Abstract
The development of the Drosophila melanogaster wing depends on the correct regulation of cell survival, growth, proliferation, differentiation, and pattern formation. These processes, and the genes controlling then, are common to the development of epithelia in many different organisms. To identify additional genes contributing to wing development we have carried out a genetic screen in mosaic wings carrying clones of homozygous mutant cells. We obtained 12 complementation groups corresponding to genes with a proven role in wing formation such as smoothened, thick veins, mothers against dpp, expanded, and fat and 71 new complementation groups affecting the pattern of veins and the size of wing. We mapped one of these groups to the mediator15 gene (med15), a component of the Mediator complex. We show that Med15 and other members of the Mediator complex are required, among other processes, for the transcription of decapentaplegic target genes.
THE Drosophila wing imaginal disc is an epithelium of undifferentiated cells that grows and becomes patterned during the larval period and that differentiates the fly wing during the pupal stage (Cohen 1993). The patterning of the wing disc involves the activities of several signaling pathways that act in collaboration with sequence-specific transcription factors to define cell fates (reviewed in de Celis 2003). First, the wing blade is specified as a domain of vestigial (vg) expression in the distal region of the wing disc by the activities of the Wingless (Wg), Epithermal growth factor receptor/Ras (EGFR/RAS), and Decapenteplegic (Dpp) signaling pathways (Williams et al. 1991; Kim et al. 1996). Later in development, the wing blade is subdivided into provein and intervein regions by the activities of the Dpp and Hedgehog (Hh) signaling pathways (reviewed in Bier 2000; de Celis 2003). Adjacent proveins are separated by broader “intervein” regions that correspond to domains of expression of the transcription factor Blistered (Bs) (Fristrom et al. 1994; Montagne et al. 1996; Roch et al. 1998). The expressions of vein-specific transcription factors in the proveins and Bs in the interveins regulate the expression of rhomboid (rho), leading to the generation of high levels of EGFR/RAS activity in these cells and to their differentiation as veins during pupal development (reviewed in Bier 2000; de Celis 2003).
Genetic screens have been instrumental in the identification of genes involved in the generation of a wing with a characteristic size and pattern of veins. In general, the imaginal discs are best suited for gain-of-function screens and for screenings carried out in sensitized genetic backgrounds, because these experiments can be done in heterozygous individuals. More conventional screens aiming to identify genes on the basis of their loss-of-function phenotype have not being frequently used, as most of the mutations of interest are likely to be homozygous lethal. These mutations can be identified only in mosaic animals, which with some exceptions (Garcia-Bellido and Dapena 1974) has prevented these experiments until the adoption of the FRT/FLP method to induce mitotic recombination (Xu and Harrison 1994). Since then, several loss-of-function screens have being reported in adult flies, using a heat-shock (hs) promoter to drive the expression of the FLP enzyme (Walsh and Brown 1998) or directing the expression of FLP to a particular domain of expression in the eye disc (Xu et al. 1995). In these cases, the mutations are identified in heterozygous animals bearing patches of homozygous cells induced by recombination between homologous FRT elements. These experiments allow the identification of genes required for imaginal development whose mutations are homozygous lethal.
We have adopted a variation of this method by combining FRT/FLP mitotic recombination with a source of FLP expressed in a broad domain of the wing blade and used a Minute mutation (M) to increase the proportion of homozygous mutants cells that otherwise might be eliminated due to their reduced viability. In our experimental setting, we induced mutations using ethyl nitrous urea (ENU) and selected heterozygous flies with a wing phenotype caused by the presence of M+ clones in the domain of expression of spalt (sal). In this experiment, carried out in an F1 generation, we isolated 140 mutations affecting the development of the wing. These mutants were classified in phenotypic classes, grouped in complementation groups, and then mapped to chromosomal intervals by complementation with a set of deficiencies covering 83% of the 2L arm. Among the complementation groups identified, 12 correspond to genes already known for their involvement in wing development, such as smoothened (smo), net, thickveins (tkv), mothers against dpp (mad), Star (S), expanded (ex), Suppressor of Hairless [Su(H)], cdc2, echinoid (ed), Protein kinase A (PKA), kuzbanian (kuz), and fat (ft), 16 are new complementation groups composed of at least two mutants, and 55 mutations appear to correspond to single alleles. We present here this screen and the mapping and characterization of one complementation group that corresponds to med15, a gene encoding a component of the Mediator complex (Lewis and Reinberg 2003; Guglielmi et al. 2004). The Mediator complex (Med) is conserved from yeast to humans and promotes the interaction of the RNApol-II with sequence-specific transcription factors (Kwon et al. 1999; Naar et al. 2001). We show that Med15 and other members of the Med complex are required, among other processes, for the transcription of dpp target genes. Interestingly, the med15 homolog in Xenopus laevis, ARC105, regulates specifically the expression of Smad3/4 and Smad2/4 target genes (Kato et al. 2002), suggesting a high degree of conservation of Med15-specific functions during evolution.
MATERIALS AND METHODS
Drosophila stocks:
Flies were cultured on standard media and crosses were carried out at 25° unless otherwise stated. We used the following stocks: hh-Gal4 (Calleja et al. 1996), salEPv-Gal4 and 638-Gal4 (Cruz et al. 2009), UAS-tauGFP (Ito et al. 1997), UAS-FLP (Duffy et al. 1998), nub1, Df(2L)vg, apHGO35, smo3 (Chen and Struhl 1996), pkaB3 (Li et al. 1995), TE35BC-GW24 (su(H)) (Morel and Schweisguth 2000), kuz1405 (Sotillos et al. 1997), Sos34Ea-6 (Rogge et al. 1991), spen5 (Kuang et al. 2000), spi1 (Freeman 1994), tkva12 (Nellen et al. 1994), Df(2L)ed-dp (a gift from S. Campuzano), net1 (Brentrup et al. 2000), aop1 (Rogge et al. 1995), fatf18 (Mahoney et al. 1991), cass2L-5 (Prout et al. 1997), Df(2L)LamB1 (Urbano et al. 2009), cdc2B47 (Clegg et al. 1993), dppd12 (St. Johnston et al. 1990), Df(2L)wgCX3 (Baker 1987), ex1 (Boedigheimer et al. 1993), lgl4 (a gift from A. Pérez), P{lacZ}bib4163 (Hao et al. 2003), the PyggyBac insertions d00080 and f06555 (Parks et al. 2004), and interference RNA lines against the genes med15 (NIG-Fly 4184R-4), med30 (VDRC 32459), med20 (NIG-Fly 18780R-3), med27 (NIG-Fly 1245R-1), med19 (NIG-Fly 5546R-1), med10 (NIG-Fly 5057R-1), med12/kto (NIG-Fly 8491R-2), med16 (NIG-Fly 5465R-1), and med25 (NIG-Fly12254R-1).
Generation of mitotic recombination clones:
We induced mitotic recombination by Flipase (FLP) at 48–72 hr after egg laying (AEL) in larvae of the following genotypes:
hsFLP1.22 f36a; M(2)z P[f+]30C FRT40A/mut al dp b pr FRT40A (f M+ clones)
hsFLP1.22 f36a; ck P[f+]30C FRT40A/mut al dp b pr FRT40A (ck/f twin clones)
salEPv-Gal4 f36a; M(2)z P[f+] FRT4A/mut FRT40A; UAS-FLP/+
salEPv-Gal4; M(2)z P[f+] FRT40A/mut FRT40A; UAS-FLP/+
w; M(2)z P[f+]30C FRT40A/mut al dp b pr FRT40A; hh-Gal4/UAS-FLP
638-Gal4; M(2)z P[f+] FRT40A/mut FRT40A; UAS-FLP/+
salEPv-Gal4; M(2)z P{arm-lacZ} FRT40A/mut FRT40A; UAS-FLP/+
638-Gal4; M(2)z P{arm-lacZ} FRT40A/mut FRT40A; UAS-FLP/+.
The salEPv-Gal4; al dp b FRT40A/FRT40A M(2)zFRT40A; UAS-FLP/+ wing disc contains homozygous al dp b M+ clones in the wing blade. The number of clones and their sizes increase during the third larval instar. We find clones covering ∼80% of the wing central domain in the corresponding adult wings. The wing blade region of 638-Gal4;al dp b FRT40A/FRT40AA M(2)zFRT40A; UAS-FLP/+ discs became entirely mutant in third instar larvae. The dp phenotype is apparent only in mosaic wings generated using the 638-Gal4 driver, suggesting that it is necessary for a large fraction of dp cells for this phenotype to develop. The presence of the dp allele does not interfere with phenotypes affecting pattern and/or size.
ENU treatment:
Groups of 50 w; al dp b pr FRT40A; UAS-FLP isogenic males 3 days old were first left starving for 4 hr and then fed during 24 hr with 0.29 mg/ml ENU in a sucrose solution. This concentration is estimated to cause one mutation per chromosomal arm (Ashburner 1989). Treated males were crossed with 100 salPE-Gal4; M(2)z FRT40A/CyO females and discarded after 3 days.
Complementation assays:
Mutants showing a similar phenotype in mosaic wings were crossed with each other, and mutations whose combination gave lethality or a visible wing phenotype were considered members of the same complementation group. We also used in these crosses the following alleles of genes localized in the 2L arm: smo1, pkaB3, biglacZ, TE35BC-GW24 (su(H)), kuz14C5, SOS34Ea-b, spen5, SPZ05671, spiEC2, madB1, tkvA12, Df(2L)ed-dp, netJ1, aop1, fatf18, cass2L-5, Df(2L)LamB1, cdc2B47, dppd5, wgCX3, ex1, and lgl4.
Complementation with deficiencies:
To map the mutants to a cytological position, they were crossed with a group of deficiencies covering the 2L chromosomal arm. The deficiencies utilized were as follows: Df(2L)net-PMF (BL3638), Df(2L)al (BL3548), Df(2L)ast2 (BL3084), Df(2L)BSC37 (BL7144), Df(2L)dppd14 (BL6648), Df(2L)C144 (BL90), Df(2L)JS17 (BL1567), Df(2L)BSC28 (BL6875), Df(2L)BSC31 (BL6965), Df(2L)drm-P2 (BL6507), Df(2L) ed1 (BL5330), Df(2L)ED250 (BL9270), Df(2L)BSC109 (BL8674), Df(2L)Exel6011 (BL7497), Df(2L)cl-h3 (BL781), Df(2L)E110 (BL490), Df(2L)BSC6 (BL6338), Df(2L)BSC7 (BL6374), Df(2L)spdj2 (BL2414), Df(2L)XE-2750 (BL4955), Df(2L)TE29Aa-11 (BL179), Df(2L)N22-14 (BL2892), Df(2L)s1402 (BL556), Df(2L)Mdh (BL1045), Df(2L)BSC32 (BL7142), Df(2L) FCK-20 (BL5869), Df(2L)Prl (BL3079), Df(2L)prd1.7 (BL3344), Df(2L)BSC30 (BL6999), Df(2L)b87e25 (BL3138), Df(2L)TE35BC-24 (BL3588), Df(2L)r10 (BL1491), Df(2L)cact 255rv64 (BL2583), Df(2L)TW137 (BL420), Df(2L)TW50 (BL3189), and Df(2L)TW161 (BL167). We first crossed one allele of each complementation group and all single mutants with these deficiencies. Subsequently, all mutations belonging to each complementation group were crossed with the corresponding deficiencies. Due to the presence of associated lethals in the treated chromosomes, we can be confident of the mapping data only for complementation groups with more than one allele (see Table S1 and Table S2).
Mapping of the complementation group affecting med15:
The 77A2 and 133A1 mutants were lethal over Df(2L)al and consequently were mapped to the 21B8–C1;21C8–D1 interval. They were then crossed with a group of smaller and molecular mapped deficiencies, Df(2L)ED5878 (BL9353), Df(2L)ED19 (BL8901), Df(2L)BSC16 (BL6608), Df(2L)BSC106 (BL8672), Df(2L)BSC107 (BL8673), and Df(2L)ast4 (BL6115), resulting in lethality in combination with Df(2L)BSC107. We then generated a new smaller deficiency by FRT recombination between the PBac insertions XP(+)d00080 and WH(+)f06555, allowing the localization of 77A2 and 133A1 to an interval including eight genes. Finally, both alleles failed to complement with the PBac insertion MED15f04180. To confirm that these mutations are med15 alleles, we amplified and sequenced the genomic region of med15 from 77A2 and 133A1 embryos using the following primers: med15-1L, TCACACTGTGCTCAGAGAAGAAGA; med15-1R, GTTGCATGGCATTTACGTT; med15-2L, AAATGCCATGCAACAGATGCCT; med15-2R, AAATGCAATAGCTGCGAAAAA; med15-3L, GATGTGGAGAAGATGACAAAG; and med15-3R, ACACTTTTTGCCCAGCGTAA.
Immunocytochemistry and in situ hybridization:
Wings discs were dissected, fixed, and stained as described in de Celis (1997). To detect apoptotic cells we used anti-activated Caspase3 (1:50; Cell Signaling), rabbit anti-Sal (1:200; Barrio and de Celis 2004), and rabbit anti-PMad (1:1000; a gift from G. Morata). Secondary antibodies were from Jackson Immunological Laboratories (used at 1/200 dilution). Pictures were taken with an Axioplan 2 Zeiss microscope and confocal images with a Microradiance–Bio-Rad microscope.
RESULTS
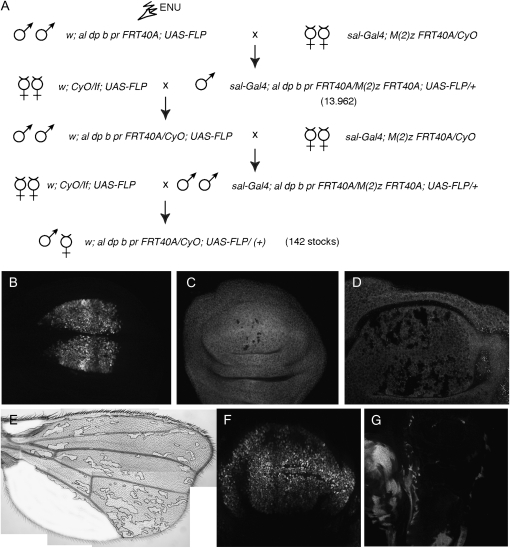
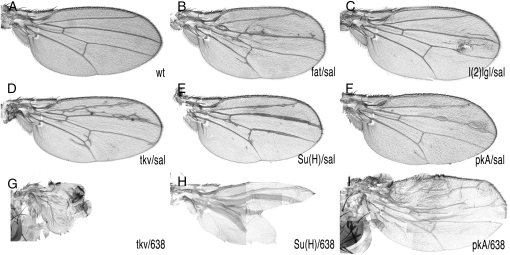
We aimed to identify loss-of-function mutations affecting the development of the Drosophila wing. Because most mutants are expected to be homozygous lethal (Ripoll and Garcia-Bellido 1979), we developed a method allowing the generation of viable and fertile heterozygous F1 animals bearing clones of cells homozygous for newly induced mutants in the wing blade (Figure 1A). In salEPv-Gal4; al dp b pr mut FRT40A/FRT40A M(2)z; UAS-FLP/+ males, the expression of FLP in the wing blade driven by salEPv-Gal4 during the third larval instar (Figure 1B) induces FRT-mediated mitotic recombination in the 2L and results in the generation of cells homozygous for the al dp b pr mut FRT40A chromosomal arm. These cells also lose the Minute mutation [M(2)z] and grow to occupy large extents of the central domain of the wing blade (Figure 1, C–E). We also used 638-Gal4 to generate mosaic wings. 638-Gal4 expression starts during the second larval instar and occurs in all wing cells (not shown and Figure 1F). In this case the entire wing blade and hinge became homozygous (Figure 1G). To estimate the visibility of phenotypes and the viability and fertility of heterozygous animals with mosaic wings, we induced clones of cells homozygous for mutations in genes whose roles in wing formation are well established. We used lethal alleles of fat (ft), lethal (2) giant larvae [l(2)gl], thick veins (tkv), Suppressor of Hairless [Su(H)], and Protein kinase A (PkA). In all cases and for both drivers, salEPv-Gal4 and 638-Gal4, the resulting mosaic animals display wing phenotypes that were easy to identify under the dissecting microscope (Figure 2). The phenotypes of combinations involving 638-Gal4 were always stronger than those of the corresponding combinations using salEPv-Gal4 (compare Figure 2D with 2G, 2E with 2H, and 2F with 2I). However, the viability of 638-Gal4 combinations was poor, and in many cases adult animals can be recovered only as escapers. For these reasons we used the salEPv-Gal4 driver for the screen and the 638-Gal4 driver to analyze the phenotype of homozygous mutant wings.
Figure 1.—
Crosses and Gal4 lines used to generate homozygous mutant wings in heterozygous flies. (A) Chromosomes and genetic crosses used to generate mosaic flies. Males of w; al dp b pr FRT40A; UAS-FLP genotype were treated with ENU and crossed in groups of 50 with salEPv-Gal4; M(2)Z FRT40A/CyO females (first row). The sal-Gal4; M(2)Z FRT40A/al dp b pr FRT40A; UAS-FLP/+ male progeny (13,962 males, second row) was screened for wing phenotypes. Selected males were crossed with w; CyO/If; UAS-FLP females, and the male progeny of w; al dp b pr FRT40A/CyO; UAS-FLP genotype were crossed with salEPv-Gal4; M(2)Z FRT40A/CyO females. We established stable w; al dp b pr FRT40A;UAS-FLP/+ stocks when the original phenotype was found in the progeny of this last cross (142 cases). (B) Expression of GFP in the wing blade region of the salEPv-Gal4/UAS-GFP wing disc. (C and D) Early (C) and late (D) third instar wing discs of salEPv-Gal4; M(2)Z FRT40A tubGFP/al dp b pr FRT40A; UAS-FLP/+ genotype, showing the clones as black spots. (E) Adult wing of f36a salEPv-Gal4; M(2)Z FRT40A Pf+30C/al dp b pr FRT40A; UAS-FLP/+ genotype, showing the area not covered by forked clones in white. (F) Third instar wing disc showing the expression of the 638-Gal4 line (638-Gal4/UAS-GFP). (G) Third instar wing disc of 638-Gal4; M(2)Z FRT40A tubGFP/al dp b pr FRT40A; UAS-FLP/+ genotype. Most of the wing blade is composed of al dp b pr FRT40A homozygous cells (shown in black).
Figure 2.—
Mosaic wings of known mutants affecting wing development. (A) Wild-type wing. (B) salEPv-Gal4; M(2)Z FRT40A/ fat18 FRT40A; UAS-FLP/+ (fat/sal). (C) salEPv-Gal4; M(2)Z FRT40A/ l(2)gl4 FRT40A; UAS-FLP/+ [l(2)gl/sal]. (D) salEPv-Gal4; M(2)Z FRT40A/tkva12 FRT40A; UAS-FLP/+ (tkv/sal). (E) salEPv-Gal4; M(2)Z FRT40A/ TE35BC-GW24 FRT40A; UAS-FLP/+ [Su(H)/sal]. (F) salEPv-Gal4; M(2)Z FRT40A/ PKAB3 FRT40A; UAS-FLP/+ (PKA/sal). (G) 638-Gal4; M(2)Z FRT40A/tkva12 FRT40A; UAS-FLP/+ (tkv/638). (H) 638-Gal4; M(2)Z FRT40A/ TE35BC-GW24 FRT40A; UAS-FLP/+ [Su(H)/638]. (I) 638-Gal4; M(2)Z FRT40A/ PKAB3 FRT40A; UAS-FLP/+ (PKA/638). Note that the phenotypes involving the Gal4 driver 638-Gal4 are much stronger than those generated with sal-Gal4.
Establishment of complementation groups:
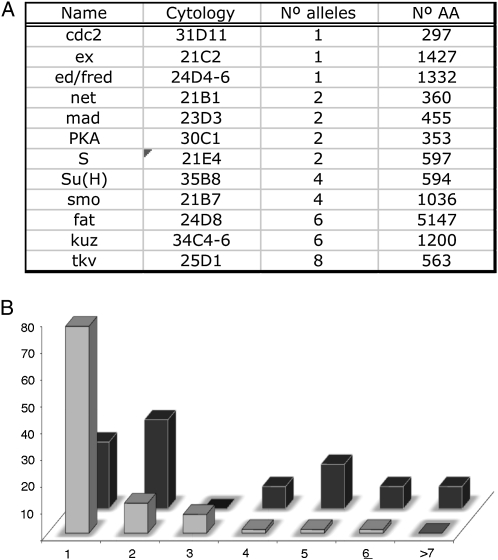
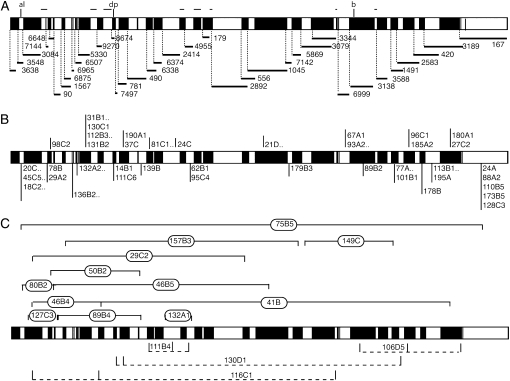
We scored ∼14,000 males of the salEPv-Gal4; al dp b pr mut FRT40A/FRT40A M(2)z; UAS-FLP/+ genotype, where mut means a recessive mutation induced in 2L. From the F1 males with abnormal wings we established 140 w; al dp b pr mut FRT40A/CyO stocks, and subsequently we used them to generate mosaic animals in which mitotic recombination is driven in the 638-Gal4 domain of expression. We grouped these alleles using the phenotypes of the combinations involving the salEPv-Gal4 and 638-Gal4 drivers and then crossed all the mutants in each group by each other to establish putative complementation groups. One member of each complementation group was crossed with alleles of known genes showing a similar phenotype in mosaics. The result of this analysis is shown in Table 1 (new complementation groups) and Figure 3A (complementation groups of known genes). As a summary, we identified 83 complementation groups, of which 12 correspond to previously known genes (Figure 3A) and 71 to mutations in other as yet unidentified genes (Figure 3B). The group of “known” genes had the higher number of alleles per complementation group (Figure 3B), whereas of the 71 novel complementation groups only 16 had more than one allele (Table 1 and Figure 3B). Although there is some correlation between the size of the coding region and the number of alleles identified for each known gene (Figure 3A), other aspects of the mutants appear more relevant to determine the probability of their identification.
TABLE 1.
Mutants identified in the screen grouped in phenotypic classes (first column), indicating the different alleles identified (“Alleles” column), the number of alleles included in each complementation group (“No.”), the phenotype in the combinations salEPv-Gal4/+; FRT40A mut al dp b pr/FRT40A M(2)z; UAS-FLP/+ (“SAL-GAL4”) and 638-Gal4/+; FRT40A mut al dp b pr/FRT40A M(2)z; UAS-FLP/+ (“638-GAL4”), and the name of the deficiency they fail to complement (“Def”)
| Phenotypic class | Alleles | No. | SAL-GAL4 | 638-GAL4 | Def |
|---|---|---|---|---|---|
| Loss of veins | MED15 | 2 | −v | +v/−v/+SO | 3548 |
| 88A2 | 1 | −s/−v/bs | −v | 167 | |
| 95C4 | 1 | −v/bs | −v/wm/CD | 4955 | |
| 106B3 | 1 | −v | +v/−v | — | |
| 128C3 | 1 | −v | −L4 | 167 | |
| Ectopic veins | kismet | 6 | +v/+s | +v/+s | 3638 |
| 111B4 | 1 | +v | — | >2 | |
| 139B | 1 | +v | +v/+v | 490 | |
| L3 duplication | 93A2, 93A6 | 1 | +v | +v/wm/N | 6999 |
| 46B4 | 1 | +L3 | +v/+v/wm | 2 | |
| 106D5 | 1 | +v | +v/+v/wm | >2 | |
| 112B3/147A2 | 2 | +v/CD | +v/wm/−L4 | 5330 | |
| Ectopic veins/loss of wing margin | 46B5 | 1 | +v | +v | 2 |
| 68B | 1 | wt | +v/wm | — | |
| 92C2 | 1 | +v/bs | wm | 6648 | |
| 111C6 | 1 | +v | +v/wm | 8674 | |
| 81C, 108D2, 112D1, 173A2, 184A3 | 5 | +v | +v/wm | 6338 | |
| 127C3 | 1 | +v | −v/wm | 2 | |
| 130D1 | 1 | +v/+s | −v/−L2/−L5/wm | >2 | |
| 132A2, 137C2, 146A2 | 3 | +v/bs/+v | +v/wm | 6507 | |
| 146C,147A1 | 2 | +v/bs | +v/wm | — | |
| 178B | 1 | +v/+v | +v/wm | 420 | |
| 185A2 | 1 | +v | +v/+v | 2583 | |
| 195A | 1 | +v/bs | +v/wm | 420/3189 | |
| Thicker veins | 20C, 39B2, 190A3 | 3 | N | N/wm | 3548 |
| 50B2 | 1 | +v | N/wm | 2 | |
| 112D7 | 1 | N | N | — | |
| 116C1 | 1 | bs/+s | N | >2 | |
| Loss of wing margin | 45C5, 149B2 | 2 | wm | +v/wm | 3548 |
| Ectopic veins/loss of wing margin | AT3, 147C2 | 2 | bs | +v/wm | — |
| 27C2 | 1 | v+ | +v/+v/wm | 3189 | |
| 37C | 1 | +v | +v/wm | 781 | |
| 41B | 1 | +v | +v/wm | 2 | |
| Larger wings | 130C1 | 1 | +s | +s | 5330 |
| 111B1 | 1 | bs | +s | — | |
| 21D, 39D3 | 2 | N | +s/+L3 | 1045 | |
| Smaller wings | 14B1 | 1 | +v/+v | +v | 8674 |
| 24C | 1 | +v | +v | 2414 | |
| 29D1 | 1 | +v/bs | +v | — | |
| 31B1, 110B4, 142C3 | 3 | +v | +v/bs | 5330 | |
| 89B2 | 1 | +v | +v | 3138 | |
| 101B1 | 1 | +v | +v/N | 1491 | |
| 119D5 | 1 | +v | +v/+v/bs | — | |
| 132A1 | 1 | +v/+SO | +v/bs | >2 | |
| 149C | 1 | +v | +v | >2 | |
| 173B5 | 1 | +v | +v | 167 | |
| 181B | 1 | +v | +v/−L2 | — | |
| 184E | 1 | +v | +v | — | |
| Blister | 18C2, 101A, 149C5 | 3 | bs | +v/bs | 3548 |
| 62B1 | 1 | bs | bs | 4955 | |
| 131B2 | 1 | wt | +v/bs | 5330 | |
| 179B3 | 1 | bs | bs | 7142 | |
| 190A1 | 1 | bs | +s | 781 | |
| Blister and pattern defects | 24A | 1 | N/CD | +v/N | 167 |
| 29A2 | 1 | +v/CD | +v/+L3/CD | 7144 | |
| 77A, 82A2, 119D2 | 3 | CD | +v/+v/wm | 1491 | |
| 80B2/80B3 | 1 | N/bs | +v/+v/wm | 2 | |
| 113B1, 136B1 | 2 | +v/−L4/CD | L | 420/3189 | |
| 66B, 127C4 | 2 | bs | +v/+v | — | |
| 89B4 | 1 | +v | +v/+v | 2 | |
| 96C1 | 1 | bs | L | 2583 | |
| 110B5 | 1 | bs | +v/+v/wm | 167 | |
| 136B3, 136B2 | 1 | bs | +v/+v/wm | 6965 | |
| 157B3 | 1 | +v/bs/CD | +v/+v/wm | 2 | |
| 180A1 | 1 | +v/bs | +v/+v/wm/CD | 3189 | |
| 78B | 1 | wt | +v | 7144 | |
| Cell differentiation | 29C2 | 1 | CD | +v/wm/CD | 2 |
| 52C1, 79A3, 101D, 130D2 | 4 | CD | +v/wm/CD | — | |
| 67A1 | 1 | +v/CD/+v | +v/wm/CD | 6999 | |
| 75B5 | 1 | +v/CD | +v/bs/CD | 2 | |
| 141C5 | 1 | +v/CD/+v | L | >2 | |
| Total | 71 | 101 |
Abbreviations in the SAL-GAL4 and 638-GAL4 columns are as follows: −v, loss of veins; +v, ectopic veins; −s, reduced size; +s, larger size; +SO, ectopic sensory organs in the wing blade; bs, loss of dorso-ventral adhesion; N, thicker veins; wm, loss of wing margin; CD, trichome differentiation or planar cell polarity; L, lethal. In some cases individual veins are also indicated. In the “Def” column, 2 and >2 indicate that the corresponding mutation fails to complement two or more than two chromosomal deficiencies, respectively.
Figure 3.—
General results of the screen. (A) Known genes identified in the screen, indicating their names (“Name”), cytological position (“Cytology”), number of alleles found (“No. alleles”), and the number of amino acids of the corresponding proteins (“No. AA”). (B) Relationship between the numbers of complementation groups (vertical values) and the number of alleles included in each complementation group (horizontal values). Data with light shading correspond to the new complementation groups identified in this screen and data with solid shading to genes that were known to play a role in wing development.
Phenotypic classes obtained in the screen:
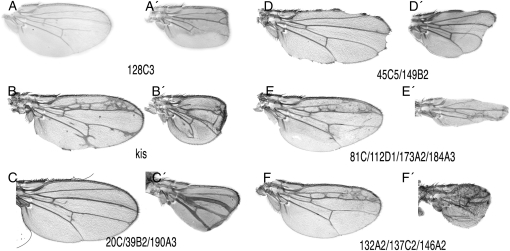
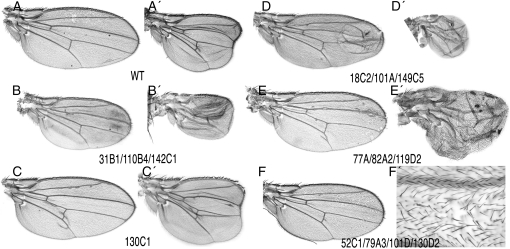
The phenotypes of alleles not corresponding to previously known genes were classified in the following groups: (1) mutants affecting primarily the development of the veins, causing the loss of vein stretches, the formation of ectopic veins, or an increase in the thickness of the veins (Figure 4, A–C); (2) mutants affecting the integrity of the wing margin, causing the loss of wing tissue around the wing margin (Figure 4D) [some of these mutants also affect the distance between the longitudinal veins (Figure 4, E and F)]; (3) mutants affecting mainly the size of the wing with no or only minor effects in the patterning of veins (Figure 5, B and C); (4) mutants affecting the adhesion between the dorsal and ventral wing surfaces (Figure 5, D and E); and (5) mutants affecting the differentiation of trichomes (Figure 5F). Although most mutants can be assigned to one of these classes, in many instances we observed phenotypes with characteristics shared between two or more groups. For example, several mutations affect the size of the wing, the patterning of veins, and the formation of the wing margin (Figures 4, D–F, and 5E). In these cases, we crossed representative alleles with members of several phenotypic classes to establish the complementation groups. The phenotypes of mosaic wings for all new alleles identified and for a representative allele of all known complementation groups isolated in the screen in combination with the Gal4 lines salEPv-Gal4 and 638-Gal4 are shown in supporting information, Figure S1, Figure S2, Figure S3, and Figure S4.
Figure 4.—
Representative wings of the phenotypic classes affecting the formation of veins. (A and A′) Loss of veins and reduced wing size in salEPv-Gal4; M(2)Z FRT40A/al dp b pr 128C3 FRT40A; UAS-FLP/+ (A) and 638-Gal4; M(2)Z FRT40A/al dp b pr 128C3 FRT40A; UAS-FLP/+ (A′). (B and B′) Loss and gain of veins in salEPv-Gal4; M(2)Z FRT40A/al dp b pr 61C FRT40A; UAS-FLP/+ (B) and 638-Gal4; M(2)Z FRT40A/al dp b pr 61C FRT40A; UAS-FLP/+ (B′). The mutant 61C is an allele of kismet (kis). (C and C′) Thick vein phenotype in salEPv-Gal4; M(2)Z FRT40A/al dp b pr 20C FRT40A; UAS-FLP/+ (C) and 638-Gal4; M(2)Z FRT40A/al dp b pr 20C FRT40A; UAS-FLP/+ (C′). (D and D′) Wing margin phenotype of salEPv-Gal4; M(2)Z FRT40A/al dp b pr 45C5 FRT40A; UAS-FLP/+ (D) and 638-Gal4; M(2)Z FRT40A/al dp b pr 45C5 FRT40A; UAS-FLP/+ (D′). (E and E′) Ectopic veins and loss of wing margin in salEPv-Gal4; M(2)Z FRT40A/al dp b pr 81C FRT40A; UAS-FLP/+ (E) and 638-Gal4; M(2)Z FRT40A/al dp b pr 81C FRT40A; UAS-FLP/+ (E′). (F and F′) Ectopic veins and reduction in wing size in salEPv-Gal4; M(2)Z FRT40A/al dp b pr 132A2 FRT40A; UAS-FLP/+ (F) and 638-Gal4; M(2)Z FRT40A/al dp b pr 132A2 FRT40A; UAS-FLP/+ (F′). The phenotypes of other mutations affecting the veins or the wing margin are shown in Figure S1 and Figure S3.
Figure 5.—
Representative wings of the phenotypic classes affecting wing size, cell adhesion, and cell differentiation. (A and A′) Wild-type wing of salEPv-Gal4; M(2)Z FRT40A/al dp b pr FRT40A; UAS-FLP/+ (A) and 638-Gal4; M(2)Z FRT40A/al dp b pr FRT40A; UAS-FLP/+ (A′) genotypes. (B and B′) Reduced wing size in salEPv-Gal4; M(2)Z FRT40A/al dp b pr 31B1 FRT40A; UAS-FLP/+ (B) and 638-Gal4; M(2)Z FRT40A/al dp b pr 31B1 FRT40A; UAS-FLP/+ (B′). (C and C′) Increased wing size in salEPv-Gal4; M(2)Z FRT40A/al dp b pr 130C1 FRT40A; UAS-FLP/+ (C) and 638-Gal4; M(2)Z FRT40A/al dp b pr 130C1 FRT40A; UAS-FLP/+ (C′). (D and D′) Failures in dorso-ventral adhesion in salEPv-Gal4; M(2)Z FRT40A/al dp b pr 18C2 FRT40A; UAS-FLP/+ (D) and 638-Gal4; M(2)Z FRT40A/al dp b pr 18C2 FRT40A; UAS-FLP/+ (D′). (E and E′) Extra veins and increased wing size with loss of wing margin in salEPv-Gal4; M(2)Z FRT40A/al dp b pr 77A FRT40A; UAS-FLP/+ (E) and 638G4-Gal4; M(2)Z FRT40A/al dp b pr 77A FRT40A; UAS-FLP/+ (E′). (F and F′) Formation of several trichomes per cell in salEPv-Gal4; M(2)Z FRT40A/al dp b pr 52C1 FRT40A; UAS-FLP/+. F′ is a higher magnification of the L3/L4 dorsal intervein. Other phenotypes affecting wing size, cell adhesion, and differentiation are shown in Figure S2 (size) and Figure S4 (adhesion and cell differentiation).
Cytological mapping of novel mutants:
To determine the cytological position of the new complementation groups, we used a collection of 36 chromosomal deficiencies that together cover the majority of the 2L arm (Figure 6A) and crossed them with 1 mutant (16 alleles) representing each complementation group and with 53 single mutants from the 55 single mutants identified in the screening. To identify gaps in the coverage of the 2L arm by these deficiencies, we also determined the viability of trans-heterozygous combinations between pairs of adjacent deletions, assuming that combinations between overlapping deficiencies are lethal. Using this criterion, we found that from the possible 35 pairwise combinations between adjacent deficiencies only 13 were lethal (Figure S5), suggesting that at least 22 small chromosomal intervals are not covered by these 36 deficiencies (Figure 6A). Using mainly lethality to define noncomplementation, we mapped to particular cytological intervals 12 of the 16 complementation groups formed by more than one allele (Figure 6B and Table 1). For individual mutations (55) we found 8 that complemented all deficiencies, 30 that failed to complement only one deficiency (Figure 6B and Table 1), and 17 that failed to complement two or more deficiencies (Figure 6C and Table 1). As we know which deficiencies overlap, those mutations that do not complement two adjacent deficiencies (2) were placed in the cytological region of overlap.
Figure 6.—
Schematic representation of the complementation data with 2L deficiencies for all the mutants isolated. (A–C) The horizontal bars represent the chromosomal arm 2L subdivided in solid and open areas that exemplified the subdivision of the arm by the overlapping of the deficiencies used for the mapping. (A) Extent of the different deletions used for the complementation tests (see material and methods). The thin lines represent the main gaps left by the deficiencies. (B) Localization of mutants (single numbers) and complementation groups (single numbers and dots) that fail to complement with only one deficiency. (C) Localization of mutants that fail to complement with more than one deficiency.
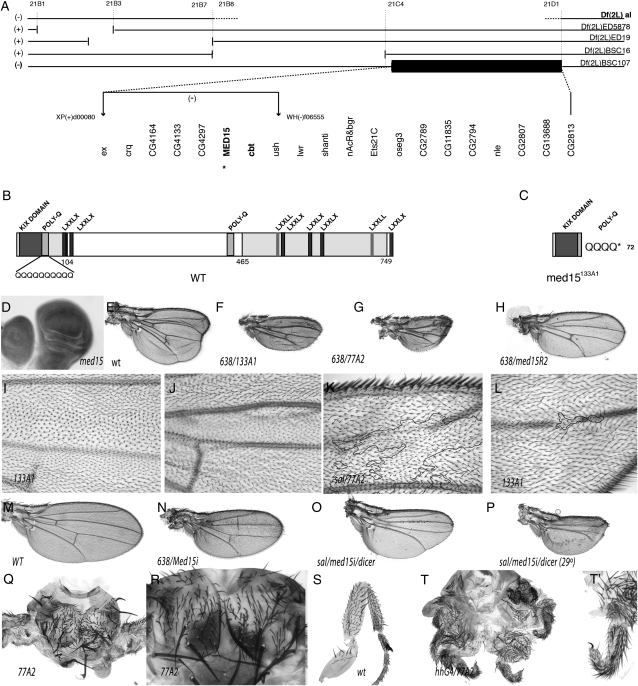
The complementation group formed by 77A2 and 133A1 corresponds to med15:
We chose the complementation group formed by the alleles 77A2 and 133A1 to carry out an in-depth analysis of the affected gene. These alleles cause in mosaic wings a strong reduction of wing size, accompanied by the loss of the L2 vein and the differentiation of some ectopic bristles along the veins L2 and L3 (Figure 7). Both 77A2 and 133A fail to complement Df(2L)al (21B8–C1;21C8–D1), and within the interval covered by this deficiency, 77A2 and 133A1 failed to complement Df(2L)BSC107, which deletes the 21C5;21D1 region (Figure 7A). There are 20 annotated genes in this interval, and we generated a smaller deficiency by FRT-mediated recombination between the Piggybac insertions XP(+)d00080 and WH(-)f06555. This deficiency includes only 8 genes and fails to complement the alleles 77A2 and 133A1 (Figure 7A). Finally, we combined 77A2 and 133A1 with mutations in the genes cabut (cbt) and med15 and found that both 77A2 and 133A1 are lethal in combination with med15f04180 and viable in combination with a cbt allele (Figure 7A). Med15 encodes a 749-amino-acid protein characterized by the presence of a Kix domain, two small poly(Q) stretches, and several LXXLL motifs (Gelbart et al. 1997; Figure 7B). We were able to map the 133A1 allele to the med15 coding region by sequencing genomic regions amplified by PCR from homozygous 133A1 embryos (see materials and methods). This mutation is associated with a C to T transition that introduces a stop codon in the region that corresponds to the N-terminal poly(Q) localized after the KIX domain (Figure 7C). Using the same approach we could not find any nucleotide change in the coding region of med15 in the 77A2 allele. Mitotic recombination clones of the med15f04180 allele, generated in 638-Gal4; med15f04180 FRT40A; M(2)z FRT40A; UAS-FLP/+ flies, result in smaller than wild-type wings with a normal pattern of veins (Figure 7H), suggesting that this allele is weaker than the novel 77A2 (Figure 7G) and 133A1 (Figure 7F) mutations. Homozygous 133A1 die during embryogenesis, and although some homozygous 77A2 embryos can hatch, they die during the first larval instar (data not shown). We have not studied the embryonic phenotypes of med15 homozygous alleles.
Figure 7.—
The complementation group formed by 77A2 and 133A1 corresponds to med15. (A) Chromosomal interval 21B3–21C4 and extent of the deficiencies used (named to the right) shown as open spaces between horizontal bars. The region included in Df(2L)BSC107, which defines the localization of the 77A2/133A1 complementation group, is shown as a solid bar, and the genes included in this deficiency are shown at the bottom. The position of two PiggyBac insertions used to generate the Df(2L)d0080-f06555 chromosome is shown by vertical arrows. (B) Protein sequence and domains of Med15. (C) Expected protein produced by the med15133A1 allele. (D) Generalized expression of med15 in the wing disc. (E) Control wing of 638-Gal4/+; FRT40A al dp b pr/FRT40A M(2)z; UAS-FLP/+ genotype showing the characteristic dp phenotype. (F and G) Homozygous wings for the med15 alleles med15133A1 (133A1) (E) and med1577A2 (77A2) (F). The genotypes of these wings are 638-Gal4/+; FRT40A al med15133A1 dp b pr/FRT40A M(2)z; UAS-FLP/+ (F) and 638-Gal4/+; FRT40A al med1577A2 dp b pr/FRT40A M(2)z; UAS-FLP/+ (G). (H) Phenotype of med15f04180 homozygous wing in 638-Gal4/+; FRT40A al med15f04180 dp b pr/FRT40A M(2)z; UAS-FLP/+ female flies (compare the wing size with its control shown in M). (I and J) Mitotic recombination clones generated in hsFLP1.22 f36a; ck P[f+]30C FRT40A/al med15133A1 dp b pr FRT40A flies. The clone in I is labeled with ck and is wild type for the med15 gene (twin spot). The clone in J is labeled with forked and is homozygous for the med15133A1 allele. Both clones are located in the L3/L4 intervein, occupying a large fraction of this dorsal (I) and ventral (J) intervein. (K) med1577A2 clone labeled with forked generated in f36a salEPv-Gal4/+; FRT40A al med1577A2 dp b pr/FRT40A P[f+]30C M(2)z; UAS-FLP/+ flies. The forked territory is enclosed by a solid line and is associated with the loss of the ventral L2 vein. (L) Small med15133A1 clone in the distal dorsal L4 vein causing the loss of this vein. (M–P) Phenotypes resulting from the expression of med15 interference RNA (med15i) in the genotypes 638-Gal4/+; UAS-med15i (N), salEPv-Gal4/UAS-med15i; UAS-dicer/+ (O), and salEPv-Gal4/UAS-med15i; UAS-dicer/+ grown at 29° (P). The wild-type control wing is shown in M. (Q and R) Two examples of thoraxes taken at different magnification showing the failure in the fusion between the left and the right hemithorax when the central region is occupied by med1577A mutant cells (labeled with forked in hsFLP1.22 f36a; P[f+]30C M(2)z FRT40A/al med1577A2 dp b pr FRT40A flies). (S) Wild-type male first leg. (T and T′) Two examples of legs taken at different magnifications (T′ is ×4 T) showing the defects in leg morphogenesis and tarsal segmentation in w; M(2)z P[f+]30C FRT40A/al med1577A dp b pr FRT40A; hh-Gal4/UAS-FLP flies.
The most characteristic phenotypes of med15 alleles in the wing are loss of the L2 vein and a reduction in the size of the wing (Figure 7, F–H). These phenotypes were observed in mosaic wings generated in 638-Gal4; med15 FRT40A/M(2)z FRT40A; UAS-FLP/+ flies. We generated mitotic recombination clones of med15 alleles in hsFLP f36a; med15 al dp b pr FRT40A/M(2)z [f+] FRT40A males, because in this case the clones are labeled with the cell marker forked (f), and this allows the study of the autonomy of the clonal phenotypes. We could generate large med15 M+ mutant clones when they were induced 48–72 h AEL, and in all cases the med15 alleles behave in a cell autonomous manner. Thus, when the clones occupy the region between the L3 and L4 veins (n = 4), the size of this territory is strongly reduced (compare Figure 7I and 7J), and when the clones include the ventral L2 vein (n = 10) or the dorsal L4 vein (n = 5), these veins are reduced or absent (Figure 7, K and L). Med15 clones running along the dorsal L3 vein diminish the pigmentation of the vein, but do not eliminate its differentiation (n = 4, data not shown). Similarly, clones affecting the proximal regions of L3, L4, and L5 do not affect vein differentiation (data not shown). We also studied the phenotypes resulting from the expression in the wing disc of interference RNA directed against med15 (med15-i). In these flies, we observe a reduction in the size of the wing, the formation of some ectopic sensory organs, and only in some cases the loss of the L2 vein (Figure 7, N–P). Finally, we also found a requirement for med15 in other tissues such as the thorax and legs. In the first case, the more frequent phenotype was a failure of the left and right hemithoraxes to fuse when one or both hemithoraxes are composed of med15 mutant cells (Figure 7, Q and R). In the legs, we found many cases of legs with severe shortening along their entire length, accompanied by fusions of tarsal segments (not shown). These phenotypes are fully penetrant when the posterior compartments of the legs are composed of med15 cells, as happens in w; med1577A2 al dp b pr FRT40A/M(2)z [f+] FRT40A; UAS-FLP/hh-Gal4 flies (Figure 7, T and T′, compare with Figure 7S). In general, the med15 phenotypes are reminiscent of those caused by reduced dpp and TGFβ signaling, which consist of loss of veins, failures in dorsal closure, leg morphogenesis defects (Dpp), and reduced wing size (TGFβ) (Posakony et al. 1990; de Celis 1997; Lecuit and Cohen 1997; Brummel et al. 1999; Harden 2002).
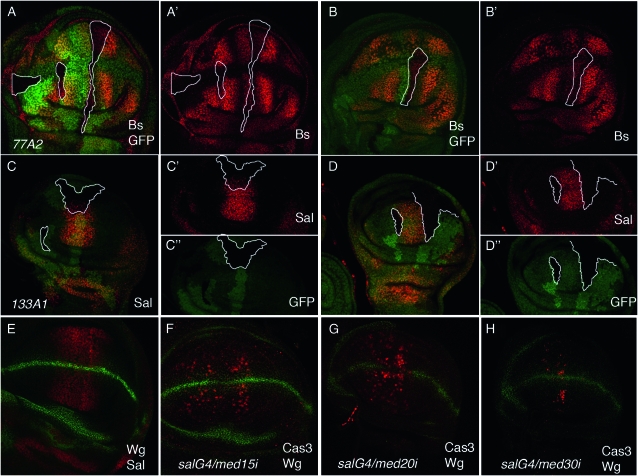
We also observed several alterations in gene expression patterns in med15 mutant cells during imaginal development. For example, the expression of Bs is reduced in intervein territories formed by med15 cells (Figure 8, A and B), suggesting that the transcriptional response to Dpp and Hedgehog signaling requires Med15 activity. Similarly, the expression of spalt (sal), a direct target of Dpp signaling (de Celis et al. 1996; Barrio and de Celis 2004), is absent in med15 clones localized in the anterior and posterior regions of the Sal domain of expression (Figure 8, C and D) and is reduced in clones localized in the central domain of sal expression (Figure 8, C and D). These effects are always cell autonomous, suggesting that Med15 does not affect dpp expression but compromises the capability of Dpp signaling to activate its targets. A reduced response to Dpp signaling might also contribute to the smaller than normal wing size of med15 mosaic wings and to the partial loss of vein stretches. We could not find changes in the expression of EGFR or Wingless target genes (argos, Delta, and distalless; data not shown), indicating that Med15 is not required as a general coactivator of transcription, but rather that its function is specific to particular enhancer–promoter interactions.
Figure 8.—
Developmental analysis of med15 function in the wing disc. (A and B) Expression of Bs (red) in med1577A2 clones induced in flies of genotype hsFLP1.22; P[tubGFP] FRT40A/med1577A2 FRT40A (twin clones). med1577A2 clones are labeled by the absence of GFP (green) and each clone is encircled in a white line in A–B′. A′ and B′ are the corresponding red channels showing reduced Bs expression in the mutant clones. (C and D) Expression of Sal (red) in med15133A1 clones induced in flies of genotype hsFLP1.22; P[tubGFP] FRT40A/med15133A1 FRT40A (twin clones). med15133A1 clones are labeled by the absence of GFP (green) and each clone is encircled in a white line in C and D, C′ and D′, and C″ and D″. ′ and ″ are the corresponding red (Sal) and green (GFP) channels, respectively. The expression of Sal is reduced or absent in mutant clones located in the center (C′) or lateral (D′) regions of the Sal domain, respectively. (E) Expression of Wingless (Wg, green) and Sal (Sal, red) in a wild-type third instar wing blade. (F–H) Induction of cell death (shown as expression of activated Caspase 3) (red in F–H) in flies expressing interference RNA in the spalt domain in salEPv-Gal4/UAS-med15-RNAi (F), salEPv-Gal4/UAS-med20-RNAi (G), and salEPv-Gal4/UAS-med30-RNAi (H).
Other members of the Med complex are required during wing development:
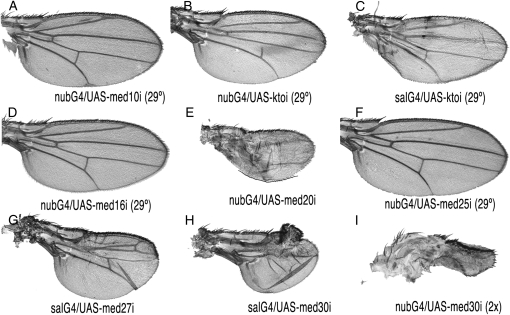
The involvement of med15 in wing disc development suggests that other members belonging to the Med complex would be required in similar processes. Alternatively, med15 functions might be independent of its participation in the Med complex. We studied the loss-of-function phenotypes of several Med complex components by driving the expression of specific interference RNAs in the wing disc. In all cases analyzed, we found that the reduction in Med expression gave rise to smaller than normal wings (med10-i, med16-i, med25-i, med27-I, and kto-i; Figure 9), which were extreme in the cases of loss of med20 and med30 (Figure 9). Only the reduction in med20 and med30 expression resulted in loss of vein phenotypes (Figure 9). In the cases of med20, med30, and med15, the expressions of their interference RNAs induce cell death (see Figure 8, E–H). In summary, and although the phenotypes observed upon a reduction in the expression of various Med components are not identical, they are similar enough to suggest that they could be caused by different degrees of loss of Mediator function.
Figure 9.—
Wing phenotypes caused by reduced expression of different components of the Mediator complex. (A–I) Adult wings of Gal4/UAS-RNAi combinations showing smaller than normal size and different defects in the patterning of veins. (A) nub-Gal4/UAS-med10i at 29°. (B) nub-Gal4/UAS-ktoi (29°). (C) salEPv-Gal4/UAS-ktoi (29°). (D) nub-Gal4/UAS-med16i (29°). (E) nub-Gal4/UAS-med20i. (F) nub-Gal4/UAS-med25i (29°). (G) salEPv-Gal4/UAS-med27i. (H) salEPv-Gal4/UAS-med30i. (I) nub-Gal4/UAS-med30i (taken at magnification ×2).
DISCUSSION
The patterning of the veins and the growth of the wing involve the activities of several conserved signaling pathways and transcription factors, and mutations in these genes result in modifications of vein positioning and wing size (Sotillos and de Celis 2005). We have carried out a mosaic screen to search for mutations in the 2L chromosomal arm that modify the pattern of veins and the growth of the wing. In this screen we generated homozygous wing regions in otherwise heterozygous flies using a combination of the FRT-FLP method and the Gal4-UAS system. We maximize the fraction of the wing occupied by homozygous mutant cells using a Minute mutation. This also allows the survival of mutants cells that otherwise could be outcompeted by the surrounding wild-type cells (Morata and Ripoll 1975). The inconvenience of using a Minute mutation is that stocks and crosses involving this allele are less healthy. In general, the flies heterozygous for newly induced mutations with mosaic wings were viable and fertile, allowing the screening of a high number of treated chromosomes.
We identified 140 mutations and grouped them in 83 complementation groups, of which 12 correspond to previously known genes and 71 to alleles in other not yet identified genes. Among the genes previously known for their roles in wing formation we found 39 alleles affecting net, mad, PKA, S, su(H), smo, kuz, ex, ed/fred, cdc2, ft, and tkv. The number of alleles in each complementation group was much higher in the case of the known genes, as only 16 of 71 novel complementation groups were formed by more than one allele. These numbers indicate that the screen is not yet at saturation and that the visibility of the phenotypes is much higher in wings homozygous for mutations in the class of known genes. We were able to map 58% of the complementation groups to individual chromosomal intervals using a collection of deficiencies that cover an estimated 80% of the 2L arm. However, these data have several caveats due to the high number of complementation groups formed by only one allele and to the presence in the treated chromosomes of associated lethals. Thus, 23% of the novel complementation groups failed to complement with more than one deficiency, and 11% of complementation groups complemented for lethality and phenotype with all deficiencies. In this manner, the cytological localization of all complementation groups composed by only one allele is still tentative. The phenotypes identified in the screen mainly affected the wing veins and wing margin, the size of the wing, the adhesion between the dorsal and ventral wing surfaces, the integrity of the epithelium, and the differentiation of trichomes by wing cells. These phenotypes correspond to alterations in processes that occur during the third larval instar (vein determination, wing disc growth, and wing margin formation) and during pupal development (dorso-ventral adhesion and trichome differentiation). Furthermore, the observed phenotypes are informative about the process, and in some cases the pathways, that might be altered in the mutants. For example, changes in wing size without effects in vein formation are expected by modifications in the insulin and TGFβ pathways (Brummel et al. 1999; Johnston and Gallant 2002), alterations in the integrity of the wing margin are a mark of loss of Notch and Wingless signaling at the dorso-ventral boundary (Couso et al. 1994; de Celis and Garcia-Bellido 1994), changes in the formation of veins are expected by modifications in the Dpp, EGFR, and Notch pathways (Sotillos and de Celis 2005), and the formation of blistered wings is typical of defects in Integrin and Laminin functions (Walsh and Brown 1998; Urbano et al. 2009). Future work will aim to unambiguously map the different complementation groups to individual genes and to identify the developmental functions they affect.
We chose to analyze in some detail the complementation group formed by the 77A and 133A1 mutants. These mutations are alleles of med15, a gene encoding one component of the Mediator complex (Guglielmi et al. 2004). Thus, they fail to complement other med15 alleles, and med15133A1 is associated with a stop codon that could truncate the protein in the N-terminal region after the KIX domain. The Mediator multiprotein complex promotes the transcription of inducible genes, acting as a link between the RNApolII holoenzyme and several sequence-specific transcription factors (Naar et al. 2001; Lewis and Reinberg 2003; Taatjes et al. 2004). The human homolog of Med15, MED105, is included in all Mediator complexes identified so far and forms part of a module named the tail that is the main target for the transcriptional activators (Guglielmi et al. 2004; Taatjes et al. 2004). Thus, Med15 homologs can bid to different transcription factors such as Gcn4 and Gal4 in Saccharomyces cerevisiae (Fishburn et al. 2005; Reeves and Hahn 2005), SREBP in Caenorhabditis elegans (Taubert et al. 2006; Yang et al. 2006), and, more interesting from the perspective of our data, to Smad2/3 and Smad4 in Xenopus (Kato et al. 2002). Other members of the Mediator complex that were previously analyzed are kohtalo and skuld (Med12 and Med13, respectively), which form part of the conserved Cdk8 module (Bourbon et al. 2004; Conaway et al. 2005; Kim and Lis 2005). Interestingly, mouse Cdk8 and Cdk9 phosphorylate Smad proteins, regulating their transcriptional activity and turnover (Alarcón et al. 2009). However, kohtalo and skuld are required for sensory organ development, for some aspects of Notch and Hedgehog signaling, and for the transcription of Wingless downstream genes (Janody et al. 2003; Carrera et al. 2008).
Med15 mutations result in smaller than normal wings and loss of mainly the L2 vein. They also affect the fusion between the left and the right hemithorax and leg morphogenesis. The reduction in the level of expression of other components of the Mediator complex, most notably med20, med27, and med30, also results in smaller than normal wings and failures in vein differentiation, in addition to causing some levels of cell death. Although these phenotypes were similar, they are not identical, which might indicate specific requirements of these subunits or, alternatively, a different degree in the effectiveness of each interference RNA used. Mutant med15 cells display specific defects in gene expression, suggesting a requirement limited to particular enhancer–promotor interactions. In particular, the expression of spalt, a direct target of Dpp signaling, is compromised in med15 mutant cells. There are no known transcriptional targets of TGFβ signaling in the wing, and consequently we could not determine directly whether the activity of this pathway is diminished in med15 mutants. A direct requirement of Med15 for the transcription of TGFβ target genes is nonetheless suggested by the similar phenotypes of wing size reduction observed in med15 mutants and in baboon mutations (Brummel et al. 1999).
Acknowledgments
We are very grateful to Rosario Hernández and Cristina Prieto for their technical assistance. This work was supported by grants BFU2006-06501 and Consolider CSD-2007-00008 from the Spanish Ministry of Research and Innovation. An institutional grant from the Ramón Areces Foundation to the Centro de Biología Molecular “Severo Ochoa” is also acknowledged.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.113670/DC1.
References
- Alarcón, C., A.-I. Zaromytidou, Q. Xi, S. Gao, J. Yu et al., 2009. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-b pathways. Cell 139 757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M., 1989. Drosophila. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Baker, N. E., 1987. Molecular cloning of sequences from wingless a segment polarity gene in Drosophila the spatial distribution of a transcript in embryos. EMBO J. 6 1765–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrio, R., and J. F. de Celis, 2004. Regulation of spalt expression in the Drosophila wing blade in response to the Decapentaplegic signaling pathway. Proc. Natl. Acad. Sci. USA 101 6021–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier, E., 2000. Drawing lines in the Drosophila wing: initiation of wing vein development. Curr. Opin. Genet. Dev. 10 393–398. [DOI] [PubMed] [Google Scholar]
- Boedigheimer, M., P. Bryant and A. Laughon, 1993. Expanded, a negative regulator of cell proliferation in Drosophila, shows homology to the NF2 tumor suppressor. Mech. Dev. 44 83–84. [DOI] [PubMed] [Google Scholar]
- Bourbon, H. M., A. Aguilera, A. Z. Ansari, F. J. Asturias, A. J. Berk et al., 2004. A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol. Cell 14 553–557. [DOI] [PubMed] [Google Scholar]
- Brentrup, D., H. P. Lerch, H. Jackle and M. Noll, 2000. Regulation of Drosophila wing vein patterning: net encodes a bHLH protein repressing rhomboid and is repressed by rhomboid dependent EGFR signalling. Development 127 4729–4741. [DOI] [PubMed] [Google Scholar]
- Brummel, T., S. Abdollah, T. E. Haerry, M. J. Shimell, J. Merriam et al., 1999. The Drosophila Activin receptor Baboon signals through dSmad2 and controls cell proliferation but not patterning during larval development. Genes Dev. 13 98–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calleja, M., E. Moreno, S. Pelaz and G. Morata, 1996. Visualization of gene expression in living adult Drosophila. Science 274 252–255. [DOI] [PubMed] [Google Scholar]
- Carrera, I., F. Janody, N. Leeds, F. Duveau and J. E. Treisman, 2008. Pygopus activates Wingless target gene transcription through the mediator complex subunits Med12 and Med13. Proc. Natl. Acad. Sci. USA 105 6644–6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., and G. Struhl, 1996. Dual roles for Patched in sequestering and transducing Hedgehog. Cell 87 553–563. [DOI] [PubMed] [Google Scholar]
- Clegg, N. J., I. P. Whitehead, J. A. Williams, G. B. Spiegelman and T. A. Grigliatti, 1993. A developmental and molecular analysis of Cdc2 mutations in Drosophila melanogaster. Genome 36 676–685. [DOI] [PubMed] [Google Scholar]
- Cohen, S. M., 1993. Imaginal Disc Development. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Conaway, R. C., S. Sato, C. Tomomorisato, T. Yao and J. W. Conaway, 2005. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem. Sci. 30 250–255. [DOI] [PubMed] [Google Scholar]
- Couso, J. P., S. A. Bishop and A. Martinez Arias, 1994. The wingless signalling pathway and the patterning of the wing margin in Drosophila. Development 120 621–636. [DOI] [PubMed] [Google Scholar]
- Cruz, C., A. Glavic, M. Casado and J. F. de Celis, 2009. A gain of function screen identifying genes required for growth and pattern formation of the Drosophila melanogaster wing. Genetics 183 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis, J. F., 1997. Expression and function of decapentaplegic and thick veins in the differentiation of the veins in the Drosophila wing. Development 124 1007–1018. [DOI] [PubMed] [Google Scholar]
- de Celis, J. F., 2003. Pattern formation in the Drosophila wing: the development of the veins. BioEssays 25 443–451. [DOI] [PubMed] [Google Scholar]
- de Celis, J. F., and A. Garcia-Bellido, 1994. Roles of the Notch gene in Drosophila wing morphogenesis. Mech. Dev. 46 109–122. [DOI] [PubMed] [Google Scholar]
- de Celis, J. F., R. Barrio and F. C. Kafatos, 1996. A gene complex acting downstream of Dpp in Drosophila wing morphogenesis. Nature 381 421–424. [DOI] [PubMed] [Google Scholar]
- Duffy, J. B., D. A. Harrison and N. Perrimon, 1998. Identifying loci required for follicular patterning using directed mosaics. Development 125 2263–2271. [DOI] [PubMed] [Google Scholar]
- Fishburn, J., N. Mohibullah, and S. Hahn, 2005. Function of a eukaryotic transcription activator during the transcription cycle. Mol. Cell 18 369–378. [DOI] [PubMed] [Google Scholar]
- Freeman, M., 1994. The spitz gene is required for photoreceptor determination in the Drosophila eye where it interacts with the EGF receptor. Mech. Dev. 48 25–33. [DOI] [PubMed] [Google Scholar]
- Fristrom, D., P. Gotwals, S. Eaton, T. Kornberg, M. A. Sturtevant et al., 1994. blistered; a gene required for vein/intervein formation in wings of Drosophila. Development 120 2661–2686. [DOI] [PubMed] [Google Scholar]
- Garcia-Bellido, A., and J. Dapena, 1974. Induction, detection and characterization of cell differentiation mutants in Drosophila. Mol. Gen. Genet. 128 117–130. [DOI] [PubMed] [Google Scholar]
- Gelbart, W. M., M. Crosby, B. Matthews, W. P. Rindone, J. Chillemi et al., 1997. FlyBase: a Drosophila database. The FlyBase consortium. Nucleic Acids Res. 25 63–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmi, B., N. L. V. Berkum, B. Klapholz, T. Bijma, M. Boube et al., 2004. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 32 5379–5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, I., R. B. Green, O. Dunaevsky, J. A. Lengyel and C. Rauskolb, 2003. The oddskipped family of zinc finger genes promotes Drosophila leg segmentation. Dev. Biol. 263 282–295. [DOI] [PubMed] [Google Scholar]
- Harden, N., 2002. Signaling pathways directing the movement and fusion of epithelial sheets: lessons from dorsal closure in Drosophila. Differentiation 70 181–203. [DOI] [PubMed] [Google Scholar]
- Ito, K., W. Awano, K. Suzuki, Y. Hiromi and D. Yamamoto, 1997. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development 124 761–771. [DOI] [PubMed] [Google Scholar]
- Janody, F., Z. Martirosyan, A. Benlali and J. E. Treisman, 2003. Two subunits of the Drosophila mediator complex act together to control cell affinity. Development 130 3691–3701. [DOI] [PubMed] [Google Scholar]
- Johnston, L. A., and P. Gallant, 2002. Control of growth and organ size in Drosophila. BioEssays 24 54–64. [DOI] [PubMed] [Google Scholar]
- Kato, Y., R. Habas, Y. Katsuyama, A. M. Naar and X. He, 2002. A component of the ARC/Mediator complex required for TGF beta/Nodal signalling. Nature 418 641–646. [DOI] [PubMed] [Google Scholar]
- Kim, J., A. Sebring, J. J. Esch, M. E. Kraus, K. Vorwerk et al., 1996. Integration of positional signals and regulation of wing formation by Drosophila vestigial gene. Nature 382 133–138. [DOI] [PubMed] [Google Scholar]
- Kim, Y. J., and J. T. Lis, 2005. Interactions between subunits of Drosophila Mediator and activator proteins. Trends Biochem. Sci. 30 245–249. [DOI] [PubMed] [Google Scholar]
- Kuang, B., S. C. Wu, Y. Shin, L. Luo and P. Kolodziej, 2000. split ends encodes large nuclear proteins that regulate neuronal cell fate and axon extension in the Drosophila embryo. Development 127 1517–1529. [DOI] [PubMed] [Google Scholar]
- Kwon, J. Y., J. M. Park, B. S. Gim, S. J. Han, J. Lee et al., 1999. Caenorhabditis elegans mediator complexes are required for developmental specific transcriptional activation. Proc. Natl. Acad. Sci. USA 96 14990–14995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit, T., and S. M. Cohen, 1997. Proximal distal axis formation in the Drosophila leg. Nature 388 139–145. [DOI] [PubMed] [Google Scholar]
- Lewis, B. A., and D. Reinberg, 2003. The mediator coactivator complex: functional and physical roles in transcriptional regulation. J. Cell Sci. 116 3667–3675. [DOI] [PubMed] [Google Scholar]
- Li, W., J. Talavera, M. Lane and D. Kalderon, 1995. Function of protein kinase A in hedgehog signal transduction and Drosophila imaginal disc development. Cell 80 553–562. [DOI] [PubMed] [Google Scholar]
- Mahoney, P. A., U. Weber, P. Onofrechuk, H. Biessmann, P. J. Bryant et al., 1991. The fat tumor suppressor gene in Drosophila encodes a novel member of the cadherin gene superfamily. Cell 67 853–868. [DOI] [PubMed] [Google Scholar]
- Montagne, J., J. Groppe, K. Guillemin, M. A. Krasnow, W. J. Gehring et al., 1996. The Drosophila serum response factor gene is required for the formation of intervein tissue of the wing and is allelic to blistered. Development 122 2589–2597. [DOI] [PubMed] [Google Scholar]
- Morata, G., and P. Ripoll, 1975. Minutes: mutants of Drosophila autonomously affecting cell division rate. Dev. Biol. 42 211–221. [DOI] [PubMed] [Google Scholar]
- Morel, V., and F. Schweisguth, 2000. Repression by Suppressor of Hairless and activation by Notch are required to define a single row of singleminded expressing cells in the Drosophila embryo. Genes Dev. 14 377–388. [PMC free article] [PubMed] [Google Scholar]
- Naar, A. M., B. D. Lemon and R. Tjian, 2001. Transcriptional coactivator complexes. Annu. Rev. Biochem. 70 475–501. [DOI] [PubMed] [Google Scholar]
- Nellen, D., M. Affolter and K. Basler, 1994. Receptor serine/threonine kinases implicated in the control of Drosophila body pattern by decapentaplegic. Cell 78 225–237. [DOI] [PubMed] [Google Scholar]
- Parks, A. L., K. R. Cook, M. Belvin, N. A. Dompe, R. Fawcett et al., 2004. Systematic generation of high resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36 288–292. [DOI] [PubMed] [Google Scholar]
- Posakony, L. G., L. A. Raftery and W. M. Gelbart, 1990. Wing formation in Drosophila melanogaster requires decapentaplegic gene function along the anterior posterior compartment boundary. Mech. Dev. 33 69–82. [DOI] [PubMed] [Google Scholar]
- Prout, M., Z. Damania, J. Soong, D. Fristrom and J. W. Fristrom, 1997. Autosomal mutations affecting adhesion between wing surfaces in Drosophila melanogaster. Genetics 146 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves, W. M., and S. Hahn, 2005. Targets of the Gal4 transcription activator in functional transcription complexes. Mol. Cell Biol. 25 9092–9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll, P., and A. Garcia-Bellido, 1979. Viability of homozygous deficiencies in somatic cells of Drosophila melanogaster. Genetics 91 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roch, F., A. Baonza, E. Martin-Blanco and A. Garcia-Bellido, 1998. Genetic interactions and cell behaviour in blistered mutants during proliferation and differentiation of the Drosophila wing. Development 125 1823–1832. [DOI] [PubMed] [Google Scholar]
- Rogge, R., P. J. Green, J. Urano, S. Hornsaban, M. Mlodzik et al., 1995. The role of yan in mediating the choice between cell division and differentiation. Development 121 3947–3958. [DOI] [PubMed] [Google Scholar]
- Rogge, R. D., C. A. Karlovich and U. Banerjee, 1991. Genetic dissection of a neurodevelopmental pathway: Son of sevenless functions downstream of the sevenless and EGF receptor tyrosine kinases. Cell 64 39–48. [DOI] [PubMed] [Google Scholar]
- Sotillos, S., and J. F. de Celis, 2005. Interactions between the Notch, EGFR, and decapentaplegic signaling pathways regulate vein differentiation during Drosophila pupal wing development. Dev. Dyn. 232 738–752. [DOI] [PubMed] [Google Scholar]
- Sotillos, S., F. Roch and S. Campuzano, 1997. The metalloprotease disintegrin Kuzbanian participates in Notch activation during growth and patterning of Drosophila imaginal discs. Development 124 4769–4779. [DOI] [PubMed] [Google Scholar]
- St. Johnston, R. D., F. M. Hoffmann, R. K. Blackman, D. Segal, R. Grimaila et al., 1990. Molecular organization of the decapentaplegic gene in Drosophila melanogaster. Genes Dev. 4 1114–1127. [DOI] [PubMed] [Google Scholar]
- Taatjes, D. J., M. T. Marr and R. Tjian, 2004. Regulatory diversity among metazoan co-activator complexes. Nat. Rev. Mol. Cell Biol. 5 403–410. [DOI] [PubMed] [Google Scholar]
- Taubert, S., M. R. V. Gilst, M. Hansen and K. R. Yamamoto, 2006. A Mediator subunit, MDT15, integrates regulation of fatty acid metabolism by NHR49 dependent and independent pathways in C. elegans. Genes Dev. 20 1137–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbano, J. M., C. Torgler, C. Molnar, A. López-Varea, N. Brown et al., 2009. Drosophila laminins act as key regulators of basement membrane assembly and morphogenesis. Development 136 4165–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, E. P., and N. H. Brown, 1998. A screen to identify Drosophila genes required for integrin mediated adhesion. Genetics 150 791–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J. A., J. B. Bell and S. B. Carroll, 1991. Control of Drosophila wing and haltere development by the nuclear vestigial gene product. Genes Dev. 5 2481–2495. [DOI] [PubMed] [Google Scholar]
- Xu, T., and S. D. Harrison, 1994. Mosaic analysis using FLP recombinase. Methods Cell Biol. 44 655–681. [DOI] [PubMed] [Google Scholar]
- Xu, T., W. Wang, S. Zhang, R. A. Stewart and W. Yu, 1995. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121 1053–1063. [DOI] [PubMed] [Google Scholar]
- Yang, F., B. W. Vought, J. S. Satterlee, A. K. Walker, Z. Y. Jim Sun et al., 2006. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 442 700–704. [DOI] [PubMed] [Google Scholar]