Abstract
Vegetative axillary bud dormancy and outgrowth is regulated by several hormonal and environmental signals. In perennials, the dormancy induced by hormonal and environmental signals has been categorized as eco-, endo- or para-dormancy. Over the past several decades para-dormancy has primarily been investigated in eudicot annuals. Recently, we initiated a study using the monoculm phyB mutant (phyB-1) and the freely branching near isogenic wild type (WT) sorghum (Sorghum bicolor) to identify molecular mechanisms and signaling pathways regulating dormancy and outgrowth of axillary buds in the grasses. In a paper published in the January 2010 issue of Plant Cell and Environment, we reported the role of branching genes in the inhibition of bud outgrowth by phyB, shade and defoliation signals. Here we present a model that depicts the molecular mechanisms and pathways regulating axillary bud dormancy induced by shade and defoliation signals in the grasses.
Key words: axillary bud, dormancy, shade, phytochrome, defoliation, shoot branching, teosinte branched1, MAX2, cell cycle, sorghum
The dormancy and outgrowth of axillary buds is regulated by several plant hormones such as auxin, cytokinins, abscisic acid and strigolactones, and by environmental factors such as light quality, quantity and duration as well as water, temperature and nutrient status.1–3 Since the fate of an axillary bud is regulated by such diverse hormonal and environmental signals and their interactions, the type of dormancy induced varies. In perennials, three types of bud dormancy have been identified.4,5 Dormancy mediated by factors within the bud is known as endo-dormancy; while dormancy induced by factors within the plant but outside the bud is called paradormancy or correlative inhibition; the best known example being apical dominance. Dormancy induced due to unfavorable environmental conditions is known as eco-dormancy. Although there is an indepth knowledge about para-dormancy in annuals,6 few studies have been conducted on eco-dormancy. Similarly, studies of endo-dormancy have largely been restricted to low-temperature mediated growth-cessation of axillary buds of perennial plants.7,8 To understand the regulation of dormancy and outgrowth of axillary buds in monocots, we initiated a study on the molecular mechanisms inhibiting bud outgrowth by shade and defoliation signals in sorghum. Our results published in the January 2010 issue of Plant, Cell & Environment indicate that different types of dormancy may be induced in axillary buds of annual grasses by various signals and there may be overlapping and independent molecular mechanisms mediating induction of axillary bud dormancy.
phyB Deficiency, Shade and Defoliation may Induce Different Types of Dormancy
The type of dormancy induced by hormonal and environmental signals has not been investigated in annuals. Our studies on the inhibition of bud outgrowth by shade and defoliation in sorghum showed that unlike shade, defoliation inhibits bud outgrowth immediately. Since defoliation causes a complete and irreversible change in the developmental status of a plant, an immediate response was anticipated. On the other hand, shade signals such as far red light reflected from neighboring plants may vary in intensity and duration throughout the day, and response to such signals may cycle between transition states of growth and dormancy of axillary buds before commitment to dormancy is established. This may be explained by the reversible nature of the photoreceptor phytochrome B (phyB) that mediates the response to shade.9,10 phyB is activated by red (R) light and inactivated by far red (FR) light, and response to shade depends on the proportion of active and inactive phyB pools (R:FR). Active phyB mediates the normal growth and development of a plant. When phyB is inactivated by shade signals, the plant initiates shade avoidance responses including increased plant height, inhibition of branching and early flowering to cope with the shade that could be detrimental to its survival.11 Since shade signals may vary in intensity and/or duration, dormancy induced by such signals could lead to different types of dormancy. In fact, in hybrid aspen, a four-week short day treatment induces an eco-dormancy of cambial cells, while a six-week short day treatment induces an endo-dormancy.12 In sorghum, the expression level of cell cycle-related genes was dramatically downregulated in axillary buds of WT repressed by shade but not in axillary buds of phyB-1 mutants repressed by phyB deficiency. Together these results suggest that, as in perennials, different types of dormancy may be induced in annuals by hormonal or developmental and environmental signals, and the type of dormancy induced by environmental signals may depend on the intensity and duration of the signal.
Inhibition of Bud Outgrowth in Annuals by Different Signals may be Integrated Through Diverse Molecular Mechanisms within the Bud
The immediate inhibition of sorghum axillary bud outgrowth by defoliation compared to shade suggests induction of different types of dormancy by those signals. This has been further revealed by the expression analysis of several branching and cell cycle-related genes in axillary buds. Branching related genes in annuals that specifically act within or close to a bud to repress its outgrowth include the TEOSINTE BRANCHED1 (TB1) and MORE AXILLARY GROWTH (MAX2).13–15 Inhibition of bud outgrowth by shade in sorghum was shown to be associated with increased expression of the sorghum TB1 (SbTB1) gene suggesting phyB controls the fate of a bud by transcriptional regulation of SbTB1.11,16 Interestingly, SbTB1 expression level was not associated with the inhibition of bud outgrowth by defoliation indicating response integration within a bud through different mechanisms. However, the expression level of SbMAX2 was upregulated to a comparable level by phyB deficiency, shade and defoliation suggesting a possible common mechanism of transcriptional regulation of SbMAX2 by those signals. The results suggest variations in the molecular mechanisms mediating response to inhibitory shade and defoliation signals possibly leading to different types or degrees of dormancy.
Signaling Pathways Regulating Bud Dormancy in the Grasses
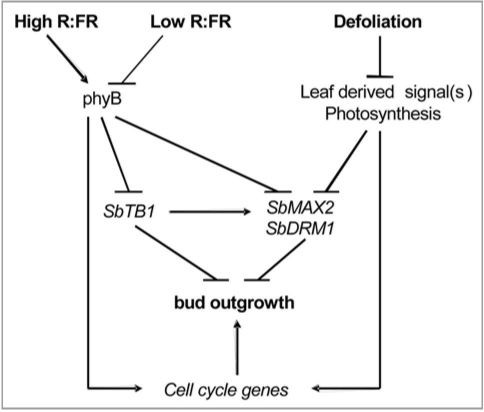
The model in Figure 1 summarizes the induction of bud dormancy in sorghum by shade through phyB, and defoliation through leaf-derived signals. phyB mediates the fate of a bud by controlling the expression level of SbTB1. However, since the inhibition of bud outgrowth by phyB deficiency is associated with changes in SbTB1 but not cell cycle-related genes, shade signals in the WT may also repress bud outgrowth by a TB1 independent pathway through transcriptional regulation of cell cycle-related genes. Inhibition of bud outgrowth by phyB deficiency, shade and defoliation was also associated with increased expression of the SbMAX2 and SbDRM1 genes. The MAX2 gene may play a role in either the perception or signal transduction of strigolactones.15 In addition it also functions in light signaling and senescence.17,18 Whether the role of MAX2 in the regulation of branching by shade or defoliation is a component of strigolactone signal transduction or signals other than strigolactones such as light or senescence needs further investigation. DRM1 was identified as one of the genes associated with dormancy in axillary buds.19 Its function has not been discovered, however, it has been used as a dormancy marker in several species. It appears both SbMAX2 and SbDRM1 act downstream of the branching inhibition signaling pathways. This model will serve as a guide toward establishing the hormonal and environmental signaling pathways regulating the fate of a bud in annual grasses.
Figure 1.
Model of the regulation of axillary bud dormancy and outgrowth by shade and defoliation signals.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/11186
References
- 1.Cline MG. Apical dominance. Bot Rev. 1991;57:319–358. [Google Scholar]
- 2.Dun EA, Brewer PB, Beveridge CA. Strigolactones: discovery of the elusive shoot branching hormone. Trends Plant Sci. 2009;14:364–372. doi: 10.1016/j.tplants.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Leyser O. The control of shoot branching: an example of plant information processing. Plant Cell Environm. 2009;32:694–703. doi: 10.1111/j.1365-3040.2009.01930.x. [DOI] [PubMed] [Google Scholar]
- 4.Lang GA. Dormancy—a New Universal Terminology. Hortscience. 1987;22:817–820. [Google Scholar]
- 5.Rohde A, Bhalerao RP. Plant dormancy in the perennial context. Trends Plant Sci. 2007;12:217–223. doi: 10.1016/j.tplants.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Ongaro V, Leyser O. Shoot branching in Arabidopsis. Comp Biochem Physiol Mol Integr Physiol. 2007;146:237–238. [Google Scholar]
- 7.McSteen P, Leyser O. Shoot branching. Ann Rev Plant Biol. 2005;56:353–374. doi: 10.1146/annurev.arplant.56.032604.144122. [DOI] [PubMed] [Google Scholar]
- 8.Poza-Carrion C, Aguilar-Martinez JA, Cubas P. Role of TCP gene BRANCHED1 in the control of shoot branching in Arabidopsis. Plant Signal Behav. 2007;2:551–552. doi: 10.4161/psb.2.6.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franklin KA, Whitelam GC. Phytochromes and shade-avoidance responses in plants. Ann Bot. 2005;96:169–175. doi: 10.1093/aob/mci165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith H, Whitelam GC. The shade avoidance syndrome: Multiple responses mediated by multiple phytochromes. Plant Cell Environm. 1997;20:840–844. [Google Scholar]
- 11.Kebrom TH, Brutnell TP. The molecular analysis of the shade avoidance syndrome in the grasses has begun. J Exp Bot. 2007;58:3079–3089. doi: 10.1093/jxb/erm205. [DOI] [PubMed] [Google Scholar]
- 12.Espinosa-Ruiz A, Saxena S, Schmidt J, Mellerowicz E, Miskolczi P, Bako L, et al. Differential stagespecific regulation of cyclin-dependent kinases during cambial dormancy in hybrid aspen. Plant J. 2004;38:603–615. doi: 10.1111/j.1365-313X.2004.02070.x. [DOI] [PubMed] [Google Scholar]
- 13.Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- 14.Hubbard L, McSteen P, Doebley J, Hake S. Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics. 2002;162:1927–1935. doi: 10.1093/genetics/162.4.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stirnberg P, Furner IJ, Leyser HMO. MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 2007;50:80–94. doi: 10.1111/j.1365-313X.2007.03032.x. [DOI] [PubMed] [Google Scholar]
- 16.Kebrom TH, Burson BL, Finlayson SA. Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol. 2006;140:1109–1117. doi: 10.1104/pp.105.074856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen H, Luong P, Huq E. The F-Box protein MAX2 functions as a positive regulator of photomorphogenesis in arabidopsis. Plant Physiol. 2007;145:1471–1483. doi: 10.1104/pp.107.107227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, et al. ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell. 2001;13:1779–1790. doi: 10.1105/TPC.010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stafstrom JP, Ripley BD, Devitt ML, Drake B. Dormancy-associated gene expression in pea axillary buds. Planta. 1998;205:547–552. doi: 10.1007/s004250050354. [DOI] [PubMed] [Google Scholar]