Abstract
SkyLine, a high-throughput homology modeling pipeline tool, detects and models true sequence homologs to a given protein structure. Structures and models are stored in SkyBase with links to computational function annotation, as calculated by MarkUs. The SkyLine/SkyBase/MarkUs technology represents a novel structure-based approach that is more objective and versatile than other protein classification resources. This structure-centric strategy provides a multidimensional organization and coverage of protein space at the levels of family, function, and genome. The concept of “modelability”, the ability to model sequences on related structures, provides a reliable criterion for membership in a protein family (“leverage”) and underlies the unique success of this approach. The overall procedure is illustrated by its application to START domains, which comprise a Biomedical Theme for the Northeast Structural Genomics Consortium (NESG) as part of the Protein Structure Initiative (PSI). START domains are typically involved in the non-vesicular transport of lipids. While 19 experimentally determined structures are available, the family, whose evolutionary hierarchy is not well determined, is highly sequence diverse, and the ligand-binding potential of many family members is unknown. The SkyLine/SkyBase/MarkUs approach provides significant insights and predicts: 1) many more family members (~4,000) than any other resource; 2) the function for a large number of unannotated proteins; 3) instances of START domains in genomes from which they were thought to be absent; and 4) the existence of two types of novel proteins, those containing dual START domain and those containing N-terminal START domains.
Keywords: Homology modeling, Structural genomics, Bioinformatics, Protein function annotation, START domain, Arabidopsis thaliana
Overview
As the number of post-genomic sequences and structures continue to grow, both high-throughput experimental and computational methodologies are needed to organize these large bodies of data, principally according to computational tools. A major goal of the Protein Structure Initiative (PSI; http://www.nigms.nih.gov/Initiatives/PSI/) has been to develop new technologies to facilitate the solution of a large number of sequence unique protein structures to provide structural coverage of protein sequence space, from different perspectives, including the biological [1–5]. These structures can then be used as templates in homology modeling with the goals of providing higher structural information content regarding biological function and the structural coverage of protein sequence space. In this paper, we introduce a structure-based approach for the coverage (or leverage) and organization of protein sequence space and illustrate how the computational methodology transforms the current view of a particular protein family, the START domains, which is the subject of a Biomedical Theme at the Northeast Structural Genomics Consortium (NESG, http://www.nesg.org).
Computational approaches to structure-based organization and coverage of protein space
We developed a computational pipeline (SkyLine) for the automated, high-throughput detection of sequence homologs and the comparative modeling of their sequences [6]. As schematically depicted in Figure 1, the SkyLine process begins with a protein structure: The sequence of the structure is used as a seed for PSI-BLAST profile searches [7] against the NCBI non-redundant protein sequence database (NRdb, [8]), and the structure itself serves as a template to construct as many “reliable” homology models for these sequence homologs. In comparison to the usual homology modeling problem of finding a template with which to model a given sequence of unknown structure, Skyline addresses the inverse problem of determining how many sequences exist for which a particular structure may serve as a template [4,9,10]. SkyLine modeling uses structure evaluation to test whether a sequence is structurally consistent with its template. A model is considered “reliable” if 1) the pG score, which is a log-transformed, length-normalized integration over the residue by residue Prosa II profile [11], is ≥ 0.7 [12], and 2) the percent coverage of the detected sequence relative to the template sequence is ≥ 75%, ensuring that the protein coded by the detected sequence contains a biologically significant number of secondary structural elements. While we recognize that there are newer measures of model reliability available, our focus has not been on model details but rather whether a reasonable model can be built. To this end, the pG score has served as a fast and effective means of evaluating modelability.
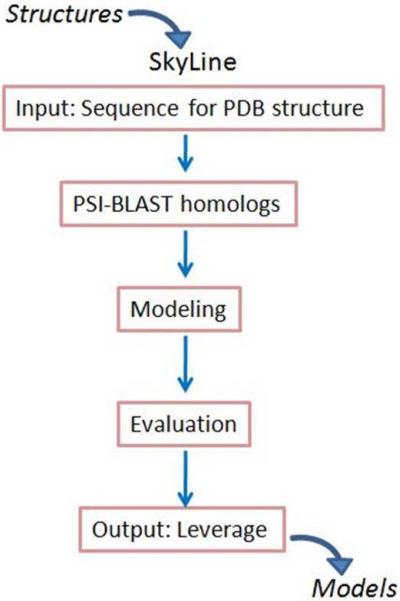
Figure 1.
Flowchart of the SkyLine pipeline. SkyLine [6] starts with a single PDB structure as input. The sequence of the structure is used as a seed in PSI-BLAST profile searches for homologs. Models are built for all non-redundant sequences detected using the input structure as the template and the sequence alignments derived from the PSI-BLAST profiles. Structure evaluation programs discern the reliable models, which comprise family members associated with the input structure. The models and associated information are stored in SkyBase, a web-accessible database.
SkyLine runs have been performed for all structures solved by NESG to date and a large fraction of the PDB defined by a 60% redundancy cutoff, whose 14 K structures likely represent virtually all protein functional types. SkyLine provides a means for defining protein families using structure-based criteria. Calculating the biophysical properties of structures and models provides a more detailed, complementary approach to analyzing families. MarkUs exploits global and local structural relationships among proteins to search for the conservation of biologically meaningful structural and functional motifs (which may be another way to view protein space) and provides access to many tools for computational function annotation ([13], http://luna.bioc.columbia.edu/honiglab/mark-us). Structures, models, and annotations derived from the MarkUs sever are stored in a publicly available database, SkyBase (http://156.145.102.40/nesg3/nesg.php).
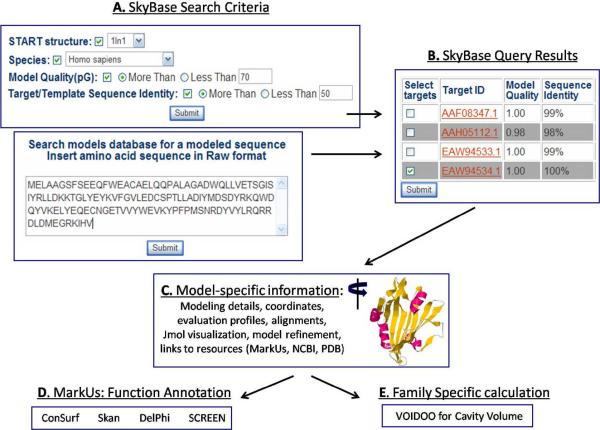
As illustrated in Figure 2, SkyBase may be queried, in a multi-factorial manner by incorporating the sequence into the online search window and/or by selecting a wide range of search criteria, such as model reliability (pG), model template, sequence identity to template, species, etc. The user of the SkyBase front-end can drill down to the level of individual model and, as listed in Figure 2, obtain a large amount of information on the model as well as access to external resources such as MarkUs [13], GenBank [8] and the PDB [14]. A Jmol window is also provided on the output page in which various user-selected representations of the model may be manipulated and rotated.
Figure 2.
Overall scheme of the computational structure-based characterization of protein families. START domain sequences and there models, calculated in the SkyLine analysis of a protein structure (PDB id 1LN1, [27]) are retrieved according to a variety of search parameters, including sequence and modeling features. Each model can be manipulated and visualized in a Jmol window, side-by-side with its Verify3D evaluation profile [37]. Links to MarkUs [13] provide access to previously calculated annotation results and the opportunity for further function analysis.
Our check for modelability/reliability (pG ≥ 0.7 and a coverage of sequence by template ≥ 75%) is independent of e-value; in most cases, there are anywhere from several to hundreds and even thousands of models for a given sequence that fit these criteria. SkyLine has a definition for discerning a “best” model per sequence, but we have found that it is most helpful to let the user have access to all of the models as well as to data such as the ProsaII structure evaluation profiles and the PSI-BLAST profiles. Different users will have different priorities, and there may be a particular region that is modeled well in one calculation versus the others. The box marked “OUTPUT” at the bottom of Figure 2 shows that any of the models retrieved according to the search criteria may be subsequently examined for more quantitative information.
The modeling step in SkyLine is key to the success of this approach, as it serves as a filter for true positives among the many false positive hits often included in PSI-BLAST results, especially if the sequence/PSI-BLAST profile inclusion e-value is relaxed beyond the typical cutoff of 0.001. Hence, this notion of a “reliability test” works just as well for models where the inclusion e-value is much larger, i.e. 0.001< e <100, and, thus, allows for the detection of true remote homolgs. We refer to this concept as “modelability”, and its application has allowed for the discovery of previously undetected protein family members and protein families, as illustrated in the next section.
START domains: An overview
Members of the steroidogenic acute regulatory (StAR)-related lipid transfer (START) domain family function in the binding and non-vesicular transport of lipid and other ligands [15–18]. Since many START domains appear in multi-domain proteins, they may serve as lipid sensors that signal biological responses. The START domain module is typically 210 residues long, and, currently, 19 experimentally determined structures are available in the PDB [19–21]. The three main classes of START domains- classical (CSD, from mammals, [15]), birch antigen (BA, from plants, [22–23]) and bacterial (BAC, [24])- share a common topology (Figure 3A): A C-terminal alpha-helix packed against a core beta-sheet provides support for a hydrophobic tunnel, which has been shown to accommodate lipid molecules in classical and BA START domains [25,26].
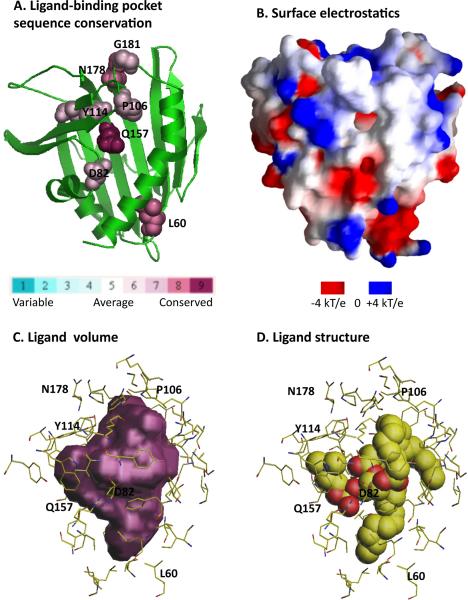
Figure 3.
Analysis of the lipid-binding pocket of the human phosphatidylcholine transfer protein (PC-TP, PDB id 1LN1, [27]). A. Conservation of residues lining the pocket is calculated and displayed by ConSurf [33]. B. Electrostatic surface potential is calculated and imaged with GRASP [34]. C. Volume plot of the ligand binding pocket is generated by VOIDOO [35]. D. The PC-TP ligand DLP (1,2-dilinoleoyl-dn-glycero-3-phosphocholine) is shown in molecular representation. Residues lining the ligand-binding pocket and predicted to be functionally important are delineated in C and D.
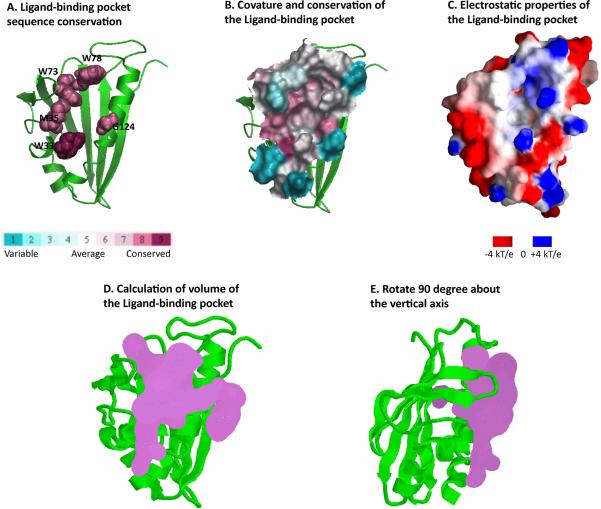
START domains are found in 15 distinct human proteins and are designated StARD1 through StARD15 [15]. Lipid specificity is known for only about half of these, and genetic disorders involved in cancers, autoimmune diseases and obesity have been found in all 15. Structures are known for four human START domains (StARD2/PCTP, StARD3/MLN64, StARD5, and StARD13) and one mouse START domains (StARD4). The phosphatidylcholine transfer protein (PCTP or StARD2; PDB id 1LN1, [27]) is depicted in Figure 3; its structure was solved in the presence of a phosphatidylcholine analog, which is accommodated in the domain's long hydrophobic groove, as illustrated in Figure 3D. The birch antigen START domains (not shown) contain similarly deep ligand-binding pockets [22]. NESG has contributed all of the bacterial START domains structure solved to date [24], and these structures are especially valuable because they provide the first structural pictures of this class of START domains about which very little is known. As illustrated in Figure 4A, the presence of the C-terminal packing helix suggests that bacterial START domains also accommodate ligands, however the absence of the three most N-terminal secondary structural elements (α1, β1 and β2) of human StarD2 (Figure 3A) suggests that the binding site is more shallow.
Figure 4.
Analysis of the proposed ligand-binding pocket of the Bacillus halodurans protein BH1534 (PDB id, 1XN5, [24]). A. Conservation of residues lining the pocket is calculated and displayed by ConSurf [33]. B. Curvature and conservation of the pocket surface is calculated with ConSurf [33]. C. Electrostatic surface potential is calculated and imaged with GRASP [34]. D and E. Volume plots of the putative ligand binding pocket were generated by SURFNET [36].
Intriguingly, the evolutionary hierarchy of START domains is not well established. Thus, the START domain functional super-family is an excellent target for our computational structure-based modeling and annotation approach.
Family analysis of START domains
SkyLine was used to search for the existence of all instances of START domains in the NCBI non-redundant sequence database. Each of the 19 available START domain structures, whose PDB identifiers and classifications are listed in the first two columns of Table 1, was used both as a sequence seed for PSI-BLAST searches against the NRdb and as the structural template for modeling the detected homologs
Table 1. START domain leverage and modelability.
PDB id's for the 19 template structures for START database analyzed by SkyLine are listed column1. BA, CSD and BAC in column 2 stand for Birch Antigen START domain, Classical START Domain, and Bacterial START domain, respectively. In column 3 through 7, the structure-based criteria for reliable models are pG ≥0.7 and query_coverage (or coverage of template by sequence) ≥ 75 %.
| START PDB id | START Sub-group | Reliable models, e-value≤0.001 | Reliable models, seqid ≥30% | Reliable models, e-value≥ 0.01 | Reliable models, e-value ≥ 0.1 | Reliable models, “unknown” |
|---|---|---|---|---|---|---|
| 1BV1 | BA | 561 | 458 | 14 | 13 | 21 |
| 1E09 | BA | 672 | 442 | 47 | 34 | 33 |
| 1EM2 | CSD | 651 | 74 | 0 | 0 | 8 |
| 1FM4 | BA | 759 | 448 | 5 | 1 | 33 |
| 1JSS | CSD | 703 | 32 | 2 | 2 | 8 |
| 1LN1 | CSD | 655 | 41 | 1 | 1 | 7 |
| 1T17 | BAC | 338 | 210 | 8 | 4 | 1 |
| 1XDF | BA | 669 | 382 | 32 | 18 | 25 |
| 1XFS | BAC | 927 | 72 | 2 | 1 | 2 |
| 1XN5 | BAC | 860 | 29 | 2 | 1 | 3 |
| 1XN6 | BAC | 867 | 24 | 1 | 0 | 2 |
| 1XUV | BA | 840 | 59 | 2 | 2 | 4 |
| 1Z94 | BA | 944 | 91 | 2 | 1 | 2 |
| 2BK0 | BA | 697 | 379 | 8 | 1 | 24 |
| 2FLH | BA | 755 | 12 | 1 | 1 | 37 |
| 2I9Y | BA | 656 | 65 | 2 | 2 | 25 |
| 2PCS | BAC | 938 | 74 | 2 | 0 | 4 |
| 2PSO | CSD | 516 | 78 | 1 | 1 | 1 |
| 2R55 | CSD | 712 | 46 | 5 | 5 | 7 |
| Total unique | 3827 | 1303 | 104 | 73 | 61 |
The results from the SkyLine runs are summarized in Table 1. The number of sequence-unique reliable models per structure is given in Column 3. The non-redundant total across all structures, i.e. the true leverage or “universe” of predicted START domains, is 3,886. Note that this number is much less than the sum across the 19 structures (13,720), which is due to the fact that more than one START domain structure may detect and serve as the template for the reliable modeling of a given sequence; these multiple models per sequence are retrievable through SkyBase (see “output” in Figure 2). This number of START domains far exceeds the number of START domains catalogued in any other available resource. This immediately raises the question of whether many of the SkyLine results may be false positive. This issue is addressed below, in the context of a specific genome (Arabidopsis thaliana) for which a detailed analysis is conducted. Of course, the most direct and only true way to test the efficacy of the prediction of novel sequences is to subject these discoveries to experimental analyses. Thus far, all of the many novel sequences tested have provided hypotheses that have been experimentally confirmed. One example involves the yeast genome, from which START domains were previously thought to be absent. SkyLine predicts the presence of four yeast START domains, one of which was recently characterized in the lab of Catherine Clarke at UCLA [28].
In order to compare our results for the number of START domains with other resources, we break down the analysis into four groupings as follows: 1) The number of predicted START domains based on SkyLine runs of CSD structures is 911; 2) the number of predicted START domains based on SkyLine runs of BA structures is 1,933; and 3) the number of predicted START domains based on SkyLine runs of BAC structures is 2,009. The sum of models from groups 1, 2 and 3 is 4,853, which provides evidence that START structure from the different classes can detect and reliably model sequences from other classes. As of Oct 2009, the number of START domains for Pfam families [29] representing classic START domains is 609, and the current similar number in the SMART database [30] in genomic mode is 314. Hence, for classical START domains, SkyLine predicts ~300 and ~600 more START domains than Pfam and SMART, respectively. The SMART database in “normal mode” reports a total of 817 START domains in 817 proteins, i.e. all instances of START domains appear once in each parent protein. A similar statement can be made based on Pfam results. SkyLine predicts a total of 33 proteins containing two instances of START domains; in each case the second, C-terminal instance is a novel discovery. Furthermore, no resources report any Archaeal START domains, and, in fact, it is stated in recent review articles that there are no START domains in Archaea. Similarly to the discovery of START domains in yeast genomes, SkyLine predicts a total of 15 sequence-unique, reliably modeled Archaeal sequences. Finally, column 8 of Table 1 shows that SkyLine produces a non-redundant total of 61 START domains in protein sequences that are annotated as “hypothetical”, “unnamed”, or “unknown” in sequence databases.
Table 1 makes additional interesting points related to the modeling procedure. The comparison of columns 3 and 4 show that utilizing an e-value ≤ 0.001 results in ~2,500 more START domains than impressing a sequence identity ≥ 30% between homolog and template sequences. Finally, columns 6 and 7 show that the approach predicts a significant number (~100) of remote homologs.
There is little overlap in the SkyLine results between classical and bacterial START domains. However, for example, human COQ10A (AAH47444), a Polyketide cyclase, is detected and reliably modeled by 1T17, a bacterial START domain. Note that this human START likely does not bind a lipid as its ligand. Bacterial START domains are thought to take polyketides as a ligand, hence these two diverse START domains may share in common the bacterial ligand, suggesting an evolutionary linkage.
START domains in Arabidopsis thaliana: A detailed modeling analysis
START domain-containing proteins are more broadly represented in plants than in animals [23]. A recent study identified 35 putative START domains in the Arabidopsis thaliana genome with a sequence-based approach (BLASTP), and, thus, these data provide an opportunity to test the efficacy of our structure-based approach in comparison [23]. SkyLine detects all 35 sequences at the PSI-BLAST stage and builds reliable models for 32 of them. Reliable models are built for the three remaining sequences upon refinement of the modeling alignments. Therefore, at least for sequences in this genome, the SkyLine modeling criteria are more conservative than sequence criteria, and while there are no false positives, it may be necessary to examine the modeling results in greater detail.
SkyLine expands the set of 35 sequences in several ways. First, reliable models are built for a group of sequences that include sequence variants of the 35 BLASTP sequences as well as two sequences that have not previously been annotated as START domains by any other resource (Table 2, rows 1 and 2). Furthermore, SkyLine predicts the presence of a START domain at the N-terminus of protein BAB01397, which contains a kinase domain at its C-terminus. According to the Conserved Domain Architecture Retrieval Tool [31], a similar architecture is also detected in a grapevine protein. This is an exciting result both because it has previously been thought that START domains appear invariably at the C-termini of multi-domain proteins and because START domains appear in concert with a kinase domain only in plant genomes. The inverse architecture (kinase-START) is found in two wheat proteins, WSK1 and WSK2 [23], where both domains are necessary to provide resistance to a devastating fungal disease. This suggests that the Arabidopsis protein BAB01397 may perform a similar defensive function. More intriguingly, SkyLine predicts, with high reliability, the presence of a second START domain in ten sequences of the reference set of 35 Arabidopsis proteins (Table 2, rows 3–12). This discovery represents the first instances of multiple START domain-containing proteins. For example, each of the 817 START domains reported by the SMART database is predicted to contain a single START domain; similar results are reported in Pfam, the NCBI Conserved Domain Database (CDD, [32]) and other available resources.
Table 2. Novel START domains in Arabidopsis thaliana predicted by SkyLine.
The models for all 12 novel sequences have high structure evaluation scores, i.e. pG ≥ 0.7, which suggests they are reliable models (column 4).
| NCBI name | Arabidopsis locus, ID | Model pG score | |
|---|---|---|---|
| 1 | AAF23188 | At3g11720 | 0.93 |
| 2 | NP_001031842 | At5g06440 | 0.95 |
| 3 | AAB71455 | At1g05230 | 0.95 |
| 4 | AAC69941 | At2g32370 | 0.90 |
| 5 | AAC80260 | At2g79840 | 0.90 |
| 6 | AAD17342 | At4g04890 | 0.95 |
| 7 | AAF97271 | At1g17920 | 0.89 |
| 8 | AAG30978 | At1g73360 | 0.83 |
| 9 | AAK59762 | At1g05230 | 0.91 |
| 10 | AAM14054 | At4g21750 | 0.83 |
| 11 | AAM91634 | At4g17710 | 0.92 |
| 12 | BAB10227 | At5g46880 | 0.92 |
All of the novel Arabidopsis sequences predicted by SkyLine are listed in Table 2. Structural models for the Arabidopsis START domains can be retrieved and analyzed using the tool MAPArT (Models of the Membrane-Associated proteins of Arabidopsis thaliana; http://156.145.102.40/araba2_ts/at_search.php), which is a SkyBase-derived resource that contains additional functional annotation performed for the NSF-funded Arabidopsis 2010 project. This example illustrates specifically how SkyBase may be used to organize protein sequence space at multiple levels, i.e. according to genome, family and function.
Calculation of biophysical properties of START domains
Because of the central role of START domains in non-vesicular lipid transport, it is of great importance both to understand the membrane associating function of START domains and to design inhibitors of START domain function. At least eight bacterial genomes contain START domains and some of these are pathogenic strains. Bacterial START domains are thought to be involved in the maintenance of membrane integrity, thus, it would be desirable to predict experimentally testable hypotheses of membrane-binding function and to find inhibitors for this class as well. SkyLine is designed to quickly detect a set of true homologs of a given structure, rather than necessarily providing the best possible models. However, experience has revealed that the models built with the SkyLine procedure are often reliable enough for accurate family classification and function annotation as predicted by calculations of 1) biophysical properties, such as surface curvature and electrostatic properties, 2) sequence conservation, and 3) ligand-binding propensities, as is performed in MarkUs. Figures 3 and 4 display the conservation of residues in the ligand-binding pocket [33], the electrostatic surface potential [34], and the calculation of the volume of the ligand-binding pocket [35,36] for human and bacterial START domains, respectively. Information obtained from the calculation of biophysical properties of START domains, such as those depicted in Figures 3 and 4, is critical in guiding and interpreting experiments. For example, highly conserved residues in the predicted ligand-binding pockets of START domains may be substituted in order to provide insight into the mechanism of ligand binding. Electrostatic potential calculations provide clues to the membrane-adsorption properties of lipid-binding START domains. And the calculation of ligand-binding volumes may be used to screen chemical libraries to detect potential ligands.
The comparison of panels C and D in Figure 3 illustrates that the ligand-binding pocket volume calculated for a START domain whose structure was solved in the presence of ligand faithfully represents the molecular shape of the ligand. Panels D and E show two views of the calculated ligand-binding pocket volume for a START domain whose structure was solved in the absence of ligand. Whether these types of calculations, for START domain structures without ligands and for high-quality homology models, have merit remains to be determined. We plan to use these calculated volumes to aid high-throughput experimental docking studies of START domain inhibitors. The integration of computational and experimental docking results into a single database will facilitate the design of more potent START domain inhibitors.
The information obtained from the computational analysis of START domain structures and models is stored in the START domain database (http://156.145.102.40/data/start/start.html) and can be used for guiding and interpreting the different kinds of experiments, for example, the cellular, molecular biological and biophysical characterization of the membrane and ligand binding functions of START domains.
Conclusions
We have outlined in this work how structural information can be used to provide novel insights about a single protein family that are well beyond what is possible with sequence alone. An essential feature of our approach is the use of modelability as a criterion to evaluate sequence relationships. Using a filtering based on modeling allows the relaxation of sequence-based criteria and thus significantly expands the number of homologs that can be identified. Together with the use of structural alignments and a range of functional information, we are now able to explore protein sequence/structure/function space in ways that were not previously possible.
Among the ~4,000 START domains identified in this work are a large number of remote homologs, including those from genomes in which START domains were previously not identified and within novel START domain-containing sequences for which our method predicts the presence of a second START domain. However, since so many of these sequences represent potential new instances of START domains, it is critical that these predictions be validated by experimental approaches, both functional studies, e.g. in the case of novel yeast START domains [28], and structure determination. To this end, NESG has instituted a target selection strategy in order to test these novel predictions and to provide comprehensive coverage of our expanded START domain universe.
The efficacy of our strategy is clearly evident in the new insights about the START domain family that we have provided. A critical aspect of the approach is the identification of a large number of putative relationships and their subsequent filtering based on modelability as done in SkyLine or functional criteria as used in MarkUs. However we recognize that the meaningfulness of a finding is often best determined by a researcher who has used computational tools with a hypothesis in mind or with the goal of identifying new hypotheses that can then be subjected to experimental verification. We believe that the computational infrastructure we have established will greatly facilitate this type of discovery process. More generally, our results highlight the fact that, combined with appropriate computational tools, structural genomics can have major impact on biological research in ways that are just now becoming evident.
Acknowledgements
B.H. acknowledges the support of National Institutes of Health Grants GM030518, GM074958, and CA121852. D.M acknowledges the support of National Institutes of Health Grants GM074958 and GM071700 and of National Science Foundation Grant NSF0738311.
Abbreviations
- BA
Birch Antigen
- BAC
Bacterial
- CSD
Classical START domain
- MLN64
metastatic lymph node 64 protein
- NCBI
National Center for Biotechnology Information
- NESG
NorthEast Structural Genomics consortium
- START
StAR-related lipid transfer
- PCTP
Phosphatidylcholine transfer protein
- PDB
Protein Data Bank
- pG
a score derived from the log-transformed, length-normalized integration over the residue by residue Prosa II structure evaluation profile
- PSI
Protein structure initiative
References
- 1.Schwede T, Sali A, et al. Outcome of a workshop on applications of protein models in biomedical research. Structure. 2009;17(2):151–9. doi: 10.1016/j.str.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dessailly BH, Nair R, et al. PSI-2: structural genomics to cover protein domain family space. Structure. 2009;17(6):869–81. doi: 10.1016/j.str.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terwilliger TC, Stuart D, Yokoyama S. Lessons from structural genomics. Annu Rev Biophys. 2009;38:371–83. doi: 10.1146/annurev.biophys.050708.133740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold K, Kiefer F, et al. The Protein Model Portal. J Struct Funct Genomics. 2009;10(1):1–8. doi: 10.1007/s10969-008-9048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berman HM, Westbrook JD, et al. The protein structure initiative structural genomics knowledgebase. Nucleic Acids Res. 2009;37(Database issue):D365–8. doi: 10.1093/nar/gkn790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mirkovic N, Li Z, et al. Strategies for high-throughput comparative modeling: applications to leverage analysis in structural genomics and protein family organization. Proteins. 2007;66(4):766–77. doi: 10.1002/prot.21191. [DOI] [PubMed] [Google Scholar]
- 7.Altschul SF, Madden TL, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson DA, Karsch-Mizrachi I, et al. GenBank: update. Nucleic Acids Res. 2004;32(Database issue):D23–6. doi: 10.1093/nar/gkh045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 10.Schwede T, Kopp J, et al. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31(13):3381–5. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sippl MJ. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17(4):355–62. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- 12.Sanchez R, Sali A. Large-scale protein structure modeling of the Saccharomyces cerevisiae genome. Proc Natl Acad Sci U S A. 1998;95(23):13597–602. doi: 10.1073/pnas.95.23.13597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrey D, Fischer M, et al. Structural relationships among proteins with different global topologies and their implications for function annotation strategies. Proc Natl Acad Sci U S A. 2009;106(41):17377–82. doi: 10.1073/pnas.0907971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berman HM, Westbrook JD. The impact of structural genomics on the protein data bank. Am J Pharmacogenomics. 2004;4(4):247–52. doi: 10.2165/00129785-200404040-00004. [DOI] [PubMed] [Google Scholar]
- 15.Alpy F, Tomasetto C. Give lipids a START: the StAR-related lipid transfer (START) domain in mammals. J Cell Sci. 2005;118(Pt 13):2791–801. doi: 10.1242/jcs.02485. [DOI] [PubMed] [Google Scholar]
- 16.Hanada K, Kumagai K, Tomishige N, Kawano M. CERT and intracellular trafficking of ceramide. Biochim Biophys Acta. 2007 Jun;1771(6):644–53. doi: 10.1016/j.bbalip.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 17.Wirtz KW. Phospholipid transfer proteins in perspective. FEBS Lett. 2006 Oct 9;580(23):5436–41. doi: 10.1016/j.febslet.2006.06.065. [DOI] [PubMed] [Google Scholar]
- 18.Strauss JF, 3rd, Kishida T, Christenson LK, Fujimoto T, Hiroi H. START domain proteins and the intracellular trafficking of cholesterol in steroidogenic cells. Mol Cell Endocrinol. 2003 Apr 28;202(1–2):59–65. doi: 10.1016/s0303-7207(03)00063-7. [DOI] [PubMed] [Google Scholar]
- 19.Tsujishita Y, Hurley JH. Structure and lipid transport mechanism of a StAR-related domain. Nat Struct Biol. 2000;7(5):408–14. doi: 10.1038/75192. [DOI] [PubMed] [Google Scholar]
- 20.Im YJ, Raychaudhuri S, et al. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 2005;437(7055):154–8. doi: 10.1038/nature03923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanno K, Wu MK, et al. Structure and function of phosphatidylcholine transfer protein (PC-TP)/StarD2. Biochim Biophys Acta. 2007;1771(6):654–62. doi: 10.1016/j.bbalip.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radauer C, Lackner P, et al. The Bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol Biol. 2008;8:286. doi: 10.1186/1471-2148-8-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schrick K, Nguyen D, et al. START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 2004;5(6):R41. doi: 10.1186/gb-2004-5-6-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen Y, Goldsmith-Fischman S, et al. 2005 [Google Scholar]
- 25.Gajhede M, Osmark P, et al. X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy. Nat Struct Biol. 1996;3(12):1040–5. doi: 10.1038/nsb1296-1040. [DOI] [PubMed] [Google Scholar]
- 26.Tsujishita Y, Hurley JH. Structure and lipid transport mechanism of a StAR-related domain. Nat Struct Biol. 2000;7(5):408–14. doi: 10.1038/75192. [DOI] [PubMed] [Google Scholar]
- 27.Roderick SL, Chan WW, et al. Structure of human phosphatidylcholine transfer protein in complex with its ligand. Nat Struct Biol. 2002;9(7):507–11. doi: 10.1038/nsb812. [DOI] [PubMed] [Google Scholar]
- 28.Barros MH, Johnson A, et al. The Saccharomyces cerevisiae COQ10 gene encodes a START domain protein required for function of coenzyme Q in respiration. J Biol Chem. 2005;280(52):42627–35. doi: 10.1074/jbc.M510768200. [DOI] [PubMed] [Google Scholar]
- 29.Finn RD, Tate J, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36(Database issue):D281–8. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schultz J, Milpetz F, et al. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95(11):5857–64. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geer LY, Domrachev M, Lipman DJ, Bryant SH. CDART: protein homology by domain architecture. Genome Res. 2002;12:1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landau M, Mayrose I, et al. ConSurf: the projection of evolutionary conservation scores of residues on protein structures. Nucl. Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicholls A, Sharp KA, Honig B. Protein Folding and Association: Insights From the Interfacial and Thermodynamic Properties of Hydrocarbons. Proteins: Stuc., Func. and Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 35.Kleywegt GJ, Jones TA. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 2):178–85. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- 36.Laskowski RA. SURFNET: “A program for visualizing molecular surfaces, cavities and intermolecular interactions. J. Mol. Graph. 1995;13:323–330. doi: 10.1016/0263-7855(95)00073-9. [DOI] [PubMed] [Google Scholar]
- 37.Eisenberg D, Lüthy R, Bowie JU. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;(277):396–404. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]