Abstract
Numerous perforin-like proteins are encoded in the genomes of apicomplexan parasites, where they are expressed in various life-cycle stages and play critical roles in pathogenesis and lifecycle progression. These ApiPLPs are characterized by the presence of a MACPF domain, responsible for pore-formation in target membranes in a number of systems, including many bacterial pathogens and effector cells of the immune response. ApiPLP MACPF domains maintain the critical structural elements but are often present in new and intriguing domain arrangements. Recent work in Toxoplasma and Plasmodium has shown that ApiPLPs are important for breaching membranes during parasite egress and cell traversal. Here we present an overview of this important protein family from a structural, functional and phylogenetic perspective across the Apicomplexa.
Keywords: pore formation, membrane, secretion, cell traversal, egress, plasmodium, toxoplasma
Introduction
Pore-forming proteins are an ancient component of the arsenal of both sides in the molecular struggle between host and pathogen. One example of this has been illustrated by recent work on a family of Membrane Attack Complex/Perforin (MACPF) domain proteins in the phylum Apicomplexa. Comprised of obligately intracellular pathogens, apicomplexan parasites are responsible for numerous diseases of medical (malaria, cryptosporidiosis, toxoplasmosis) and veterinary (coccidiosis, babesiosis, theileriosis) importance.
Originally characterized in the pore-forming effector proteins of the mammalian immune response (complement components C6–C9, perforin),1 MACPF-domain proteins bind the surface of target cells and oligomerize to form large pre-pore complexes.2 These pre-pores undergo a molecular rearrangement and insert into target membranes to form large pores (∼100 Å diameter).2,3 Exciting work over the past couple of years has provided additional insight into their molecular mechanism of action by revealing that the MACPF fold is shared with that of the cholesterol- dependent cytolysins (CDCs), an extensively studied family of bacterial pore-forming virulence factors.4,5
Here we present an overview of the Apicomplexan Perforin-Like Proteins (ApiPLPs) that have been identified in parasites for which genome sequence is available. It should be noted that apicomplexan genome sequencing efforts differ greatly in terms of completion, sequence coverage and quality of the gene predictions. Furthermore, the availability of supporting data sets such as stage specific expressing of varies greatly from the microarray, proteomics and expansive EST data available for Plasmodium6 to only a limited number of Babesia ESTs sequenced.7 While genomic sequences and gene predictions are available for six species of Plasmodium (falciparum, vivax, chabaudi, knowlesi, yoelii and berghei) and two species of Theileria (annulata and parva),8,9 intra-genus orthology is very high both with respect to sequence identity and the number of MACPF-domain proteins predicted for each species within these genera. For this reason the ApiPLPs of Plasmodium and Theileria will be discussed on the genus level using the P. falciparum and T. annulata gene predictions as representatives.
Conservation & Expression
The MACPF domain is an ancient fold found in every domain of the tree of life with the exception of Archaea (Pfam ID: PF01823,10). Among the Protista, MACPF proteins are not limited to the apicomplexa but are also expressed by free-living predatory organisms such as Paramecium and Tetrahymena.
The number of encoded MACPF domains (Fig. 1) varies across the major clades of the phylum with none in Cryptosporidium, two or three in the Coccidiae (Toxoplasma, Neospora, Eimeria), five in the Haemosporididae (Plasmodium) and nine in the Piroplasmidae (Theileria and Babesia). The expansion of ApiPLPs in Plasmodium, Theileria and Babesia may be driven by the need to adapt the functions of PLPs to both their mammalian and arthropod hosts, while the apicomplexans with fewer PLPs appear to be limited to mammalian hosts. Indeed several of the malarial PLPs are expressed in the insect stages (Table 1). Relatively little work has been done to characterize global gene expression of the tick-borne stages of either Theileria or Babesia, which may explain why the expression evidence is limited to only a few PLPs, all expressed in the mammalian, blood-borne stages.
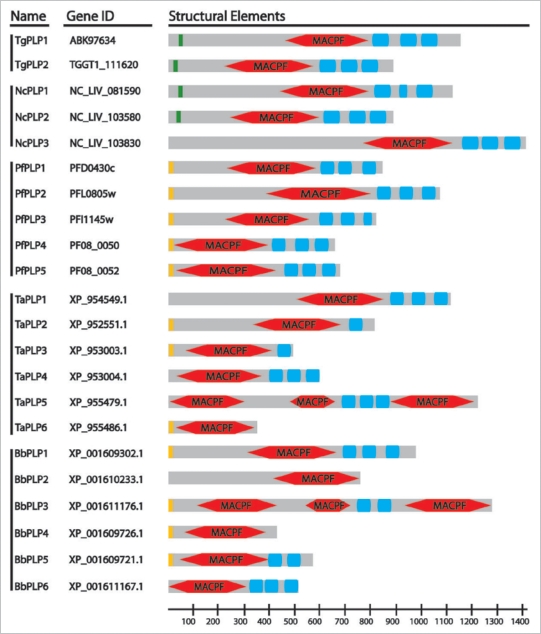
Figure 1.
ApiPLP Domain Architecture. Indicated are predicted signal peptides (orange) or signal anchors (green), MACPF domains (red) and APC-β domains (blue). TaPLP5 is predicted as three separate genes in T. parva. No gene predictions are available for Eimeria but the Eimeria tenella genome contains two PLPs with APC-β domains. P. berghei and P. chabaudi genomes each have six predicted PPLPs but in both cases two of these predictions represent the same gene.
Table 1.
ApiPLP stage-specific expression
| Genus | Gene | Stage expressed | Evidence |
| Toxoplasma | TgPLP1 | Tachyzoites | Protein (11) |
| Bradyzoites | EST (38) | ||
| Neospora | NcPLP1 | Tachyzoites | EST (38) |
| NcPLP2 | Tachyzoites | EST (38) | |
| NcPLP3 | Tachyzoites | EST (38) | |
| Eimeria | EtPLP1 | Sporozoites | EST (39) |
| EtPLP2 | Sporozoites | EST (39) | |
| Plasmodium | PPLP1 | Sporozoites | Protein (12) |
| IDC* | Proteomic (28) | ||
| PPLP2 | IDC* | Proteomic (29) | |
| PPLP3 | Ookinete | Protein (14) | |
| PPLP4 | Ookinete | Proteomic (41)/EST (42) | |
| PPLP5 | Ookinete | Protein (15) | |
| Sporozoite | Proteomic (29 | ||
| Gametocyte | Proteomic (29) | ||
| Theileria | TpPLP1 | Piroplasm | EST (8) |
| TaPLP5 | Schizont | EST (9) | |
| TaPLP6 | Macroschizont | EST (9) | |
| Babesia | BbPLP1 | Intraerthrocytic | EST (7) |
*Intra-erythrocytic development cycle.
As Cryptosporidium represents a low-branching clade in the apicomplexa it is difficult to assess whether its PLP genes were lost over time or whether it branched off before the MACPF domain was acquired. Given the presence of MACPF domains in non-apicomplexan alveolates, their ubiquity in the remaining apicomplexa and the streamlined genome of Cryptosporidium, it seems more likely that Cryptosporidium initially possessed a gene(s) encoding the MACPF domain and later lost it.
Structure
The canonical domain architecture of MACPF/CDC proteins combines the pore-forming domain with a variety of different β-pleated sheet-rich C-terminal domains that typically mediate binding of target membranes.2 The four ApiPLPs characterized to date (TgPLP1,11 PPLP1/SPECT1,12,13 PPLP3/MAOP,14 PPLP5,15) all share similar domain architectures with a N-terminal region of variable length followed by a centrally located MACPF domain and ending in a β-pleated sheet-rich domain unique to the apicomplexa (Fig. 1).
The three ApiPLPs for which intracellular localization was determined (TgPLP1,11 PPLP1/SPECT2,12,13 PPLP3/MAOP14) all localize to the micronemes, which are specialized secretory organelles that play critical roles in parasite invasion, egress and motility. Microneme secretion is a carefully controlled and directed event that is calcium-regulated and occurs only at the apical end of the parasite.16 This is analogous to the calciummediated and directed release of perforin from secretory lysosomes of natural killer cells and cytotoxic T lymphocytes and represents an effective mechanism to achieve a high local concentration of PLP monomers delivered to a limited area on the target membrane.3,17 The majority of uncharacterized ApiPLPs are also likely to be secreted based on the presence of a secretory signal sequence (Fig. 1). Given the difficult task of accurately predicting the first exon it may well be that virtually all ApiPLP2 are secreted.
The remainder of the N-terminal region of ApiPLPs preceding the MACPF is not conserved across the phylum, varying considerably in both length and sequence, suggesting that they make unique contributions to the function of each ApiPLP. TgPLP1 is proteolytically processed within this region during trafficking (Kafsack BFC and Carruthers VB, unpublished). Since TgPLP1 contains a predicted transmembrane signal anchor near its N-terminus, this processing untethers TgPLP1 from the membrane and may also be a necessary prerequisite for membraneolytic activity.
The characteristic MACPF structural elements of a twisted β-pleated sheet sitting atop two helical clusters4,5 appear to be well conserved in the ApiPLPs, based multiple sequence alignments and a structural homology model of TgPLP1.11 During pore-formation these helical clusters convert into amphipathic β-hairpins and insert into the target membrane where they form the lower lining of the pore’s barrel.18 As expected the helical clusters of ApiPLP MACPF domains exhibit the pattern of alternating hydrophobic and hydrophilic amino acids necessary for this conversion.11
A Hidden-Markov sequence conservation model based on an alignment of ApiPLP MACPF domains reveals a signature pattern of W-x(2)-[FL]-[FI]-x(2)-[FY]-G-T-H-x(7)-G-G, similar in spacing and amino acid usage to the canonical MACPF signature (PROSITE ID: PDOC00251,19). Interestingly, the ApiPLP MACPF domains contain a conserved insertion comprised of a pair of anti-parallel α-helices, which is absent from canonical MACPFs. This insertion is modeled to reside on what would be external face of the ApiPLP barrel in a spatial region that contains important residues for oligomerization of membrane-bound perforin monomers.20
A second feature unique to ApiPLP is the presence of two highly conserved pairs of cysteine residues MACPF domains, whereas the canonical MACPF domains are devoid of disulfide bridges. The first pair likely stabilizes the two anti-parallel α-helices of the above-mentioned insertion, while the second might pin together two strands of the central twisted β-pleated sheet near the bend region.
In a novel arrangement, both Theileria and Babesia are predicted to encode multiple MACPF domains within a single protein. Such an increase in valency might reduce the number of PLP monomers required to form a pore, potentially lowering the effective monomer concentration necessary for pore-formation. However, as indicated above, the gene predictions should be interpreted with some caution and it will be necessary to validate that these multiple domains are indeed in a single protein rather than in separate proteins encoded in adjacent genes.
The pore-forming MACPF-domain proteins and the CDCs all pair the MACPF/CDC domain with one or more C-terminal domains responsible for binding to target membranes via lipid or protein receptors. These C-terminal domains vary in sequence and structure, but are consistently comprised almost entirely of β-pleated sheets, as in the case of the EGF-like and C2 domains of perforin,21 the β-prism domains of the bacterial PluMACPF,4 and the β-sandwich Ig domain of the CDCs.22
Similarly, the C-termini of ApiPLPs are generally occupied by multiple copies of a predicted domain comprised of five β-strands, which we have termed ApiPLP C-terminal β-pleated sheet (APC-β) domains. Based on sequence homology and pattern searches, the ∼55aa APC-β domain is unique to the Apicomplexa and only found in association with ApiPLPs. While definitive studies are necessary, the APC-β domains are likely responsible for the initial binding of ApiPLPs to their target membranes. Given that these membranes can vary widely in their origin as well as lipid and proteins composition ranging from arthopod mid-gut epithelium to the parasitophorous vacuolar membrane of parasitized mammalian cells, it will be interesting to see if this novel domain binds a ubiquitous receptor such as cholesterol or has been adapted to specific receptors depending on the host species and target cell. As the MACPF + APC-β pairing is found across the phylum it appears likely that this union occurred early in the evolutionary history of the Apicomplexa.
The APC-β domain is generally present in three tandem copies, although some of the Theileria and Babesia PLPs are predicted to have fewer copies, which may be due in part to the difficulty of correctly predicting the last exon. Alternatively, PLPs lacking the APC-β domain altogether may interact with a second protein, which mediates the initial membrane interaction and facilitates oligomerization similar to the two-component system used by the perforin-like pleurotolysin of the edible oyster mushroom.23
Lastly, several ApiPLPs (TgPLP1, NcPLP1, PPLP3/MAOP, PPLP4) contain a patch of four or more positively charges amino acids near the C-terminus, which may facilitate interaction with negatively charged head groups of phospholipids on target membranes, perhaps in a fashion similar to the interaction of a basic patch in the HIV gag protein that facilitates for viral assembly by binding to PIP2 in the host cell plasma membrane.24
Function
Pore formation by MACPF/CDC proteins is a multi-step process that initiates with the secretion of monomers, which bind to their cognate receptor on the target membrane.2 This is followed by oligomerization into a large circular pre-pore by lateral interactions of the monomers followed by conformational change of the central MACPF domain, resulting in membrane insertion and formation of a large β-barrel pore of ∼100 Å diameter.18 Functions of these pores include: (1) killing target cells by disrupting their outer membrane as is the case for many bacterial CDCs as well as components of the membrane attack complex (C6–C9);2 (2) delivering other effector proteins across membranes as in the delivery of granzyme by perforin;3 or (3) facilitating pathogen traversal of a membrane barrier as in the case of Listeriolysin O mediated release from the phagosome.25 While pore formation by proteins containing the MACPF fold has long been described, the Apicomplexa have retooled this domain to accomplish some novel and interesting tasks. The PPLP gene family was first described by Kaiser et al.13 but the first ApiPLP to be functionally characterized was PPLP3/MAOP,14 which localizes to secretory organelles of the Plasmodium ookinete stage. The ookinete forms after fertilization in the midgut of the mosquito host and migrates from the blood meal across the midgut epithelium before transforming into an oocyst. In a PPLP3 knockout strain, ookinetes remained motile within the midgut but were unable to invade or traverse the midgut epithelial layer. Disruption of PPLP5, a second MACPF protein expressed in ookinetes, yielded an identical phenotype of ookinetes unable to invade or traverse the midgut epithelium.15 The mirrored phenotype of PPLP3 and PPLP5 knockout strains suggests that these two PPLPs may interact to accomplish midgut traversal, analogous to the interaction of multiple MACPF-domain proteins (C6–C9) to form the membrane attack complex. Proteomic evidence suggests the expression of a third MACPF-domain protein (PPLP4) in ookinetes but it has not been functionally characterized to date.8
A second instance of an ApiPLP required for traversal of a cellular barrier was revealed by the disruption of PPLP1/SPECT2, which is expressed in Plasmodium salivary gland sporozoites.12 Following the bite of an infectious mosquito, sporozoites enter the blood stream and exit circulation by traversing resident macrophages (Kupffer cells) of the hepatic sinusoids. Following this entry into the space of Disse, sporozoites will often traverse additional hepatocytes before forming a parasitophorous vacuole (PV) during productive invasion of a final hepatocyte and converting into the extra-erythrocytic form (EEF).26 While the PPLP1 knockout strain was able to form EEF normally in cultured hepatocytes, the number of liver EEF was drastically reduced in vivo during infection but could be restored to wild-type levels by destruction of the Kupffer cells within the sinusoids.12 Work by Amino et al. showed that the kinetics of cell entry strongly favored PV formation by PPLP1 knockout sporozoites, while wild-type controls were more likely to traverse several host cells before forming a PV during cell entry.27 This suggests that PPLP1 is a critical determining factor of whether or not a PV is generated during cell entry.
Recent work in our lab has implicated TgPLP1 in the egress of Toxoplasma tachyzoites from the host cell.11 Following treatment with a calcium ionophore TgPLP1 knockout tachyzoites became motile but were significantly delayed in their exit from host cells and often remaining trapped within spherical structures comprised of the parasitophorous vacuolar membrane (PVM) and host cell remnants. Further experiments revealed that TgPLP1 was necessary for the permeabilization of the PVM following ionophore treatment. The important role of TgPLP1 was underscored by a substantial attenuation of virulence in mice compared to wild-type control.
While the characterized functions of the PPLP all implicate a role in cell traversal of either the mosquito midgut epithelium by ookinetes or Kupffer cells by sporozoites, a role for PPLPs in Plasmodium egress remains possible. Proteomic studies indicate the expression of both PPLP1,28 and PPLP2,29 in the intraerythrocytic forms and PPLP5 in gametocytes.29 In neither of these stages is there an apparent need for cell traversal but egress of microgametes and macrogametes from gametocytes or merozoites from the schizont is crucial for progression of the life cycle. Similarly, one or more PPLP may act during the transition from the liver stage to the erythrocytic cycle where a large numbers of merozoites escape the PVM with in the infected hepatocyte and bud off into the liver vasculature in large vesicles termed merosomes,30 which subsequently rupture within pulmonary capillary beds.31 Thus, PPLPs may contribute both to the PVM escape and eventual release from the merosome.
Unlike Plasmodium and Toxoplasma, the Piroplasmidae (Theileria and Babesia) escape their PVM shortly after invasion and replicate within the host cell cytoplasm. In addition to facilitating their exit from the cell following replication, PLPs could also play a role in this early escape, analogous to the action of Lysteriolysin O in the escape of L. monocytogenes from the phagosome.25
While ApiPLPs have been shown to be necessary for rapid membrane disruption and appear to have all the necessary structural features, the sufficiency of ApiPLPs for pore formation has not been directly demonstrated. Further biochemical analyses will be necessary to determine not only whether ApiPLPs form pores but also to determine their cognate receptors on target membranes. The targets of the characterized PPLPs are the extracellular plasma membrane face of either mosquito midgut epithelium or a variety of mammalian cells in the skin and liver. TgPLP1 can act on both the internal as well as the cytoplasmic face of the PVM and may also act on the cytoplasmic face of the host cell plasma membrane.11 As their name indicates, cholesterol is a common receptor of the bacterial CDCs,18 and use of lipid receptor may explain how ApiPLPs can act on such diverse membranes.
Recent work by Chandramohanadas et al. showed that the host proteases calpain-1 and calpain-2 are important to egress in both Plasmodium and Toxoplasma and their depletion in cells infected with Toxoplasma lead to the same spherical structures of trapped parasites observed in the TgPLP1 knockout.32 Intriguingly, these proteases are activated by spikes in the local calcium concentration33 and the Toxoplasma PVM is in tight association with the host endoplasmic reticulum, a major sites of calcium storage.34 Secretion of TgPLP1 during egress may not only permeabilize the PVM but also act on the nearby host ER, liberating calcium locally to activate host calpains. Alternatively, ApiPLPs may directly destabilize membrane barriers to egress or act as gateways for additional effector proteins already present in the PV or released during egress, such as the SERA proteases that are activated by the protease PfSUB1 during Plasmodium merozoite egress.35 These effectors, once released into the host cytoplasm may then lead to the activation of the host calpains or breakdown of egress barriers.
The activity of pore-forming proteins must be carefully controlled to avoid inadvertent damage to non-target membranes. This is often achieved by one or more mechanisms such as regulated secretion (perforin),3 proteolytic activation (perforin),3 pH sensitivity (LLO, perforin),3,25 receptor specificity to target membranes (intermedilysin)36 or the presence of immunity proteins on non-target membranes (complement MAC).37 How apicomplexan parasites prevent autolysis by ApiPLPs following secretion remains an interesting area deserving of additional work.
The ApiPLPs studied to date are involved in two apparently distinct mechanisms: cell traversal from outside of cells and egress from within. Yet both mechanisms begin by disrupting the HPM or the PVM, itself initially derived from the HPM and topologically analogous, and end with the zoite crossing the HPM to exit the cell. A possible, unifying view of ApiPLP function is that they disrupt the PVM in both cases, either following replication during egress or immediately after its formation during invasion. The decision between traversal and invasion may be controlled by the amount of ApiPLPs secreted, with a stable PVM only forming once the amount of secreted ApiPLPs is sufficiently low. Hence, early host cell contacts of salivary gland Plasmodium sporozoites, packed with micronemes, result in large bursts of PPLP1 secretion and disruption of the nascent PVM leading to cell traversal. As available micronemes become depleted with each subsequent traversal, the amount of secreted PPLP1 drops until it fails to disrupt the newly forming PVM and leads to productive invasion. This is consistent with both the multiple cell traversal events prior to invasion and the observation of Amino et al.27 that the absence of PPLP1 heavily skews the kinetics of sporozoite invasion away from cell traversal and toward formation of a PV. For Toxoplasma tachyzoites, which feature far fewer micronemes, the depletion of micronemes during egress may be sufficient to drop the amount of secreted TgPLP1 to below the threshold for membrane traversal, thus explaining the failure to observe cell wounding by tachyzoites.
Outlook
Work in Plasmodium and Toxoplasma is beginning to reveal the importance of ApiPLPs in the life-cycle of apicomplexan parasites, thereby opening the door for a wealth of new and intriguing questions regarding ApiPLP structure and function.
Central to these efforts will be demonstrating pore-formation by ApiPLP proteins directly and defining the requirements such as the receptor for membrane binding, optimal pH, and the role of metal co-factors. Answers to these questions will also shed light in how apicomplexan parasites protect themselves from deleterious pore-formation during ApiPLP trafficking and secretion. The unusual domain arrangements potentially present in some Theileria and Babesia ApiPLPs, such as multiple MACPF domains or the lack of APC-β domains, may present a new structural twist in utilizing the MACPF/CDC fold. Similarly, work towards determining the function and fold of the uniquely apicomplexan APC-β domain may provide insight into how they direct the MACPF domain to the target membrane.
From a functional perspective, it remains unclear how the ApiPLPs enable parasites to cross the target membranes. Does insertion of ApiPLPs alone sufficiently weaken the target membranes for parasites to cross or do the pores act as gateways for additional effectors which in turn responsible for the breakdown of these barriers? Additionally, it remains unclear if ApiPLPs are only responsible for disrupting the initial target membrane or whether they also form pores in subsequent membranes such as host organelles or HPM upon parasite exit from the cell.
Finally, it will be intriguing to see what roles the many yet uncharacterized ApiPLPs play and whether these extend beyond egress and cell traversal, such as escape of Theileria and Babesia into the host cytoplasm. Similarly, looking beyond the Apicomplexa, the presence of MACPF proteins in free-living, predatory protists such as Paramecium and Tetrahymena may shed light on the structural and functional evolution this important and fascinating protein family.
Acknowledgements
PLP-related studies in the Carruthers lab are supported by grant RO1AI046675 from the National Institutes of Health.
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/9794
References
- 1.Podack ER, Deyev V, Shiratsuchi M. Pore formers of the immune system. Adv Exp Med Biol. 2007;598:325–341. doi: 10.1007/978-0-387-71767-8_23. [DOI] [PubMed] [Google Scholar]
- 2.Rosado CJ, Kondos S, Bull TE, Kuiper MJ, Law RH, Buckle AM, et al. The MACPF/CDC family of poreforming toxins. Cell Microbiol. 2008;9:1765–1774. doi: 10.1111/j.1462-5822.2008.01191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pipkin ME, Lieberman J. Delivering the kiss of death: Progress on understanding how perforin works. Curr Opin Immunol. 2007;19:301–308. doi: 10.1016/j.coi.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosado CJ, Buckle AM, Law RH, Butcher RE, Kan WT, Bird CH, et al. A common fold mediates vertebrate defense and bacterial attack. Science. 2007;317:1548–1551. doi: 10.1126/science.1144706. [DOI] [PubMed] [Google Scholar]
- 5.Hadders MA, Beringer DX, Gros P. Structure of C8alpha-MACPF reveals mechanism of membrane attack in complement immune defense. Science. 2007;317:1552–1554. doi: 10.1126/science.1147103. [DOI] [PubMed] [Google Scholar]
- 6.Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, et al. PlasmoDB: A functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:539–543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Vries E, Corton C, Harris B, Cornelissen AW, Berriman M. Expressed sequence tag (EST) analysis of the erythrocytic stages of Babesia bovis. Vet Parasitol. 2006;138:61–74. doi: 10.1016/j.vetpar.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 8.Gardner MJ, Bishop R, Shah T, de Villiers EP, Carlton JM, Hall N, et al. Genome sequence of Theileria parva, a bovine pathogen that transforms lymphocytes. Science. 2005;309:134–137. doi: 10.1126/science.1110439. [DOI] [PubMed] [Google Scholar]
- 9.Pain A, Renauld H, Berriman M, Murphy L, Yeats CA, Weir W, et al. Genome of the host-cell transforming parasite Theileria annulata compared with T. parva. Science. 2005;309:131–133. doi: 10.1126/science.1110418. [DOI] [PubMed] [Google Scholar]
- 10.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, et al. The pfam protein families database. Nucleic Acids Res. 2008;36:281–288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kafsack BF, Pena JD, Coppens I, Ravindran S, Boothroyd JC, Carruthers VB. Rapid membrane disruption by a perforin-like protein facilitates parasite exit from host cells. Science. 2009;323:530–533. doi: 10.1126/science.1165740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishino T, Chinzei Y, Yuda M. A plasmodium sporozoite protein with a membrane attack complex domain is required for breaching the liver sinusoidal cell layer prior to hepatocyte infection. Cell Microbiol. 2005;7:199–208. doi: 10.1111/j.1462-5822.2004.00447.x. [DOI] [PubMed] [Google Scholar]
- 13.Kaiser K, Camargo N, Coppens I, Morrisey JM, Vaidya AB, Kappe SH. A member of a conserved plasmodium protein family with membrane-attack complex/perforin (MACPF)-like domains localizes to the micronemes of sporozoites. Mol Biochem Parasitol. 2004;133:15–26. doi: 10.1016/j.molbiopara.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 14.Kadota K, Ishino T, Matsuyama T, Chinzei Y, Yuda M. Essential role of membrane-attack protein in malarial transmission to mosquito host. Proc Natl Acad Sci USA. 2004;101:16310–16315. doi: 10.1073/pnas.0406187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ecker A, Pinto SB, Baker KW, Kafatos FC, Sinden RE. Plasmodium berghei: Plasmodium perforin-like protein 5 is required for mosquito midgut invasion in anopheles stephensi. Exp Parasitol. 2007;116:504–508. doi: 10.1016/j.exppara.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carruthers VB, Tomley FM. Microneme proteins in apicomplexans. Subcell Biochem. 2008;47:33–45. doi: 10.1007/978-0-387-78267-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobelli J, Andres PG, Boisvert J, Krummel MF. New views of the immunological synapse: Variations in assembly and function. Curr Opin Immunol. 2004;16:345–352. doi: 10.1016/j.coi.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Tweten RK. Cholesterol-dependent cytolysins, a family of versatile pore-forming toxins. Infect Immun. 2005;73:6199–6209. doi: 10.1128/IAI.73.10.6199-6209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, et al. The 20 years of PROSITE. Nucleic Acids Res. 2008;36:245–249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baran K, Dunstone M, Chia J, Ciccone A, Browne KA, Clarke CJ, et al. The molecular basis for perforin oligomerization and transmembrane pore assembly. Immunity. 2009;30:684–695. doi: 10.1016/j.immuni.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 21.Voskoboinik I, Smyth MJ, Trapani JA. Perforinmediated target-cell death and immune homeostasis. Nat Rev Immunol. 2006;6:940–952. doi: 10.1038/nri1983. [DOI] [PubMed] [Google Scholar]
- 22.Polekhina G, Giddings KS, Tweten RK, Parker MW. Insights into the action of the superfamily of cholesterol-dependent cytolysins from studies of intermedilysin. Proc Natl Acad Sci USA. 2005;102:600–605. doi: 10.1073/pnas.0403229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomita T, Noguchi K, Mimuro H, Ukaji F, Ito K, Sugawara-Tomita N, et al. Pleurotolysin, a novel sphingomyelin-specific two-component cytolysin from the edible mushroom Pleurotus ostreatus, assembles into a transmembrane pore complex. J Biol Chem. 2004;279:26975–26982. doi: 10.1074/jbc.M402676200. [DOI] [PubMed] [Google Scholar]
- 24.Chukkapalli V, Hogue IB, Boyko V, Hu WS, Ono A. Interaction between the human immunodeficiency virus type 1 gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient gag membrane binding. J Virol. 2008;82:2405–2417. doi: 10.1128/JVI.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schnupf P, Portnoy DA. Listeriolysin O: A phagosome-specific lysin. Microbes Infect. 2007;9:1176–1187. doi: 10.1016/j.micinf.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 26.Prudencio M, Mota MM. To migrate or to invade: Those are the options. Cell Host Microbe. 2007;2:286–288. doi: 10.1016/j.chom.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Amino R, Giovannini D, Thiberge S, Gueirard P, Boisson B, Dubremetz JF, et al. Host cell traversal is important for progression of the malaria parasite through the dermis to the liver. Cell Host Microbe. 2008;3:88–96. doi: 10.1016/j.chom.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 28.Lasonder E, Ishihama Y, Andersen JS, Vermunt AM, Pain A, Sauerwein RW, et al. Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature. 2002;419:537–542. doi: 10.1038/nature01111. [DOI] [PubMed] [Google Scholar]
- 29.Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, Haynes JD, et al. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419:520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- 30.Sturm A, Amino R, van de Sand C, Regen T, Retzlaff S, Rennenberg A, et al. Manipulation of host hepatocytes by the malaria parasite for delivery into liver sinusoids. Science. 2006;313:1287–1290. doi: 10.1126/science.1129720. [DOI] [PubMed] [Google Scholar]
- 31.Baer K, Klotz C, Kappe SH, Schnieder T, Frevert U. Release of hepatic Plasmodium yoelii merozoites into the pulmonary microvasculature. PLoS Pathog. 2007;3:171. doi: 10.1371/journal.ppat.0030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandramohanadas R, Davis PH, Beiting DP, Harbut MB, Darling C, Velmourougane G, et al. Apicomplexan parasites co-opt host calpains to facilitate their escape from infected cells. Science. 2009;324:794–797. doi: 10.1126/science.1171085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goll DE, Thompson VF, Li H, Wei W, Cong J. The calpain system. Physiol Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 34.Sayer RJ. Intracellular Ca2+ handling. Adv Exp Med Biol. 2002;513:183–196. doi: 10.1007/978-1-4615-0123-7_6. [DOI] [PubMed] [Google Scholar]
- 35.Yeoh S, O’Donnell RA, Koussis K, Dluzewski AR, Ansell KH, Osborne SA, et al. Subcellular discharge of a serine protease mediates release of invasive malaria parasites from host erythrocytes. Cell. 2007;131:1072–1083. doi: 10.1016/j.cell.2007.10.049. [DOI] [PubMed] [Google Scholar]
- 36.Giddings KS, Zhao J, Sims PJ, Tweten RK. Human CD59 is a receptor for the cholesterol-dependent cytolysin intermedilysin. Nat Struct Mol Biol. 2004;11:1173–1178. doi: 10.1038/nsmb862. [DOI] [PubMed] [Google Scholar]
- 37.Davies A, Lachmann PJ. Membrane defence against complement lysis: The structure and biological properties of CD59. Immunol Res. 1993;12:258–275. doi: 10.1007/BF02918257. [DOI] [PubMed] [Google Scholar]
- 38.Gajria B, Bahl A, Brestelli J, Dommer J, Fischer S, Gao X, et al. ToxoDB: An integrated Toxoplasma gondii database resource. Nucleic Acids Res. 2008;36:553–556. doi: 10.1093/nar/gkm981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. www.ncbi.nlm.nih.gov/est?term=(txid5802[orgn])
- 40.Canuel M, Libin Y, Morales CR. The interactomics of sortilin: An ancient lysosomal receptor evolving new functions. Histol Histopathol. 2009;24:481–492. doi: 10.14670/HH-24.481. [DOI] [PubMed] [Google Scholar]
- 41.Hall N, Karras M, Raine JD, Carlton JM, Kooij TW, Berriman M, et al. A comprehensive survey of the plasmodium life cycle by genomic, transcriptomic and proteomic analyses. Science. 2005;307:82–86. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- 42.Raibaud A, Brahimi K, Roth CW, Brey PT, Faust DM. Differential gene expression in the ookinete stage of the malaria parasite Plasmodium berghei. Mol Biochem Parasitol. 2006;150:107–113. doi: 10.1016/j.molbiopara.2006.07.001. [DOI] [PubMed] [Google Scholar]