from Hilliker (1976), clearly places Nipped-B proximal to Nipped-A. Similar mapping problems have been encountered in 2Lh heterochromatin. For example, on the left arm of the second chromosome, the genetic locus E(Sd) has been mapped to different locations by different laboratories (Brittnacher and Ganetzky 1984; Sharp et al. 1985).
Our genetic and molecular analysis of existing mutants has allowed the identification of eight previously molecularly undefined essential loci in 2Rh. It is unclear how many more of the 41 currently annotated genes in 2Rh [i.e., the segment deleted by Df(2R)M41A10] are actually essential, but identification of novel essential genes within the 2Rh region through RNAi analysis indicates that the previous 2Rh EMS mutagenesis screens (Hilliker 1976; Sharp 1988; Eberl 1990; Myster et al. 2004) did not saturate the region. This finding is consistent with the observation that different genes and regions have different mutability (Hilliker et al. 1981) and that it may be difficult to isolate lethal alleles of all of the essential loci using EMS as a mutagen.
It seems reasonable to suggest that one of the limiting factors in the recovery of EMS-induced alleles of most genes is the size of their protein products, simply because genes that encode smaller products would provide smaller targets. In other words, essential genes could be missed because of their size. However, it is noteworthy in this regard that a gene with a very small protein product, RpL38 (70 amino acids; Marygold et al. 2005), yielded EMS alleles in the two previous EMS mutagenesis studies. Thus, gene size is clearly not the only variable with respect to gene mutability. Two of the genes that were knocked down with RNAi in the current study did not show a lethal phenotype using a daughterless-GAL4 driver transgene (Table 6). These genes may be truly nonessential due to functional redundancy or for some other reason. Alternatively, they may be essential, but the level of RNAi knockdown achieved using the daughterless-GAL4 driver transgene was insufficient to cause death. Thus, we may need to use GAL4 driver lines that utilize different promoters, such as Actin-5C or tubulin, to enhance knock-down.
The existence of essential, as well as nonessential loci, in 2Rh supports the view that heterochromatin is neither genetically inert nor enriched only in essential genes. There is also sufficient evidence that the set of heterochromatic genes in D. melanogaster and other organisms is not unique when compared to the set of euchromatic genes [reviewed by Corradini et al. 2003)]. Similar to any set of euchromatic genes, heterochromatic genes encode an extensive array of proteins with a wide variety of molecular functions, as has been established by a number of genetic and molecular studies (Biggs et al. 1994; Warner et al. 1998; Rollins et al. 1999; Schulze et al. 2005; Yasuhara et al. 2005; Hallson et al. 2008). This conclusion is also clearly supported by the recent annotation of heterochromatin sequence (Hoskins et al. 2007; Dimitri et al. 2009; Tweedie et al. 2009). Additionally, the expression profiles of heterochromatic genes are not especially restricted; heterochromatic genes are often expressed throughout development and in different tissues (for examples, see Biggs et al. 1994; Marygold et al. 2005; Schulze et al. 2005; Dimitri et al. 2009).
The principal difference between heterochromatic vs. euchromatic genes is the chromatin environment in which each type of gene resides. One of the main distinguishing features of heterochromatin is the high density of repeat sequences (Gatti and Pimpinelli 1992; Lohe et al. 1993; Bartolomé et al. 2002); in fact, it has been shown that some heterochromatic genes are located within regions containing nearly 90% repeats, including transposable element and repetitive satellite sequences (Smith et al. 2007). In addition, heterochromatic genes generally possess larger introns consisting mainly of transposable element remnants (Devlin et al. 1990; Uchida et al. 1993; Biggs et al. 1994; Tulin et al. 2002; Dimitri et al. 2003b; Smith et al. 2007) and are often enriched in terms of AT content (Adams et al. 2000; Díaz-Castillo and Golic 2007).
The evolution of certain heterochromatic genes argues that it is the chromatin environment, and not the genes themselves, that distinguishes heterochromatic genes from euchromatic genes. In evolutionary time frames, there appears to be flexibility between the euchromatic and heterochromatic environments in terms of where genes can reside (Schulze et al. 2006; Yasuhara and Wakimoto 2006). For example, in the evolution of the lt gene in Drosophila, the ancestral version of D. melanogaster's lt gene has been found to be euchromatic (Yasuhara et al. 2005). In the D. melanogaster version of lt, the introns have grown in size due to the accumulation of transposable element sequences compared to its euchromatic orthologs with smaller intron sizes. Contrariwise, the male-specific fertility factors in D. pseudoobscura are moving from a heterochromatic location to a euchromatic location, and in the process, the size of their introns is decreasing (Carvalho and Clark 2005). Finally, in D. melanogaster, the genes Dbp80 and RpL15 are adjacent heterochromatic genes located deep within centromeric heterochromatin, but in D. psuedoobscura these genes are euchromatic. Furthermore, in D. virilis, Dbp80 is euchromatic while RpL15 is heterochromatic (Schulze et al. 2006).
Perhaps the most important aspect of comparison between heterochromatic and euchromatic genes is their dependence on their own chromatin environment for their proper expression. To the extent that it has been examined, heterochromatic genes require heterochromatin to function properly, just as euchromatic genes require euchromatin. For heterochromatic genes, this has been most extensively studied for the rolled gene (Eberl et al. 1993), and to a lesser extent, for the light gene (Howe et al. 1995). These genes variegate when moved to distal euchromatin, with the strength of the variegation varying inversely to the size of the heterochromatic block in which the variegating gene resides. Heterochromatic gene variegation is also sensitive to the effect of the modifiers of position effect variegation, just as with euchromatic genes. For the most part, the effects of these modifiers on variegating heterochromatic genes are opposite to the effects on variegating euchromatic genes (Locke et al. 1988; Hearn et al. 1991; Weiler and Wakimoto 1995; Lu et al. 2000; Schulze and Wallrath 2007), as would be expected if genes required their own chromatin environment in which to function properly. Thus, there may be some flexibility with respect to heterochromatin and euchromatin in terms of where genes can reside over evolutionary time scales, as discussed above, but in general, any gene moving from one chromatin environment to another will tend to not function properly. Therefore, the heterochromatic dependency of the rl and lt genes in D. melanogaster is no more surprising than the presumed euchromatin dependence of rl and lt in Drosophila species where these genes reside in euchromatin. As such, constitutive heterochromatin should no longer be described as silencing DNA, which it is commonly termed, for the simple reason that euchromatin is silencing chromatin from the view point of heterochromatic genes.
Acknowledgments
This work was supported by a joint grant from the Canadian Institutes of Health Research awarded to A. J. Hilliker, B. Honda, and D. Sinclair and by a grant from the National Sciences and Engineering Research Council of Canada awarded to A. J. Hilliker.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.117259/DC1.
References
- Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne et al., 2000. The genomic sequence of Drosophila melanogaster. Science 287 2185–2195. [DOI] [PubMed] [Google Scholar]
- Bartolomé, C., X. Maside and B. Charlesworth, 2002. On the abundance and distribution of transposable elements in the genome of Drosophila melanogaster. Mol. Biol. Evol. 19 926–937. [DOI] [PubMed] [Google Scholar]
- Bellen, H. J., R. W. Levis, G. Liao, Y. He, J. W. Carlson et al., 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 162(2): 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs, W. H., III, K. H. Zavitz, B. Dickson, A. Van der Straten, D. Brunner et al., 1994. The Drosophila rolled locus encodes a MAP kinase required in the sevenless signal transduction pathway. EMBO J. 13 1628–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittnacher, J. G., and B. Ganetzky, 1984. On the components of segregation distortion in Drosophila melanogaster. III. Nature of enhancer of SD. Genetics 197 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, S.W., 1966. Heterochromatin. Science 151 417–425. [DOI] [PubMed] [Google Scholar]
- Carvalho, A. B., and A. G. Clark, 2005. Y chromosome of D. pseudoobscura is not homologous to the ancestral Drosophila Y. Science 307 108–110. [DOI] [PubMed] [Google Scholar]
- Corradini, N., F. Rossi, F. Verni and P. Dimitri, 2003. FISH analysis of Drosophila melanogaster heterochromatin using BACs and P elements. Chromosoma 112 26–37. [DOI] [PubMed] [Google Scholar]
- Coulthard, A. B., D. F. Eberl, C. B. Sharp and A. J. Hilliker, 2003. Genetic analysis of the second chromosome centromeric heterochromatin of Drosophila melanogaster. Genome 46 343–352. [DOI] [PubMed] [Google Scholar]
- Devlin, R. H., B. Bingham and B. T. Wakimoto, 1990. The organization and expression of the light gene, a heterochromatic gene of Drosophila melanogaster. Genetics 125 129–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Castillo, C., and K. G. Golic, 2007. Evolution of gene sequence in response to chromosomal location. Genetics 177 359–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl, G., D. Chen, F. Schnorrer, K. C. Su, Y. Barinova et al., 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448 151–156. [DOI] [PubMed] [Google Scholar]
- Dimitri, P., 1991. Cytogenetic analysis of the second chromosome heterochromatin of Drosophila melanogaster. Genetics 127(3): 553–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitri, P., 2004. Fluorescent in situ hybridization with transposable element probes to mitotic chromosome heterochromatin of Drosophila. Methods Mol. Biol. 260 29–39. [DOI] [PubMed] [Google Scholar]
- Dimitri, P., B. Arcà, L. Berghella and E. Mei 1997. High genetic instability of heterochromatin after transposition of the LINE-like I factor in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 94 8052–8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitri, P., N. Corradini, F. Rossi, F. Verni, G. Cenci et al., 2003. a Vital genes in the heterochromatin of chromosome 2 and 3 of Drosophila melanogaster. Genetica 117 209–215. [DOI] [PubMed] [Google Scholar]
- Dimitri, P., N. Junakovic and B. Arcà, 2003. b Colonization of heterochromatic genes by transposable elements in Drosophila. Mol. Biol. Evol. 20 503–512. [DOI] [PubMed] [Google Scholar]
- Dimitri, P., R. Caizzi, E. Giordano, M. Carmela Accardo, G. Lattanzi et al., 2009. Constitutive heterochromatin: a surprising variety of expressed sequences. Chromosoma 118 419–435. [DOI] [PubMed] [Google Scholar]
- Eberl, D. F., 1990. Genetic studies on the nature and function of autosomal heterochromatin in Drosophila melanogaster. Ph.D. Thesis, University of Guelph, Guelph, ON, Canada.
- Eberl, D. F., B. J. Duyf and A. J. Hilliker, 1993. The role of heterochromatin in the expression of a heterochromatic gene, the rolled locus of Drosophila melanogaster. Genetics 134 277–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti, M., and S. Pimpinelli, 1992. Functional elements in Drosophila melanogaster heterochromatin. Annu. Rev. Genet. 26 239–275. [DOI] [PubMed] [Google Scholar]
- Gause, M., H. A. Webber, Z. Misulovin, G. Haller, R. A. Rollins et al., 2008. Functional links between Drosophila Nipped-B and cohesin in somatic and meiotic cells. Chromosoma 117 51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallson, G., M. Syrzycka, S. A. Beck, J. A. Kennison, D. Dorsett et al., 2008. The Drosophila cohesin subunit Rad21 is a trithorax group (trxG) protein. Proc. Natl. Acad. Sci. USA 105 12405–12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah, A., 1951. Localization and function of heterochromatin Drosophila melanogaster. Adv. Genet. 4 87–125. [DOI] [PubMed] [Google Scholar]
- Hearn, M. G., A. Hedrick, T. A. Grigliatti and B. T. Wakimoto, 1991. The effect of modifiers of position-effect variegation on the variegation of heterochromatic genes of Drosophila melanogaster. Genetics 128 785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz, E., 1928. Das heterochromatin der moose. I. Jb. Wiss. Bot. 69 762–818. [Google Scholar]
- Hessler, A. Y., 1958. V-type position effects at the light locus in Drosophila melanogaster. Genetics 43 395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker, A. J., 1976. Genetic analysis of the centromeric heterochromatin of chromosome 2 of Drosophila melanogaster: deficiency mapping of EMS-induced lethal complementation groups. Genetics 83 765–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker, A. J., and D. G. Holm, 1975. Genetic analysis of the proximal region of chromosome 2 of Drosophila melanogaster. I. Detachment products of compound autosomes. Genetics 81 705–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker, A. J., R. Appels and A. Schalet, 1980. The genetic analysis of D. melanogaster heterochromatin. Cell 21 607–619. [DOI] [PubMed] [Google Scholar]
- Hilliker, A. J., A. Chovnick and S. H. Clark, 1981. The relative mutabilities of vital genes in D. melanogaster. Drosoph. Inf. Serv. 56 64–65. [Google Scholar]
- Hoskins, R. A., C. D. Smith, J. W. Carlson, A. B. Carvalho, A. Halpern et al., 2002. Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol. 3 RESEARCH0085. [DOI] [PMC free article] [PubMed]
- Hoskins, R. A., J. W. Carlson, C. Kennedy, D. Acevedo, M. Evans-Holm et al., 2007. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science 316 1625–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, M., P Dimitri, M. Berloco and B. T. Wakimoto, 1995. Cis-effects of heterochromatin on euchromatic and heterochromatic gene expression in Drosophila melanogaster. Genetics 140 1033–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, J. K., and L. A. Snyder, 1974. Cytogenetic and complementation analyses of recessive lethal mutations induced in the X chromosome of Drosophila by three alkylating agents. Genet. Res. 24 1–10. [DOI] [PubMed] [Google Scholar]
- Locke, J., M. A. Kotarski and K. D. Tartof, 1988. Dosage dependent modifiers of position effect variegation in Drosophila and a mass action model that explains their effects. Genetics 120 181–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe, A. R., A. J. Hilliker and P. A. Roberts, 1993. Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics 134 1149–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, B. Y., P. C. Emtage, G. J. Duyf, A. J. Hilliker and J. C. Eissenberg, 2000. Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila. Genetics 155 699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marygold, S. J., C. M. Coelho and S. J. Leevers, 2005. Genetic analysis of RpL38 and RpL5, two Minute genes located in the centric heterochromatin of chromosome 2 of Drosophila melanogaster. Genetics 169 683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myster, S. H., F. Wang, R. Cavallo, W. Christian, S. Bhotika et al., 2004. Genetic and bioinformatic analysis of 41C and the 2R heterochromatin of Drosophila melanogaster: a window on the heterochromatin-euchromatin junction. Genetics 166 807–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpinelli, S., M. Berloco, L. Fanti, P. Dimitri, S. Bonaccorsi et al, 1995. Transposable elements are stable structural components of Drosophila melanogaster heterochromatin. Proc. Natl. Acad. Sci USA 92 3804–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins, R. A., P. Morcillo and D. Dorsett, 1999. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics 152 577–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi, F., R. Moschetti, R. Caizzi, N. Corradini and P. Dimitri, 2007. Cytogenetic and molecular characterization of heterochromatin gene models in Drosophila melanogaster. Genetics 175 595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, J., 1936. Variegation in Drosophila and the inert chromosome regions. Proc. Natl. Acad. Sci. USA 22 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze, S. R, and L. L. Wallrath, 2007. Gene regulation by chromatin structure: paradigms established in Drosophila melanogaster. Annu. Rev. Entomol. 52 171–192. [DOI] [PubMed] [Google Scholar]
- Schulze, S. R., D. A. Sinclair, K. A. Fitzpatrick and B. M. Honda 2005. A genetic and molecular characterization of two proximal heterochromatic genes on chromosome 3 of Drosophila melanogaster. Genetics 169 2165–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze, S. R., B. F. McAllister, D. A. R. Sinclair, K. A. Fitzpatrick, M. Marchetti et al., 2006. Heterochromatic genes in Drosophila: a comparative analysis of two genes. Genetics 173 1433–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, C. B., 1988. Biometrical and genetic studies of segregation distortion in Drosophila melanogaster. Ph.D. Thesis, University of Guelph, Guelph, ON, Canada.
- Sharp, C. B., A. J. Hilliker and D. G. Holm, 1985. Further characterization of genetic elements associated with the segregation distorter phenomenon in Drosophila melanogaster. Genetics 110 671–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C. D., S. Q. Shu, C. J. Mungall and G. H. Karpen, 2007. The release 5.1 annotation of Drosophila melanogaster heterochromatin. Science 316 1586–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styhler, S., A. Nakamura and P. Lasko, 2002. VASA localization requires the SPRY-domain and SOCS-box containing protein, GUSTAVUS. Dev. Cell 3 855–876. [DOI] [PubMed] [Google Scholar]
- Tulin, A., D. Stewart and A. C. Spradling, 2002. The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev. 16 2108–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S., M. Ashburner, K. Falls, P. Leyland, P. McQuilton et al.; The FlyBase Consortium, 2009. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 37 D555–D559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida, K., S. Hanai, K. Ishikawa, Y. I. Ozawa, M. Uchida et al., 1993. Cloning of cDNA encoding Drosophila poly(ADP-ribose) polymerase: leucine zipper in the auto-modification domain. Proc. Natl. Acad. Sci. USA 90 3481–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner, T. S., D. A. Sinclair, K. A. Fitzpatrick, M. Singh, R. H. Devlin et al., 1998. The light gene of Drosophila melanogaster encodes a homologue of VPS41, a yeast gene involved in cellular-protein trafficking. Genome 41 236–243. [PubMed] [Google Scholar]
- Weiler, K. S., and B. T. Wakimoto, 1995. Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet. 29 577–605. [DOI] [PubMed] [Google Scholar]
- Yasuhara, J. C., and B. T. Wakimoto, 2006. Oxymoron no more: the expanding world of heterochromatic genes. Trends Genet. 22 330–338. [DOI] [PubMed] [Google Scholar]
- Yasuhara, J. C., C. H. DeCrease and B. T. Wakimoto, 2005. Evolution of heterochromatic genes of Drosophila. Proc. Natl. Acad. Sci. USA 102 10958–10963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P., and A. C. Spradling, 1994. Insertional mutagenesis of Drosophila heterochromatin with single P elements. Proc. Natl. Acad. Sci. USA 91 3539–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]