Abstract
Parasitic angiosperms are an ecologically and economically important group of plants. However our understanding of the basis for host specificity in these plants is embryonic. Recently we investigated host specificity in the parasitic angiosperm Orobanche minor, and demonstrated that this host generalist parasite comprises genetically defined races that are physiologically adapted to specific hosts. Populations occurring naturally on red clover (Trifolium pratense) and sea carrot (Daucus carota subsp. gummifer) respectively, showed distinct patterns of host specificity at various developmental stages, and a higher fitness on their natural hosts, suggesting these races are locally adapted. Here we discuss the implications of our findings from a broader perspective. We suggest that differences in signal responsiveness and perception by the parasite, as well as qualitative differences in signal production by the host, may elicit host specificity in this parasitic plant. Together with our earlier demonstration that these O. minor races are genetically distinct based on molecular markers, our recent data provide a snapshot of speciation in action, driven by host specificity. Indeed, host specificity may be an underestimated catalyst for speciation in parasitic plants generally. We propose that identifying host specific races using physiological techniques will complement conventional molecular marker-based approaches to provide a framework for delineating evolutionary relationships among cryptic host-specific parasitic plants.
Key words: host specificity, parasitic plant, broomrape, orobanche, speciation
Introduction
Parasitic angiosperms have evolved at least 11 times and occur in all major biomes, from arctic islands to tropical forests.1,2 In natural ecosystems, they generate profound effects on plant community structure,1,3 whereas in agroecosystems weedy species attack commercial crops and cause crop losses that amount to billions in USD annually.4 In spite of their importance from both ecological and agronomic perspectives, our understanding of parasitic angiosperms and their interactions with host plants still lags behind that of other plant-pathogen associations.3 Previously we identified patterns of host-specific genetic divergence among the morphologically cryptic taxa O. minor var. minor infecting red clovers (Trifolium pratense) and O. minor subsp. maritima infecting sea carrots (Daucus carota subsp. gummifer) respectively in northern Europe.5 Recently, to explore the potential for host specificity to drive this genetic divergence, we used reciprocal-infection experiments to quantify host specificity in terms of (1) relative parasite success during early development; (2) fitness in terms of biomass, and (3) anatomy of the host-parasite interface. The results from this investigation demonstrate that genetic races occurring naturally on red clover and sea carrots show distinct developmental pathways on their natural and alien hosts.6 Collectively these data suggest that morphologically cryptic races of O. minor may be physiologically adapted to their local hosts, which offers a template for genetic divergence and speciation.
Host-Parasite Communication by Chemical Signaling
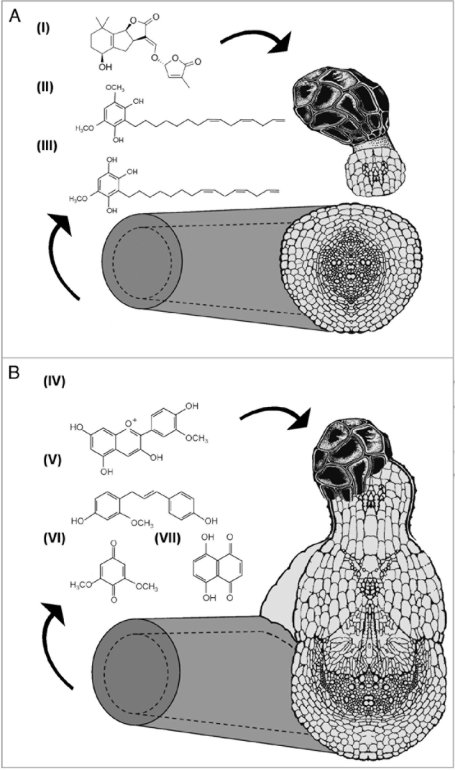
Many interactions between plants and other organisms are mediated by signal molecules that initiate developmental and physiological programs.7 Parasitic angiosperms are a key example of such interactions, in which the development of the parasite is initiated by host plant signals (Fig. 1). The isolation of these host plant signals from root exudates has been hampered by their low abundance and instability.8 Strigolactones, a group of sesquiterpene lactones, are well known germination stimulants for Orobanche and Striga, and have also been identified to be the signaling molecules that trigger hyphal branching in arbuscular mycorrhizal (AM) fungi.9 Recent research has suggested that strigolactones also pay a role in the regulation of shoot branching in plants,10,11 in addition to their role in rhizosphere communication, and may represent a novel class of plant hormones or their biosynthetic precursors.10 Variation in germination stimulant production by different host species influences the host specificity of the parasite.12 For example, different hosts induce different rates of seed germination among various Orobanche spp.,13 suggesting host specificity in this genus is elicited at this early developmental stage. We cultivated O. minor subsp. maritima and O. minor var. minor on their natural hosts reciprocally in Petri dish bioassays to observe the infection process. Orobanche minor subsp. maritima showed a higher rate of germination on sea carrot than on red clover, suggesting it is physiologically adapted to its natural host. Red clovers may therefore produce less of the stimulants to which Orobanche minor subsp. maritima seeds are responsive, or there may be qualitative differences in the stimulants produced by these host species. Interestingly, O. minor var. minor showed higher germination on sea carrot than on its natural red clover host, which may be due to the dense root architecture of sea carrot producing a greater exudate output, suggesting quantitative differences in stimulant production may also be a determining factor; later stages of parasite development and parasite biomass were however better on red clover for this race.
Figure 1.
Host specificity is elicited at the molecular level by variation in host production and parasite perception of signal molecules, including the following germination factors (A): (i) strigol; (ii) an antioxidant enhancer of SXSg, (iii) SXSg, and haustoria inducing factors (B): (iv) peonidin, (v) xenognosin A; (vi) DMBQ; (vii) 5,7-dihydroxynapthoquinone. Figure adapted from Yoder JI. Current Opinion in Plant Biology 2004; 4:359–65.
Host signals also govern later stages of the host-parasite interaction.14 The haustorium is a defining phenotypic feature of all parasitic angiosperms,12,15 and provides a physiological bridge between the two plants,16,17 through which hormonal interactions, viruses, proteins and mRNA transcripts may be transported.3,18 Chemical signals initiating haustorial development are distinct from the signals which induce germination.14 Haustorium initiation is triggered by a suite of specific exogenous molecules in the rhizosphere, called haustoria-inducing factors (HIFs), which act as signals for undifferentiated parenchymatous cells17 (Fig. 1). Qualitative differences exist in HIFs exuded by even closely related potential host species,14 suggesting that these signals are important determinants of host specificity. In our experiments, Orobanche minor subsp. maritima seedlings showed no evidence of attachment or haustoria formation on the roots of its alien host red clover, suggesting the HIF to which this race is responsive to was not exuded by this species. Conversely O. minor var. minor showed compatible development on sea carrot as well as red clover. This suggests that both differences in parasite responsiveness to host signals, and differences in signal production by the host are important elicitors of host specificity and may define host range in parasitic plants.
Host Specificity and Varietal Selection
The evolutionary shift to parasitism in flowering plants has been associated with the degeneration of morphological features traditionally used in plant classification, making systematic studies in these plants extremely challenging. Quantifying host specificity using physiological parameters combined with sequence data, may provide a robust platform for delineating morphologically cryptic host-specific races in taxonomically difficult groups such as Orobanche where traditional molecular markers alone, such as rbcL and ITS, fail to resolve closely related taxa. This is analogous to other taxonomically challenging groups such as microorganisms, in which biochemical characteristics are used alongside sequence data to delineate species relationships. Although host-driven speciation processes have been well documented in phytophagus insects,19 the possibility that host-driven speciation also occurs in parasitic plants remains relatively uncharted. However a similar pattern of host-driven speciation in parasitic plants is now becoming stronger, thanks to case studies of host specificity in, for example hemiparasite mistletoes,20–23 and holoparasitic species Cytinus spp.24,25 and Orobanche spp.5,6,26 Given the predominance of host specificity among parasitic angiosperms, and our rudimentary understanding of the evolution of host-parasite interactions, it is likely that this process may have been an underestimated driving force in the evolution of this important group of plants.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/10660
References
- 1.Press MC, Phoenix GK. Impacts of parasitic plants on natural communities. New Phytol. 2005;166:737–751. doi: 10.1111/j.1469-8137.2005.01358.x. [DOI] [PubMed] [Google Scholar]
- 2.Barkman TJ, McNeal JR, Lim S-H, Coat G, Croom HB, Young ND, et al. Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evolutionary Biology. 2007;7:248. doi: 10.1186/1471-2148-7-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Press MC, Graves JD, Stewart GR. Physiology of the interaction of angiosperm parasites and their higher plant hosts. Plant Cell Environ. 1990;13:91–104. [Google Scholar]
- 4.Parker C. Observations on the current status of Orobanche and Striga problems worldwide. Pest Manag Sci. 2009;65:453–459. doi: 10.1002/ps.1713. [DOI] [PubMed] [Google Scholar]
- 5.Thorogood CJ, Rumsey FJ, Harris SA, Hiscock SJ. Host-driven divergence in the parasitic plant Orobanche minor Sm. (Orobanchaceae) Mol Ecol. 2008;17:4289–4303. doi: 10.1111/j.1365-294x.2008.03915.x. [DOI] [PubMed] [Google Scholar]
- 6.Thorogood CJ, Rumsey FJ, Hiscock SJ. Host-specific races in the holoparasitic angiosperm Orobanche minor: implications for speciation in parasitic plants. Ann Bot. 2009a;103:1005–1014. doi: 10.1093/aob/mcp034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar SP. Signaling in plant-microbe interactions. Science. 1997;276:726–733. doi: 10.1126/science.276.5313.726. [DOI] [PubMed] [Google Scholar]
- 8.Rani K, Zwanenburg B, Sugimoto Y, Yoneyama K, Bouwmeester HJ. Biosynthetic considerations could assist the structure elucidation of host plant produced rhizosphere signalling compounds (strigolactones) for arbuscular mycorrhizal fungi and parasitic plants. Plant Physiol Biochem. 2008;46:617–626. doi: 10.1016/j.plaphy.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 9.Akiyama K, Matsuzaki K-I, Hayashi H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature. 2005;435:824–827. doi: 10.1038/nature03608. [DOI] [PubMed] [Google Scholar]
- 10.Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pages V, Dun EA, Pillot J-P, et al. Strigolactone inhibition of shoot branching. Nature. 2008;455:189–195. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- 11.Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature. 2008;455:195–200. doi: 10.1038/nature07272. [DOI] [PubMed] [Google Scholar]
- 12.Estabrook EM, Yoder JI. Plant-plant communications: rhizosphere signaling between parasitic angiosperms and their hosts. Plant Physiol. 1998;116:1–7. [Google Scholar]
- 13.Fernández-Aparicio M, Flores F, Rubiales D. Recognition of root exudates by seeds of broomrape (Orobanche and Phelipanche) species. Ann Bot. 2009;103:423–431. doi: 10.1093/aob/mcn236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoder JI. A species-specific recognition system directs haustorium development in the parasitic plant Triphysaria (Scrophulariaceae) Planta. 1997;202:407–413. doi: 10.1007/s004250050144. [DOI] [PubMed] [Google Scholar]
- 15.Hibberd JM, Jeschke WD. Solute flux into parasitic plants. J Exper Bot. 2001;52:2043–2049. doi: 10.1093/jexbot/52.363.2043. [DOI] [PubMed] [Google Scholar]
- 16.Kuijt J. The biology of parasitic flowering plants. Berkeley: University of California Press,; 1969. [Google Scholar]
- 17.Riopel JL, Timko MP. In: In Parasitic plants. Press MC, Graves JD, editors. London: Chapman and Hall,; 1995. pp. 39–79. [Google Scholar]
- 18.David-Schwartz R, Runo S, Townsley B, Machuka J, Sinha N. Long-distance transport of mRNA via parenchyma cells and phloem across the host-parasite junction in Cuscuta. New Phytol. 2008;179:1133–1141. doi: 10.1111/j.1469-8137.2008.02540.x. [DOI] [PubMed] [Google Scholar]
- 19.Funk DJ, Futuyma DJ, Ortí G, Meyer A. A history of host associations and evolutionary diversification for Ophraella (Coleoptera: Chrysomelidae): new evidence from mitochondrial DNA. Evolution. 1995;49:1008–1017. doi: 10.1111/j.1558-5646.1995.tb02335.x. [DOI] [PubMed] [Google Scholar]
- 20.Nickrent DL, Stell AL. Biochemical systematics of the Arceuthobium campylopodum complex (dwarf mistletoes, Viscaceae) II. Electrophoretic evidence for genetic differentiation in two host races of hemlock dwarf mistletoe (A. tsugense) Biochem Syst Ecol. 1990;18:267–280. [Google Scholar]
- 21.Zuber D, Widmer A. Genetic evidence for host specificity in the hemi-parasitic Viscum album L. (Viscaceae) Mol Ecol. 2000;9:1069–1073. doi: 10.1046/j.1365-294x.2000.00963.x. [DOI] [PubMed] [Google Scholar]
- 22.Jerome CA, Ford BA. The discovery of three genetic races of the dwarf mistletoe Arceuthobium americanum (Viscaceae) provides insight into the evolution of parasitic angiosperms. Mol Ecol. 2002a;11:387–405. doi: 10.1046/j.0962-1083.2002.01463.x. [DOI] [PubMed] [Google Scholar]
- 23.Jerome CA, Ford BA. Comparative population structure and genetic diversity of Arceuthobium americanum (Viscaceae) and its Pinus host species: insight into host-parasite evolution in parasitic angiosperms. Mol Ecol. 2002b;11:407–420. doi: 10.1046/j.0962-1083.2002.01462.x. [DOI] [PubMed] [Google Scholar]
- 24.Thorogood CJ, Hiscock SJ. Host Specificity in the Parasitic Plant Cytinus hypocistis. Res Lett Ecol. 2007:84234. [Google Scholar]
- 25.deVega C, Berjano R, Arista M, Ortiz PL, Talavera S, Stuessy TF. Genetic races associated with the genera and sections of host species in the holoparasitic plant Cytinus (Cytinaceae) in the western Mediterranean basin. New Phytol. 2008;178:875–887. doi: 10.1111/j.1469-8137.2008.02423.x. [DOI] [PubMed] [Google Scholar]
- 26.Thorogood CJ, Rumsey FJ, Harris SA, Hiscock SJ. Gene flow between alien and native races of the holoparasitic angiosperm Orobanche minor (Orobanchaceae) Plant Syst Evol. 2009b;282:31–42. [Google Scholar]