Abstract
Mating phenotype in the yeast Saccharomyces cerevisiae is a dynamic trait, and efficient transitions between alternate haploid cell types allow the organism to access the advantageous diploid form. Mating identity is determined by cell type-specific transcriptional regulators, but these factors must be rapidly removed upon mating-type switching to allow the master regulators of the alternate state to establish a new gene expression program. Targeted proteolysis by the ubiquitin–proteasome system is a commonly employed strategy to quickly disassemble regulatory networks, and yeast use this approach to evoke efficient switching from the α to the a phenotype by ensuring the rapid removal of the α2 transcriptional repressor. Transition to the a cell phenotype, however, also requires the inactivation of the α1 transcriptional activator, but the mechanism by which this occurs is currently unknown. Here, we report a central role for the ubiquitin–proteasome system in α1 inactivation. The α1 protein is constitutively short lived and targeted for rapid turnover by multiple ubiquitin-conjugation pathways. Intriguingly, the α-domain, a conserved region of unknown function, acts as a degradation signal for a pathway defined by the SUMO-targeted ligase Slx5–Slx8, which has also been implicated in the rapid destruction of α2. Our observations suggest coordinate regulation in the turnover of two master regulatory transcription factors ensures a rapid mating-type switch.
THE activation of biological regulatory pathways is a central event in the establishment of cellular phenotypes, but an equally important process is the dismantling of such control mechanisms to ensure proper cellular homeostasis. For example, the activation of protein effectors triggers cells to proliferate, differentiate, and adapt to changing environments, but these multistep transitions are faithfully accomplished only if the underlying activation events are coordinated into temporal cascades to precisely order key events along the pathway. Such coordination requires the activation events to be transient, so that the next step in the cascade can initiate without interference from the previous state.
Cells have evolved many pathways to inactivate regulatory proteins in spatially and temporally restricted patterns, including modification and redirected localization, but the selective proteolysis of these regulators is a particularly powerful mechanism to induce an irreversible transition (Ang and Harper 2004). The most common mechanism for the degradation of regulatory proteins involves the ubiquitin–proteasome system (UPS). Proteins are targeted for destruction via the UPS following the attachment of the small protein modifier ubiquitin (Hochstrasser 1996; Hershko and Ciechanover 1998; Pickart and Eddins 2004; Ravid and Hochstrasser 2008). The ubiquitin moiety is conjugated to substrate proteins through a series of steps, each mediated by distinct enzymes. Ubiquitin is initially activated by the E1 activating enzyme, which subsequently transfers the activated ubiquitin to an E2 ubiquitin-conjugating enzyme. Together with an E3 ubiquitin ligase, the E2 transfers ubiquitin to the substrate (Hershko and Ciechanover 1998; Pickart 2001; VanDemark and Hill 2002). Typically, a single ubiquitin moiety is not sufficient to target a protein for degradation; however, the assembly of ubiquitin chains or the independent attachment of additional ubiquitin monomers to multiple sites within the target protein directs the substrate to a large multisubunit protease called the 26S proteasome for proteolysis (Thrower et al. 2000; Pickart and Cohen 2004; Kravtsova-Ivantsiv et al. 2009).
The UPS is a common component of many biological control mechanisms. One particularly well-studied process that is dependent on the UPS for the dismantling of a preexisting regulatory pathway is the yeast mating-type determination and switching system (Laney and Hochstrasser 2004). Yeast cells exist as one of three phenotypically distinct types, the a or α haploid variants or the a/α diploid state. These distinct cell types are determined by a set of master regulatory transcription factors that establish the expression patterns of mating-type-specific genes. For example, the α cell type is defined by the transcriptional activator α1 that promotes expression of α-specific genes, as well as the transcriptional repressor α2 that blocks the expression of a-specific genes (Nasmyth and Shore 1987; Herskowitz 1989).
Interestingly, transitions between the haploid cell types occur and are an important aspect of yeast biology that likely confers distinct advantages to the organism, including the ability to access a protective spore state (Oshima and Takano 1971; Hicks and Herskowitz 1976). Such transitions not only require the establishment of a new gene regulatory program, but also depend on the dismantling of the previous state of gene expression. For transitions from the α to the a state, ubiquitin-dependent proteolysis of the α2 transcriptional repressor is a requisite step (Laney and Hochstrasser 2003). α2 is targeted to the UPS via multiple ubiquitination pathways that recognize and likely modify distinct regions of the protein (Chen et al. 1993). An amphipathic helix in the N terminus of the protein functions as a degradation signal and is targeted by the E2 conjugating enzymes Ubc6 and Ubc7 along with the RING domain E3 ligase Doa10 (Chen et al. 1993; Johnson et al. 1998; Swanson et al. 2001). Another degradation signal in the C-terminal domain has not been structurally defined, but is recognized by the E2s Ubc4 and Ubc5 and the heterodimeric E3 Slx5–Slx8 (Hochstrasser and Varshavsky 1990; Chen et al. 1993; Y. Xie and M. Hochstrasser, personal communication). Importantly, either pathway alone is sufficient to ensure an efficient α-to-a cell type transition, suggesting the evolution of redundancy in the system to protect this biologically advantageous event (Laney and Hochstrasser 2003).
The destruction of α2 alone, however, is not sufficient to evoke the α-to-a transition. In addition to α2, the α1 protein must also be rapidly removed after mating-type switching. As the transcriptional activator of α-specific genes, retention of the existing α1 protein after transition to the a state would derail the newly emerging gene expression program and phenotype of these nascent a cells. Indeed, the misexpression of α1 or its target gene STE3 causes sterility in a cells (Hirsch and Cross 1993; J. D. Laney, unpublished observations), indicating the need for a mechanism to destroy α1 activity upon mating-type switching.
Given the importance of the UPS in α2 proteolysis, we explored a role for this pathway in the inactivation of α1. Here, we establish that α1 is indeed a short-lived protein targeted for destruction via the UPS. We define the conserved α-domain of this protein as a degradation signal and genetically characterize the multiple ubiquitin-conjugation pathways essential for α1 turnover. Intriguingly, some of these pathways overlap with those regulating α2 turnover, suggesting coordinate regulation in the turnover of the two master regulatory proteins that define the α-state. As the dismantling of regulatory pathways often requires the inactivation of multiple protein effectors, the yeast mating-type determination system provides a tractable experimental model to explore such coordination.
MATERIALS AND METHODS
Yeast strains:
Strains used in this study are listed in Table 1. Specific gene deletions and modifications were made using standard methods of transforming linearized plasmid fragments or PCR products. Proper integrations and substitutions were confirmed by PCR and segregation analysis. To generate strains containing multiple disruptions, standard genetic crosses with congenic strains of opposite mating type were performed. To generate strains containing the smt3-allR and smt3-3NR alleles (JY817 and JY818), the modified smt3∷TRP1 genes from GBY5 and GBY1 (Bylebyl et al. 2003) were amplified from genomic DNA and transformed into MHY501. Direct sequencing of diagnostic PCR products from the transformants verified the presence of these mutations. To generate the mms21-ΔRING strain (JY764), the Mms21 ORF was truncated after L182 by replacing the RING domain with a stop codon, the terminator from ADH1, and a kanMX cassette.
TABLE 1.
Yeast strains
| Strain | Genotype | Source or reference |
|---|---|---|
| MHY501 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, gal2 | Chen et al. (1993) |
| JY357 | matα1Δ∷His3MX6, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1 | This study |
| JY138 | MATα, ade2-101, his3-Δ200, leu2-Δ1, lys2-801, ura3-52, adh4∷URA3-TEL-VIIL, DIA5-1, cim3-1 | J. D. Laney (unpublished data) |
| JY142 | MATα, ade2-101, his3-Δ200, leu2-Δ1, lys2-801, ura3-52, adh4∷URA3-TEL-VIIL, DIA5-1, cim5-1 | J. D. Laney (unpublished data) |
| JY560 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, rpn13Δ∷His3MX | This study |
| JY470 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, pdr5Δ∷His3MX | This study |
| MHY509 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, ubc1Δ∷HIS3 | Seufert et al. (1990) |
| MHY612 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, rad6Δ∷LEU2 | M. Hochstrasser |
| JY395 | MATα, his3Δ, ura3-52, trp1, cdc34-2 | This study |
| MHY498 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, ubc4-Δ1∷HIS3 | Seufert and Jentsch (1990) |
| MHY499 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, ubc5-Δ1∷LEU2 | Seufert and Jentsch (1990) |
| JY597 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, ubc4-Δ1∷HIS3, ubc5-Δ1∷LEU2 | This study |
| MHY495 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, ubc6-Δ1∷HIS3 | Chen et al. (1993) |
| MHY507 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, ubc7∷LEU2 | Jungmann et al. (1993) |
| MHY597 | MATα, ade2-1, ura3-1, his3-11, leu2-3,112, trp1-1, can1-100 | Qin et al. (1991) |
| MHY601 | MATα, ade2-1, ura3-1, his3-11, leu2-3,112, trp1-1, can1-100, ubc8∷URA3 | Qin et al. (1991) |
| JY385 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, ubc10Δ∷kanMX | This study |
| JY387 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, ubc11Δ∷kanMX | This study |
| JY389 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, ubc13Δ∷kanMX | This study |
| JY507 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, mms2Δ∷His3MX | This study |
| JY527 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, rad5Δ∷His3MX | This study |
| JY710 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, dma1Δ∷hphMX4 | This study |
| JY714 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, dma2Δ∷kanMX | This study |
| JY706 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, dma1Δ∷hphMX4, dma2Δ∷kanMX | This study |
| JY612 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, ufd4Δ∷hphMX | This study |
| MHY2480 | MATα, his3Δ200, leu2-3,112, ura3-52∷rsp5-2∷URA3, lys2-801, trp1-1, rsp5Δ∷HIS3 | Hoppe et al. (2000) |
| MHY1631 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, doa10Δ∷HIS3 | Swanson et al. (2001) |
| MHY3712 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, slx5Δ∷kanMX | Xie et al. (2007) |
| MHY3716 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, slx8Δ∷kanMX | Xie et al. (2007) |
| MHY3861 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, slx5Δ∷kanMX, slx8Δ∷kanMX | Xie et al. (2007) |
| JY680 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, ris1Δ∷hphMX4 | This study |
| JY751 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, ufd4Δ∷hphMX4, slx5Δ∷kanMX | This study |
| JY743 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, ufd4Δ∷hphMX4, slx8Δ∷kanMX | This study |
| JY798 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, dma1Δ∷His3MX, slx8Δ∷kanMX | This study |
| JY788 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, dma1Δ∷His3MX, slx5Δ∷kanMX | This study |
| JY794 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, dma1Δ∷His3MX, ufd4Δ∷hphMX4 | This study |
| JY800 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, dma2Δ∷CaURA3MX, slx8Δ∷kanMX | This study |
| JY790 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, dma2Δ∷CaURA3MX, slx5Δ∷kanMX | This study |
| JY792 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, dma2Δ∷CaURA3MX, ufd4Δ∷hphMX4 | This study |
| JY745 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, dma1Δ∷hphMX4, dma2Δ∷kanMX, ufd4Δ∷hphMX4 | This study |
| JY739 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, dma1Δ∷hphMX4, dma2Δ∷kanMX, slx5Δ∷kanMX | This study |
| JY768 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, dma1Δ∷hphMX4, ufd4Δ∷hphMX4, slx5Δ∷kanMX | This study |
| JY776 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, ufd4Δ∷hphMX4, dma2Δ∷CaURA3MX, slx8Δ∷kanMX | This study |
| JY774 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, ufd4Δ∷hphMX4, slx8Δ∷kanMX, dma1Δ∷His3MX | This study |
| JY796 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, dma1Δ∷His3MX, dma2Δ∷CaURA3MX, slx8Δ∷kanMX | This study |
| JY770 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, ufd4Δ∷hphMX4, dma2Δ∷CaURA3MX, slx5Δ∷kanMX | This study |
| JY772 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, ufd4Δ∷hphMX4, slx5Δ∷kanMX, dma1Δ∷His3MX, dma2Δ∷CaURA3MX | This study |
| JY778 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, dma1Δ∷His3MX, dma2Δ∷CaURA3MX, slx8Δ∷kanMX, ufd4Δ∷hphMX4 | This study |
| MHY3997 | MATα, his3-Δ200, leu2-3,112∷LEU2∷ubc9-1, ura3-52, lys2-801, trp1-1, ubc9∷TRP1 | Seufert et al. (1995) |
| MHY4664 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, uba2Δ∷kanMX, pIS1-uba2-ts9 (CEN LEU2) | Schwienhorst et al. (2000) |
| MHY2423 | MATα, his3-Δ200, leu2-3,112∷LEU2∷ulp1-333, ura3-52, lys2-801, trp1-1, ulp1∷his3∷URA3 | Li and Hochstrasser (1999) |
| JY764 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, mms21-ΔRING∷kanMX | This study |
| JY802 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, siz2Δ∷hphMX4, siz1Δ∷kanMX | This study |
| JY817 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, smt3-allR∷TRP1 | This study |
| JY818 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, smt3-R11, 15, 19∷TRP1 | This study |
| GBY5 | MATa, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-Δ1, smt3-allR∷TRP1 | Bylebyl et al. (2003) |
| GBY1 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-Δ1, smt3-R11, 15, 19∷TRP1 | Bylebyl et al. (2003) |
Plasmids:
To facilitate detection of α1, an α1 yeast expression plasmid derived from p414ADH (Mumberg et al. 1995) was constructed using a PCR product encoding the α1 ORF and a fragment containing the 3HA epitope-encoding sequence and ADH1 terminator from pFA6a-3HA-kanMX6 (Longtine et al. 1998). For the α1-Ura3 fusion protein, a similar yeast expression plasmid was constructed using a PCR product encoding the Ura3 ORF in addition to the α1 and 3HA fragments. Structure predictions from the Jpred secondary structure prediction server (Cole et al. 2008) were used to design the α1 truncation mutants; PCR products encoding these α1 fragments were subcloned into the p414ADH-α1-Ura3-3HA expression vector. Details of plasmid constructions are available upon request.
Cycloheximide-chase degradation assays:
Strains were grown to midlog phase (OD600 ∼ 0.5–1.0) in minimal media lacking the appropriate amino acid for plasmid selection. To initiate the chase, cycloheximide was added to a final concentration of 0.2 mg/ml. Immediately after cycloheximide addition, 2.5 ODs of cells were harvested and added to 10 ml of ice-cold water containing 30 mm NaN3. Cells were harvested in this way for each subsequent time point. Cells were collected by centrifugation and washed once in water and lysates were prepared as described (Kushnirov 2000). Briefly, cells were resuspended in 200 μl of 0.1 m NaOH, incubated at room temperature for 5 min, and pelleted and the cell pellet was lysed by boiling in 50 μl of 1× Laemmli loading buffer. A portion of the lysate (typically, 1 OD equivalent) was analyzed by quantitative Western blotting. The amount of α13HA present at a given chase time was expressed relative to that observed at time zero. Replicates of multiple experiments were averaged and the error is represented as the standard deviation. Error bars are included for all data points; where error bars are not visible, they have been obscured by the size of the symbol. The only exception is the experiments with ubc4Δ ubc5Δ cells, where multiple overlapping time courses with nonidentical time points were performed. Note that a wild-type cell control was assayed in parallel to the various ubiquitination mutants in most individual experiments and all of these data were compiled and averaged (thus, in instances where the wild-type control is in the same genetic background and experiments were performed at the same temperature, the wild-type data are identical: Figures 2–5, Figure 7, B and C, and Figures 1A and 7A). For experiments with an inhibitor of the proteasome, cultures of a pdr5Δ strain were treated with 25 μg/ml of MG-132 (Enzo Life Sciences) dissolved in DMSO at 30° for 30 min prior to the cycloheximide-chase assay. For experiments involving temperature-sensitive alleles, strains were grown at the permissive temperature and then shifted to 37° for 1 hr prior to the cycloheximide-chase assay. In all experiments, the α1 protein contained three copies of the HA epitope to allow detection with the anti-HA 12CA5 antibody.
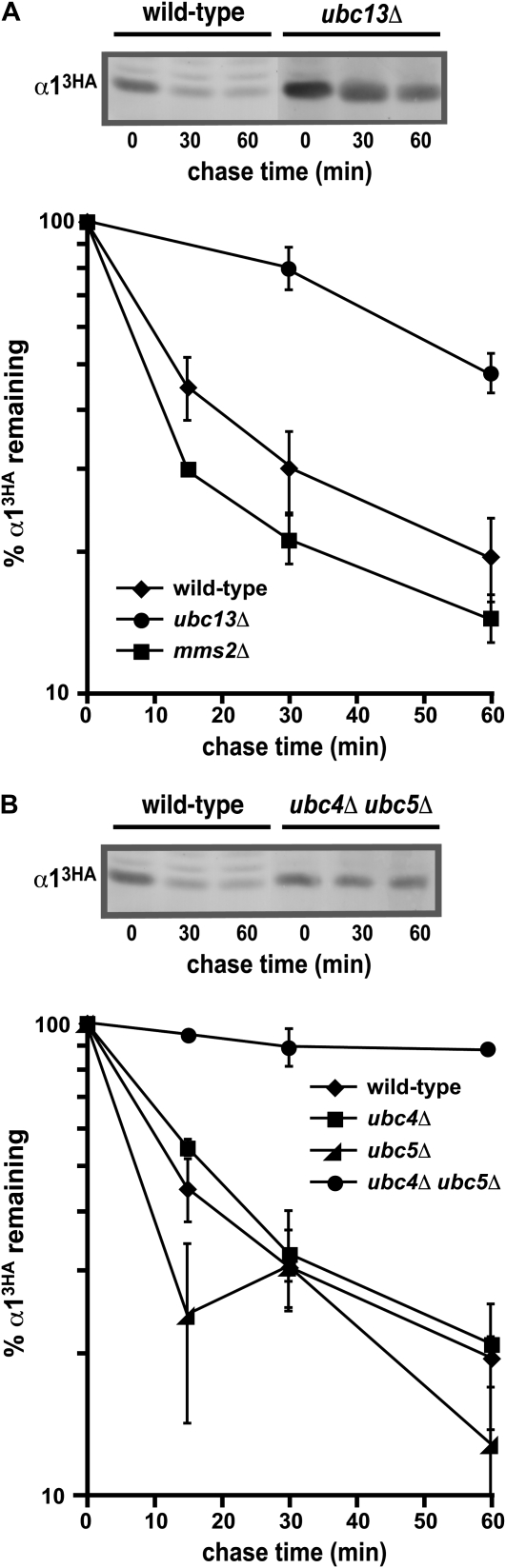
Figure 2.—
The degradation of α1 requires multiple ubiquitin-conjugating enzymes. (A) Cycloheximide-chase analysis of α13HA turnover at 30° in wild-type, ubc13Δ, and mms2Δ cells. A representative experiment is shown at the top, and the quantitative results of multiple experiments are shown below. (B) Measurement of the α13HA degradation rate at 30° in wild-type, ubc4Δ, ubc5Δ, and ubc4Δ ubc5Δ cells. A representative experiment is shown at the top, and the quantitative results of multiple experiments are shown below.
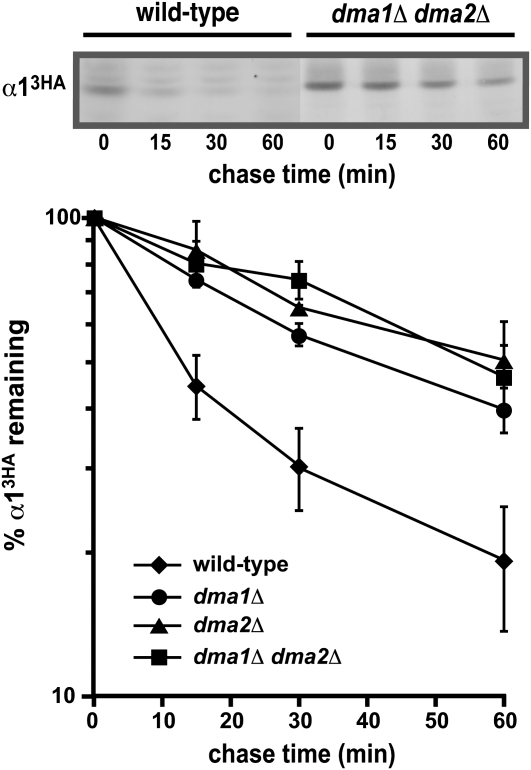
Figure 3.—
Ubiquitin ligases that interact with Ubc13 are necessary for the rapid degradation of α1. Measurement of the α13HA degradation rate at 30° is shown in wild-type, dma1Δ, dma2Δ, and dma1Δ dma2Δ cells. A representative experiment is shown at the top, and the quantitative results of multiple experiments are shown below.
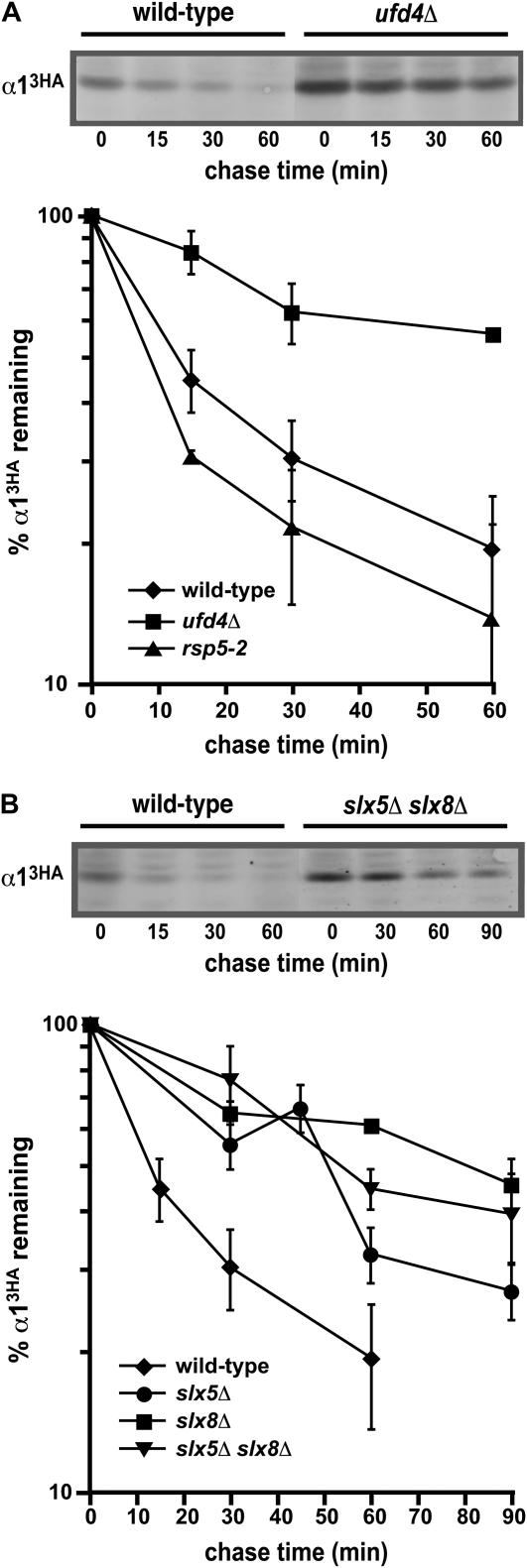
Figure 4.—
Ubc4/5-dependent ubiquitin ligases are necessary for the rapid degradation of α1. (A) Cycloheximide-chase analysis of α13HA turnover in wild-type, ufd4Δ, and rsp5-2 cells. In experiments with the rsp5-2 mutant, the temperature-sensitive strain and a congenic wild-type strain were shifted to 37° for 1 hr prior to the cycloheximide-chase assay and maintained at this temperature throughout the assay. A representative experiment is shown at the top, and the quantitative results of multiple experiments are shown below. (B) Measurement of the α13HA degradation rate at 30° in wild-type, slx5Δ, slx8Δ, and slx5Δ slx8Δ cells. A representative experiment is shown at the top, and the quantitative results of multiple experiments are shown below.
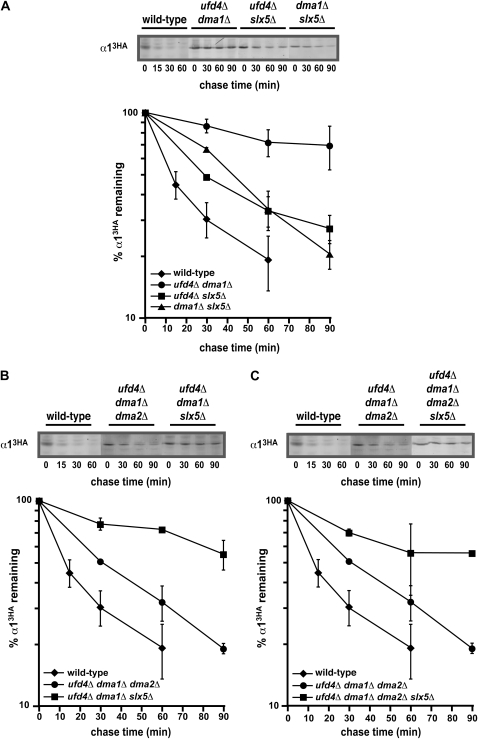
Figure 5.—
Genetic interactions among the α1 degradation pathways. (A) A representative cycloheximide-chase assay of α13HA turnover at 30° in wild-type, ufd4Δ dma1Δ, ufd4Δ slx5Δ, and dma1Δ slx5Δ cells is shown at the top, and the quantitative results of multiple experiments are shown below. (B) Cycloheximide-chase analysis of α13HA turnover at 30° in wild-type, ufd4Δ dma1Δ dma2Δ, and ufd4Δ dma1Δ slx5Δ cells. A representative experiment is shown at the top, and the quantitative results of multiple experiments are shown below. (C) A representative cycloheximide-chase assay of α13HA turnover at 30° in wild-type, ufd4Δ dma1Δ dma2Δ, and ufd4Δ dma1Δ dma2Δ slx5Δ cells is shown at the top, and the quantitative results of multiple experiments are shown below. The data for ufd4Δ dma1Δ dma2Δ cells from B are included for comparison.
Figure 7.—
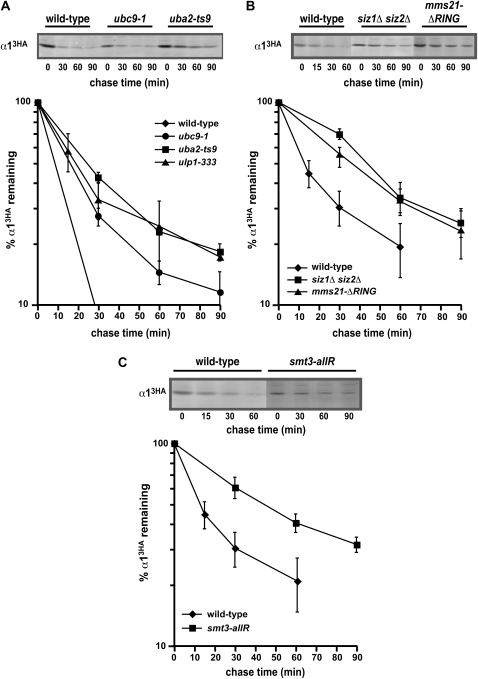
The rapid degradation of α1 requires an intact SUMO conjugation pathway. (A) Cycloheximide-chase analysis of α13HA turnover in wild-type, uba2-ts9, ubc9-1, and ulp1-333 cells. In these experiments, the temperature-sensitive strains and a congenic wild-type strain were shifted to 37° for 1 hr prior to the cycloheximide-chase assay and maintained at this temperature throughout the assay. A representative experiment is shown at the top, and the quantitative results of multiple experiments are shown below. (B) Measurement of the α13HA degradation rate at 30° in wild-type, siz1Δ siz2Δ, and mms21-ΔRING cells. A representative experiment is shown at the top, and the quantitative results of multiple experiments are shown below. In the mms21-ΔRING allele, the Mms21 ORF is truncated after L182 by replacing the RING domain with a stop codon, the terminator from ADH1, and a kanMX cassette. (C) A representative cycloheximide-chase assay of α13HA turnover at 30° in a wild-type and a smt3-allR strain is shown at the top, and the quantitative results of multiple experiments are shown below.
Figure 1.—
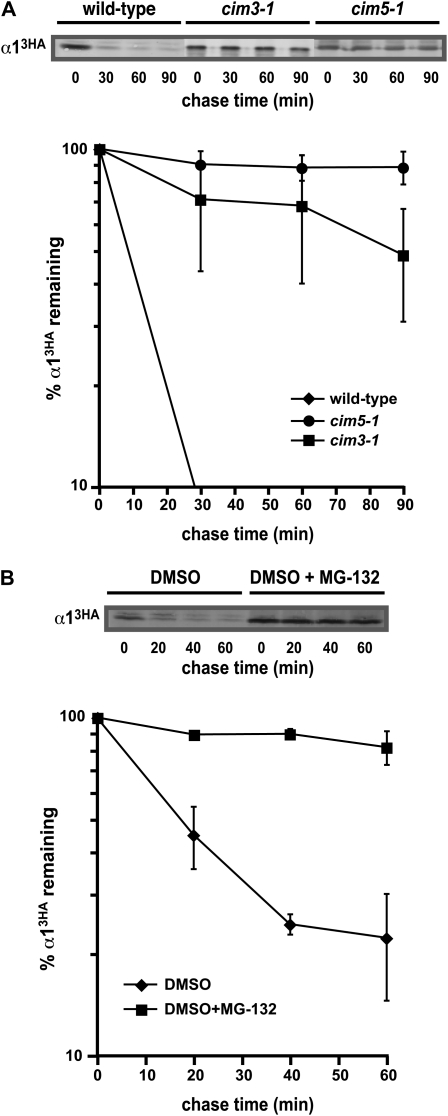
The α1 protein is short-lived and degraded by the ubiquitin–proteasome pathway. (A) A representative cycloheximide-chase assay of α13HA turnover in wild-type, cim3-1, and cim5-1 strains is shown at the top, and the quantitative results of multiple experiments are shown below. In these experiments, the strains were shifted to 37° for 1 hr prior to the cycloheximide-chase assay and maintained at this temperature throughout the assay. The amount of α13HA present at the given chase time was expressed relative to that observed at time zero and replicates of multiple experiments were averaged. (B) A representative cycloheximide-chase assay of α13HA turnover in a pdr5Δ strain treated with 25 μg/ml of MG-132 or the solvent DMSO is shown at the top, and the quantitative results of multiple experiments are shown below. These experiments were performed at 30°.
Quantitative Western blotting:
Immunoblots were probed with 1:1000 dilution of α-HA antibody (12CA5, Roche) overnight at 4° followed by an hour-long room temperature incubation with a Qdot 605-labeled α-mouse secondary antibody (Invitrogen, Carlsbad, CA) diluted 1:1000 in Seablock (Pierce, Rockford, IL). After washing, proteins on the membrane were visualized on a Typhoon 9410 variable mode imager and analyzed using ImageQuant software.
Purification of α1-His6Ub conjugates:
p414TEF-α1-3HA or p414TEF-αl and p425CUP1-His6-Ub or p425CUP1-Ub were transformed into MHY501, JY768, JY772, and JY774 as appropriate. Purification of His6Ub conjugates with Ni-NTA agarose was performed as described (Kaiser and Tagwerker 2005) with the following alterations. Strains grown at 30° in synthetic medium (0.17% yeast nitrogen base without ammonium sulfate) supplemented with 0.003% SDS, 0.1% proline, appropriate amino acids, 2% glucose as the carbon source (Liu et al. 2007), and 100 μm CuSO4 were added for 4 hr to induce expression of His6-Ub. MG-132 (Enzo Life Sciences) was added to 25 μg/ml and the cultures were incubated for an additional 60 min. Cells were lysed with glass beads in urea lysis buffer containing 10 mm imidazole and the conjugate-bound beads were washed in urea wash buffer containing 30 mm imidazole. Purified material and the unpurified lysates were analyzed by α-HA and α-Pgk1 Western blotting as described above. Similar results were observed with purifications from cells expressing α1-3HA from the ADH1 promoter.
RESULTS
The α1 protein is short-lived and degraded by the ubiquitin–proteasome pathway:
To determine if α1 is a short-lived protein targeted to the UPS for rapid proteolysis, we used cycloheximide-chase assays to examine the stability of α1 in yeast cells with loss-of-function mutations in proteasomal components. Detection of α1 was facilitated by expression from the constitutive ADH1 promoter and by the addition of three copies of the HA epitope sequence to the C terminus; this latter alteration had no effect on the functional properties of the protein since expression of α1-3HA from either the ADH1 or the α1 promoter fully complemented the mating defect of an α1Δ mutant strain. Like many transcription factors, α13HA is rapidly degraded in wild-type cells; α13HA has an in vivo half-life of ∼15 min at 30° (Figure 1 and supporting information, Figure S1). In strains containing the cim3-1 allele of the RPT6 gene that encodes a subunit of the 19S proteasomal regulatory particle, α13HA turnover was strongly impaired (Figure 1A). The α13HA protein was similarly very stable in cim5-1 strains that contain a mutation in RPT1, another 19S subunit (Figure 1A). Furthermore, α13HA turnover was inhibited ∼2-fold in rpn13Δ cells, which contain a deletion of a nonessential subunit that acts as one of multiple ubiquitin receptors for the proteasome (Figure S1). We also examined if the catalytic activity of the proteasome was necessary for α13HA degradation by treating cells with the proteasome inhibitor MG-132. Under these conditions, the α13HA protein was stable (Figure 1B). Together, these data indicate that the degradation of the α1 transcription factor requires the function of the 26S proteasome.
The degradation of α1 requires multiple ubiquitin-conjugating enzymes:
Since substrates of the 26S proteasome are typically modified by ubiquitin, we expected that α1 turnover would be impaired in strains lacking the enzymatic machinery necessary to transfer ubiquitin to a short-lived proteasomal substrate like α1. Saccharomyces cerevisiae contains 11 different ubiquitin-conjugating (Ubc) enzymes that function in ubiquitin transfer. To identify the Ubc(s) required for the rapid proteolysis of α1, we measured the rate of α13HA turnover in strains deficient for each of the UBC genes that encode a ubiquitin-conjugating enzyme. Degradation assays were performed with deletion mutant strains for the 10 nonessential UBC genes, while cells containing a temperature-sensitive allele of the sole essential UBC gene CDC34/UBC3 were examined at the nonpermissive temperature. Although the degradation of many substrates of the UPS strongly depends on a single specific UBC gene (Dohmen et al. 1991; Schwob et al. 1994; Johnson et al. 1995; Drury et al. 1997; Hampton and Bhakta 1997; Henchoz et al. 1997; Gardner et al. 2005), we found that α13HA was only partially stabilized in an ubc13Δ strain (Figure 2A) and that ubc13Δ was the only single mutant that had any detectable alteration in α1 turnover (Figure 2B and data not shown). Moreover, the identification of UBC13 was unexpected, since its best-characterized function is in a ubiquitin-dependent but protein degradation-independent process. In this functional role, Ubc13 forms a heterodimeric complex with the ubiquitin-conjugating enzyme variant (UEV) Mms2 to assemble noncanonical (lysine 63-linked) ubiquitin chains for signaling during DNA repair processes (Hofmann and Pickart 1999; Brusky et al. 2000; Ulrich and Jentsch 2000). To determine if UBC13 functions to promote α1 turnover in a manner similar to its role in DNA repair, we analyzed the degradation of α13HA in mms2Δ cells. Interestingly, no proteolytic defect was observed (Figure 2A). From these results, we conclude that the function of UBC13 in promoting α13HA proteolysis is independent of MMS2 and their role in assembling lysine 63-linked ubiquitin chains for signaling.
The approach of analyzing single mutations in UBC genes may be problematic, since important but functionally redundant ubiquitin-conjugation pathways required for α13HA turnover could be overlooked. One potential redundant pathway is that controlled by UBC4 or UBC5, since the proteins encoded by these genes have nearly identical sequence and are functionally interchangeable (Seufert and Jentsch 1990). Because of this possibility for redundancy, we analyzed α13HA degradation in ubc4Δ ubc5Δ strains and found that α13HA was strongly stabilized in the double mutant (Figure 2B). The UBC4 and UBC5 genes are transcribed in an approximately reciprocal pattern, with UBC4 expressed in the exponential phase of growth and UBC5 expressed in stationary phase. Because of this pattern of expression, the degradation of many short-lived proteins that require a UBC4/UBC5-dependent ubiquitination pathway is impaired in singly mutant ubc4Δ strains (Johnson et al. 1992; Chen et al. 1993; Rodrigo-Brenni and Morgan 2007). In contrast, α13HA turnover proceeded normally in the absence of UBC4 (Figure 2B), suggesting that the small amount of UBC5 expressed in exponentially growing cultures is sufficient to support the rapid degradation of α13HA. Thus, the cumulative results of these experiments indicate that the normal rapid rate of α1 degradation in vivo requires at least three different UBC genes: UBC4, UBC5, and UBC13.
Multiple ubiquitin ligases are necessary for the rapid degradation of α1:
By sequence criteria alone—that is, the presence of motifs characteristic of ubiquitin ligases—numerous factors exist in budding yeast that may function in this capacity. To find ubiquitin ligases that could promote α1 turnover, we surveyed the literature for factors that function with UBC4/5 or UBC13 in known ubiquitination pathways. As detailed below, a number of candidate ligases were identified for each UBC and their involvement in α13HA proteolysis was determined by examining the stability of α13HA in strains lacking these factors with cycloheximide-chase degradation experiments.
UBC13-dependent ligases:
The chromatin-associated RING domain protein Rad5 cooperates with Ubc13 during the repair of DNA damage, and the two proteins make physical interactions (Ulrich and Jentsch 2000; Torres-Ramos et al. 2002). To determine if α13HA turnover requires the function of RAD5, we analyzed the degradation of α13HA in rad5Δ cells and observed no defect (Figure S2). This result was not unexpected, given that Mms2 is an essential component of the Ubc13-Rad5 complex and our finding that MMS2 does not have a role in α13HA proteolysis (Figure 2A). In fact, the degradation rate of α13HA appears to be modestly enhanced in both mms2Δ and rad5Δ cells (Figure 2A and Figure S2), suggesting that functional quantities of Ubc13 may be limiting for α13HA turnover and that the ubiquitin ligase important for the degradation of α13HA competes with Mms2 and Rad5 for interaction with Ubc13. Consistent with this hypothesis, Ubc13 strongly interacts with Mms2, such that a pool of Ubc13 independent of the UEV is not thought to exist in vivo (Hofmann and Pickart 1999; McKenna et al. 2001, 2003).
In vertebrate cells, the RING domain ubiquitin ligase Rnf8 also interacts with Ubc13 (Plans et al. 2006), where it appears to recruit Ubc13 independently of its UEV partners to sites of DNA damage (Huen et al. 2008). Rnf8 also contains a forkhead-associated (FHA) domain, and two FHA-RING proteins exist in S. cerevisiae (encoded by DMA1/CHF1 and DMA2/CHF2) (Brooks et al. 2008). The UEV-independent activity of UBC13 in promoting α13HA turnover prompted us to determine if these genes play a role in the degradation of α13HA. Simultaneous deletion of DMA1 and DMA2 stabilized α13HA approximately threefold, indicating that these FHA-RING factors are involved in α13HA proteolysis (Figure 3). Interestingly, these two related factors do not functionally substitute for one another, since α13HA turnover is similarly inhibited approximately threefold in either a dma1Δ or a dma2Δ strain (Figure 3). Thus, DMA1 and DMA2 each make unique contributions to α13HA proteolysis. Although we identified Dma1 and Dma2 as candidates that function with UBC13, recent genetic and biochemical evidence indicates that both of these RING ligases interact with Ubc4 as well as Ubc13, suggesting that these factors may have dual specificity for different ubiquitin-conjugating enzymes (Loring et al. 2008).
UBC4/5-dependent ligases:
In addition to Dma1 and Dma2, a number of other ubiquitin ligases also utilize Ubc4/5 to promote the ubiquitination of protein substrates. Among these are two different HECT domain proteins, Ufd4 and Rsp5 (Johnson et al. 1995; Rotin et al. 2000; Horak and Wolf 2001). Ufd4 was initially identified as a component required for the ubiquitin fusion degradation (UFD) proteolytic pathway that recognizes engineered fusion proteins containing a “nonremovable” ubiquitin moiety at the N terminus (Johnson et al. 1995). In addition, Ufd4 targets unassembled Ubc7, a ubiquitin-conjugating enzyme involved in endoplasmic reticulum-associated degradation (Ravid and Hochstrasser 2007), as well as Rad4 and Mgt1, two physiological substrates involved in DNA repair (Ju and Xie 2006; Hwang et al. 2009). To examine if Ufd4 plays a similar role in promoting α1 proteolysis, we disrupted UFD4 in cells and analyzed the stability of α13HA with cycloheximide-chase assays. Figure 4A shows that α13HA turnover was inhibited approximately fourfold in ufd4Δ cells, implicating UFD4 in the degradation of α13HA. In contrast, α13HA proteolysis was not impaired in temperature-sensitive rsp5-2 loss-of-function mutants, demonstrating that the destruction of α13HA is independent of RSP5 function (Figure 4A).
Recently, a family of ubiquitin ligases that target proteins modified by the small ubiquitin-related modifier (SUMO) has been described (Perry et al. 2008). In S. cerevisiae, the SUMO-targeted ligases Ris1 and Slx5–Slx8 appear to utilize Ubc4/5 to conjugate ubiquitin to SUMO-modified protein substrates in vitro and in vivo (Uzunova et al. 2007; Xie et al. 2007; Mullen and Brill 2008; Wang and Prelich 2009). Therefore, we determined if either of these SUMO-targeted ligases was necessary for rapid turnover of α13HA. We were particularly interested in RIS1, since previous studies implicated this factor in mating-type switching (Zhang and Buchman 1997). However, disruption of RIS1 had little effect on the stability of α13HA (the degradation rate was increased less than twofold; Figure S3), whereas α13HA proteolysis was clearly defective in yeast cells lacking Slx5–Slx8 (Figure 4B). Consistent with the observation that these two RING domain proteins interact and function as a heterodimer (Yang et al. 2006), the turnover of α13HA was similarly impaired in singly mutant slx5Δ and slx8Δ strains and in the slx5Δ slx8Δ double mutant (Figure 4B). These data indicate that SLX5 and SLX8 are required for the rapid degradation of α1.
Interestingly, ubiquitin-dependent degradation of the α2 repressor also requires Slx5–Slx8 (Y. Xie and M. Hochstrasser, personal communication), suggesting that the turnover of these two determinants of the α-cell type is coordinately regulated. The rapid proteolysis of α2 also requires another ubiquitination pathway that utilizes the RING domain ligase Doa10 (Swanson et al. 2001), so we asked if DOA10 was also necessary for the degradation of α13HA. Although α13HA turnover depends on the activity of Slx5–Slx8, it does not require Doa10 (Figure S4). Thus, only some of the proteolytic pathways that act on α1 and α2 are shared.
In summary, the degradation of α1 in vivo requires the activity of at least four different ubiquitin ligases, Dma1, Dma2, Ufd4, and Slx5–Slx8.
Genetic interactions among the α1 degradation pathways:
These four ubiquitination enzymes may each define a separate degradation pathway that functions independently to promote the rapid proteolysis of α1. Alternately, two or more of these ligases may be part of the same α1 proteolytic pathway, whereby they function together to target α1 for rapid degradation. To distinguish between these ideas, we performed epistasis analysis and determined the metabolic stability of α13HA in strains containing multiple disruptions of genes encoding these ubiquitin ligases. Our expectation was that if two or more ubiquitin ligases function in separate degradation pathways for α1, then the simultaneous disruption of the corresponding genes should exhibit a more severe defect in α13HA proteolysis than any of the single mutants alone.
Consistent with this expectation, the degradation rate of α13HA was strongly impaired (more than eightfold) in cells lacking both Ufd4 and Dma1 (Figure 5A). These results contrast with the three- to fourfold stabilization of α13HA in singly mutant ufd4Δ or dma1Δ strains (Figures 3B and 4A), indicating that these two ligases function in distinct α1 degradation pathways. Interestingly, analysis of the complete set of epistasis tests with the other ligase mutations indicates more complex genetic interactions among these ubiquitination factors. For example, the inhibition of α13HA proteolysis in ufd4Δ slx5Δ cells is no greater than that observed in either single mutant (compare Figures 5A and 4). This result suggests that UFD4 and SLX5 function in the same α1 degradation pathway, in that disruption of either ligase alone is already sufficient to inactivate this pathway. However, the turnover of α13HA in a dma1Δ slx5Δ strain is also equivalent to the α13HA degradation rate in singly mutant dma1Δ or slx5Δ cells (Figures 5A, 3B, and 4B), implying that SLX5 participates in the same pathway as DMA1. Thus, SLX5 appears to function not only in the proteolytic pathway defined by UFD4, but also in the pathway defined by DMA1. Similar results were observed in epistasis tests with dma2Δ mutations (Figure 3B and data not shown), in that DMA2 seems to be a member of both the UFD4 and the DMA1 epistasis groups. Since DMA2 and SLX5–SLX8 appear to function in both degradation pathways, it is surprising that the effects of disrupting these factors on α13HA degradation are not more severe than the approximately threefold stabilization that is observed. The strength of this phenotypic defect could indicate that the function of DMA2 and SLX5–SLX8 is of secondary importance to α1 degradation, with UFD4 and DMA1 playing primary roles in promoting the turnover of α1. An alternative interpretation is that DMA2 and SLX5–SLX8 have multiple, complex functions in the α1 degradation pathway and that one of these role(s) is antagonistic to its other function(s). In this scenario, the disruption of these factors would lead to the loss of all functions, and the deficiency of the antagonistic roles could compensate for one another and mask an important overall function in α1 degradation.
Indeed, analysis of triple-mutant combinations revealed that in addition to promoting α13HA turnover, DMA2 also functions as a negative regulator of α13HA proteolysis. This conclusion follows from the surprising observation that combining the dma2Δ mutation with ufd4Δ dma1Δ resulted in strong suppression of the ufd4Δ dma1Δ proteolytic defect; the degradation of α13HA was impaired only approximately twofold in ufd4Δ dma1Δ dma2Δ cells, a modest inhibition relative to the more than eightfold effect of the ufd4Δ dma1Δ mutant (compare ufd4Δ dma1Δ dma2Δ in Figure 5B to ufd4Δ dma1Δ in Figure 5A). This effect was specific to dma2Δ mutations, since ufd4Δ dma1Δ combinations with slx5Δ or slx8Δ resulted in triply mutant cells (ufd4Δ dma1Δ slx5Δ or ufd4Δ dma1Δ slx8Δ) with comparable α13HA degradation defects to the doubly mutant ufd4Δ dma1Δ strain (Figure 5, A and B, and data not shown). One hypothesis that could explain the suppression effects of the dma2Δ mutation, and therefore the negative role of DMA2 in α13HA turnover, is that the Dma2 ubiquitin ligase not only functions to promote the degradation of α1, but also targets another member of the α1 degradation pathway. In this scenario, the other component that functions in α13HA turnover would be stabilized in dma2Δ cells, its cellular activity would accumulate, and the rate of α13HA proteolysis would increase. A candidate for such a component is Slx5–Slx8, since this ubiquitin ligase is the only characterized ligase implicated in α1 degradation remaining in the ufd4Δ dma1Δ dma2Δ triple mutant. To examine this hypothesis, we determined the rate of α13HA turnover in an ufd4Δ dma1Δ dma2Δ slx5Δ strain, since one would predict that disrupting SLX5 would ameliorate the dma2Δ suppression effect if the activity of Slx5–Slx8 were increased in the absence of DMA2. The results of cycloheximide-chase experiments are consistent with this prediction: the inhibitory role of DMA2 on α13HA turnover was no longer apparent and the strong inhibition of α13HA proteolysis was restored in quadruply mutant cells (Figure 5C). Together, these findings highlight the complexity of interactions among the ligases implicated in α1 turnover and suggest that cross-regulation among these multiple ubiquitin ligases may play an important role in regulating the α1 degradation pathways.
The α1 protein is ubiquitinated in vivo by multiple ubiquitin ligases:
The rapid degradation of α13HA requires the general function of the UPS and the activity of multiple specific ubiquitin ligase pathways, suggesting that α13HA was modified directly by ubiquitin. To examine this idea, we expressed hexahistidine-tagged ubiquitin in cells and affinity-purified ubiquitinated proteins from cell lysates and determined if α13HA was present in this purified material by immunoblotting. The results of these experiments revealed a number of discrete bands as well as smears of reactivity (Figure 6, lane 3). All of this immunoreactivity was absent in strains expressing untagged α1 or untagged ubiquitin (Figure 6, lanes 1 and 2), demonstrating the specificity of the assay for ubiquitinated α13HA. Furthermore, the immunoblotting signals representing ubiquitinated α13HA were enhanced when the activity of the proteasome is impaired (data not shown), a condition known to increase the steady-state levels of ubiquitinated proteins. Taken together, these results indicate that α1 is ubiquitinated in vivo.
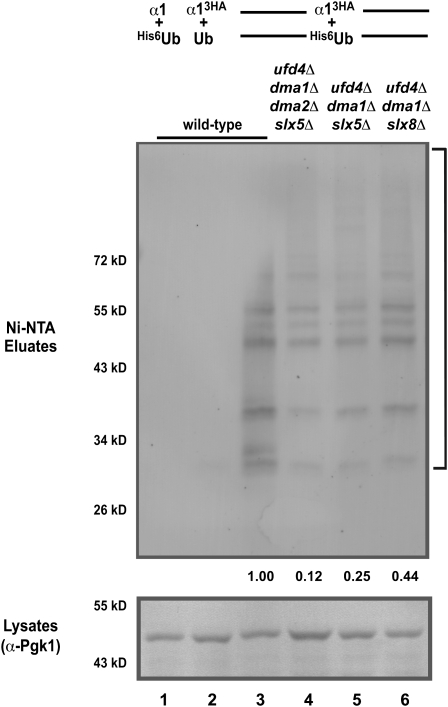
Figure 6.—
The α1 protein is ubiquitinated and this modification requires multiple ubiquitin ligases. Ubiquitinated proteins from lysates of cells of the indicated genotypes expressing His6-ubiquitin (His6-Ub) or untagged ubiquitin (Ub) and HA-tagged or untagged α1 were purified by binding to Ni-NTA beads. The affinity-purified material was analyzed for the presence of α13HA by α-HA immunoblotting (top). The unpurified lysates were analyzed by immunoblotting with α-Pgk1 (bottom) in parallel. The bracket (top) indicates ubiquitinated α13HA and the positions of molecular weight markers are given on the left side of the gel. Note the enrichment of ubiquitinated α13HA in cells expressing the tagged forms of both α1 and Ub (compare lane 3 to lanes 1 and 2) and the reduced levels of ubiquitinated α13HA in strains lacking the ubiquitin ligases Ufd4, Dma1, Dma2, and Slx5–Slx8 (compare lanes 4–6 to lane 3). Equal amounts of protein were loaded in each lane and proteins were equivalently transferred, as judged by the levels of Pgk1 in the unpurified lysates. Numbers indicate the relative amount of ubiquitinated α13HA after normalizing to the levels of Pgk1.
Since the rapid turnover of α13HA requires the activity of Ufd4, Dma1, Dma2, and Slx5–Slx8, we examined the function of these ubiquitin ligases in the ubiquitination of α13HA. In strains lacking all four ligases, α13HA ubiquitination was significantly reduced (Figure 6, compare lane 4 to lane 3). The level of ubiquitinated α13HA was also reduced in triple mutants that are deficient in Ufd4, Dma1, and Slx5–Slx8 (Figure 6, lanes 5 and 6). However, similar analyses of strains singly mutant for the various ubiquitin ligases showed little, if any, reduction in the ubiquitination of α13HA (data not shown). Thus, in cells, the stability of α13HA correlates with the level of α13HA ubiquitination: strains in which α13HA is strongly stabilized have reduced levels of α13HA-ubiquitin conjugates.
An intact SUMO pathway is necessary for the rapid degradation of α1:
Like that of α13HA, the ubiquitin-dependent turnover of the Mot1-301 mutant protein requires SLX5 and SLX8. Consistent with these genes encoding a SUMO-targeted ubiquitin ligase, the degradation of Mot1-301 also requires the components of the SUMO conjugation pathway (Wang et al. 2006; Wang and Prelich 2009). To determine if α1 turnover similarly required an intact sumoylation pathway, we analyzed the kinetics of α13HA degradation in strains containing temperature-sensitive alleles of UBA2, UBC9, and ULP1. These genes encode a subunit of the SUMO-activating enzyme, the SUMO-conjugating enzyme, and a SUMO protease, respectively. In uba2-ts9, ubc9-1, and ulp1-333 cells, α13HA turnover was modestly but reproducibly inhibited (Figure 7A). We further explored the role of the SUMO pathway in α1 degradation by examining mutants in SUMO ligases. Siz1 and Siz2 are homologous proteins that account for a large majority of the total sumoylation in yeast (Johnson and Gupta 2001). Strains that are deficient for both of these factors degrade α13HA with impaired kinetics, similar to strains lacking the Slx5–Slx8 SUMO-targeted ubiquitin ligase (Figure 7B). These results indicate that an intact sumoylation pathway is necessary for the rapid turnover of α13HA. Consistent with this finding, we have observed that mutations in MMS21, a gene encoding another SUMO ligase (Zhao and Blobel 2005), also inhibit α13HA degradation. Degradation assays with strains that contain the mms21-ΔRING allele, which lack the C-terminal RING domain essential for SUMO ligase activity, demonstrate that α13HA is also stabilized in the absence of this SUMO ligase (Figure 7B).
Like ubiquitin, SUMO can form polymeric chains on proteins, with chain assembly occurring through the C terminus of SUMO and the ɛ-amino group of lysine residues in the neighboring SUMO molecule (Ulrich 2008). Intriguingly, polysumoylated substrates are the preferred targets of Slx5–Slx8-dependent ubiquitination in vitro (Mullen and Brill 2008). These findings raise the question of whether the ability to form SUMO chains is necessary for the rapid turnover of α1. To address this question, we measured the kinetics of α13HA turnover in strains whose sole copy of SMT3, the gene encoding SUMO, contains arginine substitutions of all the lysine residues in the Smt3 protein. In these smt3-allR strains, α13HA is stabilized, indicating that polysumoylation is necessary for the rapid degradation of α1 (Figure 7C). Since the three lysine residues in the N terminus of Smt3 are used preferentially in the formation of polymeric chains (Bylebyl et al. 2003), we examined whether the smt3-3NR mutant, which contains arginine substitutions of the N-terminal lysines, also inhibited α13HA turnover. In this mutant strain, we observed that α13HA was degraded normally (Figure S5). These findings indicate that while poly-SUMO chain formation is necessary for degradation of α1, chains formed through the three N-terminal lysines are not absolutely required. These results are consistent with an elegant biochemical study using a variety of Smt3 lysine-to-arginine mutants in SUMO-dependent ubiquitination reactions catalyzed by Slx5–Slx8, which demonstrate that lysines other than the three located in the N terminus can be used to form poly-SUMO chains on a model substrate and that these “alternative” SUMO chains are productively recognized and ubiquitinated by Slx5–Slx8 (Mullen and Brill 2008).
A degradation signal in α1 targeted by Slx5–Slx8:
Most short-lived proteins contain sequence or structural determinants that are targeted by the ubiquitin-conjugation machinery (Ravid and Hochstrasser 2008). We sought to define such a degron in α1 and connect this signal to a proteolytic pathway important for the rapid turnover of this protein to ensure its biological relevance. We focused initially on identifying an Slx5–Slx8-dependent signal and took advantage of the transferability of degrons to map this signal. As a proof-of-principle for this approach, we first fused full-length α1 to the normally stable protein Ura3. Like the α13HA that is not part of a Ura3 fusion protein, α1-Ura3 is a metabolically unstable protein whose turnover is dependent upon Slx5–Slx8 (Figure 8, A and B).
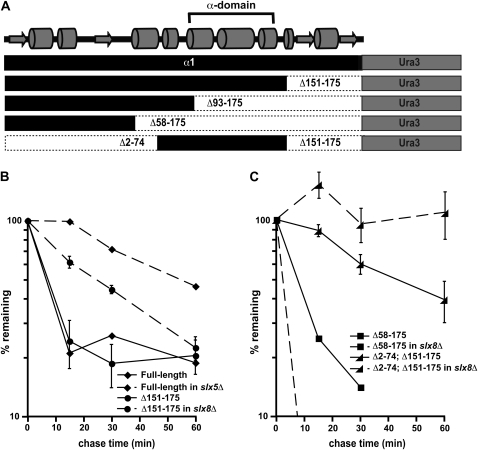
Figure 8.—
The α-domain acts as a Slx5–Slx8-dependent degradation signal. (A) Diagram of the α1-Ura3 fusion proteins analyzed for this study. Shown at the top is a predicted secondary structure of α1, including the position of the conserved α-domain. (B) Cycloheximide-chase analysis of full-length α1-Ura3 turnover at 30° in wild-type and slx5Δ cells and the Δ151–175 derivative of α1-Ura3 in wild-type and slx8Δ cells. (C) Cycloheximide-chase analysis of the Δ58–175 derivative of α1-Ura3 turnover at 30° in wild-type and slx8Δ cells and the Δ2–74; Δ151–175 derivative of α1-Ura3 in wild-type and slx8Δ cells. Note that the results of experiments with the Δ93–175 derivative of α1-Ura3 are not shown because this protein is undetectable on immunoblots of wild-type and slx8Δ cells, suggesting that the Δ93–175-Ura3 protein is very unstable in both of these strains. This protein fusion is expressed in wild-type cells, since radiolabeled protein can be detected by pulse labeling cells with Tran35S-Label. Pulse-chase analysis at 30° measuring the stability of the Δ93–175-Ura3 protein indicate a half-life of ∼2 min.
With evidence of a transferable Slx5–Slx8-dependent degradation signal in hand, we attempted to identify the degron by constructing a series of α1 deletion mutants on the basis of secondary structure predictions of the protein (Figure 8A). Removing a small 25-amino-acid region (Δ151–175), an ∼80-amino-acid region (Δ93–175), or a larger ∼120-amino-acid region (Δ58–175) from the C terminus of α1 did not alter the instability of these α1-Ura3 fusions (Figure 8, B and C, and data not shown). However, the degradation of Δ151–175 remained dependent upon Slx5–Slx8, while the turnover of Δ93–175 and Δ58–175 was not impaired by the loss of this E3 ligase (Figure 8, B and C, and data not shown) and therefore must be targeted by another ubiquitination pathway. We interpret these results to indicate that the region between amino acids 92 and 150 of α1 is necessary for Slx5–Slx8-dependent degradation. Notably, the α-domain, a region strongly conserved in fungi but of unknown function (Butler et al. 2004), lies within this ∼60-amino-acid stretch of the protein. To directly determine if the α-domain served as a Slx5–Slx8-dependent degradation signal, we expressed amino acids 75–150 of α1 as a fusion to Ura3 and determined its stability. Although the α-domain fusion is more stable than the full-length α1-Ura3 protein, it is degraded with a half-life of ∼40 min, indicative of a short-lived protein. Remarkably, the degradation of the α-domain-Ura3 fusion was wholly dependent upon Slx5–Slx8, demonstrating that the α1 domain is sufficient to target Ura3 for destruction by this ubiquitination pathway (Figure 8C).
DISCUSSION
The degradation of α1—multiple interacting proteolytic pathways, one of which overlaps with that regulating the turnover of α2:
As one of the master regulators of the α-cell type, the α1 protein must be swiftly and efficiently inactivated to allow a productive transition of cell identity following mating-type switching. Taken together, our observations implicate the UPS as the central regulator that ensures α1 inactivation by targeting this factor for degradation and indicate that multiple interacting proteolytic pathways act upon α1 to facilitate its rapid destruction. The genetic analyses presented here do not allow us to discriminate between direct and indirect effects, but an economical interpretation of our findings is that at least some of these degradation pathways function to directly catalyze the formation of α1-ubiquitin conjugates and thereby promote its proteasome-mediated degradation. Indeed, α13HA is covalently modified by ubiquitin in vivo, and multiple ubiquitin ligase pathways contribute to the levels of ubiquitinated α13HA in cells. Further insights into the mechanism of α1 turnover await a more detailed biochemical characterization of the proteolytic pathways defined here.
These α1 degradation pathways are defined by multiple ubiquitin-conjugating enzymes and ubiquitin ligases, and the connections between these factors cannot be fully discerned through epistasis analysis. Previous studies have also revealed complex effects on the kinetics of substrate degradation when distinct ubiquitination pathways interact (Chen et al. 1993; Helliwell et al. 2001; Hoppe et al. 2004; Richly et al. 2005; Rodrigo-Brenni and Morgan 2007). The mechanisms by which these interactions alter turnover rates are varied. For example, the conjugation of multiple ubiquitin moieties to a substrate is required for efficient targeting to the proteasome (Pickart 2000; Thrower et al. 2000), and this minimal ubiquitin density may be achieved through the extension of an initial ubiquitination event into a polymeric chain (Koegl et al. 1999; Petroski and Deshaies 2005; Rodrigo-Brenni and Morgan 2007) or, alternatively, through multiple distinct ubiquitination events on the same substrate (Kravtsova-Ivantsiv et al. 2009). The need to build sufficient ubiquitin density on α1 for proteasomal targeting could explain our identification of four ubiquitin ligases, which may have hierarchical effects on α1 turnover. On the other hand, ubiquitination pathways may also influence one another through interactions that are independent of the substrate, such as the direct targeting of the ubiquitination factors themselves for destruction by the UPS (Loring et al. 2008). Such a mechanism could explain our observations of a negative role for DMA2 in the turnover of α1 by the UFD4 and DMA1 pathways. Whatever the mechanisms involved, the regulated turnover of α1 must reflect the need to retain sufficient levels of the protein to define the α-state, while at the same time ensuring that α1 activity can be rapidly removed upon the transition to the a cell type following mating-type switching.
How can these complex genetic pathways that regulate the efficient turnover of α1 be deconvoluted? We took the most parsimonious interpretation of our genetic analysis of α1 degradation—that one of the identified pathways leads to direct ubiquitination of α1—to define a discrete and transferable degradation signal within the protein and reduce the genetic complexity of the targeting pathways. Using this approach, we have found that one pathway regulating the destruction of α1 recognizes the conserved α-domain of the protein. The turnover directed by this signal is genetically linked to the ubiquitin ligase Slx5–Slx8. Interestingly, Slx5–Slx8 has nonspecific DNA-binding activity and has been implicated in the ubiquitin-dependent degradation of α2, the other master regulator of the α-cell type (Yang et al. 2006; Y. Xie and M. Hochstrasser, personal communication). The commonality of this pathway in the destruction of two DNA-binding regulators that specify the α-phenotypic state raises the intriguing possibility of coordinated destruction of the functionally engaged forms of these transcription factors.
Acknowledgments
We gratefully acknowledge Yang Xie and Mark Hochstrasser for sharing data and reagents prior to their publication and for many helpful discussions. We thank Mark Hochstrasser and Erica Johnson for providing yeast strains, Mark Grabiner and Robin Brese for technical assistance, and Tricia Serio for numerous helpful discussions and her help in the preparation of the manuscript. The manuscript was improved by comments from Tricia Serio and Alec DeSimone. This work was supported by a grant from the National Institutes of Health (GM071764 to J.D.L.).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.115907/DC1.
References
- Ang, X. L., and J. W. Harper, 2004. Interwoven ubiquitination oscillators and control of cell cycle transitions. Sci. STKE 2004 pe31. [DOI] [PubMed] [Google Scholar]
- Brooks, 3rd, L., E. G. Heimsath, Jr., G. L. Loring and C. Brenner, 2008. FHA-RING ubiquitin ligases in cell division cycle control. Cell Mol. Life Sci. 65 3458–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusky, J., Y. Zhu and W. Xiao, 2000. UBC13, a DNA-damage-inducible gene, is a member of the error-free postreplication repair pathway in Saccharomyces cerevisiae. Curr. Genet. 37 168–174. [DOI] [PubMed] [Google Scholar]
- Butler, G., C. Kenny, A. Fagan, C. Kurischko, C. Gaillardin et al., 2004. Evolution of the MAT locus and its Ho endonuclease in yeast species. Proc. Natl. Acad. Sci. USA 101 1632–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylebyl, G. R., I. Belichenko and E. S. Johnson, 2003. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J. Biol. Chem. 278 44113–44120. [DOI] [PubMed] [Google Scholar]
- Chen, P., P. Johnson, T. Sommer, S. Jentsch and M. Hochstrasser, 1993. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell 74 357–369. [DOI] [PubMed] [Google Scholar]
- Cole, C., J. D. Barber and G. J. Barton, 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36 W197–W201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen, R. J., K. Madura, B. Bartel and A. Varshavsky, 1991. The N-end rule is mediated by the UBC2(RAD6) ubiquitin-conjugating enzyme. Proc. Natl. Acad. Sci. USA 88 7351–7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury, L. S., G. Perkins and J. F. Diffley, 1997. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 16 5966–5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, R. G., Z. W. Nelson and D. E. Gottschling, 2005. Degradation-mediated protein quality control in the nucleus. Cell 120 803–815. [DOI] [PubMed] [Google Scholar]
- Hampton, R. Y., and H. Bhakta, 1997. Ubiquitin-mediated regulation of 3-hydroxy-3-methylglutaryl-CoA reductase. Proc. Natl. Acad. Sci. USA 94 12944–12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell, S. B., S. Losko and C. A. Kaiser, 2001. Components of a ubiquitin ligase complex specify polyubiquitination and intracellular trafficking of the general amino acid permease. J. Cell Biol. 153 649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henchoz, S., Y. Chi, B. Catarin, I. Herskowitz, R. J. Deshaies et al., 1997. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 11 3046–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko, A., and A. Ciechanover, 1998. The ubiquitin system. Annu. Rev. Biochem. 67 425–479. [DOI] [PubMed] [Google Scholar]
- Herskowitz, I., 1989. A regulatory hierarchy for cell specialization in yeast. Nature 342 749–757. [DOI] [PubMed] [Google Scholar]
- Hicks, J. B., and I. Herskowitz, 1976. Interconversion of yeast mating types. I. Direct observations of the action of the homothallism (HO) gene. Genetics 83 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch, J. P., and F. R. Cross, 1993. The pheromone receptors inhibit the pheromone response pathway in Saccharomyces cerevisiae by a process that is independent of their associated G alpha protein. Genetics 135 943–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser, M., 1996. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30 405–439. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M., and A. Varshavsky, 1990. In vivo degradation of a transcriptional regulator: the yeast α2 repressor. Cell 61 697–708. [DOI] [PubMed] [Google Scholar]
- Hofmann, R. M., and C. M. Pickart, 1999. Noncanonical Mms2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96 645–653. [DOI] [PubMed] [Google Scholar]
- Hoppe, T., K. Matuschewski, M. Rape, S. Schlenker, H. D. Ulrich et al., 2000. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell 102 577–586. [DOI] [PubMed] [Google Scholar]
- Hoppe, T., G. Cassata, J. M. Barral, W. Springer, A. H. Hutagalung et al., 2004. Regulation of the myosin-directed chaperone UNC-45 by a novel E3/E4-multiubiquitylation complex in C. elegans. Cell 118 337–349. [DOI] [PubMed] [Google Scholar]
- Horak, J., and D. H. Wolf, 2001. Glucose-induced monoubiquitination of the Saccharomyces cerevisiae galactose transporter is sufficient to signal its internalization. J. Bacteriol. 183 3083–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen, M. S., J. Huang, J. Yuan, M. Yamamoto, S. Akira et al., 2008. Noncanonical E2 variant-independent function of UBC13 in promoting checkpoint protein assembly. Mol. Cell. Biol. 28 6104–6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, C. S., A. Shemorry and A. Varshavsky, 2009. Two proteolytic pathways regulate DNA repair by cotargeting the Mgt1 alkylguanine transferase. Proc. Natl. Acad. Sci. USA 106 2142–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, E. S., and A. A. Gupta, 2001. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106 735–744. [DOI] [PubMed] [Google Scholar]
- Johnson, E. S., B. Bartel, W. Seufert and A. Varshavsky, 1992. Ubiquitin as a degradation signal. EMBO J. 11 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, E. S., P. C. M. Ma, I. M. Ota and A. Varshavsky, 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270 17442–17456. [DOI] [PubMed] [Google Scholar]
- Johnson, P. R., R. Swanson, L. Rakhilina and M. Hochstrasser, 1998. Degradation signal masking by heterodimerization of MATalpha2 and MATa1 blocks their mutual destruction by the ubiquitin-proteasome pathway. Cell 94 217–227. [DOI] [PubMed] [Google Scholar]
- Ju, D., and Y. Xie, 2006. A synthetic defect in protein degradation caused by loss of Ufd4 and Rad23. Biochem. Biophys. Res. Commun. 341 648–652. [DOI] [PubMed] [Google Scholar]
- Jungmann, J., H.-A. Reins, C. Schobert and S. Jentsch, 1993. Resistance to cadmium mediated by ubiquitin-dependent proteolysis. Nature 361 369–371. [DOI] [PubMed] [Google Scholar]
- Kaiser, P., and C. Tagwerker, 2005. Is this protein ubiquitinated? Methods Enzymol. 399 243–248. [DOI] [PubMed] [Google Scholar]
- Koegl, M., T. Hoppe, S. Schlenker, H. D. Ulrich, T. U. Mayer et al., 1999. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96 635–644. [DOI] [PubMed] [Google Scholar]
- Kravtsova-Ivantsiv, Y., S. Cohen and A. Ciechanover, 2009. Modification by single ubiquitin moieties rather than polyubiquitination is sufficient for proteasomal processing of the p105 NF-kappaB precursor. Mol. Cell 33 496–504. [DOI] [PubMed] [Google Scholar]
- Kushnirov, V. V., 2000. Rapid and reliable protein extraction from yeast. Yeast 16 857–860. [DOI] [PubMed] [Google Scholar]
- Laney, J. D., and M. Hochstrasser, 2003. Ubiquitin-dependent degradation of the yeast Matα2 repressor enables a switch in developmental state. Genes Dev. 17 2259–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laney, J. D., and M. Hochstrasser, 2004. Ubiquitin-dependent control of development in Saccharomyces cerevisiae. Curr. Opin. Microbiol. 7 647–654. [DOI] [PubMed] [Google Scholar]
- Li, S. J., and M. Hochstrasser, 1999. A new protease required for cell-cycle progression in yeast. Nature 398 246–251. [DOI] [PubMed] [Google Scholar]
- Liu, C., J. Apodaca, L. E. Davis and H. Rao, 2007. Proteasome inhibition in wild-type yeast Saccharomyces cerevisiae cells. Biotechniques 42 158, 160, 162. [DOI] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, 3rd, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 953–961. [DOI] [PubMed] [Google Scholar]
- Loring, G. L., K. C. Christensen, S. A. Gerber and C. Brenner, 2008. Yeast Chfr homologs retard cell cycle at G1 and G2/M via Ubc4 and Ubc13/Mms2-dependent ubiquitination. Cell Cycle 7 96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna, S., L. Spyracopoulos, T. Moraes, L. Pastushok, C. Ptak et al., 2001. Noncovalent interaction between ubiquitin and the human DNA repair protein Mms2 is required for Ubc13-mediated polyubiquitination. J. Biol. Chem. 276 40120–40126. [DOI] [PubMed] [Google Scholar]
- McKenna, S., J. Hu, T. Moraes, W. Xiao, M. J. Ellison et al., 2003. Energetics and specificity of interactions within Ub.Uev.Ubc13 human ubiquitin conjugation complexes. Biochemistry 42 7922–7930. [DOI] [PubMed] [Google Scholar]
- Mullen, J. R., and S. J. Brill, 2008. Activation of the Slx5-Slx8 ubiquitin ligase by poly-small ubiquitin-like modifier conjugates. J. Biol. Chem. 283 19912–19921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg, D., R. Muller and M. Funk, 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156 119–122. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K., and D. Shore, 1987. Transcriptional regulation in the yeast life cycle. Science 237 1162–1170. [DOI] [PubMed] [Google Scholar]
- Oshima, Y., and I. Takano, 1971. Mating types in Saccharomyces: their convertibility and homothallism. Genetics 67 327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, J. J., J. A. Tainer and M. N. Boddy, 2008. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem. Sci. 33 201–208. [DOI] [PubMed] [Google Scholar]
- Petroski, M. D., and R. J. Deshaies, 2005. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell 123 1107–1120. [DOI] [PubMed] [Google Scholar]
- Pickart, C. M., 2000. Ubiquitin in chains. Trends Biochem. Sci. 25 544–548. [DOI] [PubMed] [Google Scholar]
- Pickart, C. M., 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70 503–533. [DOI] [PubMed] [Google Scholar]
- Pickart, C. M., and R. E. Cohen, 2004. Proteasomes and their kin: proteases in the machine age. Nat. Rev. Mol. Cell Biol. 5 177–187. [DOI] [PubMed] [Google Scholar]
- Pickart, C. M., and M. J. Eddins, 2004. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695 55–72. [DOI] [PubMed] [Google Scholar]
- Plans, V., J. Scheper, M. Soler, N. Loukili, Y. Okano et al., 2006. The RING finger protein RNF8 recruits UBC13 for lysine 63-based self polyubiquitylation. J. Cell Biochem. 97 572–582. [DOI] [PubMed] [Google Scholar]
- Qin, S., B. Nakajima, M. Nomura and S. M. Arfin, 1991. Cloning and characterization of a Saccharomyces cerevisiae gene encoding a new member of the ubiquitin-conjugating protein family. J. Biol. Chem. 266 15549–15554. [PubMed] [Google Scholar]
- Ravid, T., and M. Hochstrasser, 2007. Autoregulation of an E2 enzyme by ubiquitin-chain assembly on its catalytic residue. Nat. Cell Biol. 9 422–427. [DOI] [PubMed] [Google Scholar]
- Ravid, T., and M. Hochstrasser, 2008. Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 9 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly, H., M. Rape, S. Braun, S. Rumpf, C. Hoege et al., 2005. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 120 73–84. [DOI] [PubMed] [Google Scholar]
- Rodrigo-Brenni, M. C., and D. O. Morgan, 2007. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell 130 127–139. [DOI] [PubMed] [Google Scholar]
- Rotin, D., O. Staub and R. Haguenauer-Tsapis, 2000. Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J. Membr. Biol. 176 1–17. [DOI] [PubMed] [Google Scholar]
- Schwienhorst, I., E. S. Johnson and R. J. Dohmen, 2000. SUMO conjugation and deconjugation. Mol. Gen. Genet. 263 771–786. [DOI] [PubMed] [Google Scholar]
- Schwob, E., T. Böhm, M. D. Mendenhall and K. Nasmyth, 1994. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79 233–244. [DOI] [PubMed] [Google Scholar]
- Seufert, W., and S. Jentsch, 1990. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 9 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufert, W., J. P. McGrath and S. Jentsch, 1990. UBC1 encodes a novel member of an essential subfamily of yeast ubiquitin-conjugating enzymes involved in protein degradation. EMBO J. 9 4535–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufert, W., B. Futcher and S. Jentsch, 1995. A ubiquitin-conjugating enzyme involved in the degradation of both S- and M-phase cyclins. Nature 373 78–81. [DOI] [PubMed] [Google Scholar]
- Swanson, R., M. Locher and M. Hochstrasser, 2001. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 15 2660–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower, J. S., L. Hoffman, M. Rechsteiner and C. M. Pickart, 2000. Recognition of the polyubiquitin proteolytic signal. EMBO J. 19 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Ramos, C. A., S. Prakash and L. Prakash, 2002. Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 22 2419–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich, H. D., 2008. The fast-growing business of SUMO chains. Mol. Cell 32 301–305. [DOI] [PubMed] [Google Scholar]
- Ulrich, H. D., and S. Jentsch, 2000. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 19 3388–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzunova, K., K. Gottsche, M. Miteva, S. R. Weisshaar, C. Glanemann et al., 2007. Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 282 34167–34175. [DOI] [PubMed] [Google Scholar]
- VanDemark, A. P., and C. P. Hill, 2002. Structural basis of ubiquitylation. Curr. Opin. Struct. Biol. 12 822–830. [DOI] [PubMed] [Google Scholar]
- Wang, Z., and G. Prelich, 2009. Quality control of a transcriptional regulator by SUMO-targeted degradation. Mol. Cell. Biol. 29 1694–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., G. M. Jones and G. Prelich, 2006. Genetic analysis connects SLX5 and SLX8 to the SUMO pathway in Saccharomyces cerevisiae. Genetics 172 1499–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Y., O. Kerscher, M. B. Kroetz, H. F. McConchie, P. Sung et al., 2007. The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J. Biol. Chem. 282 34176–34184. [DOI] [PubMed] [Google Scholar]
- Yang, L., J. R. Mullen and S. J. Brill, 2006. Purification of the yeast Slx5-Slx8 protein complex and characterization of its DNA-binding activity. Nucleic Acids Res. 34 5541–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., and A. R. Buchman, 1997. Identification of a member of a DNA-dependent ATPase family that causes interference with silencing. Mol. Cell. Biol. 17 5461–5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., and G. Blobel, 2005. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA 102 4777–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]