Abstract
An ever increasing amount of transcriptomic data and analysis tools provide novel insight into complex responses of biological systems. Given these resources we have undertaken to review aspects of transcriptional regulation in response to the plant hormone gibberellic acid (GA) and its second messenger guanosine 3′,5′-cyclic monophosphate (cGMP) in Arabidopsis thaliana, both wild type and selected mutants. Evidence suggests enrichment of GA-responsive (GARE ) elements in promoters of genes that are transcriptionally upregulated in response to cGMP but downregulated in a GA insensitive mutant (ga1-3). In contrast, in the genes upregulated in the mutant, no enrichment in the GARE is observed suggesting that GARE motifs are diagnostic for GA-induced and cGMP-dependent transcriptional upregulation. Further, we review how expression studies of GA-dependent transcription factors and transcriptional networks based on common promoter signatures derived from ab initio analyses can contribute to our understanding of plant responses at the systems level.
Key words: gibberellic acid; gibberellic acid response elements (GARE); guanosine 3′,5′-cyclic monophosphate (cGMP); plant homeostasis
It is critical for growth and differentiation that plants can rapidly and systemically respond to environmental changes such as light, temperature, CO2, water and nutrient availability as well as biotic and abiotic stresses. Many of the responses are, at least in part, mediated and or modulated by either the classical amino acid derived hormones,1 peptidic hormones,2 plant steroid hormones3 or nitric oxide (NO).4 The link between environment and hormones and the physiological response are second messengers that form a network of molecular transducers. Second messengers relay information and directly or indirectly affect biological processes such as gating of ion channels5 or the regulation of transcription. 6 To do this effectively their cellular activities must be transient and specific.7
In the last decade, an increasing number of second messengers and signaling mechanisms have been discovered which either act on their own or in conjunction with Ca2+. These messengers include cyclic ADP-ribose,8,9 phospholipids such as phosphatidic acid10 as well as cyclic nucleotides, notably adenosine 3′,5′-cyclic monophosphate (cAMP)11–13 and guanosine 3′,5′-cyclic monophosphate (cGMP).14,15
Here the emphasis is on cGMP and its role as second messenger in responses to the plant hormone gibberellic acid (GA) and in particular GA-dependant transcription. GAs are a large family of tetracyclic diterpenoid plant hormones that affect a wide range of plant growth, developmental and environmental responses. These include seed development and germination, leaf expansion, stem elongation and flowering.16 While cGMP is not the only second messenger reported in GA signal transduction, Ca2+,17,18 the cytosolic pH19 and phosphoinositides20,21 responses have been described too, we shall concentrate on cGMP-dependent processes.
GA has been shown to cause increases in cytosolic cGMP which in turn have been proven to be essential and sufficient for α-amylase synthesis and secretion in barley aleurone22 thus suggesting the existence of a cGMP-dependent transcriptome. Experimental evidence for the latter has been obtained6 and shows an enrichment for transporters such as non-selective ion channels and cation:proton antiporters which are affected by cGMP in roots. This together with the fact that NaCl leads to cGMP increases23 and cGMP in turn was found to modulate influx and efflux of the monovalent cations Na+ and K+, would suggest a complex cGMP-dependent response pattern.
We use a recently described method for the inference of biological function at the systems level.24 This method consists of a sequential analysis of genome and expression data as well as on-line web tools for functional genomics at the systems level and is particularly well suited to research in model species such as Arabidopsis.24,25 Here we apply this method to GA- and cGMP-dependent trascription with a view to uncover novel functional aspects of these signaling molecules in a systems context.
Analyses of Genes Transcriptionally Induced by cGMP
The point of departure of the analysis was the identification of genes that are upregulated in response to cGMP treatment. We used supplementary microarray data6 where A. thaliana root tissue was exposed to 10 μM membrane permeable cGMP (8-Br-cGMP) for 30 minutes, and plant tissue was collected after two hours and five hours respectively to identify the effect of cGMP on transcription. Fifty nine genes with average ratios ≥2 from replicate arrays were considered (Suppl. Table 1). This group of genes was analyzed in FatiGO+,26 in order to identify possible bias in distribution that is indicative of a common functional role. FatiGO+ identified a bias in GO functional annotation terms in the cGMP upregulated genes compared to the remainder of the A. thaliana genome. In the GO search category of biological process there is a significant [Family Wise Error Rate—(FWER) adjusted p-value] enrichment in genes involved in ion transport and homeostasis at a number of levels. The most notable bias is found at level 7 with a significant (adjusted p-value-3.81E−02 + 0.0000038) enrichment in genes involved in monovalent inorganic cation transport as well as cellular metal ion homeostasis (Table 1). The GO analysis for the cellular component and molecular function category revealed no significant difference in biologically relevant labels between the two lists. The results of the Swiss-Prot keyword search also identified a significant enrichment in genes involved in ionic channels (adjusted p value = 8.17E−04), ion transport (adjusted p value = 8.17E−04) and associated with the membrane (adjusted p value = 7.94E−03) in the cGMP upregulated genes (appended in Suppl. data file). In the 59 most downregulated genes (Suppl. Table 1) in response to two hours of cGMP exposure we also identified significant enrichment in genes involved in ion transport and homeostasis at a number of levels. The most notable bias is found at level 7 with a significant (adjusted p-value-2.06E−03) enrichment in genes involved in monovalent inorganic cation transport (appended in Suppl. data file). It is thus noteworthy that cGMP may affect ion homeostasis by both up and downregulation of ion transporter encoding genes.
Table 1.
FatiGO+ analysis of cGMP upregulated genes
| List 1 | List 2 | |||
| GO term | Level | # of genes | % | p-value |
| Chemical homeostasis | 3 | 4 | 0.52 | 1.68E−02 |
| Establishment of localization | 3 | 14 | 13.71 | 1.85E−02 |
| Cellular Metabolic Process | 3 | 9 | 64.42 | 1.97E−02 |
| Ion Homeostasis | 4 | 4 | 0.47 | 1.40E−02 |
| Transport | 4 | 14 | 14.19 | 1.85E−02 |
| Ion Transport | 5 | 7 | 3.63 | 1.85E−02 |
| Cation Transport | 6 | 6 | 3.60 | 3.81E−02 |
| Cell. Metal Ion Homeostasis | 7 | 3 | 0.72 | 3.81E−02 |
| Monov. Inorg. Cation Trans. | 7 | 4 | 1.97 | 3.81E−02 |
Genes in list 1 used in analysis: 59; Genes in list 2 used in analysis: 28751.
Identification of Transcription Factors with a Role in cGMP Regulated Genes
Since transcription factors are a key to the regulation of gene expression,27 the identification of common transcription factors in the cGMP upregulated gene set, is likely to indicate collective functional features within this set of genes. Hence we searched promoter regions of the cGMP upregulated genes 1 kb upstream of the predicted transcription start site (TSS) for the presence of known cis-elements to identify common elements and infer transcriptional activation characteristics.
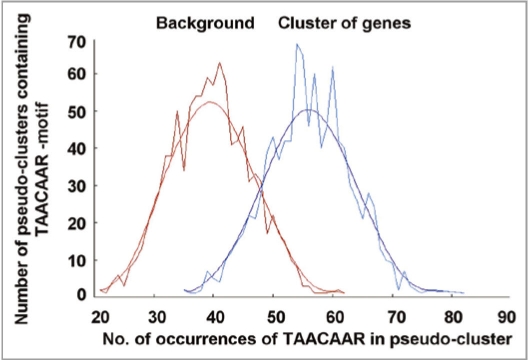
The Athena database (www.bioinformatics2.wsu.edu/cgi-bin/Athena/cgi/home.pl)28 was used to detect common transcription factors and the search revealed that the TAACAA(A/G) gibberellic acid response element GARE was present in 43/59 genes a total of 74 times at an average of 1.25 copies/promoter (p-value <10E−4). A Bonferroni correction was automatically used in Athena to account for multiple hypotheses testing and the p-value represents an enrichment of the genes according to the stringent enrichment threshold of <10−4 and indicates that a high percentage of our cGMP upregulated genes contain multiple copies of the TAACAA(A/G) motif. The subsequent POBO analysis (ekhidna.biocenter.helsinki.fi/poxo/pobo)29 of the cGMP upregulated genes shows that the TAACAAR motif was significantly enriched (t-test p-value <0.0001), being present in 40/59 genes a total number of 64 times at an average of 1.08 copies/promoter compared to the average of 0.78 across all A. thaliana promoters (Fig. 1).
Figure 1.
Frequency of occurrence of the GARE (TAACAAR) core motif in artificial clusters generated in POBO (ekhidna.biocenter.helsinki.fi/poxo/pobo)29 for A. thaliana background promoters compared to the promoters (1 kb upstream of the transcription start site) of the 59 genes upregulated >2 fold after cGMP treatment6 was analyzed. The default settings were used (number of sequences to pick-out = 50, number of samples to generate = 1000, sequence length = 1000 bps) and a two-tailed p-value was calculated in the linked online GraphPad Web site using the generated t-value and degrees of freedom to determine the statistical differences between input sequences and background. This analysis determined that compared to the background (0.78 copies/promoter), there was a significant (t-test: p-value >0.0001) enrichment in the frequency of the TAACAAR motif in our dataset (1.08 copies/promoter).
Functional Analysis of Expression Data in Arabidopsis Wild-Type and Selected Mutants
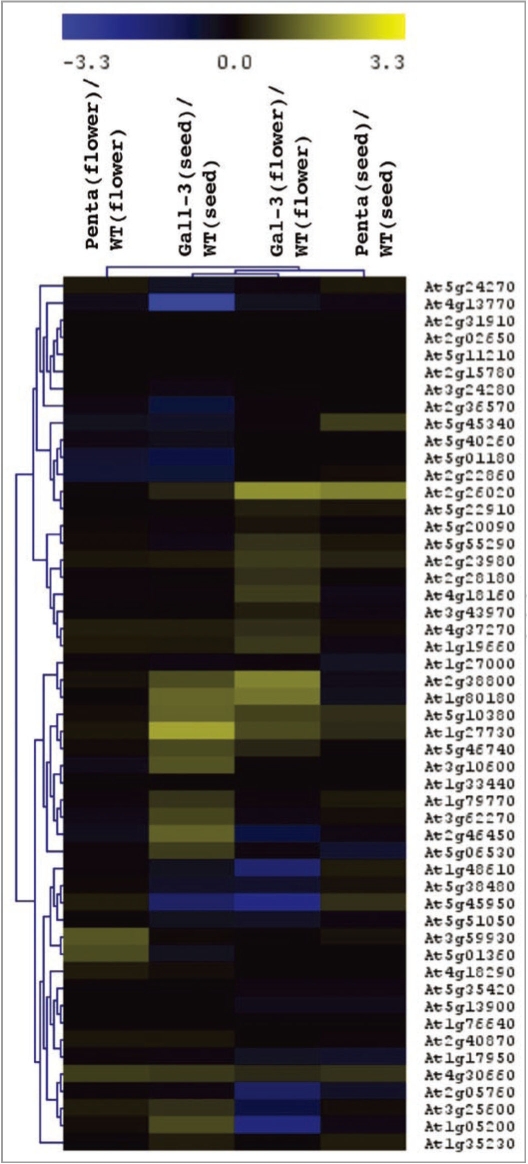
In order to elucidate the role of the GARE motif in cGMP- and GA-dependent transcription, we examined responses in GA mutants in A. thaliana. Expression data was extracted from publicly available supplementary microarray data for an experiment involving GA Arabidopsis thaliana mutants (Accession no: E-MEXP-849)30 and a heatmap was generated. The GA mutant used for the analysis was ga1-3, a null mutant that is severely GA deficient since GA1 encodes ent-copalyl diphosphate synthase, an enzyme that catalyzes the first committed step in GA biosynthesis.31 Mutant plants show extreme dwarfism with delayed flowering and male sterility. A heatmap was also generated from the expression data of cGMP upregulated genes in the ga1-3 in Arabidopsis flowers and seeds (Supplementary micro-array data,30 NASCarray experiment, reference number −385; Roots treated with +/− 2 µM GA for two hours). The results show that of the 59 cGMP upregulated genes, the expression of the genes were divided, with some of the genes having the same expression in both ga1-3 and the wildtype (indicated in black), while the remainder of the genes were either upregulated (yellow) or downregulated (blue) when compared to the wildtype (Fig. 2). FatiGO+ analysis of cGMP genes upregulated in ga1-3 mutant showed enrichment in K+ ion transport proteins (level 8; p-value = 1.2E−02), while genes that are downregulated in the mutant, showed enrichment in proteins with a role in ion homeostasis (level 6; p-value = 2.52E−02).
Figure 2.
Heatmap illustrating fold change (log2) in expression of cGMP-induced genes in the Arabidopsis GA-deficient mutants ga1-3 and penta. In order to reveal the transcriptional relationship between cGMP-induced genes and GA, expression of the top 59 cGMP upregulated genes was investigated in a publicly available microarray data set (Accession no: E-MEXP-849) containing the GA-deficient mutants ga1-3 and penta (a gal-3 della mutant lacking all four DELLA proteins).30 Gene expression M values [log2(mutant/wildtype)] were determined for seed and flower tissues in both mutants as previously described. The heatmap indicates clusters of up-and downregulated GA-responsive genes, most noticeably in the gal-3 mutant with evidence of tissue-specific expression between seeds and leaves.
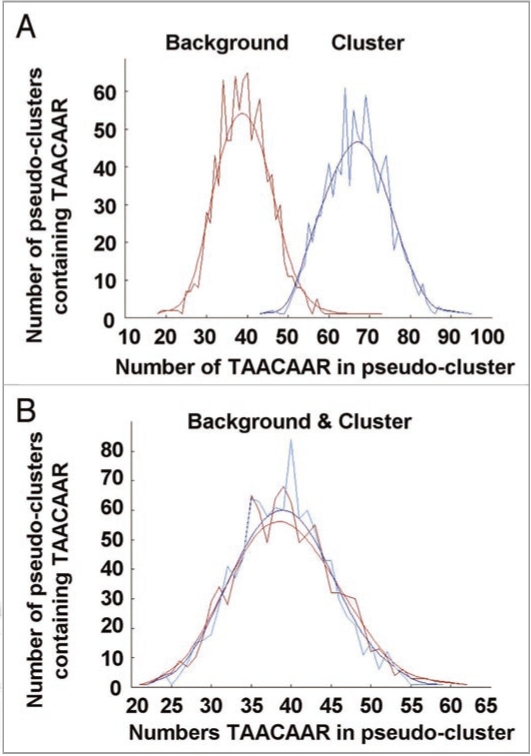
To test whether the GARE motif (TAACAAR), is present in both the up and downregulated genes we analyzed the distribution in POBO. The results showed that the genes downregulated in the mutant, had multiple copies of the GARE element (1.33 copies/promoter) with the TAACAAR motifs being significantly enriched compared to expected frequencies in the A. thaliana genome (0.78 copies/promoter) (Fig. 3A). This GARE frequency of occurrence is even higher than in the entire selection of cGMP upregulated genes (1.08 copies/promoter) while POBO showed that the TAACAAR motif was not enriched in the genes upregulated (0.77 copies/promoter) in the mutant compared to the expected frequencies in the A. thaliana genome (0.78 copies/promoter) (Fig. 3B). Furthermore the 50 most significantly upregulated genes in A. thaliana root tissue treated with 2 µM GA for 2 hours (NASCarray experiment: −385) were analyzed with promoter analysis tool Athena and showed no enrichment in the GARE motif and no enrichment in any ontology category. Furthermore, there are no genes shared between the cGMP upregulated data set and the GA upregulated data set (top 50) and there is also no overlap between the cGMP upregulated data set and all of the GA upregulated genes detected in the microarray experiment, thus suggesting no similarity between the two gene selections.
Figure 3.
Frequency of occurrence of the GARE (TAACAAR) core motif in artificial clusters generated in POBO for A. thaliana background promoters compared to the promoters of cGMP upregulated genes that were downregulated (A) or upregulated (B) in the ga1-3 mutant. (A) The 1 kb upstream promoter sequences of the selected genes were analyzed in POBO to determine the frequency of occurrence of the TAACAAR GARE core motif. The analysis determined that compared to the A. thaliana background (0.78 copies/promoter), there was a significant (t-test: p-value >0.0001) enrichment in the frequency of the TAACAAR motif in our dataset (1.33 copies/promoter). (B) The analysis determined that compared to the A. thaliana background (0.78 copies/promoter), there was no significant (t-test: p-value >0.0001) enrichment in the frequency of the TAACAAR motif in our dataset (0.77 copies/promoter).
In a next step of the analysis we looked at expression in the ga1-3 mutant in seeds and flower buds.30 Firstly, 541 genes upregulated in seeds and 828 genes upregulated in flower buds were extracted and the 60 genes that had the highest average ratios (log2 values) were subjected to a promoter analysis in Athena. The GARE motif is present in 43/60 GA upregulated genes in seeds a number of 64 times giving an average of 1.06 times copies/promoter (p-value ≤10E−4). This p-value indicates a significant enrichment of the GARE motif in the GA upregulated genes in seeds. GA upregulated genes in flower buds do not show an enrichment of the GARE motif.
The GA upregulated genes containing the GARE motif were further subjected to FatiGO+ analysis and the genes in flowers showed enrichment in genes involved in monovalent inorganic cation transport as well as cellular metal ion homeostasis at level 7 (adjusted p-value-3.81E−02 + 0.0000038). Distribution of the GARE motif was also identified across the entire A. thaliana genome. The results showed that the TAACAAR motif is present in 55% of the promoters in the genome. Subsequently, a FatiGO+ analysis was performed with genes where the GARE motif is present 5 or more times within their promoters. The result yielded no significant distribution of any GO annotation.
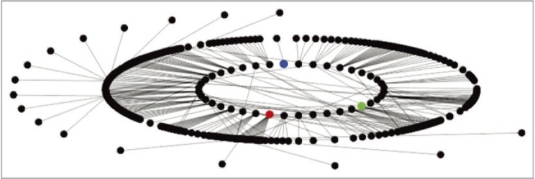
In order to gain further insight into gene function at the systems level we have analyzed the promoter content of the 59 cGMP upregulated genes using an ab initio method and then built a putative transcription regulatory network (Fig. 4) based on promoter content similarity using the Dragon Motif Builder.32 In the resuting network, a hub gene with 24 common edges, CHX8 (Cation Hydrogen Exchanger 8), was identified. CHX8 is a cation/hydrogen exchanger gene that belongs to the putative monovalent cation:proton antiporter family 2 (CPA2).33 Expression correlation analyses of the top co-expressed 100 genes (Suppl. Table 2) shows high r-values ranging from 0.999611 to 0.973635 and the Fatigo+ analysis reveals a highly significant enrichment (2.94E−09) for “Phosphorous Metabolic Process” at level 4. Other highly significant terms at higher levels include “Phosphorylation” and “Protein Modification” at level 6, “Posttranslational Protein Modification” at level 7 and “Protein Amino Acid Phosphorylation” at level 8, thus pointing to a role in signaling and regulation of this hub-linked co-expressed group of genes. This is further supported by the significant terms of “Protein Amino Acid Acylation” at level 7 and “Protein Myristoylation” at level 9 (Table 2).
Figure 4.
Transcriptional regulatory network diagram of cGMP upregulated genes generated by promoter analysis in order to predict A. thaliana co-expressed genes. The 59 upregulated gene promoters of a length 2,200 nt covering 2,000 nt upstream and 200 nt downstream of the 5′ end of the respective genes. The data was created with the Dragon Motif Builder (apps.sanbi.ac.za/MotifBuilder/index.php)32 and displayed with Cytoscape. (Dragon Motif Builder parameters: Algorithm EM2, motif length 9, threshold 0.9, no background, number of motif families = 30). The red dot denotes a hub gene (AtCHX8) and signifies the cGMP upregulated gene that shares the highest number of common promoter motifs with other A. thaliana genes. The green dot indicates At5G36240, a zinc knuckle (CCHC-type) family protein which contains the highest number of shared motifs per edge. At5g38480 general regulatory factor, a 14-3-3 gene, is represented by the blue dot on the figure. This gene is only linked to one gene and thus shares notable common motifs with a single gene only.
Table 2.
FatiGO+ analysis of CHX8 correlated genes
| List 1 | List 2 | |||
| GO term | Level | # of genes | % | p-value |
| Pi Metabolic Process | 4 | 22 | 7.75 | 2.94E−09 |
| Phosphate Metabolic Process | 5 | 21 | 8.38 | 4.36E−11 |
| Biopolymer Mod. Peptide | 5 | 22 | 30.49 | 9.97E−08 |
| Amino-terminal Blocking | 5 | 7 | 3.32 | 1.26E−03 |
| Cell. Prot. Metabolic Process | 5 | 26 | 30.49 | 1.26E−03 |
| Phosphorylation | 6 | 21 | 9.21 | 6.48E−12 |
| Protein Modification | 6 | 21 | 15.06 | 11.0E−08 |
| Lipoprot. Metabolic Process | 6 | 7 | 4.0 | 2.69E−03 |
| Post-translational Prot. Mod. | 7 | 21 | 13.52 | 1.21E−10 |
| Peptidyl Amino Acid Mod. | 7 | 7 | 5.12 | 3.47E−03 |
| Lipoprotein Biosynth. Proc. | 7 | 7 | 5.05 | 3.47E−03 |
| Prot. a.a. Acylation | 7 | 7 | 4.92 | 3.47E−03 |
| Prot. a.a. Phosphorylation | 8 | 21 | 15.6 | 3.54E−10 |
| Protein Myristoylation | 9 | 7 | 14.19 | 4.43E−03 |
Genes in list 1 used in analysis: 102; Genes in list 2 used in analysis: 28708.
For 16 out of the 24 hub group genes co-expression coefficients could be determined and three genes, a DNA repair protein RAD54 (AT3G19210, r = 0.806444), pentatricopeptide (PPR) repeat-containing protein with strong similarity to PCMP-H2 (AT3G61170, r = 0.750457) and one un-annotated expressed protein (AT1G17830, r = 0.74996) show a positive correlation and are all expressed in the male gametophyte. The remaining 13 are not expression correlated with “r” values between 0.020641 and −0.156385.
Analysis of tissue expression patterns of CHX8 shows that this hub gene is very highly expressed [linear value = 49460 (standard error = 455)/log2 value = 15.59 (s.e = 0.01)] in pollen.34 Supplementary microarray data of Arabidopsis genes expressed in pollen, leaves, seedlings and siliques,35 verified that CHX8 is enriched in pollen expression compared to other tissues. In addition, genes that were identified in Arabidopsis Co-Expression Tool (ACT; www.arabidopsis.leeds.ac.uk)36 as CHX8 co-expressed also showed a higher expression in pollen than in any of the other tissue and this finding was confirmed by a Genevestigator37 analysis using the anatomy tool for CHX8 and the co-expressed genes.
The promoter analysis of CHX8 and the most highly correlated genes also revealed an enrichment in the CARGW8GAT motif (p-value = 10−3) as well as the MYB1AT motif (p-value = 10−3), MYB2AT motif (p-value = 10−5) and the GARE motif (p-value = 0.0373) (Suppl. Table 3). The CARGW8GAT motif was present in 70/100 genes at a total number of 252 times and the GARE motif was present in 57/100 genes a total of 97 times. Another cGMP-induced gene in the network, AT5G36240, is the zinc knuckle (CCHC-type) family protein. It contains the highest number of shared motifs per edge and notably 20 with AT1G6770 and AT5G36250, a protein phosphatase 2C. Another cGMP-induced gene, AT5G38480.1, is only linked to AT5G38480.3 and is annotated as a GENERAL REGULATORY FACTOR 3 (GRF3).
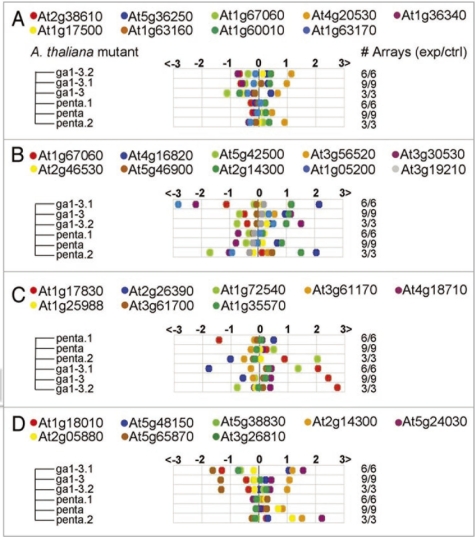
If we consider the expression of the CHX8 node genes and CCHC-type family protein node genes in the GA mutant display (ga1-3.1, ga1-3, ga1-3.2, penta, penta.1 and penta.2), we note different response patterns with regard to the number of common elements shared by the node members (Fig. 5). CCHC-type family protein node genes which contains the highest number of shared motifs per edge (20) show a less varied response pattern (Fig. 5A) than in the case of the CHX8 node genes (Fig. 5B and C), which only shared 13/14 motifs per edge. As a control we used a gene belonging to the K+ channel AtTPK/KCO family of proteins (At4g18160), which has only 13 common elements shared with other node members. Analysis of its node genes also showed a more varied response pattern than observed in the case of CCHC-type family protein node genes (Fig. 5D). This indicates that the higher the number of common elements shared by different genes the more similar we would expect their expression patterns to be, a hypothesis that has been supported by experimental evidence in rice LEA (Late Embryogenesis Abundant) genes.38
Figure 5.
Genevestigator analysis (www.genevestigator.com/gv/index.jsp)37 of GA mutant genes. Expression of zinc knuckle (CCHC-type) family protein (AT5G36240) node members, which shares 20 common elements, show a less varied response pattern (A) than in the case of the ATCHX8 linked genes (B and C), which only shares 13/14 motifs per edge. As a control, genes linked to AT4g18160, K+ channel ATTPK/KCO family of proteins were used (D), since all its node members only shared 13 common motifs. The results also showed a response pattern that was more varied than the one of the zinc knuckle (CCHC-type) family protein.
Cis-Elements and Transcription Factors in GA Rersponses
Isolated GA-related mutants have helped to determine both the physiological role of GA and to elucidate its biosynthetic pathways. Several negative regulators of GA signaling have been isolated by characterization of the recessive slender mutants and the dominant GA-unresponsive dwarf mutants. In Arabidopsis these mutants include the spindly (spy) and SHORT INTERNODES (SHI). GA-unresponsive dwarf mutants have pointed to a number of positive regulators of GA signaling including the dwarf1 (d1) (79) mutants in rice and the sleepy1 (sly1) (115) mutants in Arabidopsis.39 GID1 (GA-INSENSITIVE DWARF1) is a receptor mutant that shows similarity to hormone-sensitive lipases and has been shown to mediate GA perception. The GID1 receptor binds to GA and the GID1-GA complex then interacts with DELLA proteins—which are negative regulators of GA action—resulting in degradation of DELLA protein.40
Early attempts to characterise GA transcriptional responses lead to the discovery of a promoter element for GA-driven expression in barley α-amylase and this element was termed GARE (Gibberellic Acid Response Element).41 Here we undertook a promoter analysis of cGMP upregulated genes and showed a significant enrichment of these elements suggesting firstly, that they are likely to play an important role as regulatory elements in the expression of these genes and that there may be a causal link between strong GA and cGMP-dependent transcriptional regulation. Analysis of the GA signaling pathway in cereal aleurone has revealed that a MYB gene, GAMYB, binds specifically to the TAACAAA motif and activates the transcription of a high-pI α-amylase promoter which implied that GAMYB is part of the GA signaling pathway that leads to α-amylase gene expression in aleurone cells.42 There is compelling evidence that cGMP is required for barley aleurone layers to act in response to GA.22 Treatment of aleurone layers with GA led to a transient increase in cGMP. This transient increase in cGMP was reduced after application of LY-83583, an inhibitor of guanylyl cyclase, which in turn led to an inhibition of α-amylase and GAMYB mRNA accumulation as well as α-amylase secretion. Dibutyryl-cGMP, a membrane permeant cGMP analogue, almost restored α-amylase secretion to control levels with LY 83583 present.22 Taken together, cGMP operates as an essential second messenger in some GA dependent responses.
GAMYB-like genes have also been shown to function in tissues other than the aleurone and in different plant species. In barley, HvGAMYB, involved in GA signaling in the aleurone, also plays a role in endosperm development43 and was found to be strongly expressed in anthers.44 GAMYB has also been implicated in the elongation of the first internode of Triticum aestivum (wheat)45 and flowering in Lolium temulentum.46 MYB33, a GAMYB-like gene has been identified in A. thaliana47 and is expressed at the shoot apex when plants were treated with GA or when endogenous GA levels increased. The expression of MYB33 in the shoot apex is correlated with the onset of flowering47 and expression at the shoot apex also leads to an increase in LFY expression. LFY is a LEAFY gene that is a potential inducer of flowering in dicots48 and is activated by GA application.49,50 The LFY promoter contains a putative GARE,46 that may serve as binding site for MYB33.47 The GARE motif in turn is necessary for GA induction of LEAFY47,51 to promote flowering thus hinting at a role of GARE as a factor in GA-regulated signaling pathways that depend on cGMP as second messenger.
In promoters of genes transcriptionally upregulated in response to cGMP and downregulated in the GA insensitive mutant (ga1-3), we have observed an enrichment in the GARE motifs. In contrast, in the genes upregulated in the mutant, there was no such enrichment and we argue that this is consistent with a role for GARE and hence GAMYB in GA-induced cGMP-dependent transcriptional activation. Interestingly, the GARE enrichment was seen only in the GA upregulated genes in seeds and flower buds and not in root tissue which is the tissue used for the cGMP analysis presented here. We identified all GARE motifs across all promoters of the entire A. thaliana genome, and genes where this motif is present in more than one copy were analyzed in FatiGO+. The result (not shown) has no significant enrichment for any GO category. It must therefore be concluded that GARE on its own is insufficient to drive a regulatory program independent of its dosage.
The finding that the cGMP upregulated genes are significantly enriched in monovalent inorganic cation transport as well as cellular metal ion homeostasis in the FatiGO+ analysis lends strength to previous reports of the role of cGMP in ion homeostasis. The enrichment is also corroborated by the the Swiss-Prot keyword search that identifies a significant enrichment of genes involved in ionic channels and ion transport. A previous electro-physiological investigation has demonstrated that cGMP can play a direct role in ion channel modulation and hence ion homeostasis.5 Cyclic GMP binding modulates the voltage/current relationship in plant voltage gated K+ channels by inducing a shift of the current-voltage curve to more negative voltages and this effect is concentration-dependent.5 Furthermore, it was also reported that plants contain cyclic-nucleotide gated low affinity cation channels where binding of cAMP and cGMP to the intracellular portion leads to direct gating.52 In A. thaliana, voltage independent channels (VIC) without selectivity for particular monovalent cations have been characterised and these channels have opening probabilities that are affected by µM concentrations of cAMP and cGMP on the cytoplasmic side of the plasma membrane.53 Short-term unidirectional Na+ influx is reduced in the presence of cyclic nucleotides and essentially, membrane permeable cyclic nucleotides can reduce net Na+ uptake and thereby improve salinity tolerance under particular experimental conditions.
GO analysis of cGMP downregulated genes also revealed an enrichment of genes involved in monovalent inorganic cation transport at level 7. This is an indication that the transcriptional cGMP response is bi-polar in the sense that cation transport regulation is affected also through the reduction of cation channels and that requires complex transcriptional regulation of channel encoding genes with a role in long-term adaptation to the environment.
New Insights from Transcription Regulatory Networks
We have used an ab initio method to analyze promoter content of the cGMP upregulated genes to generate a transcriptional regulatory network based on numbers of shared motifs families (Fig. 4). Analysis of the regulatory network of the cGMP upregulated genes revealed a hub gene that is linked to the lagest number genes and this gene was identified as CHX8 which encode cation/hydrogen exchangers belonging to the putative monovalent cation:proton antiporter family 2 (CPA2).33 A number of CHX genes, including CHX8 are specifically expressed during male gametophyte development and tissue expression pattern analysis of CHX8 also indicate a higher expression in pollen compared to other plant tissues.34 It is therefore safe to conclude CHX genes have a role in male gametogenesis. Pollen development and tube growth are associated with large fluctuations in water and ion homeostasis and a number of responses involving transport which include: vacuole formation, ion and metabolite transport and osmotic adjustments during dehydration and rehydration as well as vesicular trafficking.54 Since dehydration sets in as male gametophytes reach maturity, several genes upregulated in vegetative tissues by salt or dehydration stress are also expressed in pollen of unstressed plants, suggesting a need to make major osmotic adjustments during microgametogenesis.55 CHX8 peaks in the tricellular or mature pollen presumably to contribute to homoeostatic adjustment during dehydration and/or pollen germination following rehydration.
Microarray data35 where Arabidopsis genes were expressed in various plant tissues were also analyzed and showed that CHX8 as well as all of the top 100 most highly co-expressed genes were markedly higher expressed in pollen as compared to other tissue. Further analysis with the anatomy tool from Genevestigator37 confirmed pollen specific upregulation of CHX8 and the co-expressed genes over a number of microarrays. CHX8 is a target gene of MIKC*proteins that are expressed exclusively in pollen and predominantly from the tricellular stage onwards56 and is assumed to play an important role in the regulation of transcription during late pollen development.57 MIKC*proteins bind to MEF2 type CArG motifs [C(A/T)8G] and these motifs are over-represented in late pollen specific promoters, coinciding with MIKC* expression. Our Athena analysis of CHX8 promoter confirmed the presence of a CARGW8GAT motif, also referred to as the MEF2 type CArG motifs, and thus being consistent with CHX8 being a target for MIKC*proteins. Promoter analysis of the CHX8 co-expressed genes in Athena also indicated a significant enrichment of the CARGW8GAT motif within this set of genes. The motif was found in 70 of the genes at a number of 252 times. These findings imply that CHX8 and its most highly expression correlated genes are most likely involved during the latter stages of pollen development since they are predominantly expressed in pollen tissue and contain the overrepresented MEF2 type CArG motifs diagnostic for late pollen specific promoters.
Activation of protein kinases that modify the phosphorylation state of proteins is an important mechanism in cellular responses to extracellular signals. Protein kinases have been identified in pollen and have been shown to be associated with pollen germination and tube growth in various plant species.58,59 Activation of protein synthesis during hydration of pollen is accompanied by protein phosphorylation.60 A maize pollen specific calcium-dependent calmodulin-independent protein kinase (CDPK) also implicates phosphorylation based mechanisms in post-translational control.58 In Arabidopsis, several protein kinases have been shown to be involved in pollen germination and tube growth. These kinases include inositol polyphosphate kinase, AtIPK2α,61 where AtIPK2α regulates pollen germination and tube growth. Further, a phosphatidylinositol-4-monophosphate 5-kinase (PIP5K4) has been implicated in regulating pollen tube growth and polarity.62 The fact that protein phosphorylation is prominently involved in pollen germination and tube growth, is also in line with the FatiGO+ analysis of CHX8 and its co-expressed genes. This analysis revealed a significant enrichment of genes involved in protein amino acid phosphorylation, where 21 genes were implicated in this GO annotation with a highly significant p-value of 3.54E−10. These CHX8 expression-correlated genes were also shown to be implicated in protein myristoylation with a significant p-value of 4.43E−03 while evidence for protein myristoylation in pollen tissues has been reported63 and, in particular, an Arabidopsis phosphatase, AtPTEN1 was predicted to be myristoylated.63 AtPTEN1 is a dual specific enzyme that can act on either phosphotyrosine residues or lipid phosphotidylinositol (3,4,5)-triphosphate in proteins.64 AtPTEN1 is exclusively expressed in pollen grain during the later stages of development and was proven to be important for pollen development.64
Since CHX8 was identified as a hub gene within a cGMP upregulated gene network, we have set out to discover further links between pollen function, CHX8 and co-expressed genes, and cGMP upregulated genes, particularly so, since cGMP has been shown to mediate NO function in Lily pollen tube, where NO levels regulate the rate and orientation of pollen tube growth with cGMP as a second messenger.65 In ferns, cGMP and NO have also been identified as playing a role in linking the gravity stimulus to polarized growth in spores by acting as downstream effectors.66
Promoter analysis of CHX8 and the co-expressed genes showed that the genes are enriched in the GARE motif while GA is an essential component in pollen tube development, further strengthening the association with CHX8 and co-expressed genes. The spy mutation 5 (an Arabidopsis mutant which increases GA responses) dislays reduced pollen tube growth and a GA biosynthesis inhibitor, uniconazole, also affects pollen tube function implicating the hormone in normal pollen tube growth.67 GA signaling alleles gar2-1 and rga were also shown to play a role in pollen tube elongation in GA-deficient pollen tubes in Arabidopsis.68 Further evidence of a critical role for GA in pollen was reported in rice, where GA-deficient or GA-insensitive mutants show reduced viability and pollen tube growth.69 Expression of GA signaling genes occurs at the premeiosis stage of the pollen developmental process, while GA synthesis genes are expressed after meiosis, suggesting that GA synthesis has a role in gametophytic processes.69
In summary, the ab initio analysis of cGMP-induced genes identifies hub genes one of which is the GARE containing cation/hydrogen exchanger gene (CHX8) that is highly expressed in pollen. Further, CHX8 co-expressed genes show a GARE enrichment as well as the highest expression levels during male gametogenesis and pollen tube growth implementing them in functions critical for gametophyte development, in particular homoestatic adaptations to rapidly changing microenvironments. Finally, we propose that this type of integrated application of on-line data mining tools for functional annotation and systems analysis can uncover novel biological mechanisms and inform experimental strategies particularly so in model systems like Arabidopsis or rice.
Acknowledgements
This project has been supported by the South African National Research Foundation.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/10718
Supplementary Material
References
- 1.Kende H, Zeevaart J. The Five “Classical” Plant Hormones. Plant Cell. 1997;9:1197–1210. doi: 10.1105/tpc.9.7.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindsey K. Plant peptide hormones: The long and the short of it. Curr Biol. 2001;11:741–743. doi: 10.1016/s0960-9822(01)00435-3. [DOI] [PubMed] [Google Scholar]
- 3.Schaller H. The role of sterols in plant growth and development. Prog Lipid Res. 2003;42:163–175. doi: 10.1016/s0163-7827(02)00047-4. [DOI] [PubMed] [Google Scholar]
- 4.Neill SJ, Desikan R, Hancock JT. Nitric oxide signalling in plants. New Phytol. 2003;159:11–35. doi: 10.1046/j.1469-8137.2003.00804.x. [DOI] [PubMed] [Google Scholar]
- 5.Hoshi T. Regulation of voltage dependence of the KAT1 channel by intracellular factors. J Gen Physiol. 1995;105:309–328. doi: 10.1085/jgp.105.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maathuis FJ. cGMP modulates gene transcription and cation transport in Arabidopsis roots. Plant J. 2006;45:700–711. doi: 10.1111/j.1365-313X.2005.02616.x. [DOI] [PubMed] [Google Scholar]
- 7.Plieth C. Calcium: just another regulator in the machinery of life? Ann Bot. 2005;96:1–8. doi: 10.1093/aob/mci144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galione A, Cui Y, Empson R, Iino S, Wilson H, Terrar D. Cyclic ADP-ribose and the regulation of calcium-induced calcium release in eggs and cardiac myocytes. Cell Biochem Biophys. 1998;28:19–30. doi: 10.1007/BF02738307. [DOI] [PubMed] [Google Scholar]
- 9.McAinsh MR, Gray JE, Hetherington AM, Leckie CP, Ng C. Ca2+signalling in stomatal guard cells. Biochem Soc Trans. 2000;28:476–481. [PubMed] [Google Scholar]
- 10.Munnik T. Phosphatidic acid: an emerging plant lipid second messenger. Trends Plant Sci. 2001;6:227–233. doi: 10.1016/s1360-1385(01)01918-5. [DOI] [PubMed] [Google Scholar]
- 11.Moutinho A, Hussey PJ, Trewavas AJ, Malho R. cAMP acts as a second messenger in pollen tube growth and reorientation. Proc natl Acad Sci USA. 2001;98:10481–10486. doi: 10.1073/pnas.171104598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemtiri-Chlieh F, Berkowitz GA. Cyclic adenosine monophosphate regulates calcium channels in the plasma membrane of Arabidopsis leaf guard and mesophyll cells. J Biol Chem. 2004;279:35306–35312. doi: 10.1074/jbc.M400311200. [DOI] [PubMed] [Google Scholar]
- 13.Suita K, Kiryu T, Sawada M, Mitsui M, Nakagawa M, Kanamaru K, et al. Cyclic GMP acts as a common regulator for the transcriptional activation of the flavonoid biosynthetic pathway in soybean. Planta. 2009;229:403–413. doi: 10.1007/s00425-008-0839-5. [DOI] [PubMed] [Google Scholar]
- 14.Newton RP, Smith CJ. Cyclic nucleotides. Phytochem. 2004;65:2423–2437. doi: 10.1016/j.phytochem.2004.07.026. [DOI] [PubMed] [Google Scholar]
- 15.Meier S, Gehring C. Emerging roles in plant biotechnology for the second messenger cGMP—guanosine 3′,5′-cyclic monophosphate. Afr J Biotech. 2006;5:1687–1692. [Google Scholar]
- 16.Davies PJ. Plant Hormones. The Netherlands: Kluwer Academic Publishers; 2004. [Google Scholar]
- 17.Gilroy S, Jones RL. Gibberellic acid and abscisic acid coordinately regulate cytoplasmic calcium and secretory activity in barley aleurone protoplasts. Proc natl Acad Sci USA. 1992;89:3591–3595. doi: 10.1073/pnas.89.8.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilroy S. Signal transduction in barley aleurone protoplasts is calcium dependent and independent. Plant Cell. 1996;8:2193–2209. doi: 10.1105/tpc.8.12.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gehring C, Irving H, Parish R. Intracellular pH changes precede gibberellic acid induced elongation in maize coleoptiles. Planta. 1994;194:532–540. [Google Scholar]
- 20.Murthy PP, Renders JM, Keranen LM. Phosphoinositides in barley aleurone layers and gibberellic acid-induced changes in metabolism. Plant Physiol. 1989;91:1266–1269. doi: 10.1104/pp.91.4.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fleet CM, Ercetin ME, Gillaspy GE. Inositol phosphate signaling and gibberellic acid. Plant Signal Behav. 2009;4:73–74. doi: 10.4161/psb.4.1.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penson SP, Schuurink RC, Fath A, Gubler F, Jacobsen JV, Jones RL. cGMP is required for gibberellic acidinduced gene expression in barley aleurone. Plant Cell. 1996;8:2325–2333. doi: 10.1105/tpc.8.12.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donaldson L, Ludidi N, Knight MR, Gehring C, Denby K. Salt and osmotic stress cause rapid increases in Arabidopsis thaliana cGMP levels. FEBS Lett. 2004;569:317–320. doi: 10.1016/j.febslet.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 24.Meier S, Bastian R, Donaldson L, Murray S, Bajic V, Gehring C. Co-expression and promoter content analyses assign a role in biotic and abiotic stress responses to plant natriuretic peptides. BMC Plant Biol. 2008;8:24. doi: 10.1186/1471-2229-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meier S, Gehring C. A guide to the integrated application of on-line data mining tools for the inference of gene functions at the systems level. Biotechnol J. 2008;3:1375–1387. doi: 10.1002/biot.200800142. [DOI] [PubMed] [Google Scholar]
- 26.Al-Shahrour F, Minguez P, Tarraga J, Medina I, Alloza E, Montaner D, et al. FatiGO+: a functional profiling tool for genomic data. Integration of functional annotation, regulatory motifs and interaction data with microarray experiments. Nucl Acids Res. 2007;35:91–96. doi: 10.1093/nar/gkm260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong W, Shen YP, Ma LG, Pan Y, Du YL, Wang DH, et al. Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol. 2004;135:773–782. doi: 10.1104/pp.104.042176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Connor TR, Dyreson C, Wyrick JJ. Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics. 2005;21:4411–4413. doi: 10.1093/bioinformatics/bti714. [DOI] [PubMed] [Google Scholar]
- 29.Kankainen M, Holm L. POBO, transcription factor binding site verification with bootstrapping. Nucl Acids Res. 2004;32:222–229. doi: 10.1093/nar/gkh463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao D, Cheng H, Wu W, Soo HM, Peng J. Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol. 2006;142:509–525. doi: 10.1104/pp.106.082289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun TP, Kamiya Y. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell. 1994;6:1509–1518. doi: 10.1105/tpc.6.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang E, Yang L, Chowdhary R, Kassim A, Bajic V. An algorithm for ab initio DNA motif detection. Information Processing and Living Systems. 2005:611–614. [Google Scholar]
- 33.Maser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, et al. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001;126:1646–1667. doi: 10.1104/pp.126.4.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sze H, Padmanaban S, Cellier F, Honys D, Cheng NH, Bock KW, et al. Expression patterns of a novel AtCHX gene family highlight potential roles in osmotic adjustment and K+ homeostasis in pollen development. Plant Physiol. 2004;136:2532–2547. doi: 10.1104/pp.104.046003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pina C, Pinto F, Feijo JA, Becker JD. Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control and gene expression regulation. Plant Physiol. 2005;138:744–756. doi: 10.1104/pp.104.057935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manfield IW, Jen CH, Pinney JW, Michalopoulos I, Bradford JR, Gilmartin PM, Westhead DR. Arabidopsis Co-expression Tool (ACT): web server tools for microarray-based gene expression analysis. Nucl Acids Res. 2006;34:504–509. doi: 10.1093/nar/gkl204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meier S, Gehring C, MacPherson CR, Kaur M, Maqungo M, Reuben S, et al. The promoter signatures in rice LEA genes can be used to build a coexpressing LEA gene network. Rice. 2008;1:177–187. [Google Scholar]
- 39.Sun TP, Gubler F. Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol. 2004;55:197–223. doi: 10.1146/annurev.arplant.55.031903.141753. [DOI] [PubMed] [Google Scholar]
- 40.Hirano K, Ueguchi-Tanaka M, Matsuoka M. GID1-mediated gibberellin signaling in plants. Trends Plant Sci. 2008;13:192–199. doi: 10.1016/j.tplants.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 41.Skriver K, Olsen FL, Rogers JC, Mundy J. cis-acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proc natl Acad Sci USA. 1991;88:7266–7270. doi: 10.1073/pnas.88.16.7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gubler F, Kalla R, Roberts JK, Jacobsen JV. Gibberellin-regulated expression of a myb gene in barley aleurone cells: evidence for Myb transactivation of a high-pI alpha-amylase gene promoter. Plant Cell. 1995;7:1879–1891. doi: 10.1105/tpc.7.11.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diaz I, Vicente-Carbajosa J, Abraham Z, Martinez M, Isabel-La Moneda I, Carbonero P. The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endospermspecific genes during seed development. Plant J. 2002;29:453–464. doi: 10.1046/j.0960-7412.2001.01230.x. [DOI] [PubMed] [Google Scholar]
- 44.Murray F, Kalla R, Jacobsen J, Gubler F. A role for HvGAMYB in anther development. Plant J. 2003;33:481–491. doi: 10.1046/j.1365-313x.2003.01641.x. [DOI] [PubMed] [Google Scholar]
- 45.Chen L, Nishizawa T, Higashitani A, Suge H, Wakeui H, Takeda K, et al. A variety of wheat tolerant to deep-seedling conditions: Elongation of the first internode depends on the response to gibberellin and potassium. Plant Cell Environ. 2001;24:469–476. [Google Scholar]
- 46.Gocal GF, Poole AT, Gubler F, Watts RJ, Blundell C, King RW. Long-day upregulation of a GAMYB gene during Lolium temulentum inflorescence formation. Plant Physiol. 1999;119:1271–1278. doi: 10.1104/pp.119.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gocal GF, Sheldon CC, Gubler F, Moritz T, Bagnall DJ, MacMillan CP, et al. GAMYB-like genes, flowering and gibberellin signaling in Arabidopsis. Plant Physiol. 2001;127:1682–1693. [PMC free article] [PubMed] [Google Scholar]
- 48.Weigel D, Nilsson O. A developmental switch sufficient for flower initiation in diverse plants. Nature. 1995;377:495–500. doi: 10.1038/377495a0. [DOI] [PubMed] [Google Scholar]
- 49.Blazquez MA, Soowal LN, Lee I, Weigel D. LEAFY expression and f lower initiation in Arabidopsis. Development. 1997;124:3835–3844. doi: 10.1242/dev.124.19.3835. [DOI] [PubMed] [Google Scholar]
- 50.Blazquez MA, Green R, Nilsson O, Sussman MR, Weigel D. Gibberellins promote flowering of arabidopsis by activating the LEAFY promoter. Plant Cell. 1998;10:791–800. doi: 10.1105/tpc.10.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blazquez MA, Weigel D. Integration of floral inductive signals in Arabidopsis. Nature. 2000;404:889–892. doi: 10.1038/35009125. [DOI] [PubMed] [Google Scholar]
- 52.Leng Q, Mercier RW, Yao W, Berkowitz GA. Cloning and first functional characterization of a plant cyclic nucleotide-gated cation channel. Plant Physiol. 1999;121:753–761. doi: 10.1104/pp.121.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maathuis FJ, Sanders D. Sodium uptake in Arabidopsis roots is regulated by cyclic nucleotides. Plant Physiol. 2001;127:1617–1625. [PMC free article] [PubMed] [Google Scholar]
- 54.Hepler PK, Vidali L, Cheung AY. Polarized cell growth in higher plants. Annu Rev Cell Dev Biol. 2001;17:159–187. doi: 10.1146/annurev.cellbio.17.1.159. [DOI] [PubMed] [Google Scholar]
- 55.Yoshiba Y, Nanjo T, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Stress-responsive and developmental regulation of Delta(1)-pyrroline-5-carboxylate synthetase 1 (P5CS1) gene expression in Arabidopsis thaliana. Biochem Biophys Res Commun. 1999;261:766–772. doi: 10.1006/bbrc.1999.1112. [DOI] [PubMed] [Google Scholar]
- 56.Honys D, Twell D. Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 2004;5:85. doi: 10.1186/gb-2004-5-11-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Verelst W, Saedler H, Munster T. MIKC* MADS-protein complexes bind motifs enriched in the proximal region of late pollen-specific Arabidopsis promoters. Plant Physiol. 2007;143:447–460. doi: 10.1104/pp.106.089805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Estruch JJ, Kadwell S, Merlin E, Crossland L. Cloning and characterization of a maize pollen-specific calcium-dependent calmodulin-independent protein kinase. Proc natl Acad Sci USA. 1994;91:8837–8841. doi: 10.1073/pnas.91.19.8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moutinho A, Trewavas AJ, Malho R. Relocation of a Ca2+-dependent protein kinase activity during pollen tube reorientation. Plant Cell. 1998;10:1499–1510. doi: 10.1105/tpc.10.9.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hiscock SJ, Doughty J, Dickinson HG. Synthesis and phosphorylation of pollen proteins during the pollen-stigma interaction in self-compatible Brassica napus L. and self-incompatible Brassica oleraceae L. Sex Plant Reprod. 1995;8:345–353. [Google Scholar]
- 61.Xu J, Brearley CA, Lin WH, Wang Y, Ye R, Mueller-Roeber B, et al. A role of Arabidopsis inositol polyphosphate kinase, AtIPK2alpha, in pollen germination and root growth. Plant Physiol. 2005;137:94–103. doi: 10.1104/pp.104.045427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sousa E, Kost B, Malho R. Arabidopsis phosphatidylinositol- 4-monophosphate 5-kinase 4 regulates pollen tube growth and polarity by modulating membrane recycling. Plant Cell. 2008;20:3050–3064. doi: 10.1105/tpc.108.058826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Podell S, Gribskov M. Predicting N-terminal myristoylation sites in plant proteins. BMC Genomics. 2004;5:37. doi: 10.1186/1471-2164-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gupta R, Ting JT, Sokolov LN, Johnson SA, Luan S. A tumor suppressor homolog, AtPTEN1, is essential for pollen development in Arabidopsis. Plant Cell. 2002;14:2495–2507. doi: 10.1105/tpc.005702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prado AM, Porterfield DM, Feijo JA. Nitric oxide is involved in growth regulation and re-orientation of pollen tubes. Development. 2004;131:2707–2714. doi: 10.1242/dev.01153. [DOI] [PubMed] [Google Scholar]
- 66.Salmi ML, Morris KE, Roux SJ, Porterfield DM. Nitric oxide and cGMP signaling in calcium-dependent development of cell polarity in Ceratopteris richardii. Plant Physiol. 2007;144:94–104. doi: 10.1104/pp.107.096131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh DP, Jermakow AM, Swain SM. Gibberellins are required for seed development and pollen tube growth in Arabidopsis. Plant Cell. 2002;14:3133–3147. doi: 10.1105/tpc.003046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Swain SM, Muller AJ, Singh DP. The gar2 and rga alleles increase the growth of gibberellin-deficient pollen tubes in Arabidopsis. Plant Physiol. 2004;134:694–705. doi: 10.1104/pp.103.031666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chhun T, Aya K, Asano K, Yamamoto E, Morinaka Y, Watanabe M, et al. Gibberellin regulates pollen viability and pollen tube growth in rice. Plant Cell. 2007;19:3876–3888. doi: 10.1105/tpc.107.054759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.