Abstract
Adventitious roots are distinct from primary and lateral roots in that adventitious roots develop ectopically from aboveground organs. Whole-genome microarrays in poplar provided the first glimpse into the gene networks that are remodeled in cells prior to the development of adventitious roots. In the first 24 hr after removal of stem cuttings, over one-half of the transcripts encoded in the genome showed evidence of differential abundance in the cells that will eventually give rise to adventitious roots. Major processes that were regulated appear related to physiological adaptation of the cutting to acute loss of water and nutrients as well as hormone signaling. Comparative transcriptome analysis of genotypes that differ in their competence to form adventitious roots may be a generally useful strategy to identify genes that regulate adventitious rooting efficiency.
Key words: adventitious root, populus, transcriptome, vegetative propagation
Roots can be divided into several categories depending on their origin and developmental history. Primary roots originate from the embryo in seeds, while lateral roots develop as branches of primary roots. Adventitious roots are distinct in that they develop ectopically from aboveground organs such as stems, often in situations where there is environmentally imposed stress. Research on adventitious rooting has a long history in woody perennial species that are clonally propagated for forestry and horticulture applications.1–3 Within the genus Populus there is evidence that genetic competence to form adventitious roots is ecologically and evolutionarily significant, perhaps as an alternative or supplement to seed propagation in ecosystems where soil disturbance is frequent.4,5 Identifying the genes that distinguish easy-to-root genotypes from difficult-to-root genotypes therefore has many practical applications, and also promises new insights into how natural selection has sculpted variation in adventitious rooting. Some recent discoveries in Populus suggest it is feasible to identify genes that regulate adventitious rooting and also generate new insights into the genes and pathways that are remodeled in cells prior to adventitious root development.6
Microarrays derived from the reference genome sequence of Populus trichocarpa7 are useful to glimpse the genes and pathways associated with adventitious rooting. Adventitious root primordia primarily arise from ray cells, pericycle or callus formed at the base of stem cuttings.1 In the initial 48 hr after removal of a stem cutting from a donor plant, prior to development of root primordia, the base of the cutting shows altered endogenous hormone pools—while auxin and ethylene levels are increased, cytokinin content is reduced.8–10 Whole-transcriptome monitoring in the base of stem cuttings revealed significant shifts during this same time period.6 By contrasting expression data from discrete sampling times (0–6 hr, 6–24 hr and 24–48 hr) a total of 15,134 transcripts (representing 27% of the predicted gene models) were differentially regulated between 0 and 6 hr; 20,111 (36%) between 6 and 24 hr, and 2,474 (4%) between 24 and 48 hr.
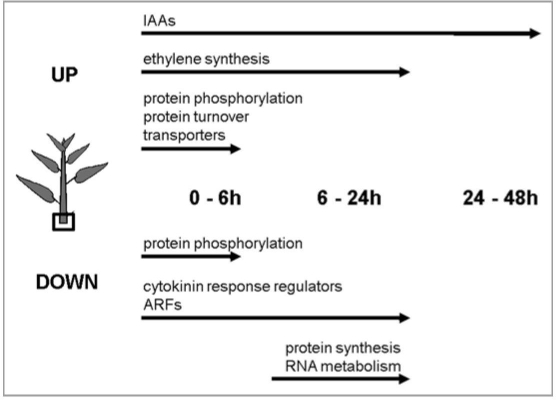
Individual genes annotated with roles in ethylene biosynthesis, auxin signaling and cytokinin signaling showed differential regulation during the initial 24 hr period6 (Fig. 1) likely related to shifts in abundance of these hormones observed in previous studies.8–10 To evaluate the processes differentially regulated during this time, genes were grouped based on Gene Ontology terms using ErmineJ software (http://bioinformatics. ubc.ca/ermineJ/). A total of 115, 46 and 15 gene sets showed enrichment in the 0–6 hr, 6–24 hr and 24–48 hr contrasts, respectively. Individual genes with contrasts significant at a false discovery rate11 (FDR) of <5% were included in the gene-set enrichment analysis, whereas the gene-sets themselves were retained if they were significant at FDR <10%.
Figure 1.
Gene families and pathways differentially regulated at the level of transcript abundance in the base of stem cuttings after removal from the donor plant.
During the initial 6 hr following excision, the three most significantly regulated gene sets were amino acid ligase activity, substrate-specific transport activity and protein phosphorylation activity (corrected p-values of 1.97E-07, 1.17E-07 and 1.36 E-07, respectively; Fig. 1). Overall, most of the genes in the amino acid ligation gene-set were upregulated at 6 hr and included components of the 26S proteasome protein degradation pathway such as ubiquitin conjugating enzymes (E2) and ubiquitin ligases (E3). In this gene-set the most significantly upregulated was a close relative of Arabidopsis U-box E3 ubiquitin ligase PUB22, a protein involved in drought signaling.12 In the substrate-specific transporter activity gene-set, the most significantly regulated genes were related to potassium, sodium, calcium, sugar, amino acid and water transport. Although most of the genes in this gene-set were upregulated, the most significantly altered transcript was downregulated and is closely related to ATHKT1, a sodium and potassium transporter involved in salt stress tolerance. 13 The most significantly regulated transcripts in the protein phosphorylation activity gene-set encode mitogen-activated protein kinases (MAPKs) and leucine rich repeat kinases. Half of these genes (5 out of 10) were upregulated, while the other half were downregulated. The most significantly upregulated gene in this category encodes a protein similar to MITOGEN-ACTIVATED PROTEIN KINASE-3 (MAPK3) from Arabidopsis that phosphorylates the ETHYLENE-INSENSITIVE-3 (EIN3) transcription factor in the presence of ethylene.14 These results suggest that during the initial 6 hr after excision transcriptional regulation of post-translational regulators may be important for the (rootless) base of the cutting to adapt to loss of water and nutrients delivered from the root system.
In the 6 to 24 hr interval, gene-sets for structural constituents of ribosomes were the two most significantly enriched (corrected p-values of 3.24E-08 and 1.24E-06), followed by ATP-dependent RNA helicases (corrected p-value of 5.41E-06; Fig. 1). In all of these gene-sets, the prevailing pattern was for reduced abundance at 24 hr compared to 6 hr. In the structural ribosome gene-sets the ten most significantly altered genes encoded for constituents of ribosomes with the most significantly reduced transcript related to the 60S ribosomal protein L14. In the ATP-dependent RNA helicase activity gene-set, all top ten transcripts encode DEAD/DEAH box (Asp-Glu-Ala-Asp/His) helicases that unwind structured double-stranded RNA molecules or RNA-protein complexes.15,16 The overall pattern of regulation may reflect high demand on translation machinery early after shoot excision that returns to steady-state at 24 hr. Transcriptome shifts between 24 hr and 48 hr were comparatively modest relative to the earlier contrasts.
Whereas the patterns of transcript abundance shifts are complex, comparison of genotypes that differ in their competence to form adventitious roots should aid in identifying the genes that regulate adventitious rooting efficiency. This strategy was used to suggest a role for a transcription factor in the cytokinin signaling pathway (the type-B response regulator PtRR13)6 in the competence of stem bases to form adventitious roots.
Acknowledgements
Supported by the Department of Energy, Office of Science, Office of Biological and Environmental Research (DE-AC05-00OR22725).
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/10705
References
- 1.Lovel PH, White J. Anatomical changes during adventitious root formation. In: Jackson MB, editor. New root formation in plants and cuttings. Martinus Nijhoff Publishers; 1986. pp. 111–140. [Google Scholar]
- 2.Stelzer HE, Goldfarb B. Implementing clonal forestry in the southeastern United States: SRIEG satellite workshop summary remarks. Can J For Res. 1997;27:442–446. [Google Scholar]
- 3.Davis JM, Becwar MR. Developments in tree cloning. Developments in Fibres and Fibre Treatment Series, PIRA International. 2007:69. [Google Scholar]
- 4.Dech JP, Maun MA. Adventitious root production and plastic resource allocation to biomass determine burial tolerance in woody plants from central Canadian coastal dunes. Ann Bot. 2006;95:1095–1105. doi: 10.1093/aob/mcl196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rood SB, Kalischuk AR, Polzin ML, Braatne JH. Branch propagation, not cladoptosis, permits dispersive, clonal reproduction of riparian cottonwoods. For Ecol Man. 2003;186:227–242. [Google Scholar]
- 6.Ramirez-Carvajal GA, Morse AM, Dervinis C, Davis JM. The cytokinin type-B response regulator PtRR13 is a negative regulator of adventitious root development in Populus. Plant Physiol. 2009;150:759–771. doi: 10.1104/pp.109.137505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuskan GA, DiFazio SP, Hellsten U, Jansson S, Rombauts S, Putnam N, et al. The genome of western black cottonwood, Populus trichocarpa. Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 8.Maldiney R, Pelese F, Pilate G, Sotta B, Sossountzov L, Miginiac E. Endogenous levels of abscisic-acid, indole-3-acetic-acid, zeatin and zeatin-riboside during the course of adventitious root-formation in cuttings of Craigella and Craigella lateral suppressor tomatoes. Physiol Plant. 1986;68:426–430. [Google Scholar]
- 9.de Klerk GJ, Keppel M, Terbrugge J, Meekes H. Timing of the phases in adventitious root-formation in apple microcuttings. J Exp Bot. 1995;46:965–972. [Google Scholar]
- 10.de Klerk G-J, Van Der Krieken W, De Jong JC. The formation of adventitious roots: New concepts, new possibilities. In Vitro Cell Dev Biol Plant. 1999;35:189–199. [Google Scholar]
- 11.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho SK, Ryu MY, Song C, Kwak JM, Kim WT. Arabidopsis PUB22 and PUB23 are homologous U-box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell. 2008;20:1899–1914. doi: 10.1105/tpc.108.060699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rus A, Yokoi S, Sharkhuu A, Reddy M, Lee B, Matsumoto TK, et al. AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. Proc Natl Acad Sci USA. 2001;98:14150–14155. doi: 10.1073/pnas.241501798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoo S-D, Cho Y-H, Tena G, Xiong Y, Sheen J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature. 2008;451:789–796. doi: 10.1038/nature06543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De la Cruz J, Kressler D, Linder P. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem Sci. 1999;24:192–198. doi: 10.1016/s0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y, Potratz JP, Tijerina P, Del Campo M, Lambowitz AM, Russell R. DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc Natl Acad Sci USA. 2008;105:20203–20208. doi: 10.1073/pnas.0811075106. [DOI] [PMC free article] [PubMed] [Google Scholar]