Abstract
Definitive identification of convergent evolution, the acquisition of the same biological trait in unrelated lineages, provides one of the most compelling sources of evidence for natural selection. Although numerous examples of convergent morphological evolution are well known (such as the independent development of wings in birds and mammals), cases of convergent evolution at the molecular-genetic level appear to be quite rare. We recently discovered a remarkable case of convergent molecular evolution involving more than 100 parallel amino-acid changes across all 13 mitochondrially-encoded proteins of snakes and agamid lizards. Just a few of these convergent substitutions were sufficient to positively mislead the inference of phylogeny, even with thousands of sites providing latent support for the correct underlying relationships. Since this example demonstrates that molecular convergence can happen en masse in nature, affecting multiple genes, it is important to consider the threat this poses to molecular systematics, and careful genome-wide assays for convergent molecular evolution are warranted. This result implies that the protein adaptive landscape is sometimes highly constrained.
Key words: adaptive evolution, positive selection, homoplasy, parallel evolution, phylogenetic bias
The outcome of convergent evolution driven by natural selection (or ‘adaptive convergence’) has been frequently recognized at the morphological level in nature. Documented cases of adaptive convergence at the molecular level are fairly unusual, however, and until now examples have involved only single proteins at just a few amino acid positions. This makes sense if the protein adaptive landscape is vast, with many different possible ways to achieve most binding, catalytic, or other protein biochemical functions (Fig. 1). This reasoning has led to the opinion that convergent molecular evolution is rare and usually of limited scope. As such, it is often considered an insignificant and largely ignorable phenomenon for the fields of molecular evolution and molecular systematics, and the study of protein evolution in general.
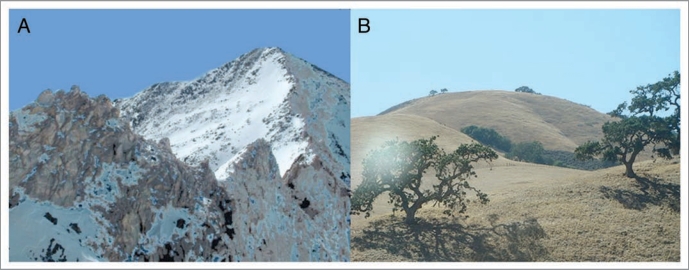
Figure 1.
Alternative views of potential protein adaptive landscapes. In (A), the protein adaptive landscape is viewed as being like an arrêt ridge, with only a single narrow path leading from the current adaptive peak in the foreground to a new adaptive peak in the distance. This landscape is conducive to convergence. In (B), the adaptive landscape is viewed as being like rolling hills, with many alternative routes to nearby adaptive hilltops that are not substantially different from one another. With so many alternative paths and alternative similar hilltops, under this scenario sequences would be unlikely to converge (i.e., follow the same path) even under similar adaptive pressure.
Contrary to these expectations, in a recent study1 we showed that widespread convergent molecular evolution occurred at an unprecedented scale across key parts of mitochondrial proteins in snakes and a distantly related group of lizards (agamids). In total, at least 44 out of 113 predicted convergent changes distributed across all 13 mitochondrial protein-coding genes appear to have been driven by selection,1 suggesting a remarkably strong directional adaptive pressure. We had also shown in another study that some of the same mitochondrial proteins had endured remarkably strong selection for radical changes at otherwise highly conserved sites early in snake evolution.2 This makes sense if the protein adaptive landscape is narrow rather than vast, with only one or a few pathways to reach a neighboring adaptive peak (Fig. 1).
Together, these studies suggest that natural selection has driven the evolution of core snake metabolic proteins at a scale far beyond what was previously thought plausible. Some of the proteins most enriched for convergent changes, such as COI, are among the most highly conserved proteins across all domains of life. Furthermore, the sites that experienced adaptive changes were otherwise highly conserved relative to other sites, implying that they were likely to have affected protein structure and function in a fundamental way. We have previously labeled this phenomenon “evolutionary redesign” because of the dramatic extent to which core functions of these proteins were evolutionarily targeted for directional change. Interestingly, the reason why such core metabolic redesign might have taken place is not yet clear, although a leading candidate is that the molecular changes support the extreme fluctuations in metabolic demand experienced by snakes. It is also intriguing to speculate that they may be related to adaptation in response to early venom gene evolution in squamate reptiles.
Although form is often said to follow function, it is not clear that protein form (the primary amino acid sequence) usually follows modification of function in any predictable way. If there are many ways to achieve the same function, common adaptive pressures will not necessarily lead to the same changes in protein sequence. Our findings, however, suggest that in the case of these core oxidative phosphorylation subunits encoded by the mitochondrial genome, there may be a limited number of ways in which sequence can be modified in response to functional pressures. Given the central nature of this issue to understanding protein evolution, it is a fair question to ask if adaptive convergence may therefore be more common than we realize. Our opinion is that in fact it may be. In the case we examined, although the shared selective pressure is uncertain, its common form is likely to have arisen through the common function of these proteins in oxidative metabolism. Although these different mitochondrial subunits have completely different structures and functions, similar adaptive pressure on the snake and agamid lineages produced many convergent molecular evolutionary events. This indicates that this type of convergent response is not a one-time event peculiar to the vagaries of a particular protein structure and function, but represents a more general phenomenon reflecting a common quality of the protein adaptive landscape (Fig. 1).
It is also conceptually consistent that adaptive convergent changes should tend to occur at otherwise conserved positions. Protein positions with amino acid residues critical to function are usually conserved over evolutionary time because the most likely effect of a change is to alter function for the worse. Nevertheless, if there is a sustained selective pressure to alter function, it is these same residues that are most likely to have an effect, and thus they are most likely to be the targets of adaptation. The only remaining ingredients are that similar adaptive pressures must occur in divergent lineages, and there must be only one or a few ways to achieve the same functional innovation (Fig. 1).
One reason that we have not yet found many cases of adaptive convergence may be that we are only beginning to acquire appropriately large data sets. Although we have entered the age of complete genomes, available vertebrate mitochondrial genome data sets have only become densely sampled (including many hundreds of species) in the last 5–7 years. Furthermore, complete vertebrate nuclear genomes still number in the tens, not hundreds; with fewer species, branch lengths are longer, and it is much harder to detect and localize adaptive and convergence events. For the same reasons, bacterial genomes, although densely sampled as a group, are less likely to yield many great examples of adaptive convergence because the amounts of time, and therefore the numbers of substitutions separating their proteins, are large. Our expectation is that as more vertebrate genomes are sequenced, we will acquire more examples of suites of genes responding to similar physiological pressures with convergent amino acid changes.
Another reason why adaptive convergence could be more common than believed is that very few systematic surveys for molecular convergence have been performed. When they have been made, such studies have typically depended on reconstructed ancestral sequences,3 which can be error prone in deep evolutionary comparisons. In addition, there have traditionally been no objective ways to distinguish adaptive convergence from random, non-adaptive (or neutral) convergence. Neutral convergence can be especially prevalent in highly divergent datasets, where convergent changes accumulate randomly between long lineages and cause the well-known ‘long branch attraction’ (LBA) artifact in phylogenetic reconstruction. Importantly, the likelihood that such convergent changes will occur on pairs of long lineages is even further increased when there is functional constraint. Functional constraint reduces the number of possible states at important sites, and thus increases the probability of random convergence events.
One of the contributions made by our study is the introduction of statistical methods that can be used to distinguish neutral phenomena from adaptive convergence. Although we were able to detect and validate our example in part because it involved such a massive adaptive burst involving multiple proteins, the use of the new statistical techniques should make it easier to detect other, perhaps smaller events in the future. Application of these methods to other data could thus reveal that suspected cases of LBA were more likely due to adaptive convergence, or worse, that strong phylogenetic support for unexpected relationships could have artificially resulted from previously unsuspected adaptive events.
For phylogenetic reconstruction, the question of how common adaptive convergent events actually are, even “microconvergent” events involving a small number of amino acids on a small number of proteins, is far from academic. The nature of the convergent sites is such that they tend to lend strong phylogenetic support to incorrect conclusions. The first known case of convergent molecular evolution in ruminant lysozymes,4,5 for example, was shown to lend parsimony support to a dramatically wrong phylogeny that placed cows within the primates, even though only a small number of convergent substitutions took place.6 In our study, an incorrect sister-relationship between snakes and agamids was recovered with extremely high statistical support using modern phylogenetic methods. This occurred despite an otherwise strong, latent phylogenetic signal in the rest of the large (>10 k.b.) mitochondrial dataset that supported the expected relationships among snakes and lizards.
Such observations highlight a previously unappreciated weakness of modern, otherwise robust, likelihood methods for phylogenetic inference: slowly-evolving sites and sites involving transversion substitutions rather than transition substitutions usually provide the most trusted evidence for clustering lineages in phylogenies. It appears that even extremely small amounts of adaptive convergent molecular evolution at such sites can be sufficient to strongly bias inferences of relationships, however, particularly if overall phylogenetic signal is low in the data. The slow evolution of such sites means they are less likely to exhibit neutral convergence, but if they respond similarly to changes in selective pressure, as in adaptive convergence, they may provide an extremely misleading degree of support for a wrong phylogeny.
Estimates of phylogeny play a central role in modern biological research, from interpreting the human genome, to inferring the tree of life and identifying new species. Regardless of its frequency in nature, even small amounts of adaptive convergence can strongly mislead phylogenetic inferences. In our experience, single adaptive convergent sites were observed to outweigh support at hundreds of other sites, which contained accurate phylogenetic signal. It is therefore imperative that new phylogenetic methods be developed that are robust to convergent changes directed by adaptation. It is also important that when phylogenetic analyses are interpreted, that adaptive convergence is considered as an explanation for incongruent or suspect phylogenetic results, just as horizontal gene transfer or incomplete lineage sorting are already considered.
Future work is required to understand in more detail what may have driven the convergence event between snakes and agamids, and what functional changes it caused in mitochondrial proteins. Such work will help to explain not just how these proteins happen to function, but, in an evolutionary sense, why they function the way they do. By understanding this case of adaptive convergence better, we should improve our ability to detect adaptive convergence in molecular evolution in other cases when it occurs, and correct for it in phylogenetic reconstruction. Greater detection of adaptive convergence events in nature may also provide important insight into the diversity of protein adaptive landscapes in nature, and help to explain why in some cases these landscapes appear highly constrained.
Acknowledgements
We are thankful for the support of the National Institutes of Health (NIH R01 GM083127) to D.D.P., and an NIH training grant (LM009451) to T.A.C.
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/10174
References
- 1.Castoe TA, de Koning AP, Kim HM, Gu W, Noonan BP, Naylor G, et al. Evidence for an ancient adaptive episode of convergent molecular evolution. Proc Natl Acad Sci USA. 2009;106:8986–8991. doi: 10.1073/pnas.0900233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castoe TA, Jiang ZJ, Gu W, Wang ZO, Pollock DD. Adaptive evolution and functional redesign of core metabolic proteins in snakes. PLoS ONE. 2008;3:2201. doi: 10.1371/journal.pone.0002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rokas A, Carroll SB. Frequent and widespread parallel evolution of protein sequences. Mol Biol Evol. 2008;25:1943–1953. doi: 10.1093/molbev/msn143. [DOI] [PubMed] [Google Scholar]
- 4.Stewart CB, Schilling JW, Wilson AC. Adaptive evolution in the stomach lysozymes of foregut fermenters. Nature. 1987;330:401–404. doi: 10.1038/330401a0. [DOI] [PubMed] [Google Scholar]
- 5.Stewart CB, Wilson AC. Sequence convergence and functional adaptation of stomach lysozymes from foregut fermenters. Cold Spring Harbor Symp Quant Biol. 1987;52:891–899. doi: 10.1101/sqb.1987.052.01.097. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J, Kumar S. Detection of convergent and parallel evolution at the amino acid sequence level. Mol Biol Evol. 1997;14:527–536. doi: 10.1093/oxfordjournals.molbev.a025789. [DOI] [PubMed] [Google Scholar]