Abstract
PVS-RIPO is a genetically recombinant, non-pathogenic poliovirus chimera with a tumor-specific conditional replication phenotype. Consisting of the genome of the live attenuated poliovirus type 1 (Sabin) vaccine with its cognate IRES element replaced with that of human rhinovirus type 2, PVS-RIPO displays an inability to translate its genome in untransformed neuronal cells, but effectively does so in cells originating from primary tumors in the central nervous system or other cancers. Hence, PVS-RIPO unleashes potent cytotoxic effects on infected cancer cells and produces sustained anti-tumoral responses in animal tumor models. PVS-RIPO presents a novel approach to the treatment of patients with glioblastoma multiforme, based on conditions favoring an unconventional viral translation initiation mechanism in cancerous cells. In this review, we summarize advances in the understanding of major molecular determinants of PVS-RIPO oncolytic efficacy and safety and discuss their implications for upcoming clinical investigations.
1. Introduction
The idea to enlist the help of human pathogens in the fight against cancer first emerged from early anecdotal reports of spontaneous tumor remissions following immunization with live attenuated viruses or co-incidental infection (reviewed in [1]). More than 100 years later, a multitude of viral agents have been proposed to serve such purposes and are at various stages of pre-clinical or clinical testing. The unifying principle underlying each of these is conditional replication and cytotoxicity in cancerous cells combined with reduced propagation in normal tissues and, hence, pathogenic potential [2]. Strategies to devise oncolytic viral agents can be roughly divided in two groups according to diverse mechanisms of tumor selectivity. Certain viruses exhibit inherent selective cytopathogenicity for transformed human cells, because viral replication/pathogenesis is naturally restricted in normal human tissues. Examples for this group include human orphans, e.g. reovirus [3], or animal viruses with lacking human tropism, e.g. vesicular stomatitis virus [4] or myxoma virus [5]. A proposed common mechanistic basis for tumor selectivity of these agents is that virus propagation in normal human tissues is tightly controlled via innate immune responses, accounting for their low inherent pathogenic potential. In contrast, tumor cells, which commonly exhibit deficient innate immunity [6], may permit replication and cell killing of human orphan viruses, e.g. reovirus. Viruses with lacking human host range may exhibit similar specificity because viral mechanisms to counter innate host defenses in their respective host species are ineffective in normal human cells [7]. However, these viruses may exhibit significant cytotoxicity in tumor cells with defective innate defenses, due to removal of the primary barrier to viral replication in cells of a foreign host.
Other viruses under consideration for use as oncolytic agents require sophisticated genetic manipulations in order to achieve tumor selectivity because of inherent pathogenic potential. Examples include adenovirus [8], herpes simplex virus (HSV) [9], or poliovirus [10]. A plethora of genetic manipulations have been introduced to viral genomes to achieve conditional replication phenotypes. For oncolytic HSVs these include deletion of genes deemed essential for replication in normal cells but dispensable in cancerous cells (e.g. the HSV γ34.5 and ribonucleotide reductase genes [11]), insertion of tumor-specific promoters [12], or manipulation of genetic elements involved in translation control [13].
2. Limitations of genetic manipulation of oncolytic viruses
Intuitively, basing oncolytic virus strategies on pathogenic viruses has certain advantages. For example, the documented inherent capacity of neuroinvasive HSVs or poliovirus to disseminate in the human brain may aid in targeting regionally diffuse glioma. Also, the availability of specific chemotherapy (e.g. against HSV) or vaccines (e.g. against poliovirus) can help to mitigate concerns about the potential unintended public health consequences of virus administration to cancer patients. However, the need for genetic manipulation to achieve tumor selectivity with such viruses comes at a price.
DNA viruses prima facie offer better opportunities for targeted genetic manipulation than RNA viruses, due to their relative genetic stability and dependence on a comparatively large set of genes. However, empirical evidence with adenoviruses and HSVs revealed certain obstacles to achieving true conditional replication in cancerous cells through genetic alteration. The γ34.5 gene in HSV-1 was shown to be essential for neurovirulence [14] because of its role in reversing innate host defenses mediated by activation of protein kinase R (PKR) in HSV-infected host cells [15]. PKR counteracts viral propagation and dissemination by phosphorylation of eukaryotic translation factor (eIF) 2α, leading to protein synthesis shut-off in infected cells. The HSV-1 γ34.5 gene product associates with protein phosphatase 1α and induces eIF2α de-phosphorylation, maintaining a pool of functional eIF2α and preventing a block of protein synthesis in infected cells [16]. Reminiscent of cell type-specificity of orphan viruses due to defects of innate host defenses in cancer, γ34.5-deleted HSVs were proposed to exhibit tumor-specific conditional replication due to cancer-specific defects of PKR function [11]. HSVs with γ34.5 deletions have highly attenuated neurovirulence in authentic primate models [17], but viral replication and cytotoxicity was also severely impaired in certain tumor cell lines [18]. Several factors may explain such observations. Deletion of viral genes rarely produces narrowly defined pinpoint defects, because the loss of a gene product is likely to affect viral functions in more than one way. More significantly, a myriad of biochemical anomalies in cancer can cause variable PKR pathway defects. These may produce different degrees of functional deficits, may be distributed unevenly within individual cancers, or may mediate sufficient PKR activity in the absence of γ34.5 to thwart HSV-1 propagation. It is of concern that no clear molecular correlates of PKR dysfunction have been identified and, thus, patient tumor samples cannot be tested for potential susceptibility to γ34.5-deleted HSVs.
Genetic manipulation of RNA viruses such as poliovirus [10] is problematic for different reasons. The genetic austerity of RNA viruses greatly limits the possibility for targeted deletion. The use of polyproteins, multifunctional gene products, or cis-acting RNA elements in open reading frames, hinder simple strategies of removing select viral functions without affecting propagation and cytotoxicity in all cells, including the intended tumor targets. Most worryingly, due to the inherent genetic infidelity of RNA viruses, any genetic alteration of their genomes may be subject to reversion in patient tumors. Although genetic variants of the viral RNA-dependent RNA polymerase with improved fidelity have been reported [19, 20], there are no simple strategies to circumvent rapid genetic adaptation. Our strategy to address this problem and the potential mechanistic basis for our approach are discussed below.
3. The rationale for oncolytic polioviruses
The principal reason to consider poliovirus for cancer targeting is simple: the poliovirus receptor, nectin-like molecule 5 (Necl-5), is broadly up-regulated in malignancy. Receptor tropism is an Achilles heel for many oncolytic virus strategies, because tropism for intended tumor targets may be inherently poor or difficult to determine empirically (e.g. due to multiple molecular entities with complex control over host cell binding/entry or in cases where receptor identity is unknown). Poliovirus uniquely depends on Necl-5 for host cell binding and entry and, based on all available empirical evidence, the molecule is sufficient for all binding and entry functions leading to uncoating of the viral genome [21]. Necl-5 is the founding member of the nectin/nectin-like family of genes [22] (Table 1). The family comprises several cell surface molecules implicated in cell-to-cell adhesion functions in various physiologic contexts [22] (Table 1). Interestingly, Nectins 1/2 serve as cellular receptors for a-HSVs [23].
Table 1.
The nectin/nectin-like family of genes.
| Member | Alternative nomenclature | Proposed functions | References |
|---|---|---|---|
| Nectin-1 | PRR1, HVEC | Cell-cell adhesion molecule | [23] |
| α-HSV receptor | [22] | ||
| Involved in dysraphism (cleft lip/palate-ectodermal dysplasia syndrome) (cleft lip/palate-ectodermal dysplasia syndrome) | [24] | ||
| Nectin-2 | PRR2, HVEB | Cell-cell adhesion molecule | [25] |
| α-HSV receptor | [22] | ||
| Nectin-3 | PRR3 | Cell-cell adhesion molecule | [26] |
| Nectin-4 | Cell-cell adhesion molecule | [27] | |
| Necl-1 | TSLL1, SynCAM3 | Cell-cell adhesion molecule in the CNS | [28,29] |
| Necl-2 | IGSF4, RA175, SgIGSF, TSLC1, SynCAM1 | Cell-cell adhesion molecule | [29] |
| Necl-3 | SynCAM2 | Putative cell-cell adhesion molecule | [30] |
| Necl-4 | TSLL2, SynCAM4 | Cell-cell adhesion molecule in the CNS | [31] |
| Necl-5 | Tage4, PVR, CD155 | Poliovirus receptor | [20] |
| Associated with malignancy | [32-36] |
There is abundant empirical evidence to suggest that Necl-5 is ectopically expressed in malignancy. Virtually all explant human tumor cell lines tested, with the exception of certain lymphoma cell lines lacking Necl-5, support poliovirus infection, implying Necl-5 expression. Association with colo-rectal [33], breast [34], or glioblastoma multiforme (GBM) [35] has been documented in patients’ tissues. An association of Necl-5 with GBM is particularly relevant for the approach discussed in this review [10]. Following simple expression studies [35], Necl-5 has been implicated in GBM cell invasion and migration [36, 37]. Ectopic expression of Necl-5 has been linked with activation of the morphogen sonic hedgehog via its effectors, the gli transcription factors [38] or Ras signaling [39]. However, the precise circumstances for abnormal regulation of nectin family members in cancer remain unknown. An interesting study in explant rodent liver cells showed de-regulation of the Necl-5 gene upon diverse types of disrupted histoarchitecture, e.g. acute injury, wound repair and transformation [40]. Lacking tumor cell tropism is a major obstacle to oncolytic virus efficacy and has prompted efforts to genetically engineer adenoviruses [41] or HSVs [42] with re-targeted receptor affinities. Thus, natural tropism for a widespread and abundant molecular signature on cancer cell surfaces is a most useful property for developing poliovirus as an anti-cancer agent. However, tropism for Necl-5 is also a major determinant for the serious neuropathogenic properties of poliovirus, mediating specific targeting of lower motor neurons [43].
4. A poliovirus recombinant with tumor-specific replication/cytotoxicity
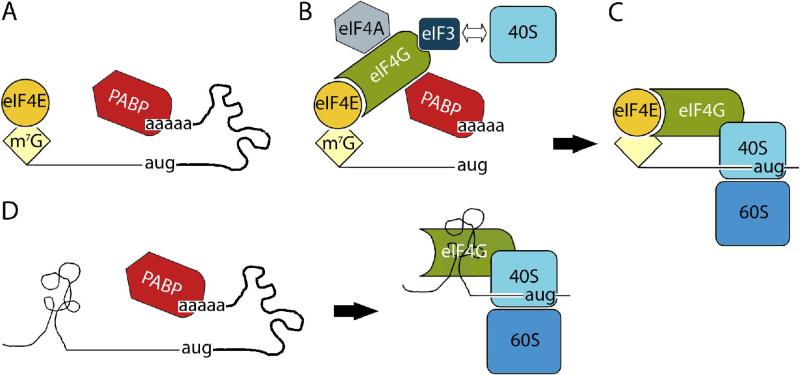
Inherent neuropathogenicity of wild-type poliovirus prevents its consideration for therapeutic purposes. Conditional replication of poliovirus in cancerous cells was achieved by manipulating the viral 5’UTR and a cis-acting genetic element involved in translation regulation, the internal ribosomal entry site (IRES), in particular. The prototype oncolytic poliovirus, termed PVS-RIPO, is the live attenuated poliovirus type 1 (Sabin) containing a heterologous IRES derived of human rhinovirus type 2 (HRV2) [44]. Poliovirus (+)strand genomic RNA is naturally devoid of a 5’ 7-methyl-guanosine (m7G) ‘cap’ structure [45] and, therefore, lacks the capacity to initiate translation via the canonical m7G-cap-dependent mechanism exemplified by eukaryotic mRNAs (Fig. 1). Conventional translation initiation occurs upon binding of the cap-binding translation factor, eIF4E, to the m7G-cap (Fig. 1A), and recruitment of 40S ribosomal subunits to mRNAs via the scaffolding factor eIF4G (Fig. 1B). Scanning of the 5’ untranslated region (UTR) leads to 60S subunit joining at the initiation codon and translation elongation (Fig. 1C).
Fig. 1.
Cap-dependent and IRES-mediated translation initiation. (A) - (C) eIF4E binding to the m7G-cap and binding of eIF4G result in recruitment of 40S ribosomal subunits to eukaryotic mRNAs. (D) Alternatively, IRES-mediated translation occurs without involvement of the 5’ terminus or eIF4E and, at least for poliovirus [47], likely occurs via direct eIF4G binding to the IRES.
Poliovirus translation initiation cannot occur via eIF4E, because the m7G-cap is absent. Instead, recent investigations suggest that poliovirus initiates viral protein synthesis via direct interaction of eIF4G with the viral RNA [46, 47] (Fig. 1D). This permits recruitment of 40S ribosomal subunits independent of the m7G-cap and eIF4E. Instead, ribosome recruitment depends on the ability of the IRES, a highly structured cis-acting genetic element of ~500 nucleotide length in the poliovirus 5’UTR [48, 49], to attract eIF4G. Exchanging the cognate poliovirus IRES for its HRV2 counterpart had no significant influence on viral replication kinetics in HeLa cells or other transformed cells routinely used for poliovirus propagation [10, 44, 50]. However, this manipulation caused substantially reduced neurovirulence in mice transgenic for the human Necl-5 gene [10]. Attenuated neurovirulence correlates with drastically reduced propagation potential in cells of neuronal derivation, e.g. neuroblastoma cells [10] or HEK-293 neuroblastic cells [51, 52]. Neuronal incompetence of PVS-RIPO is associated with reduced translation initiation capability at the recombinant HRV2 IRES, possibly due to association with a heterodimeric RNA-binding protein, DRBP76:NF45, with neuron-specific cytoplasmic distribution [53, 54]. The non-pathogenic, non-neurovirulent phenotype of PVS-RIPO was thoroughly documented in a recent, IND-directed dose-range finding, toxicology and biodistribution study by intrathalamic inoculation in a confirmed relevant primate species, Cynomolgus macaques (unpublished data).
A key question emerging from these findings is the mechanism of PVS-RIPO conditional replication in cancerous cells. Since the IRES may participate in genome replication functions involving the 3’UTR [55, 56], it may interact with viral as well as host proteins and depends on host translation initiation factors (at a minimum eIF4G, eIF4A and eIF3; Fig. 1D). There are infinite possibilities for variable performance in a cell type-specific manner. Our investigations indicate that cell type-specific cytotoxicity is the result of both repressive factors in the CNS and functions specifically favoring viral, cap-independent translation in cancer. Important hints towards possible mechanistic explanations for PVS-RIPO selective propagation and cytotoxicity in cancer cells stem from unrelated functional studies of the IRES:eIF4G relationship [46], and from safety studies in experimental animal models of GBM.
6. Genetic stability of PVS-RIPO
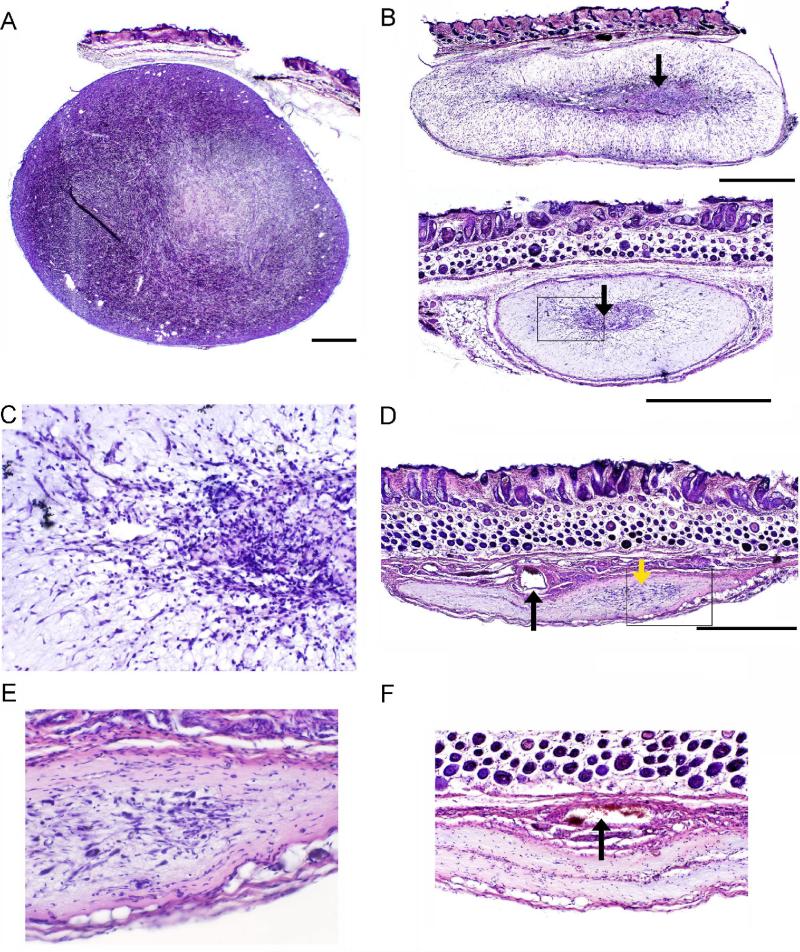
Plans for clinical tests of PVS-RIPO demanded rigorous investigation of the major biosafety concern, genetic instability (see above). We observed the remarkable genetic stability of PVS-RIPO upon long-term serial passage in HeLa cells in the laboratory in preliminary pre-clinical studies in experimental animals [10] and during good-manufacturing-practice production of the agent in VERO cells (unpublished data). However, to systematically address concerns that PVS-RIPO, while proliferating in tumor tissue, might acquire genetic alterations resulting in a changed pathogenic phenotype, serial in vivo passage in GBM xenografts were performed [57]. Intratumoral virus administration produced rapid histological changes in xenografts (Fig. 2). Ten days after virus administration, xenografts had shrunk considerably (Fig. 2B). A remaining central core containing potentially viable tumor cells was surrounded by fibrotic periphery, which was presumably free of tumor cells (Fig. 2B). The central core was filled with invading inflammatory cells extending into the periphery (Fig. 2C). Eighteen days later, there was no evidence of significant viable tumor cells in the majority of the animals; instead, the former xenograft appeared to have been replaced by a scar (Fig. 2D-F). Attempts to isolate virus from lesions 28 days after PVS-RIPO administration failed, possibly correlating with the absence of viable tumor cells. Virus was recovered, however, from intermediate lesions 10 days after PVS-RIPO administration. It was estimated that virus recovered from two consecutive serial passages in GBM xenografts, including amplification of virus in U-118 cells in vitro, was the result of at least 15 rounds of replication [57]. The final isolate was tested for tumor-specific replication in an assay based on cell type-specific growth deficits in HEK-293 cells [51] vs. efficient viral propagation in U-87 GBM cells [58]. Moreover, the material was subjected to full-length genome sequencing. These tests revealed that PVS-RIPO remained genetically stable and maintained the cell type-specific conditional replication phenotype upon prolonged serial passage in GBM cells in vitro and in vivo [57].
Fig. 2.
Histopathological analysis of PVS-RIPO-induced oncolysis of HTB-15 xenografts in athymic mice [57]. The bar shown with low-magnification images represents 1 mm. (A) Crosssection of a vehicle-injected tumor in an animal sacrificed 40 days after tumor implantation. The xenograft displays typical dense, hyper-cellular morphology. (B) Representative xenografts 10 days post PVS-RIPO inoculation. Only a minor, central portion of the xenograft retains the characteristic appearance of tumor (arrows). The boxed region is shown at higher magnification in (C). (C) Detail of (B) (bottom panel). Dense infiltrates surrounding and invading the central part of the xenograft can be distinguished at higher magnification. (D) Overview of a HTB-15 xenograft 28 days after PVS-RIPO inoculation. A scar had replaced the tumor, but a minor focus of remaining active tissue re-arrangement is visible (yellow arrow). A large patent vessel (black arrow) and surrounding collapsed minor vessels may represent the former tumor's vascular supply. The boxed region is shown at higher magnification in (E). (E) Detail of (D) showing the area of active tissue re-modeling at higher magnification. (F) Two out of 4 tumors had entirely been eliminated 28 days post PVS-RIPO and replaced by a scar. The remnants of several large vessels are visible at the center of the scar's base (arrow).
Although scientifically rather unexciting, the genetic stability tests provided important information about possible mechanisms responsible for efficient PVS-RIPO propagation and cytotoxicity in tumors. It had been suggested that neuro-attenuation of PVS-RIPO may simply be the result of generally impaired viral propagation potential due to the heterologous HRV2 IRES. Efficient viral propagation in GBM could be the result of the broadly more permissive environment for viral propagation in these cells. If this were the case, we would almost certainly have observed genetic adaptation in GBM cells to remove any obstacle to viral growth. Polioviruses react with immediate genetic rearrangement to any possible impediment to efficient replication [55]. We interpret the absence of genetic adaptation upon sustained growth in GBM cells as an indication of specific conditions in GBM cells that favor optimal function of the genetically engineered 5’UTR in PVS-RIPO. Since the major genetic distinction of PVS-RIPO is the presence of the heterologous HRV2 IRES, unraveling mechanisms of alternative translation initiation in GBM became a priority for pre-clinical studies aimed at identifying molecular correlates for PVS-RIPO efficacy in GBM.
7. PVS-RIPO and translation control in cancer
Despite its reduced requirements for protein synthesis initiation, even picornavirus IRES-mediated translation depends on initiation factors mediating ribosome recruitment to RNAs. Therefore, an obvious level of regulation of PVS-RIPO translation and growth, or picornavirus propagation in general, is controlled activity of translation factors. This concept is supported by the fact that both central initiation factors critically involved in ribosome recruitment at eukaryotic mRNAs, eIF4E and −4G, are at the receiving end of multiple signal transduction pathways with widespread activity in GBM [59]. Accordingly, activation of major oncogenic signaling pathways in GBM has been implicated in differential ribosome recruitment to select host cell transcripts [60]. Based on the observed genetic stability of PVS-RIPO in GBM cells, it is compelling to assume that conditions for translation initiation at the heterologous HRV2 IRES are optimal in such cells. Although other factors cannot be excluded, the most immediate candidates for a role determining the rate of alternative, cap-independent translation in GBM are eIF4E and −4G.
Major signaling pathways via phospho-inosite-3-kinase/mammalian target of rapamycin (PI3K/mTOR) and RAS/mitogen activated protein kinase (MAPK) are universally active in GBM [59]. Both pathways are intricately involved in translation regulation via modification of eIF4E, −4G or the eIF4E-binding proteins (4E-BPs), which regulate their interaction. Activation of mTOR produces 4E-BP hyperphosphorylation and dissociation from eIF4E, thus stimulating cap-dependent translation initiation [61, 62]. Activation of Erk 1/2 MAPK signaling by growth factors via oncogenic RAS culminates in activation of the MAPK-integrating kinase-1 (Mnk1) [63, 64], which phosphorylates eIF4E at Ser209. eIF4G is phosphorylated at multiple sites, including Ser1108, which is sensitive to the mTOR inhibitor rapamycin [65]. Involvement of other signal transduction pathways is likely, but control of eIF4G function remains poorly understood. In contrast to eIF4E, which only interacts with eIF4G and the 4E-BPs, eIF4G engages in complex interactions with translation factors eIF4E, −4A, poly(A) binding protein (PABP), and eIF3 [66], with 40S ribosomal subunits (indirectly) and, at a minimum, with kinases of the Mnk family [67]. Therefore, the potential functional consequences of post-translational modification of eIF4G are significant, in particular when considering eIF4G's role in direct association with viral IRESes [46, 47].
While several distinct signaling effects on translation factors were investigated for their role in translation regulation, e.g. the role of eIF4E phosphorylation [68], there is no unifying hypothesis explaining the effects of the combined, converging oncogenic signal to the protein synthesis machinery. The matter is complicated by the simultaneous confluence of multiple signaling events and the enormous heterogeneity of eukaryotic translation templates, which may enable alternative means of translation initiation to varying degrees. Therefore, we speculate that signal transduction to the protein synthesis machinery produces a range of discrete changes in translation rate at diverse templates rather than uniform shifts of translation activity across the transcriptome. The question of pre-eminent interest in the context of PVSRIPO is control of the balance of cap-dependent vs. viral, strictly cap-independent translation in cells with active oncogenic signaling. This issue has been addressed empirically before in a variety of experimental systems [62, 69, 70], but remains inconclusive. PVS-RIPO is a unique sensor of this balance because it categorically depends on eIF4G and cap-independent translation via its IRES. Also, since the viral RNA is self-amplifying, even modest changes favoring viral translation of few viral RNAs immediately upon uncoating can be conveniently detected as enhanced viral propagation, translation and cytotoxicity. Intense investigations of the effects of the converging oncogenic signal to the protein synthesis machinery on specific modes of translation initiation are underway.
8. Summary and outlook
Ten years of pre-clinical investigation of PVS-RIPO, the molecular mechanisms responsible for neuro-attenuation and GBM-specific cytotoxicity, have revealed a surprisingly complex interplay of extracellular factors, e.g. receptor expression and/or tissue type-specific receptor function, and intracellular factors, e.g. the protein synthesis machinery and its regulation in malignancy. These studies suggest that conditional replication phenotypes associated with oncolytic viruses can rarely be attributed to single factors simultaneously explaining viral propagation and cytotoxicity in multiple target tissues (e.g. inherent ‘normal’ targets vs. cancerous cells). This assertion is supported by the increasingly complex empirical record for molecular mechanisms involved in cell type-specific replication of oncolytic adenoviruses or HSVs.
Deciphering the intricate regulatory networks that dictate viral replication and cytotoxicity appears cumbersome in light of the urgent need for innovative cancer therapy, but it may be essential to successful application of oncolytic viruses. Moreover, correlative mechanistic analyses of oncolytic viruses may shed surprising insight into basic properties of malignancy, e.g. translation regulation and its response to oncogenic signals, which have broad implications for the understanding of malignancy in general or other types of experimental therapies. PVSRIPO is scheduled to enter phase-I clinical investigation against recurrent GBM. As plans for introduction into the clinic materialize, we will continue to rigorously investigate the molecular mechanisms underlying viral oncolysis.
Biographies
 Christian Goetz is a PhD candidate in the Dept. of Molecular Cancer Biology at Duke University. He received his undergraduate degree at the Eberhard Karls Universitaet in Tuebingen, Germany and at the University of Michigan. He joined Dr. Gromeier's group in 2007 and his studies have focused on the tumor cell killing mechanism of oncolytic polioviruses. He has been particularly interested in oncogenic signaling pathways controlling translation of the viral genome.
Christian Goetz is a PhD candidate in the Dept. of Molecular Cancer Biology at Duke University. He received his undergraduate degree at the Eberhard Karls Universitaet in Tuebingen, Germany and at the University of Michigan. He joined Dr. Gromeier's group in 2007 and his studies have focused on the tumor cell killing mechanism of oncolytic polioviruses. He has been particularly interested in oncogenic signaling pathways controlling translation of the viral genome.
Matthias Gromeier, MD, is Associate Professor in of Surgery, Div. of Neurosurgery at Duke Univ. Medical School. He is a former recipient of a Burroughs Wellcome Career Award in the Biomedical Sciences. His research interests are in regulation of protein synthesis in cancer and novel ways to target abnormal translation regulation, e.g. via the use of oncolytic viruses.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sinkovics JG, Horvath JC. Virus therapy of human cancers. Melanoma Res. 2003;13:431–2. doi: 10.1097/00008390-200308000-00015. [DOI] [PubMed] [Google Scholar]
- 2.Gromeier M. Viruses for treating cancer. ASM News. 2002;68:438–45. [Google Scholar]
- 3.Coffey MC, Strong JE, Forsyth PA, Lee PW. Reovirus therapy of tumors with activated Ras pathway. Science. 1998;282:1332–4. doi: 10.1126/science.282.5392.1332. [DOI] [PubMed] [Google Scholar]
- 4.Stojdl DF, Lichty B, Knowles S, Marius R, Atkins H, Sonenberg N, Bell JC. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat Med. 2000;6:821–5. doi: 10.1038/77558. [DOI] [PubMed] [Google Scholar]
- 5.Wang G, Barrett JW, Stanford M, Werden SJ, Johnston JB, Gao X, Sun M, Cheng JQ, McFadden G. Infection of human cancer cells with myxoma virus requires Akt activation via interaction with a viral ankyrin-repeat host range factor. Proc Natl Acad Sci U S A. 2006;103:4640–5. doi: 10.1073/pnas.0509341103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balachandran S, Barber GN. Defective translational control facilitates vesicular stomatitis virus oncolysis. Cancer Cell. 2004;5:51–65. doi: 10.1016/s1535-6108(03)00330-1. [DOI] [PubMed] [Google Scholar]
- 7.McFadden G. Poxvirus tropism. Nat Rev Microbiol. 2005;3:201–13. doi: 10.1038/nrmicro1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bischoff JR, Kirn DH, Williams A, Heise C, Horn S, Muna M, Ng L, Nye JA, Sampson-Johannes A, Fattaey A, McCormick F. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science. 1996;274:373–6. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- 9.Martuza RL, Malick A, Markert JM, Ruffner KL, Coen DM. Experimental therapy of human glioma by means of a genetically engineered virus mutant. Science. 1991;252:854–6. doi: 10.1126/science.1851332. [DOI] [PubMed] [Google Scholar]
- 10.Gromeier M, Lachmann S, Rosenfeld MR, Gutin PH, Wimmer E. Intergeneric poliovirus recombinants for the treatment of malignant glioma. Proc Natl Acad Sci U S A. 2000;97:6803–8. doi: 10.1073/pnas.97.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andreansky SS, He B, Gillespie GY, Soroceanu L, Markert JM, Chou J, Roizman B, Whitley RJ. The application of genetically engineered herpes simplex viruses to the treatment of experimental brain tumors. Proc Natl Acad Sci U S A. 1996;93:11313–8. doi: 10.1073/pnas.93.21.11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kambara H, Okano H, Chiocca EA, Saeki Y. An oncolytic HSV-1 mutant expressing ICP34.5 under control of a nestin promoter increases survival of animals even when symptomatic from a brain tumor. Cancer Res. 2005;65:2832–9. doi: 10.1158/0008-5472.CAN-04-3227. [DOI] [PubMed] [Google Scholar]
- 13.Campbell SA, Mulvey M, Mohr I, Gromeier M. Attenuation of herpes simplex virus neurovirulence with picornavirus cis-acting genetic elements. J Virol. 2007;81:791–9. doi: 10.1128/JVI.00714-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou J, Kern ER, Whitley RJ, Roizman B. Mapping of herpes simplex virus-1 neurovirulence to gamma 134.5, a gene nonessential for growth in culture. Science. 1990;250:1262–6. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- 15.Chou J, Roizman B. Herpes simplex virus 1 gamma(1)34.5 gene function, which blocks the host response to infection, maps in the homologous domain of the genes expressed during growth arrest and DNA damage. Proc Natl Acad Sci U S A. 1994;91:5247–51. doi: 10.1073/pnas.91.12.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He B, Gross M, Roizman M. The gamma(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci U S A. 1997;94:843–8. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunter WD, Martuza RL, Feigenbaum F, Todo T, Mineta T, Yazaki T, Toda M, Newsome JT, Platenberg RC, Manz HJ, Rabkin SD. Attenuated, replication-competent herpes simplex virus type 1 mutant G207: safety evaluation of intracerebral injection in nonhuman primates. J Virol. 1999;73:6319–26. doi: 10.1128/jvi.73.8.6319-6326.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taneja S, MacGregor J, Markus S, Ha S, Mohr I. Enhanced antitumor efficacy of a herpes simplex virus mutant isolated by genetic selection in cancer cells. Proc Natl Acad Sci U S A. 2001;98:8804–8. doi: 10.1073/pnas.161011798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–8. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfeiffer JK, Kirkegaard K. Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog. 2005;1:e11. doi: 10.1371/journal.ppat.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendelsohn CL, Wimmer E, Racaniello VR. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989;56:855–65. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 22.Takai Y, Miyoshi J, Ikeda W, Ogita H. Nectins and nectin-like molecules: roles in contact inhibition of cell movement and proliferation. Nat Rev Mol Cell Biol. 2008;9:603–15. doi: 10.1038/nrm2457. [DOI] [PubMed] [Google Scholar]
- 23.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–20. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi K, Nakanishi H, Miyahara M, Mandai K, Satoh K, Satoh A, Nishioka H, Aoki J, Nomoto A, Mizoguchi A, Takai Y. Nectin/PRR: an immunoglobulin-like cell adhesion molecule recruited to cadherin-based adherens junctions through interaction with Afadin, a PDZ domain-containing protein. J Cell Biol. 1999;145:539–49. doi: 10.1083/jcb.145.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki K, Hu D, Bustos T, Zlotogara J, Richieri-Costa A, Helms JA, Spritz RA. Mutations of PVRL1, encoding a cell-cell adhesion molecule/herpesvirus receptor, in cleft lip/palate-ectodermal dysplasia. Nat Genet. 2000;25:427–30. doi: 10.1038/78119. [DOI] [PubMed] [Google Scholar]
- 26.Aoki J, Koike S, Asou H, Ise I, Suwa H, Tanaka T, Miyasaka M, Nomoto A. Mouse homolog of poliovirus receptor-related gene 2 product, mPRR2, mediates homophilic cell aggregation. Exp Cell Res. 1997;235:374–84. doi: 10.1006/excr.1997.3685. [DOI] [PubMed] [Google Scholar]
- 27.Reymond N, Borg JP, Lecocq E, Adelaide J, Campadelli-Fiume G, Dubreuil P, Lopez M. Human nectin3/PRR3: a novel member of the PVR/PRR/nectin family that interacts with afadin. Gene. 2000;255:347–55. doi: 10.1016/s0378-1119(00)00316-4. [DOI] [PubMed] [Google Scholar]
- 28.Reymond N, Fabre S, Lecocq E, Adelaide J, Dubreuil P, Lopez M. Nectin4/PRR4, a new afadin-associated member of the nectin family that trans-interacts with nectin1/PRR1 through V domain interaction. J Biol Chem. 2001;276:43205–15. doi: 10.1074/jbc.M103810200. [DOI] [PubMed] [Google Scholar]
- 29.Kakunaga S, Ikeda W, Itoh S, Deguchi-Tawarada M, Ohtsuka T, Mizoguchi A, Takai Y. Nectin-like molecule-1/TSLL1/SynCAM3: a neural tissue-specific immunoglobulin-like cell-cell adhesion molecule localizing at non-junctional contact sites of presynaptic nerve terminals, axons and glia cell processes. J Cell Sci. 2005;118:1267–77. doi: 10.1242/jcs.01656. [DOI] [PubMed] [Google Scholar]
- 30.Shingai T, Ikeda W, Kakunaga S, Morimoto K, Takekuni K, Itoh S, Satoh K, Takeuchi M, Imai T, Monden M, Takai Y. Implications of nectin-like molecule 2/IGSF4/RA175SgIGSF/ TSLC1/SynCAM1 in cell-cell adhesion and transmembrane protein localization in epithelial cells. J Biol Chem. 2003;278:35421–7. doi: 10.1074/jbc.M305387200. [DOI] [PubMed] [Google Scholar]
- 31.Pellissier F, Gerber A, Bauer C, Ballivet M, Ossipow V. The adhesion molecule Necl- 3/SynCAM-2 localizes to myelinated axons, binds to oligodendrocytes and promotes cell adhesion. BMC Neurosci. 2007;8:90. doi: 10.1186/1471-2202-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maurel P, Einheber S, Galinska J, Thaker P, Lam I, Rubin MB, Scherer SS, Murakami Y, Gutmann DH, Salzer JL. Nectin-like proteins mediate axon Schwann cell interactions along the internode and are essential for myelination. J Cell Biol. 2007;178:861–74. doi: 10.1083/jcb.200705132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masson D, Jarry A, Baury B, Blanchardie P, Laboisse C, Lustenberger P, Denis MG. Overexpression of the CD155 gene in human colorectal carcinoma. Gut. 2001;49:236–40. doi: 10.1136/gut.49.2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ochiai H, Moore SA, Archer GE, Okamura T, Chewning TA, Marks JR, Sampson JH, Gromeier M. Treatment of intracerebral neoplasia and neoplastic meningitis with regional delivery of oncolytic recombinant poliovirus. Clin Cancer Res. 2004;10:4831–8. doi: 10.1158/1078-0432.CCR-03-0694. [DOI] [PubMed] [Google Scholar]
- 35.Merrill MK, Bernhardt G, Sampson JH, Wikstrand CJ, Bigner DD, Gromeier M. Poliovirus receptor CD155-targeted oncolysis of glioma. Neuro-oncol. 2004;6:208–17. doi: 10.1215/S1152851703000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sloan KE, Stewart JK, Treloar AF, Matthews RT, Jay DG. CD155/PVR enhances glioma cell dispersal by regulating adhesion signaling and focal adhesion dynamics. Cancer Res. 2005;65:10930–7. doi: 10.1158/0008-5472.CAN-05-1890. [DOI] [PubMed] [Google Scholar]
- 37.Sloan KE, Eustace BK, Stewart JK, Zehetmeier C, Torella C, Simeone M, Roy JE, Unger C, Louis DN, Ilag LL, Jay DG. CD155/PVR plays a key role in cell motility during tumor cell invasion and migration. BMC Cancer. 2004;4:73. doi: 10.1186/1471-2407-4-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Solecki DJ, Gromeier M, Mueller S, Bernhardt G, Wimmer E. Expression of the human poliovirus receptor/CD155 gene is activated by sonic hedgehog. J Biol Chem. 2002;277:25697–702. doi: 10.1074/jbc.M201378200. [DOI] [PubMed] [Google Scholar]
- 39.Hirota T, Irie K, Okamoto R, Ikeda W, Takai Y. Transcriptional activation of the mouse Necl-5/Tage4/PVR/CD155 gene by fibroblast growth factor or oncogenic Ras through the Raf-MEK-ERK-AP-1 pathway. Oncogene. 2005;24:2229–35. doi: 10.1038/sj.onc.1208409. [DOI] [PubMed] [Google Scholar]
- 40.Erickson BM, Thompson NL, Hixson DC. Tightly regulated induction of the adhesion molecule necl-5/CD155 during rat liver regeneration and acute liver injury. Hepatology. 2006;43:325–34. doi: 10.1002/hep.21021. [DOI] [PubMed] [Google Scholar]
- 41.Waehler R, Russell SJ, Curiel DT. Engineering targeted viral vectors for gene therapy. Nat Rev Genet. 2007;8:573–87. doi: 10.1038/nrg2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grandi P, Wang S, Schuback D, Krasnykh V, Spear M, Curiel DT, Manservigi R, Breakefield XO. HSV-1 virions engineered for specific binding to cell surface receptors. Mol Ther. 2004;9:419–27. doi: 10.1016/j.ymthe.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 43.Gromeier M, Solecki D, Patel DD, Wimmer E. Expression of the human poliovirus receptor/CD155 gene during development of the central nervous system: implications for the pathogenesis of poliomyelitis. Virology. 2000;273:248–57. doi: 10.1006/viro.2000.0418. [DOI] [PubMed] [Google Scholar]
- 44.Gromeier M, Alexander L, Wimmer E. Internal ribosomal entry site substitution eliminates neurovirulence in intergeneric poliovirus recombinants. Proc Natl Acad Sci USA. 1996;93:2370–5. doi: 10.1073/pnas.93.6.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nomoto A, Lee YF, Wimmer E. The 5' end of poliovirus mRNA is not capped with m7G(5')ppp(5')Np. Proc Natl Acad Sci U S A. 1976;73:375–80. doi: 10.1073/pnas.73.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaiser C, Dobrikova EY, Bradrick SS, Shveygert M, Herbert JT, Gromeier M. Activation of cap-independent translation by variant eukaryotic initiation factor 4G in vivo. RNA. 2008;14:2170–82. doi: 10.1261/rna.1171808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CU. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc Natl Acad Sci U S A. 2009;106:9197–202. doi: 10.1073/pnas.0900153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5' nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–43. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–5. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 50.Gromeier M, Bossert B, Arita M, Nomoto A, Wimmer E. Dual stem loops within the poliovirus internal ribosomal entry site control neurovirulence. J Virol. 1999;73:958–64. doi: 10.1128/jvi.73.2.958-964.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Campbell SA, Lin J, Dobrikova EY, Gromeier M. Genetic determinants of cell typespecific poliovirus propagation in HEK 293 cells. J Virol. 2005;79:6281–90. doi: 10.1128/JVI.79.10.6281-6290.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shaw G, Morse S, Ararat M, Graham FL. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 2002;16:869–71. doi: 10.1096/fj.01-0995fje. [DOI] [PubMed] [Google Scholar]
- 53.Merrill MK, Gromeier M. The double-stranded RNA binding protein 76:NF45 heterodimer inhibits translation initiation at the rhinovirus type 2 internal ribosome entry site. J Virol. 2006;80:6936–42. doi: 10.1128/JVI.00243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Merrill MK, Dobrikova EY, Gromeier M. Cell-type-specific repression of internal ribosome entry site activity by double-stranded RNA-binding protein 76. J Virol. 2006;80:3147–56. doi: 10.1128/JVI.80.7.3147-3156.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Florez de Sessions P, Dobrikova E, Gromeier M. Genetic adaptation to untranslated region-mediated enterovirus growth deficits by mutations in the nonstructural proteins 3AB and 3CD. J Virol. 2007;81:8396–405. doi: 10.1128/JVI.00321-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dobrikova E, Florez P, Bradrick S, Gromeier M. Activity of a type 1 picornavirus internal ribosomal entry site is determined by sequences within the 3' nontranslated region. Proc Natl Acad Sci U S A. 2003;100:15125–30. doi: 10.1073/pnas.2436464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobrikova EY, Broadt T, Poiley-Nelson J, Yang X, Soman G, Giardina S, Harris R, Gromeier M. Recombinant oncolytic poliovirus eliminates glioma in vivo without genetic adaptation to a pathogenic phenotype. Mol Ther. 2008;16:1865–72. doi: 10.1038/mt.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang X, Chen E, Jiang H, Muszynski K, Harris RD, Giardina SL, Gromeier M, Mitra G, Soman G. Evaluation of IRES-mediated, cell-type-specific cytotoxicity of poliovirus using a colorimetric cell proliferation assay. J Virol Methods. 2009;155:44–54. doi: 10.1016/j.jviromet.2008.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holland EC, Celestino J, Dai C, Schaefer L, Sawaya RE, Fuller GN. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet. 2000;25:55–7. doi: 10.1038/75596. [DOI] [PubMed] [Google Scholar]
- 60.Rajasekhar VK, Viale A, Socci ND, Wiedmann M, Hu X, Holand EC. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol Cell. 2003;12:889–901. doi: 10.1016/s1097-2765(03)00395-2. [DOI] [PubMed] [Google Scholar]
- 61.Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5'- cap function. Nature. 1994;371:762–7. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 62.Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15:658–64. [PMC free article] [PubMed] [Google Scholar]
- 63.Waskiewicz AJ, Flynn A, Proud CG, Cooper JA. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997;16:1909–20. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fukunaga R, Hunter T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997;16:1921–33. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Raught B, Gingras AC, Gygi Sp, Imataka H, Morino S, Gradi A, Aebersold R, Sonenberg N. Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. EMBO J. 2000;19:434–44. doi: 10.1093/emboj/19.3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–83. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 67.Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 1999;18:270–9. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scheper GC Proud CG. Does phosphorylation of the cap-binding protein eIF4E play a role in translation initiation? Eur J Biochem. 2002;269:5350–9. doi: 10.1046/j.1432-1033.2002.03291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Knauf U, Tschopp C, Gram H. Negative regulation of protein translation by mitogenactivated protein kinase-interacting kinases 1 and 2. Mol Cell Biol. 2001;21:5500–11. doi: 10.1128/MCB.21.16.5500-5511.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Svitkin YV, Herdy B, Costa-Mattioli M, Gingras AC, Raught B, Sonenberg H. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol Cell Biol. 2005;25:10556–65. doi: 10.1128/MCB.25.23.10556-10565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]