Abstract
Background
Apolipoprotein E is polymorphic in the human population. APOE4 has previously been reported to correlate with symptomatic oral and genital herpes disease.
Methods
We genotyped APOE in 182 subjects with HSV-2 and in 62 subjects with HSV-1, including 44 subjects with both viral types for a total of 200 adults. HSV shedding was measured by PCR from swab samples obtained daily from mucosa for at least 30 days. Participants also maintained a dairy of oral or genital lesions.
Results
Observed APOE genotypes reflected the US Caucasian population and the Hardy-Weinberg equilibrium. Genital and oral HSV shedding was detected on 17.2% and 3.7% of overall days, respectively, while genital and oral lesion rates were 10.1% and 2.9%. Using Poisson regression and adjusting for known correlates of HSV shedding, we did not observe a significant association between APOE genotype and genital or oral HSV shedding, or genital HSV lesions. However, the presence of the APOE4 allele was associated with a higher rate of oral herpetic lesions, with a relative risk of 4.64 (1.32–15.05, P=0.016).
Conclusions
Variation at the APOE locus may be associated with clinical manifestations of HSV-1 infection but does not to correlate with herpes simplex viral reactivation in humans.
Keywords: Apoliprotein E, herpes simplex virus, genotype, viral shedding
Introduction
Herpes simplex virus (HSV) types 1 and 2 are ubiquitous human pathogens, infecting about 60% and 20% of US adults, respectively 1. Both viral types have a tropism for epithelial and neuronal cells. Their ability to undergo latency and reactivation in neurons leads to a phenotype of recurrent clinical disease in some persons and detectable, intermittent viral shedding in infected individuals 2. HSV-1 clinical disease typically effects the orolabial and ocular areas and HSV-2 is usually detected in the anogenital region 3. While most persons who have antibody to HSV-1 or HSV-2 do not have a clinical history of mucosal ulcerations, the current consensus is that all seropositive persons have lifelong latent infection in neural ganglia and at least occasional shedding of virus. One possible explanation for this is variation in host genes that interact with HSV replication, the immune response to HSV, or the generation of clinically apparent signs or symptoms or infection.
Apolipoprotein E is a protein involved in the transport of lipids that is strongly linked to Alzheimer’s disease (AD) pathogenesis. It is encoded by the autosomal APOE gene. Amongst the three common alleles (E2, E3, and E4), which vary in amino acid sequence at residues 112 and 158, the E4 allele has a pathogenic association with the prevalence and age of onset of AD. HSV-1 DNA can be present in both normal and AD brain tissues. Investigations into the interaction between HSV-1 and APOE alleles have suggested an association between HSV-1 DNA detection in AD tissues and the presence of the E4 allele 4. An association between the E4 genotype and a history of symptomatic HSV-1 was also detected 4. Mouse models also support an interaction between HSV infection and APOE genotype 5–8.
We have used daily home collection of anogenital and oral swabs from persons with HSV infection to define objectively the frequency of mucosal viral shedding, and prospective diaries to characterize the frequency of oral and genital lesions. The relationship between the virologic and clinical severity of genital HSV shedding and APOE variants was studied in 200 genotyped persons.
Material and Methods
Study participants and procedures
All participants were 18 years old or older, had serologic evidence of HSV-1 and/or HSV-2 infection, and no clinical or laboratory evidence of HIV infection, and were enrolled into prospective studies at the University of Washington Virology Research Clinic in Seattle or Westover Heights Clinic in Portland. Participants provided signed informed consent. Study protocols were approved by the University of Washington (Seattle site) or Western (Portland site) institutional review boards. Subjects did not receive anti-HSV therapy during the study observation period. A standardized history of herpes simplex virus infection, and general health was obtained. Subjects received gender-specific instruction on how to obtain swabs of the oral and/or genital and rectal regions as previously described 9 10. Participants kept a diary of genital and oral symptoms and were seen in the clinic every 2 weeks for collection of samples and diary review. The analysis included participants with data for 30 or more days at genital, oral, or both sites. Blood for DNA was collected by venipuncture
Laboratory testing
Antibodies to HSV-1 and HSV-2 were detected by the University of Washington immunoblot 11. Antibodies to HIV-1 were detected by screening ELISA. Swabs were placed into vials containing 1 mL of transport medium and refrigerated until laboratory processing. HSV DNA was detected by polymerase chain reaction (PCR) 12. The PCR assay uses type-common primers to the HSV gene encoding glycoprotein B 13 14. An internal control was included in the PCR reaction to ensure that HSV-negative findings were not due to inhibition. Samples were considered positive for HSV if we detected >3 copies of HSV DNA per 20 µL of specimen (i.e.,>150 copies of HSV DNA per mL of transport media) 15. Results were analyzed both dichotomously and, among positive specimens, as HSV copies/swab. Laboratory personnel were blinded to clinical data. Genomic DNA was isolated from blood collection tubes (Paxgene, Becton Dickinson, Franklin Lakes, NJ) and measured with a spectrophotometer. APOE genotyping was performed by standard PCR restriction fragment length polymorphism-based methods 16.
Statistical Analysis
We examined the association between APOE alleles and the outcomes: viral shedding and mucosal lesions, at both the oral and genital sites, using Poisson regression. The rate ratios due to specific APOE alleles were adjusted for known influences on HSV shedding, such as gender, HSV-1 infection among HSV-2 infected persons, and time since HSV acquisition 2 17. The duration of infection (categorized as great than one year, less than one year, or unknown) as also considered, as viral shedding is higher in the 1st year after primary HSV-2 infection 17. To analyze the relationship of APOE genotype with HSV DNA copy number, linear mixed models were used, both without and with adjustment for the factors mentioned above. Genotype frequencies were examined for independent assortment (Hardy-Weinberg equilibrium 18) with the polynomial
(a + b + c)2 = a2 + b2 +c2 + 2ab + 2bc + 2ac.
Results
Virological and clinical outcomes
We studied 200 participants. Among these, 78 had both HSV-1 and HSV-2 infection, 107 had only HSV-2 infection, and 15 had only HSV-1 infection. Overall, 44% were male and 86% were Caucasian (racial data were available on 87% of participants). Genital swabs were obtained from 182 HSV-2 seropositive persons (Table 1). Among these, 143 (79%) had a history of genital herpes. We note that this rate is higher than the typical rate for HSV-2-seropositive persons, and likely reflects the recruiting pathway for our research clinics that encourages participation by persons with symptomatic genital herpes. 75 (41%) were also HSV-1 seropositive. Oral swabs were obtained from 62 HSV-1 seropositive persons, including 47 (76%) who were also HSV-2-seropositive. Both genital and oral swabs and history were obtained from 44 of these dually-seropositive persons.
Table 1.
Participants enrolled in the study.
| anatomic site for swab collection | ||
|---|---|---|
| genitala | oral | |
| total subjects | 182 | 62 |
| HSV-1 only seropositive | 0 | 15 |
| HSV-2 only seropositive | 107 | 0 |
| HSV-1 and HSV-2 dually seropositive |
75 | 47 |
| men | 78 (43%) | 34 (55%) |
| history of genital herpes | 143 (79%) | 30/44 (68%) |
| Caucasian | (85%) | (82%) |
44 subjects collected swabs from both sites, giving a total of 200 unique participants.
The virological endpoints were mucosal HSV shedding rates. Overall, HSV shedding data were available from 13,417 swab specimens. Among the 182 persons and 10,425 days with genital swabs collected, 1,788 (17.2%) were positive for HSV. The median observed shedding rates was 13% (range 0 to 80%) of days. Among the 62 persons participating in the oral shedding cohort, 112 of 2,992 oral swabs (3.7%) were positive. The median observed shedding rate was 0% (ranged from 0% to 28% of days).
The clinical endpoints were the rates of oral and genital lesions as noted in participant diaries. The number of days with oral lesion reports was fewer than the number of days with genital lesion reports, reflecting the smaller oral lesion cohort. Of the 11,770 days containing diary entries, genital lesions were noted on 1,192 (10.1%) days. Among the 3,431 days containing diary entries, oral lesions were noted on 99 (2.9%) days. The median reported lesion rate was 5%, ranging from 0 to 67% of days for genital sites, with the corresponding median rate for oral sites of 0%, ranging from 0 to 29% of days
APOE genotypes
APOE allele frequencies (Table 2) were similar to those reported in the US population. Amongst the 400 alleles from 200 persons, the allele frequencies were 7.5, 74.0, and 18.5% for APOE2, APOE3, and APOE4, respectively. It is well known that APOE4 has high allele frequencies in African-heritage populations (up to 22%) 19. As expected from our high proportion of Caucasian subjects, our observe APOE4 frequency was similar to those reported from predominantly Caucasian groups 19. APOE genotype frequencies reflected the allele frequencies, and displayed independent assortment per the Hardy-Weinberg equilibrium and were similar to those reported 20 for Caucasian populations (not shown). Amongst the participants in the genital herpes study, each of the six possible APOE genotype was represented, with between 1 and 99 persons having each genotype. Amongst the persons with oral herpes data, three genotypes were not represented (APOE 2/2, APOE 2/4, and APOE 4/4), while between 11 and 31 persons per genotype were present in the other three genotypes (APOE 2/3, APOE 3/3, and APOE 3/4).
Table 2.
APOE allele frequencies in the study population.
Correlation between virological and clinical endpoints and APOE genotype
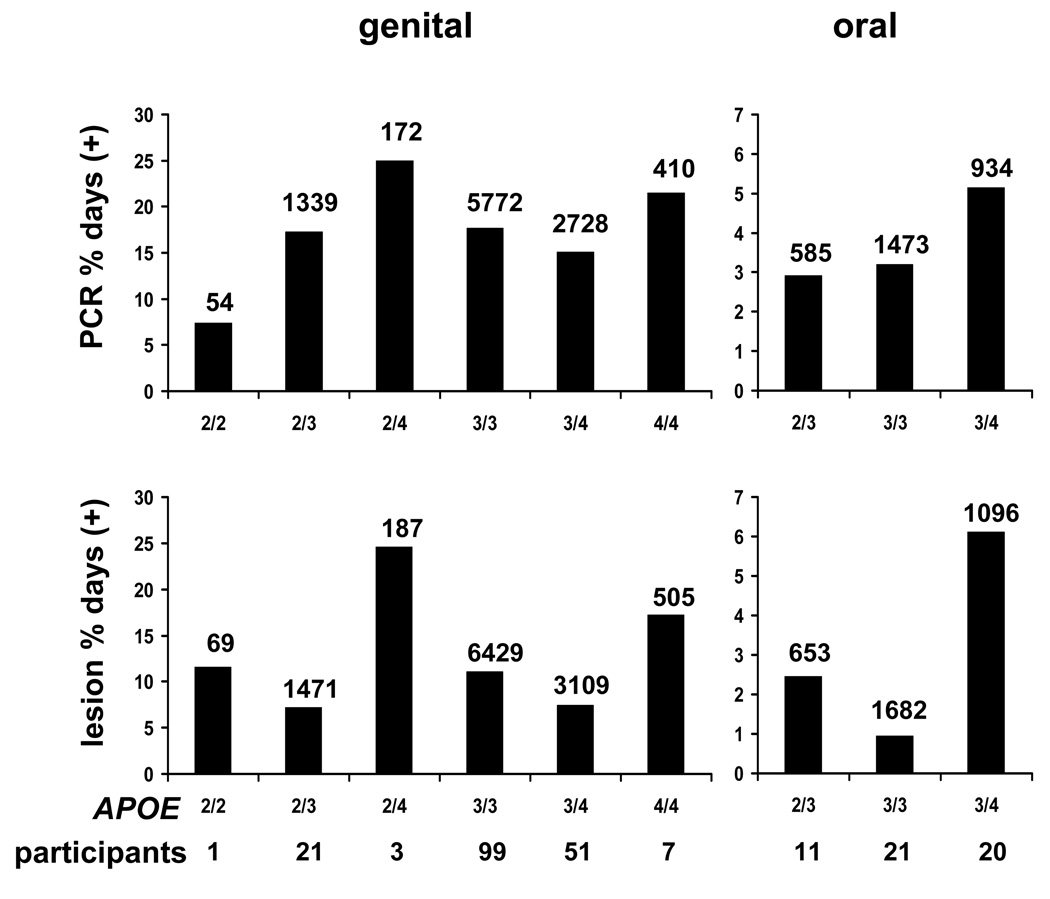
We examined the relationship between APOE genotype and four distinct HSV outcomes: genital HSV shedding and lesions, and oral HSV shedding and lesions (Table 3). Bar graph representations of shedding and lesion data for (Fig. 1) show that genotypes containing APOE4 generally had the highest levels of HSV activity. The genital shedding rates for genotypes 2/2, 2/3, 2/4, 3/3, 3/4, and 4/4 were 7.4%, 17.3%, 25%, 17.7%, 15.1%, and 21.4%, respectively. The corresponding lesion rates were 11.6%, 7.2%, 24.6%, 11.1%, 7.5%, and 17.2%, respectively. For participants with the 2/3, 3/3, and 3/4 in the oral herpes study, the oral shedding rates were 2.9%, 3.2%, and 5.1%, respectively. The corresponding oral lesion rates were 2.5%, 0.95%, and 6.1%. Within each genotype, large ranges were noted for shedding and lesion rates. Therefore, when the individual participants’ shedding or lesion rates are analyzed, overlap is present in both shedding rates and lesion rates between groups of participants with distinct APOE genotypes (not shown).
Table 3.
Poisson regressions for shedding and lesions.
| genital shedding | oral shedding | |||
|---|---|---|---|---|
| RR (95% C.I.a) | P | RR (95% C.I.a) | P | |
| any APOE4 versus none | 0.94 (0.68–1.29) | 0.69 | 1.65 (0.69–3.97) | 0.26 |
| any APOE4 versus noneb | 0.93 (0.68–1.29) | 0.68 | 1.61 (0.68–3.79) | 0.28 |
| male gender | 0.87 (0.64–1.18) | 0.37 | 1.41 (0.58–3.46) | 0.45 |
| HSV-1 and HSV-2 infection | 1.23 (0.91–1.67) | 0.17 | 3.35 (0.52–21.77) | 0.21 |
| genital lesions | oral lesions | |||
| RR (95% C.I.a) | P | RR (95% C.I.a) | P | |
| any APOE4 versus none | 0.93 (0.61–1.41) | 0.72 | 4.64 (1.32–15.05) | 0.016 |
| any APOE4 versus noneb | 0.86 (0.57–1.29) | 0.45 | 4.04 (1.09–15.01) | 0.037 |
| male gender | 1.02 (0.69–1.52) | 0.91 | 1.57 (0.37–6.59) | 0.54 |
| HSV-1 and HSV-2 infection | 1.08 (0.73–1.60) | 0.70 | 0.74 (0.16–3.39) | 0.70 |
| history of genital herpes | 5.13 (2.20–11.98) | <0.001 | -- | -- |
C.I. stands for confidence interval.
Risk ratio adjusted for gender, co-infection, time since infection, and symptoms at site. Data set on oral symptoms is incomplete so no adjustment is made for oral shedding.
Fig. 1.
Association of APOE genotypes and oral HSV shedding and oral lesions. The left bars refer to genital shedding rates (top) and lesion rates (bottom), while the right bars depict oral shedding (top) and lesions (bottom). The abscissa values differ between genital and oral sites. The numbers of days with data available are indicated over the bars for each genotype, anatomic site, and study endpoint. The number of persons with each genotype is listed below the APOE genotypes across the ordinates.
Poisson regression was performed to examine what factors might be associated with shedding or lesions. APOE genotype was not associated with genital HSV shedding or lesions. As expected, a history of symptomatic genital herpes was associated with genital lesion rates (Table 3). For oral disease, the presence of any number of copies of APOE4 was associated with increased rates of oral lesions (relative risk 4.64, 95% confidence interval 1.32–15.05, p = 0.016), but not with oral shedding. Adjustment of the risk ratio for oral lesions based on gender, HSV dual infection, and time since infection (if known) yielded a relative risk for any APOE4 allele any oral lesions of 4.04 (95% confidence interval 1.09–15.01, p = 0.037). Amongst participants with oral lesion data, each person with any copies of APOE4 also had a copy of APOE3. Therefore, it was not possible to examine whether the mechanism of association was dominant, recessive, additive or heterogeneous.
We also examined the relationship between APOE genotype and the amount of HSV DNA detected using quantitative HSV DNA copy number data from 112 oral swabs and 1,788 genital swabs. There was no significant difference in HSV copy numbers between the most prevalent APOE genotype, 3/3, and any other APOE 3/3 genotype, and there was no significant difference between persons with and without any copies of APOE 4. These observations were true for both oral and genital swabs.
Discussion
We investigated the association between host APOE genotype and the virological and clinical severity of infection by two prevalent pathogens, HSV-1 and HSV-2. Data were collected separately at oral and genital sites. We obtained 244 clinical and virological data sets for the 200 study participants. We found no association between APOE genotype and genital HSV shedding or lesion rates. There was also no association with oral HSV shedding, but we did observe an association between the APOE4 allele and higher rates of oral herpetic lesions.
Our study had several strengths. We obtained viral shedding data in addition to subjective clinical information. Viral shedding represents the outcome of both neuronal reactivation and epithelial replication, and may occur with or without lesion formation. Shedding is important as it accounts for transmission of HSV. We asked our subjects to prospectively rather than retrospectively report lesions and provided guidance in the assessment of lesions. We have previously shown this educational intervention can improve on the accuracy of HSV diagnosis 2. Our primary endpoints, HSV lesion and shedding rates, were both studied as continuous rather than as dichotomous variables. Potential weaknesses of our study include the inclusion, amongst HSV-2 seropositive persons, of mostly subjects with a history of genital herpes. Previous studies have shown that the majority of HSV-2 seropositive persons have asymptomatic or unrecognized infection 21. Results could have differed in such persons, although previous reports from our group have found that the virologic characterstics of genital HSV-2 shedding are similar between symptomatic and asymptomatic persons 2. An inherent weakness is our limited power to exclude a relationship between APOE genotype and shedding rates, related to the size of the cohorts. In the genital shedding cohort, we had 80% power to detect associations of either shedding rate or lesions rate with APOE4, as long as the magnitude of the risk ratio was at least as high as 1.4-fold, or smaller than 0.7-fold. In the oral shedding cohort, with fewer subjects, we had 80% power to detect risk ratios either larger than 1.9-fold or smaller than 0.6-fold. Associations of smaller magnitude cannot be ruled out by this analysis.
Our cohort differed from those of Itzahki et al. 4 22 and Jayasuriya et al. 23 in previous studies. Entry into our clinic population is frequently the result of a diagnosis of genital herpes. Itzhaki et al. did not provide details on how participants were recruited for oral herpes studies. When comparing HSV-2 studies, the most obvious difference between our cohort and that of Jayasuriya et al. is HIV infection status. HIV infection is accelerates HSV-2 pathogenesis, and subtle effects of host genotype could be overwhelmed by immune changes. Inclusion of shedding studies could clarify this issue. The allele frequencies we observed generally matched those previously reported in the US Caucasian population. As expected, our APOE4 allele frequency was lower than that of Jayasuriya et al., as our cohort was largely Caucasian, and APOE4 is more prevalent in African populations 24.
APOE is a reasonable “target gene” candidate for HSV genotypic association research. In 1997, Itzhaki et al. measured APOE genotypes in adult subjects with and without cold sores. HSV-1 serology was performed on 70% of the 77 subjects, and the vast majority were HSV-1 seropositive. The odds ratio for having any copies of APOE4 amongst persons with cold sores was 5.7 compared to persons without cold sores 4. A follow-up report in 2008 analyzed 146 adults, again split roughly 50:50 into those with and without cold sores. A similarly raised odds ratio for APOE4 for cold sores was reported, as was a trend to earlier onset of symptomatic oral herpes amongst a separate cohort of children who had the APOE4 allele 22.
The APOE studies of Itzhaki et al. were begun in the context of Alzheimer’s disease (AD). APOE4 is a risk factor for early-onset AD. Brain infections are a rational candidate etiologic co-factor in this largely idiopathic condition, and Itzhaki et al. detected HSV-1 DNA in the brain in AD 4. In contrast to focusing on a candidate gene such as APOE or the toll-like receptor genes, Hobbs et al. used a genome-wide scan to look for human chromosomal loci associated with symptomatic oral herpes (defined as 2 or more episodes per year) 25. They found a susceptibility allele on chromosome 21 but, using a nearest marker about 0.8 Mb from APOE on chromosome 19, did not detect a signal from the region of APOE.
There are several genetic loci such as toll-like receptors that are rational candidates for linkage with HSV severity. Previously, we have reported a relationship between toll-like receptor 2 (TLR2) alleles and their associated haplotypes, and genital HSV shedding and lesion rates 26. The presence of a G nucleotide at 15,607 base pairs 5’ to the ATG start of TLR2 had an allele frequency of 33%. This minor allele (TLR2-15607A/G) was associated with higher genital HSV shedding rate and lesion rates 26. Amongst the persons with APOE and HSV data in the current report, 133 of 182 (73.1%) studied at the genital site have had TLR genotyping, while 48 of 62 (77.4%) with oral herpes data have TLR genotypes available. TLR2 and APOE are on different chromosomes and are expected to assort independently. There was no observed association between presence of TLR2-15607A/G and APOE4 in either the genital or oral cohort (p=0.59, p=0.84). Polymorphisms in TLR2 and APOE could also interact biologically. Adjustment for presence of TLR2-15607A/G did not alter the findings regarding the lack of association between genital shedding or genital lesions and presence of APOE4, though TLR2-15607A/G remained predictive. TLR2-15607A/G was not associated with either oral shedding (p=0.98) or oral lesions (p=0.75) after adjustment for APOE genotype, gender, HSV dual infection, and time since infection, although the size of the oral HSV cohort (62 persons) may have limited our power of observe an effect.
HSV-1 and HSV-2 have important similarities and differences. While their genomic structures are similar, HSV-1 reactivates more efficiently from craniofacial ganglia while HSV-2 reactivates better from sacral ganglia. Host genetic loci could be associated with the severity of both viral types, or distinct for HSV-1 versus HSV-2. Jayasuriya et al. studied 96 HSV-2/HIV co-infected persons in an African immigrant-dominated clinic in England 23. The presence or absence of any history of prior genital ulceration was recorded. Among participants with a history of genital ulcers, the number of episodes of genital ulceration in the preceding 12 months was recorded. Genital ulceration severity was dichotomously classified as recurrent (two or more episodes in the prior 12 months) or non-recurrent (0 or 1 episodes in the prior 12 months). Analyses of groups with or without any lifetime history of genital ulcerations, or recurrent vs. non-recurrent disease as defined above, both showed a significant correlation of the presence of APOE4 and symptomatic genital ulceration. In multivariate analyses, ethnicity, CD4 count, and anti-HIV therapy were not associated with recurrent ulceration. In contrast to Jayasuriya et al., we did not find any association between APOE genotype and genital herpes history.
Mechanistic biologic links between APOE and HSV have not been rigorously described. APOE has biological activities linked to HSV replication 27 28. Burgos et al. studied HSV-1 pathogenesis in mice deleted for endogenous APOE, or in which murine APOE was replaced by specific human APOE alleles 5–8. Murine APOE generally enhanced HSV infection compared to APOE deletion. Higher levels of acute and latent infection were detected in APOE4 transgenic mice, including in the brain and trigeminal ganglia, than in APOE3 mice. We observed an association between APOE genotype and oral lesions, most of which are due to HSV-1, but not with recurrent genital lesions, most of which are caused by HSV-2. HSV-2 pathogenesis has not yet been studied in APOE-recombinant mice. HSV-1 and HSV-2-encoded proteins differ at myriad amino acid-coding loci, including loci in genes reported to interact with host immune or inflammatory molecules and cells such as glycoprotein D 29, glycoprotein B 30, the Us5, Us3, and γ34.5 gene products 31 32, and ICP47 33. Spontaneous recurrences, the topic of this report, are difficult to study in murine models. This limits the potential for currently available murine models to address the mechanism for the type-specific association between APOE genotype and lesional recurrences that we have observed.
In summary, we found that the APOE4 allele was significantly associated with a higher rate of oral lesions reported by HSV-1 seropositive study participants. Neither genital lesion rate, nor oral or genital HSV DNA detection, were correlated with APOE genotype. Because HSV shedding is strongly correlated with transmission, it is unlikely that APOE genotype has had a significant effect on shaping HSV dissemination in the population. It remains possible that other genetic loci in linkage disequilibrium with APOE determine oral lesion formation, or that APOE cooperates with other host genes to modulate HSV-1 or HSV-2, requiring further research in genetic host-pathogen relationships for these common infections.
Additional Information:
DM Koelle conceived the project, purified the DNA from blood samples, and wrote the paper. A Magaret performed the biostatistical analyses. T Warren recruited subjects and obtained specimens. GD Schellenberg performed APOE genotyping. A Wald recruited subjects, obtained specimens and provided biostatistical and writing expertise.
Competing Interest: None declared
Key Messages:
Human APOE genotypes were not associated with HSV-1 or HSV-2 shedding rates as measured by daily swabs and a sensitive PCR assay
Human APOE genotypes were not associated with the rate of self-reported genital lesions
The human APOE4 allele was associated with increased rate of self-reported oral lesions
Acknowledgements
The authors thank Dr. Rhoda Morrow and the University of Washington Clinical Virology Lab for performing HSV serology, Dr. Meei-li Huang and the University of Washington Molecular Virology lab for performing HSV PCR, Christopher McClurkan for overseeing DNA processing, and the staff of the Virology Research Clinic and the Westover Heights Clinics.
Footnotes
Funding was provided by NIH grants U19 AI031448 and PO1 AI30731.
The work was presented in part at the International Society for STD Research meeting in London, England in June 2009.
Competing Interest: None declared
References
- 1.Xu F, Sternberg MR, Kottiri BJ, McQuillan GM, Lee FK, Nahmias AJ, et al. Trends in herpes simplex virus type 1 and type 2 seroprevalence in the United States. JAMA. 2006;296(8):964–973. doi: 10.1001/jama.296.8.964. [DOI] [PubMed] [Google Scholar]
- 2.Wald A, Zeh J, Selke S, Warren T, Ryncarz AJ, Ashley R, et al. Reactivation of genital herpes simplex virus type 2 infection in asymptomatic seropositive persons. N Engl J Med. 2000;342(12):844–850. doi: 10.1056/NEJM200003233421203. [DOI] [PubMed] [Google Scholar]
- 3.Corey L. Herpes simplex viruses. In: Kasper DL, Braunwald E, Fauci AS, Hauser SL, Jameson JL, editors. Harrison's Principles of Internal Medicine. 16th ed. New York: McGraw-Hill Book Company; 2004. pp. 1035–1042. [Google Scholar]
- 4.Itzhaki RF, Lin WR, Shang D, Wilcock GK, Faragher B, Jamieson GA. Herpes simplex virus type 1 in brain and risk of Alzheimer's disease. Lancet. 1997;349(9047):241–244. doi: 10.1016/S0140-6736(96)10149-5. [DOI] [PubMed] [Google Scholar]
- 5.Burgos JS, Ramirez C, Sastre I, Bullido MJ, Valdivieso F. Involvement of apolipoprotein E in the hematogenous route of herpes simplex virus type 1 to the central nervous system. J Virol. 2002;76(23):12394–12398. doi: 10.1128/JVI.76.23.12394-12398.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burgos JS, Ramirez C, Sastre I, Bullido MJ, Valdivieso F. ApoE4 is more efficient than E3 in brain access by herpes simplex virus type 1. Neuroreport. 2003;14(14):1825–1827. doi: 10.1097/00001756-200310060-00013. [DOI] [PubMed] [Google Scholar]
- 7.Burgos JS, Ramirez C, Sastre I, Valdivieso F. Effect of apolipoprotein E on the cerebral load of latent herpes simplex virus type 1 DNA. J Virol. 2006;80(11):5383–5387. doi: 10.1128/JVI.00006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burgos JS, Ramirez C, Sastre I, Valdivieso F. Apolipoprotein E genotype influences vertical transmission of herpes simplex virus type 1 in a gender specific manner. Aging Cell. 2007;6(6):841–842. doi: 10.1111/j.1474-9726.2007.00332.x. [DOI] [PubMed] [Google Scholar]
- 9.Mark KE, Corey L, Meng TC, Magaret AS, Huang ML, Selke S, et al. Topical resiquimod 0.01% gel decreases herpes simplex virus type 2 genital shedding: a randomized, controlled trial. J Infect Dis. 2007;195(9):1324–1331. doi: 10.1086/513276. [DOI] [PubMed] [Google Scholar]
- 10.Kim HN, Meier A, Huang ML, Kuntz S, Selke S, Celum C, et al. Oral herpes simplex virus type 2 reactivation in HIV-positive and -negative men. J Infect Dis. 2006;194(4):420–427. doi: 10.1086/505879. [DOI] [PubMed] [Google Scholar]
- 11.Ashley RL, Militoni J, Lee F, Nahmias A, Corey L. Comparison of Western blot (immunoblot) and glycoprotein G-specific immunodot enzyme assay for detecting antibodies to herpes simplex virus types 1 and 2 in human sera. J Clin Microbiol. 1988;26(4):662–667. doi: 10.1128/jcm.26.4.662-667.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magaret AS, Wald A, Huang ML, Selke S, Corey L. Optimizing PCR positivity criterion for detection of herpes simplex virus DNA on skin and mucosa. J Clin Microbiol. 2007;45(5):1618–1620. doi: 10.1128/JCM.01405-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jerome KR, Huang ML, Wald A, Selke S, Corey L. Quantitative stability of DNA after extended storage of clinical specimens as determined by real-time PCR. J Clin Microbiol. 2002;40(7):2609–2611. doi: 10.1128/JCM.40.7.2609-2611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corey L, Huang ML, Selke S, Wald A. Differentiation of herpes simplex virus types 1 and 2 in clinical samples by a real-time taqman PCR assay. J Med Virol. 2005;76(3):350–355. doi: 10.1002/jmv.20365. [DOI] [PubMed] [Google Scholar]
- 15.Magaret A, Johnston C, Wald A. Use of the designation "shedder" in mucosal detection of herpes simplex virus DNA involving repeated sampling. Sex Transm Infect. 2009 doi: 10.1136/sti.2008.034751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bekris LM, Millard SP, Galloway NM, Vuletic S, Albers JJ, Li G, et al. Multiple SNPs within and surrounding the apolipoprotein E gene influence cerebrospinal fluid apolipoprotein E protein levels. J Alzheimers Dis. 2008;13(3):255–266. doi: 10.3233/jad-2008-13303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koelle DM, Benedetti J, Langenberg A, Corey L. Asymptomatic reactivation of herpes simplex virus in women after first episode of genital herpes. Ann Intern Med. 1992;116:433–437. doi: 10.7326/0003-4819-116-6-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy GH. Mendelian proportions in a mixed population. 1908. Yale J Biol Med. 2003;76(2):79–80. [PMC free article] [PubMed] [Google Scholar]
- 19.Gerdes LU. The common polymorphism of apolipoprotein E: geographical aspects and new pathophysiological relations. Clin Chem Lab Med. 2003;41(5):628–631. doi: 10.1515/CCLM.2003.094. [DOI] [PubMed] [Google Scholar]
- 20.Fullerton SM, Clark AG, Weiss KM, Nickerson DA, Taylor SL, Stengard JH, et al. Apolipoprotein E variation at the sequence haplotype level: implications for the origin and maintenance of a major human polymorphism. Am J Hum Genet. 2000;67(4):881–900. doi: 10.1086/303070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corey L, Wald A. Genital Herpes. In: Holmes KK, Sparling PF, Stamm WE, Piot P, Wasserheit JN, Corey L, Cohen MS, Watts DH, editors. Sex. Trans. Dis. New York: McGraw Hill Medical; 2008. pp. 399–437. [Google Scholar]
- 22.Itzhaki R, Wozniak M. Susceptibility to herpes simplex labialis conferred by the gene encoding apolipoprotein E. J Infect Dis. 2008;198(4):624–625. doi: 10.1086/590213. author reply 25-6. [DOI] [PubMed] [Google Scholar]
- 23.Jayasuriya AN, Itzhaki RF, Wozniak MA, Patel R, Smit EJ, Noone R, et al. Apolipoprotein E-epsilon 4 and recurrent genital herpes in individuals co-infected with herpes simplex virus type 2 and HIV. Sex Transm Infect. 2008;84(7):516–517. doi: 10.1136/sti.2008.032367. [DOI] [PubMed] [Google Scholar]
- 24.Singh PP, Singh M, Mastana SS. APOE distribution in world populations with new data from India and the UK. Ann Hum Biol. 2006;33(3):279–308. doi: 10.1080/03014460600594513. [DOI] [PubMed] [Google Scholar]
- 25.Hobbs MR, Jones BB, Otterud BE, Leppert M, Kriesel JD. Identification of a herpes simplex labialis susceptibility region on human chromosome 21. J Infect Dis. 2008;197(3):340–346. doi: 10.1086/525540. [DOI] [PubMed] [Google Scholar]
- 26.Bochud PY, Magaret AS, Koelle DM, Aderem A, Wald A. Polymorphisms in TLR2 are associated with increased viral shedding and lesional rate in patients with genital herpes simplex type 2 infection. J. Infect. Dis. 2007;196:505–509. doi: 10.1086/519693. [DOI] [PubMed] [Google Scholar]
- 27.Miller RM, Federoff HJ. Isoform-specific effects of ApoE on HSV immediate early gene expression and establishment of latency. Neurobiol Aging. 2008;29(1):71–77. doi: 10.1016/j.neurobiolaging.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 28.Corder EH, Paganelli R, Giunta S, Franceschi C. Differential course of HIV-1 infection and APOE polymorphism. Proc Natl Acad Sci U S A. 2008;105(46):E87. doi: 10.1073/pnas.0808164105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mauri DN, Ebner R, Montgomery RI, Kochel KD, Cheung TC, Yu G-L, et al. LIGHT, a new member of the TNF superfamily, and lymphotoxin alpha are ligands for herpesvirus entry mediator. Immunity. 1998;8:21–30. doi: 10.1016/s1074-7613(00)80455-0. [DOI] [PubMed] [Google Scholar]
- 30.Satoh T, Arii J, Suenaga T, Wang J, Kogure A, Uehori J, et al. PILRalpha is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell. 2008;132(6):935–944. doi: 10.1016/j.cell.2008.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jerome KR, Fox R, Chen Z, Sears AE, Lee H, Corey L. Herpes simplex virus inhibits apoptosis through the action of two genes, US3 and US5. J. Virol. 1999;73:8950–8957. doi: 10.1128/jvi.73.11.8950-8957.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chou J, Kern ER, Whitley RJ, Roizman B. Mapping of herpes simplex virus 1 neurovirulence to gamma 1 34.5, a gene nonessential for growth in cell culture. Science. 1990;252:1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- 33.Hill A, Jugovic P, York I, Russ G, Bennink J, Yewdell J, et al. Herpes simplex virus turns off the TAP to evade host immunity. Nature. 1995;375:411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- 34.Tsuang DW, Bird TD. Genetics of dementia. Med Clin North Am. 2002;86(3):591–614. doi: 10.1016/s0025-7125(02)00003-2. [DOI] [PubMed] [Google Scholar]