Abstract
The spliceosome is a complex small nuclear (sn)RNA–protein machine that removes introns from pre–mRNAs via two successive phosphoryl transfer reactions. The chemical steps are isoenergetic, yet splicing requires at least eight RNA–dependent ATPases responsible for substantial conformational rearrangements. To comprehensively monitor pre–mRNA conformational dynamics, we developed a strategy for single molecule FRET (smFRET) that utilizes a small, efficiently spliced yeast pre–mRNA, Ubc4, in which donor and acceptor fluorophores are placed in the exons adjacent to the 5′ and 3′ splice sites. During splicing in vitro we observe a multitude of generally reversible, time– and ATP–dependent conformational transitions of individual pre–mRNAs. The conformational dynamics of branchpoint and 3′ splice site mutants differ from one another and from wild–type. Because all transitions are reversible, spliceosome assembly appears to be occurring close to thermal equilibrium.
INTRODUCTION
Introns in eukaryotic pre–mRNAs are removed by RNA splicing. This process is carried out by the spliceosome, a large supra–molecular assembly consisting of five small nuclear RNAs (snRNAs) and more than 100 proteins1,2. The spliceosome lacks a pre–formed catalytic core. Rather, spliceosomes are assembled on intron–containing substrates in a stepwise manner that requires both binding and release of small nuclear ribonucleoproteins (snRNPs; reviewed in Ref. 2). Notably, splicing entails two successive phosphoryl transfer reactions that are isoenergetic. Nonetheless, spliceosome assembly requires at least eight RNA–dependent ATPases of the so–called DEAD–box (DExD/H) family3. These enzymes, which catalyze successive rearrangements of RNA and protein pairing partners, are believed to enhance the fidelity of splicing by acting as proofreading clocks to allow the discard of mutant substrates at multiple steps in the assembly and catalytic pathway4–7.
In the budding yeast Saccharomyces cerevisiae, assembly (reviewed in Ref. 2) is initiated by the ATP–independent formation of a commitment complex in which the U1 snRNP interacts with the 5′ splice site (5′SS) while BBP and Mud2 interact with the branchpoint sequence (Fig. 1). If the branchpoint is mutated (BP mutant) this complex cannot form and spliceosome assembly is blocked. With a wild–type precursor (WT), the commitment complex is converted to the pre–spliceosome by the ATP–dependent binding of U2 to the branchpoint, followed by association of the U4/U6.U5 triple snRNP. Before the first transesterification can occur, U1 and U4 snRNPs must be released from the assembled spliceosome, again in ATP–dependent reactions. Importantly, the first chemical step, which results in formation of the lariat–intermediate and free 5′ exon, can occur even in the presence of a mutation at the 3′ splice site (3′SS). This mutation specifically blocks the second transesterification. Finally, the spliceosome undergoes ATP–dependent disassembly from the lariat intron and mature mRNA (Fig. 1).
Figure 1.

Canonical spliceosome assembly pathway.
Despite this level of understanding of the pathway, major questions about the specific conformational rearrangements and particularly their kinetics (and splicing signal dependence) remain unanswered. This gap in our knowledge is in large part due to the fact that to date splicing must be studied in a crude extract in vitro, where multi–step reversible processes are obscured by asynchronous progression along the pathway8. In addition, splicing in a yeast extract is generally inefficient, leading to only a fraction of molecules undergoing one or both steps of splicing. To overcome these severe limitations and monitor in real–time the conformational states of the pre–mRNA in spliceosome assembly, we have developed an in vitro assay based on single molecule fluorescence resonance energy transfer (smFRET) that obviates the need to isolate or synchronize reaction intermediates.
We first identified a natural yeast pre–mRNA with a small intron, Ubc4, which is efficiently spliced in vitro. This made possible the chemical synthesis of the pre–mRNA with donor (Cy 3) and acceptor (Cy 5) fluorophores in the exons adjacent to the 5′– and 3′– splice sites, respectively. The Förster radius of the Cy3–Cy5 fluorophore pair is ~54 Å so that smFRET can effectively monitor changes in inter–fluorophore distance between ~20–100 Å. Cryo–EM micrographs of snRNPs and spliceosomal complexes range in size from 200–300 Å9,10, yet cross–linking and footprinting indicate proximity of the two exons throughout splicing11,12. Using total internal reflection fluorescence microscopy (TIRFM) of immobilized Ubc4 we show here that the pre–mRNA reversibly folds into conformations with proximal exons even in the absence of splicing extract. Spliceosome assembly further accelerates and diversifies these conformational dynamics. We establish four criteria that suggest that we are indeed monitoring spliceosome assembly at the level of single molecules; an in situ assay verifies that the surface immobilized pre–mRNA is efficiently spliced; the observed complex smFRET transitions are time– and ATP–dependent; and they are dependent on appropriate intron signals. Our results suggest that the spliceosome is a cellular machine that operates close to equilibrium like the ribosome.
RESULTS
Identification and functional characterization of a suitable pre–mRNA
In Sacchromyces cerevisiae, most in vitro splicing assays have utilized the ~1,400 nucleotide (nt) actin pre–mRNA (with an intron of 308 nt)13. To develop our smFRET approach, however, we sought to identify a smaller pre–mRNA efficiently spliced in vitro that would be amenable to chemical synthesis. To this end, we analyzed pre–mRNAs from the temperature sensitive yeast mutant prp2–114.
In this mutant, splicing is blocked before the first catalytic step when cells are shifted to the non–permissive temperature, resulting in the efficient accumulation of most yeast pre–mRNAs15,16. Total RNA was extracted from prp2–1 cells grown first at permissive temperature (30°C), and then shifted for 30 min to the non–permissive temperature (37°C). Previous experiments had shown that this RNA preparation contains pre–mRNAs from most genes containing introns and a small background of mature mRNAs, spliced during the permissive growth period16. This RNA mixture was subjected to in vitro splicing conditions in yeast cell extract, isolated, and analyzed using a splicing microarray that could detect mature, spliced mRNAs16. Several efficiently spliced candidates with small introns were identified (Supplementary Table 1). We ultimately chose the pre–mRNA for UBC4 as the most suitable substrate for our smFRET analysis.
The entire UBC4 gene is 542 nt long with a short intron of 95 nt. We synthesized a number of Ubc4 pre–mRNAs with different exon lengths and assayed splicing activity. In a trade–off between ease of synthesis and splicing activity of the substrate we chose to develop a variant of the Ubc4 pre–mRNA for smFRET consisting of 20–nt long exons and the 95–nt intron for a total size of 135 nt. This truncated Ubc4 pre–mRNA was prepared through a two–piece ligation of synthesized RNAs (see Methods). We post–synthetically coupled the donor fluorophore Cy3 to the 5′ exon and the acceptor Cy5 to the 3′ exon, seeking to place the fluorophores as close as possible to the 5′ and 3′ splice sites, respectively, for maximum distance sensitivity without interfering with splicing activity. We prepared pre–mRNAs with dyes located at uridines in multiple positions, all of which are spliced with a similar efficiency of ~30–40% (Fig. 2a). The substrate used in this work carries Cy3 in exon 1, 7 nt from the 5′ splice site, and Cy5 in exon 2, 4 nt from the 3′ splice site (Fig. 2a). Introduction of 3′SS and BP mutations into this smFRET substrate was found to have the expected effects17 of blocking splicing prior to the second and first steps of splicing, respectively (Fig. 2b).
Figure 2.
Splicing activity of different Ubc4 pre–mRNA variants. (a) Positioning the dyes in different sites in the 20–nt exon of Ubc4 pre–mRNA results in similar splicing efficiency. In this experiment splicing was assayed at room temperature as described in Methods. The three lanes for each pre–mRNA are (−)ATP, (+)ATP for 15 min and (+)ATP for 30 min. All pre–mRNAs examined have an apparent splicing efficiency of first plus second step [(mRNA+lariat intermediate)/(pre–mRNA+ mRNA+lariat intermediate)] of between 30–40%. The Cy5 fluorescence scan is shown. (b) The splicing assay of wildtype (WT), 3′ splice site mutant (3′SS) and branchpoint mutant (BP) Ubc4 pre–mRNAs shows that the 3′SS mutant RNA only carries out step 1, leading to a lariat–intron intermediate. The BP mutant is inactive in splicing. The experiment was carried out as described above for panel A. The Cy5 fluorescence scan is shown. (c) Design of an in situ assay to probe for the presence of introns in immobilized (pre–)mRNAs. (d) Results of RNase H probing for the presence of the intron after incubation of WT, BP, and 3′SS pre–mRNA in yeast splicing extract. Control experiments on WT pre–mRNA and mature mRNA (leftmost 4 columns) show that an intron containing substrate strongly loses Cy3 signal after incubation with a complementary DNA oligonucleotide and RNase H, but not the intron–free mRNA. Loss of the intron to splicing is thus indicated by a small difference in signal before and after RNase H treatment. Error bars indicate 1 s.d. from the mean.
In our smFRET assays the 3′ end of the labeled pre–mRNA was immobilized through hybridization to a short 2′–O–methyl capture RNA with a 5′ biotin, which was bound to a streptavidin coated and PEG passivated quartz slide (Fig. 3a). Hybridization of the capture RNA to Ubc4 pre–mRNA does not affect the efficiency of splicing in ensemble assays (data not shown). The Cy3 and Cy5 fluorophores were detected by total internal reflection fluorescence microscopy (TIRFM) in real–time to determine the smFRET efficiency as described previously8,18–21. By utilizing TIRFM we minimized the background fluorescence of the splicing extract, while capturing all sufficiently long–lived conformational states of the pre–mRNA (even in a fully extended pre–mRNA of ~50 nm length the distal Cy3 dye is still well within the ~100 nm deep evanescent light field of TIRFM8). An enzymatic oxygen scavenger system (OSS) was used to limit photobleaching, and Trolox was added to suppress fluorophore blinking (see Methods). Ensemble splicing assays showed that the presence of OSS and Trolox in the reaction mix does not inhibit splicing (data not shown). Under these conditions, we observed limited quenching/blinking and photobleaching with rate constants of ~0.01 s−1. These effects are the same for each of the three pre–mRNAs, and we found no appreciable change in the quenching/blinking and photobleaching upon addition of ATP.
Figure 3.
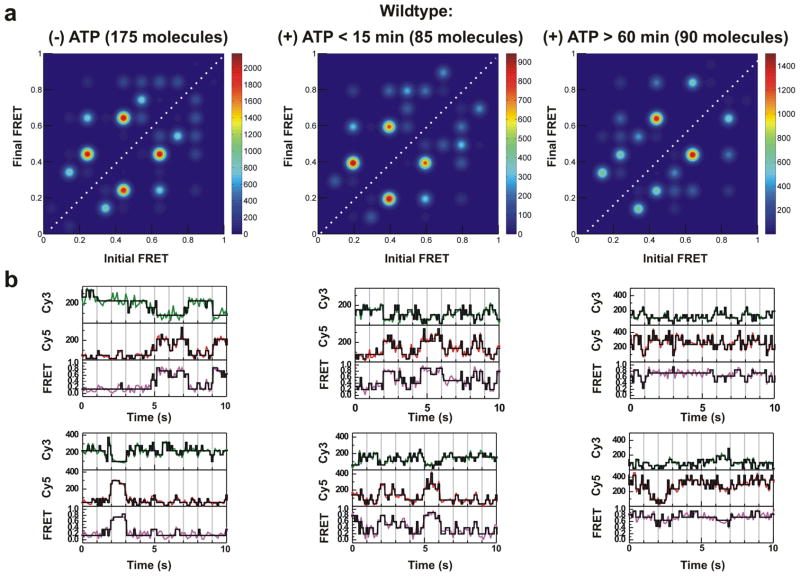
Data analysis and examples of analysis. (a) Synthetic Ubc4 pre mRNA is hybridized via a 17–nt 2′–O–methyl RNA tether to the 3′ exon, and attached via biotin to a streptavidin coated quartz slide. (b) Raw Cy3 (green), Cy5 (red), FRET (Magenta) time trajectories are analyzed using Hidden Markov Modeling (HMM) algorithms to yield idealized trajectories (black) as described in Methods. (c) The first 10 s of the raw FRET trajectories of a subset of (36 out of 175) WT molecules incubated in ATP–depleted cell extract were analyzed to determine the ensemble distribution of FRET states within the population of molecules analyzed. (d) Transition Density Plots (TDPs) utilize the idealized FRET trajectories to determine the entire set of transitions for a given set of molecules. The number of times a transition occurs is represented as a heat map whose index is defined by the color bar. The fact that this analysis, of a subset (36 of 175 molecules as in panel c) of the data shown in Figs. 5a and 6a, provides qualitatively the same result obtained for the full data set attests to the convergence of the analysis.
To verify that the spliceosome assembles on the immobilized pre–mRNA to remove its intron we performed an in situ RNase H sensitivity assay under our TIRFM conditions. Briefly, a DNA oligonucleotide complementary to the intron was added together with RNase H to slides on which WT pre–mRNA had been immobilized and incubated in either ATP–depleted [(−)ATP] or ATP supplemented [()ATP] yeast splicing extract for an hour (Methods and Fig 2c). Only pre–mRNA is expected and experimentally found to be susceptible to RNase H degradation, leading to the specific loss of the 5′ exon and thus Cy3 signal from the slide surface, while the mature mRNA is RNase H resistant (first four columns of Fig 2d). We find that after incubation in (−)ATP cell extract 75% of all molecules lose Cy3 but not Cy5 upon RNase H challenge, while in ()ATP this fraction is negligible ( Fig. 2d). Performing the same assay with the BP mutant results in extensive loss of Cy3 signal upon addition of RNase H both before and after incubation in ()ATP extract (Fig. 2d), consistent with the expected block of the first step of splicing. The 3′SS mutant displays an intermediate picture as much of its Cy3 signal is lost during the high salt washes used to remove the cell extract (Fig. 2d), consistent with a block after the first step of splicing when the 5′ exon is no longer covalently attached to the intron. These results strongly support the notion that the immobilized WT pre–mRNA is splicing active to at least the extent observed in standard ensemble splicing assays in solution (~30–40%, see above). The apparent increase in splicing efficiency of the immobilized WT substrate may be the result of the more favorable stoichiometric ratio of limiting splicing factors to pre–mRNA under our single molecule conditions. Additionally, effects such as RNA aggregation and precipitation are known to inhibit reactions under in vitro conditions that favor intramolecular and thus inevitably intermolecular interactions22. Immobilization of isolated pre–mRNAs for single molecule studies is expected to suppress such inhibition and may thus provide ideal conditions for studying splicing.
Dissecting conformational states and dynamics in complex smFRET trajectories
Our smFRET approach monitors the relative dynamic positioning of the two exons within the spliceosomal complex. Results are presented in three ways. First, we show smFRET time trajectories with their corresponding donor and acceptor traces, which reveal multiple discrete FRET values or states that reflect the complexity of pre–mRNA conformational rearrangements during spliceosome assembly (Fig. 3b). Second, FRET occupancy histograms display the probability of observing a particular FRET value (Fig. 3c). In these histograms we plot frame–by–frame FRET values for a set of molecules collected over the first 100 frames (10 s of real–time) of all FRET trajectories in a given data set, which is proportional to the probability that any single molecule will have that FRET value.
Third, we use traditional Transition Density Plots (TDPs), weighted by the number of times a particular FRET transition occurs, as well as novel POpulation–weighted and Kinetically–Indexed Transition density (POKIT) plots to comprehensively represent all observed discrete FRET states, their transitions and kinetics. To first reliably identify genuine FRET states and their transition kinetics in naturally noisy data sets we use Hidden Markov Modeling (HMM), which traces the most probable sequence of FRET states and derives their dwell times23 (Methods and Fig. 3b). To eliminate from our analysis possible effects of the high concentrations of proteins directly on the fluorophores we additionally evaluate the donor and acceptor signals by HMM and introduce a scoring function selecting for anti–correlated donor and acceptor changes that reflect changes in FRET (see Methods and Supplementary Fig. 2 online). As a further control, we monitored the donor and acceptor signals from a mature (synthetic) mRNA under the same conditions used for the pre–mRNA and found a largely steady FRET signal (Supplementary Fig. 3 online). These observations indicate that any smFRET dynamics we observe for pre–mRNA are not caused by spurious photophysical artifacts but by the expected changes in fluorophore distance.
To determine the most likely number of states in our smFRET trajectories we analyzed whole data sets of 50–200 single molecule traces by our scored HMM approach. The best global fit to the entire trace set for each condition generally found 10 or 11 distinct FRET states (to be evaluated as distinct two states needed to exhibit a FRET difference of at least 0.1, see Methods), with each molecule adopting only a subset of states (Supplementary Fig. 4 online). With knowledge of the number of states in each data set we identified the transitions between these states, summarized as TDPs24 and POKIT plots (see below). The transition from an initial FRET state of 0.8, for example, to a final FRET state of 0.3 is one point in these plots. There can be a maximum of N(N−1) different transitions between N states. We observed many fewer than the possible 110 transitions.
To test the robustness of our analysis we randomly selected a subset of 36 out of a total of 175 WT substrate molecules in (−)ATP extract and found this subset to yield a similar FRET occupancy histogram and TDP as the entire data set (compare Figs. 3c and 3d with Figs. 5a and 6a, respectively), suggesting that our molecule sample is of sufficient size for the HMM model to converge.
Figure 5.
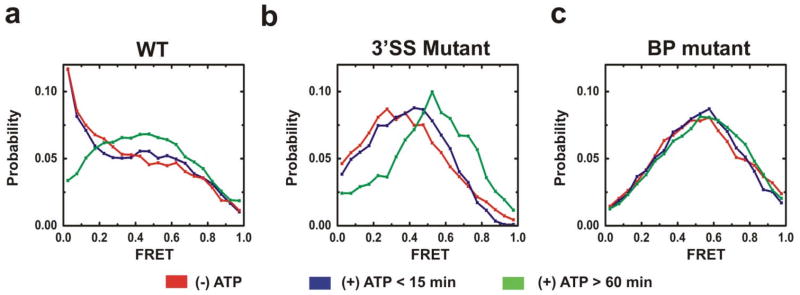
ATP–dependent conformational dynamics of the wildtype (WT), 3′ splice site mutant (3′SS), and branchpoint mutant (BP) pre–mRNAs in yeast cell extract. (a) Probability distributions of FRET states for the WT substrate for each experimental condition. (b) Probability distributions of FRET states for the 3′SS substrate for each experimental condition. (c) Probability distributions of FRET states for the BP substrate for each experimental condition.
Figure 6.
Mapping conformational changes of the wildtype (WT) pre–mRNA during spliceosome assembly and splicing in vitro. (a) TDPs for WT substrate; in ATP–depleted extract, within 15 min after ATP addition to cell extract, and after 60 min incubation in (+)ATP cell extract. (b) Representative trajectories (donor, green; acceptor, red; FRET, magenta) and idealized HMM models (black) for WT substrate.
The pre–mRNA substrate dynamically folds and unfolds in splicing buffer
When incubated in standard splicing buffer alone, all three pre–mRNAs exhibit substantial global dynamics with fluctuations between high and low FRET states (Fig. 4a). The several second long dwell times in high FRET states indicate that the pre–mRNAs must transiently form secondary structures that bring the distal 5′ and 3′ splice sites into close proximity. The distribution of FRET states is not identical between the pre–mRNAs, as the WT and BP mutant favor high FRET states (shorter distances) relative to the 3′SS mutant. By comparison, the 3′SS spends relatively more time in low FRET states as evident from the cumulative FRET occupancy histograms in Fig. 4b. These differences are also apparent in the corresponding TDPs, where the WT and BP pre–mRNAs exhibit more transitions in the high (>0.7) and fewer in the low (<0.4) FRET range than the 3′SS. Notably, there are some transitions with long dwell times found in all pre–mRNAs (Figs. 4a,c). Computational predictions suggest that in the ensembles of lowest free energy states distinct secondary structures separate the 5′ and 3′ splice sites in the WT and BP mutant less than in the 3′SS mutant, qualitatively correlating with their experimentally observed relative bias towards higher FRET values (Supplementary Fig. 6 online).
Figure 4.
Conformational dynamics of wildtype (WT), 3′ splice site mutant (3′SS), and branchpoint mutant (BP) pre–mRNA substrates in splicing buffer. (a) Sample traces of all three substrates in splicing buffer, showing raw donor (Cy3, green), acceptor (Cy5, red), and FRET (blue) trajectories and their idealized HMM models (black). (b) FRET histograms of the three pre–mRNAs in splicing buffer. (c) TDPs for all three pre–mRNAs in splicing buffer.
Since all RNA molecules assume transient secondary structures25, it is not surprising that the Ubc4 pre–mRNA exhibits transient folding yielding high FRET states. In the case of Ubc4 pre–mRNA even single base changes affect these structures. That we can observe these single base differences by smFRET attests to the power of this method. Furthermore, this kinetic analysis of the substrate RNA in buffer is necessary to distinguish the pre–mRNA dynamics caused by the formation of transient secondary structures and those induced by spliceosome assembly. To achieve a maximal dynamic time range for observing spliceosome related kinetics, in the following we performed 100–ms time resolution smFRET experiments at various instances after addition of splicing extract.
Sequence and time dependent dynamics of single pre–mRNAs in splicing extract
The addition of yeast splicing extract depleted of ATP [(−)ATP] results in a profound difference from the FRET histograms observed in buffer alone (compare Figs. 4b and 5). Lower FRET values are relatively favored for all three pre–mRNAs. The effect is most pronounced for the WT, which becomes biased toward particularly low FRET values. These changes to lower FRET distributions show that the relatively stable high FRET states observed in buffer alone are disfavored in extract.
The addition of ATP to extract [(+)ATP] leads to a time dependent change in the FRET distributions of the WT and 3′SS mutant pre–mRNAs, but not that of the BP mutant (Fig. 5c). In the case of the 3′SS mutant, the overall FRET distribution changes within 15 min and after 60 min of incubation in (+)ATP extract to increasingly more prevalent mid and high FRET states (Fig. 5b). In the case of the WT, the prevalent low FRET states are lost over 60 min of incubation (Fig. 5a), while the BP mutant shows no substantial change in the overall FRET distribution (Fig. 5c), in agreement with the requirement of an intact branch point for ATP–dependent spliceosome assembly17. Thus, the time dependent changes in FRET distribution upon addition of ATP require a pre–mRNA substrate that can undergo the first step (3′SS) or both steps (WT) of splicing.
The time–, ATP– and mutant dependent changes in the FRET histograms observed upon incubation in extract strongly suggest that we are observing splicing dependent changes at the single molecule level, but FRET histograms cannot tell us about the dynamics of those changes. To track even subtle changes in these conformational dynamics upon addition of splicing extract we turned to HMM and TDP plot analysis.
Spliceosome dependent dynamics of pre–mRNA substrates
The TDP of the WT pre–mRNA shows that upon addition of (−)ATP cell extract fewer transitions are observed when compared to the same substrate in buffer and the most prevalent transitions are on average lower on the FRET scale (compare Figs. 4c and 6a). Within the first 15 min after the addition of (+)ATP cell extract we find that the distribution of transitions changes yet again, with more transitions originating from the 0.8 FRET state. After a 60 min incubation in (+)ATP cell extract, FRET transitions appear in an even higher FRET region (0.75–0.95), indicating that conformations are favored where the two exons are in close proximity (Fig. 6a). Most observed FRET transitions are reversible, leading to the relative mirror symmetry of the TDPs with respect to their main diagonal (Fig. 6a). Reversible FRET transitions imply conformational reversibility, a property that is characteristic of all substrates and extract conditions (Figs. 6–8).
Figure 8.
Mapping conformational changes of the branchpoint mutant (BP) pre–mRNA spliceosome assembly and splicing in vitro. (a) TDPs for BP substrate; in ATP–depleted extract, within 15 min after ATP addition to cell extract, and after 60 min incubation in (+)ATP cell extract. (b) Representative trajectories (donor, green; acceptor, red; FRET, magenta) and idealized HMM models (black) for BP substrate.
Upon addition of (−)ATP extract the 3′SS pre–mRNA, like the WT, displays fewer, still symmetric (i.e., reversible) transitions than in buffer (compare Figs. 4c and 7a). After a 15 min incubation in (+)ATP cell extract (compare Figs. 6 and 7) there is a shift towards higher FRET states. Most notably, transitions that become more prevalent after 60 min incubation in (+)ATP extract generally fall into a mid FRET regime (Fig. 7a), unlike for the WT where a shift occurs towards high FRET transitions (Fig. 6a).
Figure 7.
Mapping conformational changes of the 3′ splice site mutant (3′SS) pre–mRNA spliceosome assembly and splicing in vitro. (a) TDPs for 3′SS substrate; in ATP–depleted extract, within 15 min after ATP addition to cell extract, and after 60 min incubation in (+)ATP cell extract. (b) Representative trajectories (donor, green; acceptor, red; FRET, magenta) and idealized HMM models (black) for 3″SS substrate.
The BP mutant does not enter the spliceosome assembly pathway and while we observe transitions between a substantial number of FRET states, unlike the WT and 3′ SS substrates there are few changes when comparing the FRET states in (−)ATP extract and (+)ATP extract. The prominent set of symmetric transitions between 0.45 and 0.65 FRET are present before and following the addition of (+)ATP extract; Figs. 8a–c). After 60 min in (+)ATP extract the number and diversity of transitions sampled by each BP mutant molecule is only slightly changed.
To assess the kinetic differences between substrates we turn to POKIT plot analysis. Supplementary Figure 5 online provides a side–by–side comparison of these plots with the more common TDPs to illustrate their close relationship; however, POKIT plots provide two additional, comprehensive pieces of information for each transition. First, they present as a number of concentric circles the fraction of molecules in the entire trace set that exhibits a specific FRET transition at least once (Supplementary Fig. 5 online). This feature complements TDPs that are weighted by number of times a specific transition is observed overall24, which emphasizes fast (frequent) transitions exhibited by possibly only a small fraction of molecules (Supplementary Fig. 5 online). Second, our POKIT plots provide the average dwell time for each transition, or time spent in the initial state before transition to the final state, in the form of circle colors (Supplementary Fig. 5 online), facilitating the rapid visual comparison of the kinetics of our various smFRET data sets as a function of ATP concentration, incubation time, and intron signals.
The POKIT plots of the WT pre–mRNA in buffer and (−)ATP extract highlight the fact that incubation with cell extract induces longer dwell times in low rather than high FRET states (Fig. 9). Within the first 15 min after the addition of (+)ATP cell extract the distribution of transitions then changes to include a higher fraction of molecules with FRET transitions in the 0.6–0.9 range (Fig. 9). After a 60 min incubation in (+)ATP cell extract, this trend continues toward even longer dwell times in an even higher FRET region (0.75–0.95, Fig. 9), indicating that relatively stable conformations are favored where the two exons are proximal. These long dwell times in the highest FRET states are observed only for the WT substrate and require an extended incubation with (+)ATP cell extract (Fig. 9). The fact that similar transitions are strongly dominant for the mature (synthetic) mRNA control (Supplementary Fig. 3 online), but are found in only ~20% of all WT molecules (Fig. 9), suggests that only a fraction of the total WT pre–mRNA observed after 60 min in (+)ATP cell extract adopts a conformation similar to that of the synthetic mRNA. These findings indicate that most pre–mRNA molecules are spliced reversibly, possibly because our 2′–O–methyl RNA tether hybridized to the short 3′ exon blocks the binding of Prp22 and disrupts spliceosome disassembly25; and/or that the mRNA derived from activity of bound splicing factors exhibits a FRET signature distinct from that of the synthetic mRNA, which is not expected to bind splicing factors.
Figure 9.
Detailed comparison of kinetic and conformational profiles of pre–mRNAs during spliceosome assembly and splicing in vitro. (a) POKIT plots for WT substrate; in ATP–depleted extract, within 15 min after ATP addition to cell extract, and after 60 min incubation in (+)ATP cell extract. (b) POKIT plots for 3′SS substrate; in ATP–depleted extract, within 15 min after ATP addition to cell extract, and after 60 min incubation in (+)ATP cell extract. (c) POKIT plots for BP substrate; in ATP–depleted extract, within 15 min after ATP addition to cell extract, and after 60 min incubation in (+)ATP cell extract.
In summary, our data suggest that in vitro the WT pre–mRNA samples both low and high FRET conformational states even in the absence of splicing extract. The addition of extract leads to a change in the distribution and kinetics of these FRET states, first by assembling with spliceosomal components in the absence of ATP and, second, by extended incubation with ATP–enriched cell extract over a time period where it undergoes both steps of splicing (Fig. 2). Additionally, WT pre–mRNA after 60 min in splicing extract adopts relatively stable high FRET conformations found in an mRNA control but not in mutant substrates when placed under the same conditions (Figs. 6–9 and Supplementary Fig. 3 online). The BP mutant largely lacks these time and ATP–dependent changes. The 3′SS mutant is generally more similar to the WT than the BP mutant, but does not achieve the stable high FRET states after 60 min incubation observed for the WT substrate. Most conformational changes of the pre–mRNAs are readily reversible and their relative abundance and kinetics both ATP and intron signal dependant, implying that they are relevant for splicing activity.
DISCUSSION
Establishment of smFRET to monitor spliceosome assembly
In this study we have employed smFRET to observe the conformational dynamics of a pre–mRNA during spliceosome assembly. We identified Ubc4 as a natural intron–containing transcript that is both small – allowing ease of chemical synthesis – and very efficiently spliced in vitro as well as in situ during our smFRET experiments (Fig. 2). We placed the donor fluorophore Cy3 in exon 1 near the 5′ splice site and the acceptor Cy5 in exon 2 near the 3′ splice site. With this substrate under standard in vitro splicing conditions we observed a splicing efficiency of 30–40%, which in situ may be even higher (Fig. 2). This result is not unique to the Ubc4 substrate; recently Crawford et al. demonstrated that a different fluorescently labeled pre–mRNA bound to a slide for TIRFM studies undergoes splicing (measured as the release of a fluorescent intron) upon addition of splicing extract26. The authors did not, however, monitor the structural dynamics of the pre–mRNA during spliceosome assembly and action as we have done here.
Several criteria establish that the smFRET changes we observe in splicing extract reflect dynamic changes in the substrate during spliceosome assembly. These criteria are that the dynamic changes be time– and ATP–dependent, and that they be affected as expected by mutations in the substrate. The FRET occupancy histograms in Fig. 5 illustrate that time and ATP–dependent changes indeed are observed for the WT substrate and the 3′SS mutant but, as expected, less so for the BP mutant (see also below). These results demonstrate that we monitor conformational changes associated with genuine spliceosome assembly processes.
The pattern of WT conformational dynamics is altered by intron mutations
When ATP–depleted extract is added, substantial differences in FRET histograms and FRET transitions of all three pre–mRNAs are observed. More specifically, the smFRET transitions observed in splicing buffer alone are replaced by a somewhat smaller set of transitions with on average shorter dwell times (Figs. 4, 6–9). This initial shift in FRET distribution of all pre–mRNAs upon extract addition is consistent with the presence of a large number of highly concentrated RNA binding proteins in yeast extract, e.g., the RNP protein NPL327. These proteins are expected to alter transient secondary structures, but they are not expected to distinguish between RNA molecules differing by a single base change. Notably, the WT adopts a set of low FRET conformations with slow transitions that are not found in the 3′SS and BP mutants. As evident from Fig. 1 the behavior of WT may reflect the ATP–independent interaction of the U1 snRNP with the 5′SS, BBP (SF1 in humans) and Mud2 (U2AF in mammals) with the branchpoint and polypyrimidine tract to form CC, the commitment complex. In any event, since an intact branchpoint is required for the resistance of a labeled pre–mRNA template to challenge by excess cold substrate28, we predicted that the BP mutant would be blocked at the earliest stage of spliceosome assembly. This is fully consistent with our observation of substantial differences in the TDP and POKIT plot patterns of WT and 3′SS mutant on the one hand and BP mutant on the other (Figs. 6–9).
The 3′SS mutant was predicted to largely mimic the WT pattern, given that the 3′ splice site sequence is thought to be required only for the second chemical step. This prediction is consistent with the greater similarity of the TDP and POKIT plots of the WT and 3′SS mutant compared to that of the BP mutant. Curiously the 3′SS mutant, unlike the WT, does not adopt stable low FRET transitions in the absence of ATP (Fig. 9). These stable transitions appear in the 3′SS mutant only after addition of (+)ATP supplemented extract, and are never observed in the BP mutant. It is tempting to speculate that our detailed kinetic smFRET analysis reveals differences between WT and 3′SS mutant that are not detectable by conventional assays. Other discrepancies to the canonical model of spliceosome assembly (Fig. 1) have been noted previously and will likely expand with the emergence of new experimental tools29,30.
The spliceosome, like the ribosome, functions near thermal equilibrium
As described in the introduction, a dominant and well–known feature of splicing is the highly dynamic nature of the spliceosome, which is driven forward by (in yeast) eight RNA–dependent ATPases of the DEAD–box family2. It is thought that each of the eight DEAD–box dependent steps in splicing provide fidelity check–points6,7. Evidence for this model is found in our observation that within the first 15 min of incubation of WT pre–mRNA in ATP–enriched extract the molecule population shifts towards higher FRET transitions compared to the transitions in ATP–depleted extract (Figs. 6 and 9). This indicates that the assembly and rearrangements of spliceosomal components initiated by ATP bring the two exons closer to each other, as envisioned in the model to achieve splicing–specific conformations. The effect is more pronounced after a long incubation (60 min) with ATP–enriched extract (Figs. 6 and 9), when a substantial fraction of mRNA product has been generated (Fig. 2).
Historically, it has been assumed that the spliceosomal ATPases would drive the pathway unidirectionally. Our observation of the dynamics of single pre–mRNAs indicates that the substrate does not follow a simple unidirectional pathway. Instead, pre–mRNAs sample a variety of reversible conformational states guided by the assembly of ATP dependent spliceosomal components. Recently, it has been demonstrated that the chemical steps are themselves reversible, at least under certain in vitro conditions31,32. A reversal of the chemical steps of splicing requires that the conformation of the substrate and spliceosome assembly be reversible as well. Such a model is supported by our observation of reversible smFRET transitions. To our knowledge, our data provide the first direct glimpse of such reversible conformational changes of the pre–mRNA throughout the splicing process. This realization prompts a provocative re–evaluation of the role of ATP in splicing, in which the function of hydrolysis is largely to improve the accuracy of the pathway, driving the “correct” substrate to completion while “incorrect” alternatives are rejected. Interestingly, this picture fits well with recent evidence from the ribosome, where spontaneous intersubunit rotation has been observed in the absence of GTP33. Thus it is tempting to speculate that both macromolecular machines carry out complex sets of conformational changes close to thermal equilibrium.
Outlook
We have shown that during spliceosome assembly a pre–mRNA’s conformations are highly dynamic and largely reversibly interchanging. These changes are reflected in a large number of complex transitions. By locating the dyes at different pre–mRNA positions, by placing the dyes in snRNAs or proteins, and by examining the effects of mutant extracts it now seems possible to assign particular sets of FRET transitions to specific steps in spliceosome assembly. Ultimately approaches such as these will yield a comprehensive dynamic model of pre–mRNA splicing.
METHODS
Microarray analysis of prp2–1 versus wild–type
To find a yeast pre–mRNA that is active in vitro and that has a small intron we performed the experiment described in detail in the Supplementary Methods. The presence of spliced mRNA was monitored by hybridization of a fluorescent cDNA copy of the mRNA to an immobilized DNA fragment whose sequence spans the spliced junction. Characterization of this microarray assay is described in Pleiss et.al16. Pleiss et. al. show that the splicing of most yeast introns is inhibited in the mutant prp2–1 at the non–permissive temperature. UBC4 was selected from among the top candidates from this microarray, see Supplementary Table 1.
Synthesis and activity of Ubc4 pre–mRNA substrates
The two oligonucleotides that comprise the Ubc4 pre–mRNA substrate; the 5′ segment of 76 nt and the 3′ segment of 59 nt, each containing a single 5–amino–allyl–uridine, were synthesized by Dharmacon. Mutant versions of these oligonucleotides were also synthesized. (Supplementary Table 2) Half of each synthesis at a time was deprotected and then purified by electrophoresis on a denaturing, 7 M urea, 6% (w/v) polyacrylamide gel. The bands were identified by UV shadowing and eluted by overnight rotation in 0.3 M NaOAc pH 5.5, 1 mM EDTA, 0.1% SDS. Following elution the gel fragments were removed by centrifugation, the supernatant extracted with phenol chloroform, and the RNA ethanol precipitated. It is important to remove all Tris in this step because it can interfere with dye conjugation. For coupling, 2–5 nanomoles of each fragment were resuspended in 40 μl of 0.1 M sodium bicarbonate buffer, pH 9.0, and incubated for 60 min at 60 °C with one dye pack of the N–hydroxysuccinimidyl ester dissolved in DMSO. The 5′ and 3′ fragments were reacted with Cy3 and Cy5 N–hydroxysuccinimidyl ester, respectively. After dye coupling, the conjugated fragments were ethanol precipitated, washed until ethanol soluble dye was removed and then purified on a denaturing, 7 M urea, 20% (w/v) polyacrylamide gel. The conjugated oligonucleotide was eluted from the gel by overnight rotation and subsequently purified as described above. The overall yield of coupled oligonucleotide was generally ~2–5 nanomoles. The purified and conjugated oligonucleotides were ligated as described34 using T4 RNA ligase I and a DNA splint (dSplint) from Integrated DNA Technologies. The ligated fragments were separated by gel electrophoresis and eluted as described above. The overall yield in this step is only 10–20%, but the resulting 200–500 picomoles suffice for a large number of smFRET experiments.
The 135 nt version of the Ubc4 pre–mRNA with a 95 nt intron and 20 nt 5′ and 3′ exons has the 5-allyl-amine U residues in the 5′ exon and in the 3′ exon (positions, −7 and +4 depicted in bold font in Supplementary Table 2) In the 3′SS mutant, the bold underlined guanine was replaced with a cytosine. In the BP mutant the italicized underlined adenosine is replaced by a cytosine. The splicing activity of this fluorescently labeled Ubc4 pre–mRNA was determined using standard in vitro assays, wherein 5–10 fmoles of Ubc4 pre–mRNA at a final concentration of 0.2 nM was incubated in 40% (v/v) yeast whole cell extract containing 8 mM Hepes–KOH, pH 7.0, 2 mM MgCl2, 0.08 mM EDTA, 60 mM Ki(PO4), 20 mM KCl, 8% (v/v) glycerol, 3% (w/v) PEG, 0.5 mM DTT, supplemented with 2 mM ATP, and the products were resolved via denaturing polyacrylamide gel electrophoresis and scanned on a Typhoon variable mode imager. All sequences for substrates and oligonucleotides used in this study are available in Supplementary Table 2.
Preparation of yeast cell extract
Splicing active whole cell extract was prepared as described previously from BJ2168 yeast cells35 with the following exceptions. Cells were grown in 8 to 16 L of YPD medium to an OD600 of 2.5–3. The cells were then harvested and washed. Pellets of a thick slurry of the extract were obtained by dripping the slurry into liquid nitrogen. These pellets can be frozen at −70 °C for further use. The cell pellets were disrupted by grinding in a ball mill grinder (RMM301, Retsch) for 5 cycles of 3 min at 10 Hz. Between each cycle the canisters were cooled in liquid nitrogen. The frozen powder was thawed in an ice bucket and centrifuged at 34,950 g in a Ja–25 Beckman rotor; the supernatant was then centrifuged at 100,000g in a Ti–60 rotor for 1 h. The clear middle layer was removed using a syringe. This extract was dialyzed for 4 h against 20 mM Hepes–KOH, pH 7.0, 0.2 mM EDTA, 0.5 mM DTT, 50 mM KCl, 20% (v/v) glycerol with one buffer exchange. 40% (v/v) cell extract was used in all assays and in all single molecule studies. ATP–depletion was achieved by adding 1 mM glucose to the cell extract and incubating at room temperature prior to each experiment.
Single molecule FRET
Slides were prepared using a protocol modified from previous published methods36,37. Quartz slides were reacted with aminopropyltriethoxysilane (APTES) in acetone for 30 min to generate an amino functionalized surface, which was reacted overnight with a 10:1 mixture of succinimidyl ester functionalized O–methyl–PEG and biotin–PEG to PEGylate the surface. Sulfo–disuccinaimidlytartarate (sulfo–DST) was reacted for 30 min with the remaining unreacted amines, thus ensuring the surface does not carry a positive charge. Slide coverslips undergo a similar procedure. The slides were then rinsed, dried and a single flow channel per slide was assembled. A solution of 0.2 mg ml−1 streptavidin in buffer (50 mM Tris–HCl, pH 7.5, 50 mM NaCl) was added to the channel and incubated for 10 min at room temperature.
The doubly labeled pre–mRNA was heat annealed to a 2′–O–methyl–RNA capture (tether) strand complementary to the 17 3′ terminal nucleotides of the 3′ exon by incubating at 70 °C for 2 min and cooling to room temperature for 10 min. The capture strand carries a 5′ biotin, which binds to the streptavidin on the surface of the slide to immobilize the pre–mRNA. Following tether annealing, the hybrid was diluted to a concentration of ~50 pM in 100 μl splicing buffer and flowed into the slide and incubated for 10 min. Using a prism–based total internal reflection fluorescence (TIRF) microscope as described38 data were collected from single molecules in splicing buffer (8 mM Hepes–KOH, pH 7.0, 2 mM MgCl2, 0.08 mM EDTA, 60 mM Ki(PO4), 20 mM KCl, 8% (v/v) glycerol, 3% (w/v) PEG, 0.5 mM DTT), 40% (v/v) cell extract depleted of ATP or 40% (v/v) cell extract supplemented with 2 mM ATP, both in splicing buffer. An oxygen scavenger system composed of protocatechuate dioxygenase (PCD) and protocatechuate (PCA) was added to splicing buffer and cell extracts to limit photobleaching26, Trolox was also added to the solution to limit fluorophore blinking39. The Cy3 donor was exited using a 532 nm laser and emission by the Cy3 and Cy5 fluorophores was recorded at 100 ms time resolution using an intensified CCD camera (Princeton Instruments, I–Pentamax). A FRET value was then calculated by dividing the intensity of acceptor emission by the total emission from both donor and acceptor. It should be noted that our ability to observe FRET transitions has a lower boundary imposed by our TIRFM time resolution of 100 ms and an upper boundary derived from the time window before either of the fluorophores photobleaches (~100 s). This dynamic range of ~4 orders of magnitude resolves many, but probably not all kinetic differences among the pre–mRNAs, especially in light of the slow splicing kinetics in vitro. To further extend our observation time window, we therefore performed experiments involving extended pre–incubation with splicing active extract before commencing high–resolution (100 ms) smFRET detection.
Anti–correlation was detected in two ways. First, trajectories to be studied were pre–filtered by searching for the presence of any substantial anti–correlation by visual inspection. Second, transition events found within the pre–filtered FRET index trajectories were screened for localized anti–correlation in the donor and acceptor traces. The post—filtering system required the use of the Hidden Markov Modeling tools in the QuB software suite on the pre—filtered data (available at http://www.qub.buffalo.edu/soft.php). The raw donor, acceptor, and FRET indices were independently modeled to determine transition boundaries using a global fitting routine that simultaneously analyzes all data points in a given experimental condition (Supplementary Fig. 2 online). The post—filter algorithm was coded and executed in a MATLAB environment. Alternatively, we carried out a preliminary analysis of individual trajectories in which we explicitly optimized the rates of FRET transitions as in previous single—molecule FRET analysis23,24. Consistently, this analysis identified similar sequence and ATP–dependent changes. However, the large number of FRET states and trajectories in our data sets and the apparent kinetic heterogeneity ultimately resulted in a lower level of confidence in the resulting transition models. In depth discussion of HMM analysis in Supplementary Methods.
Verification of splicing in situ by RNase H probing
Each Ubc4 construct (WT, BP, and 3′SS, mRNA) was immobilized on a quartz slide, incubated with splicing buffer, ATP depleted extract, or ATP supplemented extract and monitored by smFRET as described above. Extract was washed out with 100 μl 1 M NaCl solution and incubation over 2 min, repeated three times. Following the final wash, the sample chamber was equilibrated with 100 μl RNase H buffer (20 mM HEPES–KOH, pH 7.8 at 25°C, 1 mM DTT, 50 mM KCl, 50% (v/v) glycerol, 0.2 mg ml−1 BSA) for 5 min, followed by incubation for 20 min with 100 μl of 250 μM Deoxyoligonucleotide dRH which is complementary to the Ubc4 intron in RNase H buffer. Next, a movie was taken for 50 frames with direct excitation of both Cy3 and Cy5 (‘before RNase H’). The slide was then washed with 100 μl 250 μM dRH and 10 U RNase H (Promega) in RNase H buffer, was incubated for 30 min, and washed again with 100 μl 250 μM dRH in RNase H buffer. A second movie was taken as above (‘after RNase H’). Movies collected before and after RNase H treatment were quantified using an in–house spot finder algorithm, which utilizes a second–order non–linear polynomial equation to correctly align the Cy3 and Cy5 channels, and quantified using a reference bead slide for calibration39.
Supplementary Material
Acknowledgments
The authors wish to thank Reinhard Lührmann and Patrizia Fabrizio (Max Planck Institute for biophysical Chemistry – Göttingen, Germany), Jonathan Staley (University of Chicago – Chicago, IL) and Beate Schwer (Weill Cornell Medical College – New York City, NY) for providing splicing active yeast cell extracts at a moment, common in this field, in which we were having difficulty in making active extracts. The oligonucleotides synthesized at Dharmacon were sometimes a lot longer than they usually make and their excellent quality was essential for this project. The work at the University of Michigan was supported by NIH grant GM062357 to N.G.W., NIH Cellular & Molecular Biology and Molecular Biophysics Training Grant fellowships to M.B. and M.A.D., respectively, as well as a Rackham Merit Fellowship to M.B. The work at UCSF was supported by an American Cancer Society Research Professor of Molecular Genetics award to C.G., by NIH grant GM021119 to C.G., by NIH postdoctoral fellowship GM077844 to C.M., and by a grant from the Agouron Institute to J.A. We acknowlege helpful comments on the manuscript from Greg Whitworth, Jonathan Staley and Haralambos Hadjivassiliou.
Footnotes
AUTHOR CONTRIBUTIONS
J.A. worked at the bench and led the development of the Ubc4 system at UCSF. J.A.P. and J.A. performed the microarray analysis in Supplementary Table 1, while D.R., T.V. and C.M. participated in various phases of the biochemistry at UCSF. M.B., M.A.D, F.F., and M.W. performed the smFRET experimentation and data analysis, whereas P.A. performed the secondary structure analysis at the University of Michigan. J.A., M.A.D., M.B., C.M., C.G. and N.G.W. wrote the manuscript.
References
- 1.Jurica MS, Moore MJ. Pre–mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 2.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–18. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–26. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 4.Burgess SM, Guthrie C. A mechanism to enhance mRNA splicing fidelity: the RNA–dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell. 1993;73:1377–91. doi: 10.1016/0092-8674(93)90363-u. [DOI] [PubMed] [Google Scholar]
- 5.Couto JR, Tamm J, Parker R, Guthrie C. A trans–acting suppressor restores splicing of a yeast intron with a branch point mutation. Genes Dev. 1987;1:445–55. doi: 10.1101/gad.1.5.445. [DOI] [PubMed] [Google Scholar]
- 6.Mayas RM, Maita H, Staley JP. Exon ligation is proofread by the DExD/H–box ATPase Prp22p. Nat Struct Mol Biol. 2006;13:482–90. doi: 10.1038/nsmb1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu YZ, Query CC. Competition between the ATPase Prp5 and branch region–U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol Cell. 2007;28:838–49. doi: 10.1016/j.molcel.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walter NG, Huang CY, Manzo AJ, Sobhy MA. Do–it–yourself guide: how to use the modern single–molecule toolkit. Nat Methods. 2008;5:475–89. doi: 10.1038/nmeth.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furman E, Glitz DG. Purification of the spliceosome A–complex and its visualization by electron microscopy. J Biol Chem. 1995;270:15515–22. doi: 10.1074/jbc.270.26.15515. [DOI] [PubMed] [Google Scholar]
- 10.Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ. Purification and characterization of native spliceosomes suitable for three–dimensional structural analysis. RNA. 2002;8:426–39. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kent OA, MacMillan AM. Early organization of pre–mRNA during spliceosome assembly. Nat Struct Biol. 2002;9:576–81. doi: 10.1038/nsb822. [DOI] [PubMed] [Google Scholar]
- 12.Newman AJ, Teigelkamp S, Beggs JD. snRNA interactions at 5′ and 3′ splice sites monitored by photoactivated crosslinking in yeast spliceosomes. RNA. 1995;1:968–80. [PMC free article] [PubMed] [Google Scholar]
- 13.Lin RJ, Newman AJ, Cheng SC, Abelson J. Yeast mRNA splicing in vitro. J Biol Chem. 1985;260:14780–92. [PubMed] [Google Scholar]
- 14.Vijayraghavan U, Company M, Abelson J. Isolation and characterization of pre–mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev. 1989;3:1206–16. doi: 10.1101/gad.3.8.1206. [DOI] [PubMed] [Google Scholar]
- 15.Clark TA, Sugnet CW, Ares M., Jr Genomewide analysis of mRNA processing in yeast using splicing–specific microarrays. Science. 2002;296:907–10. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- 16.Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Transcript specificity in yeast pre–mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 2007;5:e90. doi: 10.1371/journal.pbio.0050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vijayraghavan U, et al. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 1986;5:1683–95. doi: 10.1002/j.1460-2075.1986.tb04412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rueda D, et al. Single–molecule enzymology of RNA: essential functional groups impact catalysis from a distance. Proc Natl Acad Sci USA. 2004;101:10066–71. doi: 10.1073/pnas.0403575101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ditzler MA, Rueda D, Mo J, Hakansson K, Walter NG. A rugged free energy landscape separates multiple functional RNA folds throughout denaturation. Nucleic Acids Res. 2008;36:7088–99. doi: 10.1093/nar/gkn871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pereira MJ, et al. Single VS Ribozyme Molecules Reveal Dynamic and Hierarchical Folding Toward Catalysis. J Mol Biol. 2008;382:496–509. doi: 10.1016/j.jmb.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Silva C, Walter NG. Leakage and slow allostery limit performance of single–drug sensing aptazyme molecules based on the hammerhead ribozyme. RNA. 2009;15:76–84. doi: 10.1261/rna.1346609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartel DP, Szostak JW. Isolation of new ribozymes from a large pool of random sequences [see comment] Science. 1993;261:1411–8. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- 23.Munro JB, Altman RB, O’Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25:505–17. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKinney SA, Joo C, Ha T. Analysis of single–molecule FRET trajectories using hidden Markov modeling. Biophys J. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwer B. A conformational rearrangement in the spliceosome sets the stage for Prp22–dependent mRNA release. Mol Cell. 2008;30:743–54. doi: 10.1016/j.molcel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crawford DJ, Hoskins AA, Friedman LJ, Gelles J, Moore MJ. Visualizing the splicing of single pre–mRNA molecules in whole cell extract. RNA. 2008;14:170–9. doi: 10.1261/rna.794808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–41. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 28.Seraphin B, Rosbash M. Identification of functional U1 snRNA–pre–mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989;59:349–58. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- 29.Stevens SW, et al. Composition and functional characterization of the yeast spliceosomal penta–snRNP. Mol Cell. 2002;9:31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- 30.Maroney PA, Romfo CM, Nilsen TW. Functional recognition of 5′ splice site by U4/U6. U5 tri–snRNP defines a novel ATP–dependent step in early spliceosome assembly. Mol Cell. 2000;6:317–28. doi: 10.1016/s1097-2765(00)00032-0. [DOI] [PubMed] [Google Scholar]
- 31.Smith DJ, Konarska MM. Mechanistic insights from reversible splicing catalysis. RNA. 2008;14:1975–8. doi: 10.1261/rna.1289808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tseng CK, Cheng SC. Both catalytic steps of nuclear pre–mRNA splicing are reversible. Science. 2008;320:1782–4. doi: 10.1126/science.1158993. [DOI] [PubMed] [Google Scholar]
- 33.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–88. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stark MR, Pleiss JA, Deras M, Scaringe SA, Rader SD. An RNA ligase–mediated method for the efficient creation of large, synthetic RNAs. RNA. 2006;12:2014–9. doi: 10.1261/rna.93506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevens SW, Abelson J. Yeast pre–mRNA splicing: methods, mechanisms, and machinery. Methods Enzymol. 2002;351:200–20. doi: 10.1016/s0076-6879(02)51849-8. [DOI] [PubMed] [Google Scholar]
- 36.Ha T, et al. Initiation and re–initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–41. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 37.van Oijen AM, et al. Single–molecule kinetics of lambda exonuclease reveal base dependence and dynamic disorder. Science. 2003;301:1235–8. doi: 10.1126/science.1084387. [DOI] [PubMed] [Google Scholar]
- 38.Zhuang X, et al. Correlating structural dynamics and function in single ribozyme molecules. Science. 2002;296:1473–6. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 39.Rasnik I, McKinney SA, Ha T. Nonblinking and long–lasting single–molecule fluorescence imaging. Nat Methods. 2006;3:891–3. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.