Abstract
Protein quality control plays an important role in the photosynthetic apparatus because its components receive excess light energy and are susceptible to photooxidative damage. In chloroplasts, photodamage is targeted to the D1 protein of Photosystem II (PSII). The coordinated PSII repair cycle (PSII disassembly, D1 degradation and synthesis, and PSII reassembly) is necessary to mitigate photoinhibition. A thylakoid protease FtsH, which is formed predominantly as a heteromeric complex with two isoforms of FtsH2 and FtsH5 in Arabidopsis, is the major protease involved in PSII repair. A mutant lacking FtsH2 (termed var2) shows compromised D1 degradation. Furthermore, var2 accumulates high levels of chloroplastic reactive oxygen species (cpROS), reflecting photooxidative stress without functional PSII repair. To examine if the cpROS produced in var2 are connected to a ROS signaling pathway mediated by plasma membrane NADPH oxidase (encoded by AtRbohD or AtRbohF), we generated mutants in which either Rboh gene was inactivated under var2 background. Lack of NADPH oxidases had little or no impact on cpROS accumulation. It seems unlikely that cpROS in var2 activate plasma membrane NADPH oxidases to enhance ROS production and the signaling pathway. Mutants that are defective in PSII repair might be valuable for investigating cpROS and their physiological roles.
Key words: reactive oxygen species (ROS), photosystem II repair cycle, chloroplast, FtsH, NADPH oxidase, D1 protein, protein turnover
Photosynthetic apparatus components receive excess light energy that can ultimately engender photoinhibition.1,2 Chloroplasts are therefore equipped with molecular systems to minimize accumulation of photodamaged proteins.3 Photosystem II, a large pigment-protein complex located in the thylakoid membrane, transfers electrons to plastoquinone and drives oxidation of water molecules using light energy.4,5 Because PSII is an initial and rate-limiting step of electron flow, photosynthetic organisms have evolved a unique mechanism of protein quality control (PSII repair cycle), in which the damage is centralized into the reaction center D1 protein, and in which PSII is recycled efficiently.6,7 Several lines of evidence from genetic and biochemical studies indicate that a prokaryotic ATP-dependent metalloprotease FtsH plays a critical role in D1 turnover of the PSII repair.8–12
In chloroplasts, FtsH forms a heteromeric complex with two major isoforms.13,14 Mutants lacking either major isoform (var2 lacking FtsH2 or var1 lacking FtsH5) show leaf variegation forming white sectors that contain cells with aberrant plastids.8,9,11,15 The variegated phenotype implies that FtsH is involved not only in D1 degradation but also in thylakoid development.16,17 We conducted in vivo D1 degradation assays using “non-variegated” var1 and var2 mutants (owing to a trans-acting suppressor mutation fug1).18,19 Results showed that both D1 degradation and PSII electron transport rates were impaired in these non-variegated lines.19 Collectively, our results corroborate the important role of chloroplast FtsH in the PSII repair cycle. We also infer that the variegation phenotype in var mutants is separable from the defect in the PSII repair.
Two important observations concomitant with impaired D1 degradation were made in our recent study.19 One is the accumulation of PSII partial complexes in var2. Two-dimensional blue-native SDS-PAGE analysis demonstrated that thylakoid-membrane fractions from var2 chloroplasts contained fewer PSII supercomplexes (representing functional PSII) and more partial PSII complexes (representing disassembled intermediates in the PSII repair cycle). These results indicate, although indirectly, that the impaired D1 degradation affects the disassembly/ reassembly step of the PSII repair cycle. The other important observation is the accumulation of reactive oxygen species (ROS), such as superoxide radical (O2−) and hydrogen peroxide (H2O2) in var2. Results of NBT staining indicated that O2− is specially localized in chloroplasts of var2 green sectors in a light-dependent manner. No NBT staining was detected in wild type under identical conditions. Similarly, DAB staining indicated that H2O2 is detectable in var2 green sectors. High ROS in var2 therefore demonstrates that chloroplasts suffer from photooxidative stress without a functional PSII repair system. Where these ROS are generated within chloroplasts remains unclear. We raise one possibility: that PSII partial complexes in var2 contribute to ROS production because they potentially accumulate excitation energy that might not be used for water oxidation.
A considerable amount of chloroplastic O2− in var2 might be converted rapidly to H2O2, which can then be exported to cytosol or to other organelles for detoxification. Simultaneously, H2O2 in cytosol might act as a signaling molecule and consequently affect responses to environmental stress.20 We raised one possibility: cpROS in var2 are influenced by an apoplastic oxidative burst that is mediated by plasma membrane-bound NADPH oxidases and which further activates downstream signaling cascades. For example, cpROS produced in guard cells of ozonetreated Arabidopsis were shown to activate certain NAPDH oxidases through the action of heterotrimeric G protein signaling. 21 Although Gα subunit activated by cpROS is primarily involved in oxidative bursts, Gβγ complexes appear to act on further production of cpROS.21 These observations led us to examine whether high cpROS in var2 are regulated by NADPH oxidases.
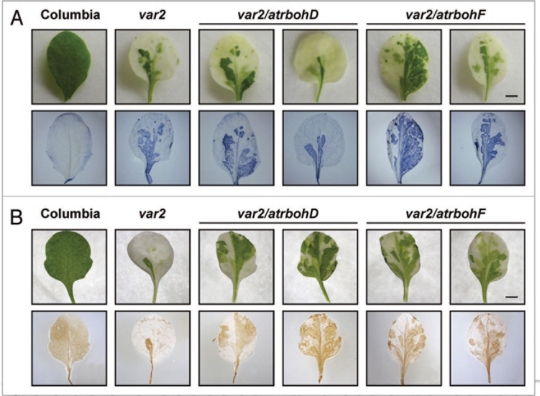
Ten genes for NADPH oxidases (AtRbohA to AtRbohJ) are reported in Arabidopsis.22 Among these, AtRbohD and AtRbohF are expressed in mesophylls and are involved in cpROS signaling in guard cells.22–24 To investigate the effect of these NADPH oxidases on cpROS accumulation in var2, we generated double mutants (var2/atrbohD and var2/atrbohF). The degrees of leaf variegation were similar in var2 and the double mutants. Single atrbohD and atrbohF mutants did not accumulate detectable ROS (not shown). We observed strong signals in var2/ atrbohD and var2/atrbohF double mutants both in NBT and DAB stains (Fig. 1). Overall, results showed no significant difference in the accumulation of cpROS between var2 and the double mutants. Furthermore, results of our microarray analyses demonstrated that expression levels of AtRbohD and AtrbohF are similar between var2 green sectors and wild type (unpublished data). Taken together, these results suggest that no apparent NADPH oxidase activities in plasma membranes contribute to cpROS detected in var2.
Figure 1.
ROS accumulation in var2 and var2/atrboh mutants. (A) In situ detection of superoxide by staining with NBT (blue, bottom panels) in four-week-old wild type (Columbia), var2, var2/atrbohD, var2/atrbohF leaves. Bar = 1 mm. (B) In situ detection of hydrogen peroxide by DA B staining (dark brown, bottom panels) in four-week-old wild type (Columbia), var2, var2/rbohD, var2/rbohF leaves. Bar = 1 mm.
Actually, ROS transiently generated by apoplastic NADPH oxidases are known to regulate a cell-death signaling pathway such as a hypersensitive response against pathogen infection.25 Based on results of our current genetic and microarray analyses, we reason that, in var2, constitutive cpROS do not activate the ROS-mediated signaling pathway. Nevertheless, involvement of cpROS in signaling cascades has been suggested in other experimental systems. The mutants described in this report might be valuable for use in future studies.
Acknowledgements
We thank Dr. Jeffery L. Dangl for providing atrboh mutants. This work was supported by a Grant-in-Aid for Scientific Research from MEXT (16085207), the Asahi Glass Foundation, and the Oohara Foundation. E.M. is supported by a postdoctoral fellowship from JSPS.
Abbreviations
- cpROS
chloroplastic reactive oxygen species
- DAB
3,3′-deaminobenzidine
- NBT
nitroblue tetrazolium
- PSII
photosystem II
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/10604
References
- 1.Barber J, Andersson B. Too much of a good thing: light can be bad for photosynthesis. Trends Biochem Sci. 1992;17:61–66. doi: 10.1016/0968-0004(92)90503-2. [DOI] [PubMed] [Google Scholar]
- 2.Aro EM, Virgin I, Andersson B. Photoinhibition of Photosystem II. Inactivation, protein damage and turnover. Biochim Biophys Acta. 1993;1143:113–134. doi: 10.1016/0005-2728(93)90134-2. [DOI] [PubMed] [Google Scholar]
- 3.Niyogi KK. Photoprotection revisited: genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:333–559. doi: 10.1146/annurev.arplant.50.1.333. [DOI] [PubMed] [Google Scholar]
- 4.Barber J. Photosystem II: an enzyme of global significance. Biochem Soc Trans. 2006;34:619–631. doi: 10.1042/BST0340619. [DOI] [PubMed] [Google Scholar]
- 5.Nelson N, Yocum CF. Structure and function of photosystems I and II. Annu Rev Plant Biol. 2006;57:521–565. doi: 10.1146/annurev.arplant.57.032905.105350. [DOI] [PubMed] [Google Scholar]
- 6.Edelman M, Mattoo AK. D1-protein dynamics in photosystem II: the lingering enigma. Photosynth Res. 2008;98:609–620. doi: 10.1007/s11120-008-9342-x. [DOI] [PubMed] [Google Scholar]
- 7.Baena-González E, Aro EM. Biogenesis, assembly and turnover of photosystem II units. Phil Trans R Soc Lond B Biol Sci. 2002;357:1451–1460. doi: 10.1098/rstb.2002.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen M, Choi Y, Voytas DF, Rodermel S. Mutations in the Arabidopsis VAR2 locus cause leaf variegation due to the loss of a chloroplast FtsH protease. Plant J. 2000;22:303–313. doi: 10.1046/j.1365-313x.2000.00738.x. [DOI] [PubMed] [Google Scholar]
- 9.Takechi K, Sodmergen, Murata M, Motoyoshi F, Sakamoto W. The YELLOW VARIEGATED (VAR2) locus encodes a homologue of FtsH, an ATPdependent protease in Arabidopsis. Plant Cell Physiol. 2000;41:1334–1346. doi: 10.1093/pcp/pcd067. [DOI] [PubMed] [Google Scholar]
- 10.Bailey S, Thompson E, Nixon PJ, Horton P, Mullineaux CW, Robinson C, Mann NH. A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J Biol Chem. 2002;277:2006–2011. doi: 10.1074/jbc.M105878200. [DOI] [PubMed] [Google Scholar]
- 11.Sakamoto W, Tamura T, Hanba Tomita Y, Murata M. The VAR1 locus of Arabidopsis encodes a chloroplastic FtsH and is responsible for leaf variegation in the mutant alleles. Genes Cells. 2002;7:769–780. doi: 10.1046/j.1365-2443.2002.00558.x. [DOI] [PubMed] [Google Scholar]
- 12.Nixon PJ, Barker M, Boehm M, de Vries R, Komenda J. FtsH-mediated repair of the photosystem II complex in response to light stress. J Exp Bot. 2005;56:357–363. doi: 10.1093/jxb/eri021. [DOI] [PubMed] [Google Scholar]
- 13.Sakamoto W, Zaltsman A, Adam Z, Takahashi Y. Coordinated regulation and complex formation of YELLOW VARIEGATED1 and YELLOW VARIEGATED2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell. 2003;15:2843–2855. doi: 10.1105/tpc.017319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu F, Park S, Rodermel SR. The Arabidopsis FtsH metalloprotease gene family: interchangeability of subunits in chloroplast oligomeric complexes. Plant J. 2004;37:864–876. doi: 10.1111/j.1365-313x.2003.02014.x. [DOI] [PubMed] [Google Scholar]
- 15.Kato Y, Miura E, Matsushima R, Sakamoto W. White leaf sectors in yellow variegated2 are formed by viable cells with undifferentiated plastids. Plant Physiol. 2007;144:952–960. doi: 10.1104/pp.107.099002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaltsman A, Feder A, Adam Z. Developmental and light effects on the accumulation of FtsH protease in Arabidopsis chloroplasts—implications for thylakoid formation and photosystem II maintenance. Plant J. 2005;42:609–617. doi: 10.1111/j.1365-313X.2005.02401.x. [DOI] [PubMed] [Google Scholar]
- 17.Sakamoto W, Uno Y, Zhang Q, Miura E, Kato Y, Sodmergen Arrested differentiation of proplastids into chloroplasts in variegated leaves characterized by plastid ultrastructure and nucleoid morphology. Plant Cell Physiol. 2009;50:2069–2083. doi: 10.1093/pcp/pcp127. [DOI] [PubMed] [Google Scholar]
- 18.Miura E, Kato Y, Matsushima R, Albrecht V, Laalami S, Sakamoto W. The balance between protein synthesis and degradation in chloroplasts determines leaf variegation in Arabidopsis yellow variegated mutants. Plant Cell. 2007;19:1313–1328. doi: 10.1105/tpc.106.049270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato Y, Miura E, Ido K, Ifuku K, Sakamoto W. The variegated mutants lacking chloroplastic FtsHs are defective in D1 degradation and accumulate reactive oxygen species. Plant Physiol. 2009;151:1790–1801. doi: 10.1104/pp.109.146589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 21.Joo JH, Wang S, Chen JG, Jones AM, Fedoroff NV. Different signaling and cell death roles of heterotrimeric G protein alpha and beta subunits in the Arabidopsis oxidative stress response to ozone. Plant Cell. 2005;17:957–970. doi: 10.1105/tpc.104.029603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 23.Torres MA, Onouchi H, Hamada S, Machida C, Hammond Kosack KE, Jones JD. Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91phox) Plant J. 1998;14:365–370. doi: 10.1046/j.1365-313x.1998.00136.x. [DOI] [PubMed] [Google Scholar]
- 24.Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, et al. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 2003;22:2623–2633. doi: 10.1093/emboj/cdg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torres MA, Dangl JL, Jones JD. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci USA. 2002;99:517–522. doi: 10.1073/pnas.012452499. [DOI] [PMC free article] [PubMed] [Google Scholar]