Abstract
Plants consist of distinct cell types distinguished by position, morphological features and metabolic activities. We recently developed a method to extract cell-type specific mRNA populations by immunopurification of ribosome-associated mRNAs. Microarray profiles of 21 cell-specific mRNA populations from seedling roots and shoots comprise the Arabidopsis Translatome dataset. This gene expression atlas provides a new tool for the study of cell-specific processes. Here we provide an example of how genes involved in a pathway limited to one or few cell-types can be further characterized and new candidate genes can be predicted. Cells of the root endodermis produce suberin as an inner barrier between the cortex and stele, whereas the shoot epidermal cells form cutin as a barrier to the external environment. Both polymers consist of fatty acid derivates, and share biosynthetic origins. We use the Arabidopsis Translatome dataset to demonstrate the significant cell-specific expression patterns of genes involved in those biosynthetic processes and suggest new candidate genes in the biosynthesis of suberin and cutin.
Key words: cell-type specific expression, polysome immunopurification, translatome, suberin, cutin, endodermis, epidermis, arabidopsis
Introduction
Plants consist of numerous specialized cell types with defined functions. For example, leaf mesophyll cells perform photosynthesis, whereas phloem cells transport sugars. Some cell types establish barriers within an organ or to the external environment, such as the root endodermis and leaf epidermis, respectively. To obtain insight into the genes involved in cell-specific specialization and function, plant scientists have labored to isolate specific cell-types to observe the regulated expression of cell-specific mRNAs. The methods that enable this are based on mechanical separation (i.e., epidermal peels,1 phloem isolation2), laser microdissection of organs3–5 or fluorescence-activated sorting of GFP-marked protoplasts.6–10 However, these methods may not enable monitoring of rapid responses to chemicals or environmental cues. To successfully profile dynamic responses to hypoxia, we developed a non-invasive method to immunopurify polysome-bound mRNAs from specific cells by expressing a FLAG-tagged ribosomal protein under control of cell- and region-specific promoters.11 This technique uses flash-frozen material to isolate the subset of cellular mRNAs associated with polysomes, the majority of which are likely to be undergoing translation. The profile of the ribosome-associate mRNAs or “translatome” is distinct from that of the transcriptome,12,13 because mRNA abundance and translation are regulated by distinct processes. Hence, translatome profiling can provide a more precise readout of gene expression.
The Arabidopsis Translatome atlas can be used to study specific processes in one of the targeted cell types of control grown and hypoxia-treated seedlings. The data are accessible for individual genes via an eFP (electronic fluorescent pictograph) platform (efp.ucr.edu/).11,14 This atlas of translatome profiles can be readily extended to the analysis of other transformable organisms or additional Arabidopsis cell types, such as the root pericycle and epidermis, cell types of the developing embryo, root, shoot and floral meristems and pollen. We demonstrate here how the Arabidopsis Translatome dataset can be used to study genes involved in the synthesis of lipid monomers that are precursors for the formation of the polymers suberin and cutin by the root endodermis and leaf epidermis, respectively.
Recent Advances in Suberin and Cutin Biosynthesis
Suberin and cutin are extracellular lipid polymers that provide barriers against water, ions and gases.15,16 While cutin is present in the outer layers of the leaves, suberin is present at the outside of the roots (rhizodermis), where it influences water uptake and provides protection from pathogen and insect attack. Suberin also forms within organs, such as the border between bundle sheath cells and vasculature in C4 leaves and the endodermis in roots, controlling water and ion flux between the cortex and the vasculature.
The suberin polymer is composed of oxygenated fatty acid derivatives, alcohols and unsubstituted fatty acids, as well as glycerol and ferulic acid.15 Suberin barriers differ in composition due to intricacies in the biosynthetic pathway that are not yet known in detail.17 Cutin contains similar monomers, but also displays variations in composition.16 Cuticular waxes are a third group of lipid polymers, produced in part by the monomer biosynthesis pathways common to cutin and suberin. Recent progress has been made in elucidation of single steps involved in oxygenated fatty acid synthesis, elongation and modification through recognition of the participant enzymes. A number of genes have been associated with suberin and/or cutin biosynthesis by use of Arabidopsis mutants. Here, we use the Arabidopsis Translatome dataset to confirm the localized expression of genes involved in suberin and cutin biosynthesis and identify genes that are likely to be involved in lipid monomer biosynthesis associated with production of these lipid biopolymers in the endodermis or epidermis of Arabidopsis.
The Arabidopsis Translatome dataset consists of 21 mRNA profiles including root and root tip endodermis (pSCR) and shoot epidermis (pCER5) mRNA populations. The dataset includes two readouts:
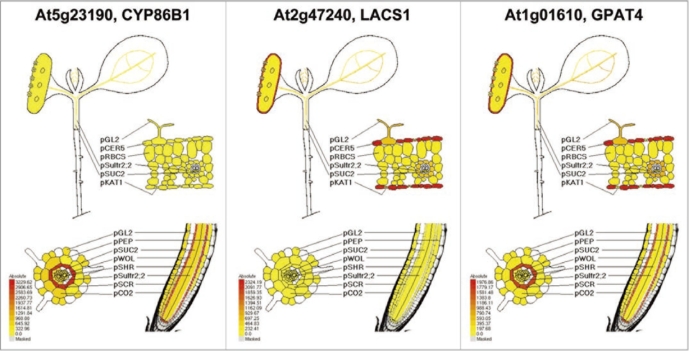
(1) the absolute signal value for a given mRNA, or mRNA abundance; (2) the fold enrichment of an mRNA in a cell type/population relative to non-overlapping cell types or cell populations evaluated for the same organ. The data can be displayed for individual genes, as shown for three endodermal and shoot epidermal-enriched mRNAs (Fig. 1). The mRNAs enriched in specific cell types are likely to be involved in cell specific processes. By use of GO term annotations, we recognized that mRNAs involved in suberin or cutin biosynthesis were significantly overrepresented in the endodermis (pSCR, i.e., peroxidase activity, adj. p-value 9.76E-09; lipid binding, 6.89E-03; fatty acid metabolic process, 1.47E-09; lipid transport, 3.17E-03; acetyl-CoA biosynthetic process from pyruvate, 3.68E-03) and the shoot epidermis (pCER5, i.e., carboxy-lesterase activity, 3.82E-09; fatty acid (omega-1)-hydroxylase activity, 7.15E-03; lipid metabolic process, 4.60E-09; cuticle development, 2.91E-05; cellulose and pectin-containing cell wall modification, 6.08E-03) (reviewed in ref. 11). This motivated us to further explore the cell-specific expression of known genes of suberin biosynthesis.
Figure 1.
Three genes involved in suberin and cutin biosynthesis and their mRNA in translatomes of different cell populations of Arabidopsis, visualized via the eFP platform (efp.ucr.edu/). Absolute signal values of transcripts in translatomes isolated from cell populations.
One major reaction in early suberin and cutin biosynthesis is the fatty acid elongation via a multi-enzyme complex (FAE).15 The rate-limiting step of this enzyme complex is the β-ketoacyl-CoA synthase (KCS), encoded by a family of 21 genes in Arabidopsis. Of those, six are involved in suberin and cutin biosynthesis (KCS1,18 KCS2,19,20 KCS6 (CUT1),21 KCS10 (FDH),22 KCS18 (FAE1),23 KCS20,20). Of the remaining KCS genes, several show highly specific expression in either the root endodermis or the leaf epidermis, and are therefore potential targets for future studies (Table 1). Another enzyme of this complex is the β-ketoacyl-CoA reductase (KCR), encoded by two genes in Arabidopsis. Whereas KCR1 is essential for suberin and wax biosynthesis24 and is enriched in the root endodermis (Table 1), KCR2 is neither essential nor found to be expressed in our dataset.
Table 1.
Summary of selected gene families associated with suberin and cutin biosynthesis—known members and potential new candidates
| AGI ID | Gene name | Described function | Cell type enriched11 | SLR R SCR (endodermis) | SLR S CER (epidermis) |
| 3-ketoacyl CoA synthase | |||||
| At1g01120 | KCS1 | cutin18 | R endo/S epi | 2.73 | 2.32 |
| At1g04220 | KCS2 (DAISY) | suberin,19 cutin20 | n.s. | 0.97 | 0.88 |
| At1g07720 | KCS3 | potential new target | S epi | 0.06 | 2.68 |
| At1g25450 | KCS5 | potential new target | S epi | −0.16 | 2.47 |
| At1g68530 | KCS6 (CUT1, CER6) | cutin21 | n.s. | 0.07 | 1.75 |
| At2g16280 | KCS9 | potential new target | S epi | 0.85 | 2.46 |
| At2g26250 | KCS10 (FDH) | cutin22 | R endo | 2.29 | 2.27 |
| At2g46720 | KCS13 (HIC) | potential new target | S epi | 0.03 | 1.79 |
| At4g34250 | KCS16 | potential new target | S epi | 0.27 | 2.15 |
| At4g34510 | KCS17 | potential new target | R endo | 1.68 | 0.08 |
| At5g04530 | KCS19 | potential new target | S epi | 0.04 | 2.98 |
| At5g43760 | KCS20 | cutin20 | n.s. | 1.19 | 1.93 |
| ketoacyl CoA reductase | |||||
| At1g67730 | KCR1 | cutin, suberin24 | R endo | 1.71 | 1.58 |
| long-chain acyl-CoA synthetases | |||||
| At2g47240 | LACS1 (CER8) | cutin35 | S epi | 0.11 | 3.36 |
| t1g49430 | LACS2 | utin36 | endo | .34 | .02 |
| At1g64400 | LACS3 | potential new target | R endo | 1.55 | 1.81 |
| fatty acid omega-hydroxylase | |||||
| At5g58860 | CYP86A1 (HORST) | suberin26 | R endo | 2.33 | −0.01 |
| At4g00360 | CYP86A2 (ATT1) | cutin27 | R endo/S epi | 2.68 | 2.62 |
| At1g01600 | CYP86A4 | cutin28 | S epi | 1.53 | 2.37 |
| At2g45970 | CYP86A8 (LCR) | cutin29 | R endo/S epi | 1.97 | 2.65 |
| At5g23190 | CYP86B1 | suberin30 | R endo | 2.71 | −0.02 |
| At5g08250 | CYP86B2 | suberin30 | R endo | 1.90 | 0.02 |
| At5g63450 | CYP94B1 | potential new target | S epi | 0.79 | 2.04 |
| other CYP450 enzymes | |||||
| At4g15330 | CYP705A1 | potential new target | R endo | 1.94 | 0.34 |
| At4g22710 | CYP706A2 | potential new target | S epi | −0.52 | 2.10 |
| At2g34490 | CYP710A2 | potential new target | S epi | 0.15 | 2.52 |
| At5g24900 | CYP714A2 | potential new target | R endo | 1.24 | −0.12 |
| At3g53280 | CYP71B5 | potential new target | S epi | 0.08 | 2.24 |
| At2g26710 | CYP734A1 | potential new target | S epi | −0.19 | 2.54 |
| At3g10570 | CYP77A6 | cutin28 | S epi | 0.07 | 1.82 |
| At1g11600 | CYP77B1 | potential new target | S epi | −0.02 | 1.92 |
| At5g09970 | CYP78A7 | potential new target | S epi | 0.13 | 1.31 |
| At5g52320 | CYP96A4 | potential new target | S epi | −0.33 | 2.11 |
| alcohol dehydrogenase | |||||
| At1g72970 | HTH | cutin32 | S epi | 0.37 | 2.64 |
| aldehyde dehydrogenase | |||||
| At4g36250 | ALDH | potential new target | S epi | −0.15 | 2.47 |
| glycerol-acyl-transferase | |||||
| At4g01950 | GPAT3 | potential new target | S epi | 0.32 | 2.12 |
| At1g01610 | GPAT4 | cutin25 | R endo | 3.48 | 2.88 |
| At3g11430 | GPAT5 | suberin33 | R endo | 2.80 | −0.18 |
| At2g38110 | GPAT6 | cutin28 | R endo | 3.01 | 0.41 |
| At4g00400 | GPAT8 | cutin25 | R endo/S epi | 2.37 | 2.83 |
| feruloyl-acyl-transferase | |||||
| At5g41040 | ASFT (ACT) | suberin31, 34 | R endo | 3.43 | 0.14 |
SLR, mean signal log ratio of pairwise comparisons of a specific cell type (here: pSCR or pCER 5) to all other non-overlapping cell types of the same organ;11 n.s., not significant.
Following the elongation step, the fatty acids are hydroxylated at the ω-position by NADPH-dependent cytochrome P450 monooxygenases of the CYP86 and CYP94 families.15 Six of the 11 putative CYP86 genes were found to modify suberin or cutin composition when knocked out in Arabidopsis, of which all have mRNAs specifically enriched in either the endodermis or epidermis in the Translatome atlas (CYP86A1,25,26 CYP86A2,27 CYP86A4,28 CYP86A8,29 CYP86B1/CYP86B2,30,31). These data affirm the cell-specific importance of CYP450 genes. The CYP94 gene family has only one out of six members, which is specific for the leaf epidermis, but has not yet been characterized in respect to fatty acid hydroxylation. Two additional CYP450 genes, CYP705A1 (At4g15330) and CYP714A2 (At5g24900) are enriched in the endodermal mRNA population, and could be also involved in suberin biosynthesis, while eight additional CYP450 genes are specifically enriched in the leaf epidermis (Table 1). One of those, CYP77A6 was recently found to be involved in cutin biosynthesis.28
An alternative pathway to modify very long chain fatty acids involves alcohol (ω-hydroxyacid) dehydrogenases and aldehyde (-oxo-acid) dehydrogenases.30 The first enzyme of this pathway was characterized in Arabidopsis,32 but the second is unknown. Our data suggest a potential candidate for this enzymatic step (Table 1).
Suberin and cutin are oligomers of fatty acids, esterified with either glycerol or ferulic acid. This oligomer formation requires the activity of acyl transferases (ACTs), such as acyl-CoA:glycerol-3-phosphate acyltransferase (GPAT) and aliphatic suberin feruloyl transferase (ASFT). Arabidopsis contains nine putative GAPT and two putative ASFT genes. Of the GAPTs, four are involved in suberin and cutin synthesis (GAPT4,25 GAPT5,33 GAPT6,28 GAPT8,25). GAPT3 is also enriched among shoot epidermis mRNAs and could therefore be involved in cutin biosynthesis (Table 1). The recently described ASFT is involved in modification of suberin composition,31,34 and is specifically expressed in root endodermis (Table 1). A second ASFT candidate, At5g63560, is also slightly enriched in this cell type.11
Outlook
In addition to biosynthetic pathways, several other processes are required for the formation and deposition of suberin, including fatty acyl or wax exporters (ABC transporters, i.e., CER5, WBC11), extracellular lipid transfer proteins, and signaling components (i.e., WIN1). With the available dataset, confirmed and potential new candidates can be selected for further in-depth studies. The 260 endodermis-specific and the 511 epidermis-specific genes identified in the Translatome atlas provide a large resource for the further exploration of genes associated with suberin and cutin synthesis, transport and deposition.11 Of the 15 genes highly enriched in both mRNA populations, only 11 have assigned functions. The epidermal data is also likely to yield leads in wax biosynthesis. Furthermore, the 26 and 24 transcription factors enriched in the root endodermis (pSCR) and shoot epidermis (pCER5), respectively, deserve examination in experiments that couple genetic, biochemical and enzymatic approaches. In conclusion, the highly cell-specific biosynthesis of lipid polymers provides one example of the data potential of the Arabidopsis Translatome atlas.
Acknowledgements
This work was supported by the National Science Foundation (IBN-0420152, IOS-0750811) and German Academic Exchange Service (DAAD). We thank John B. Ohlrogge for critical reading of the manuscript.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/11187
References
- 1.Suh MC, Samuels AL, Jetter R, Kunst L, Pollard M, Ohlrogge J, et al. Cuticular lipid composition, surface structure and gene expression in Arabidopsis stem epidermis. Plant Physiol. 2005;139:1649–1665. doi: 10.1104/pp.105.070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao C, Craig JC, Petzold HE, Dickerman AW, Beers EP. The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis roothypocotyl. Plant Physiol. 2005;138:803–818. doi: 10.1104/pp.105.060202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spencer MW, Casson SA, Lindsey K. Transcriptional profiling of the Arabidopsis embryo. Plant Physiol. 2007;143:924–940. doi: 10.1104/pp.106.087668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai S, Lashbrook CC. Stamen abscission zone transcriptome profiling reveals new candidates for abscission control: enhanced retention of floral organs in transgenic plants overexpressing Arabidopsis ZINC FINGER PROTEIN2. Plant Physiol. 2008;146:1305–1321. doi: 10.1104/pp.107.110908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiao Y, Tausta SL, Gandotra N, Sun N, Liu T, Clay NK, et al. A transcriptome atlas of rice cell types uncovers cellular, functional and developmental hierarchies. Nat Genet. 2009;41:258–263. doi: 10.1038/ng.282. [DOI] [PubMed] [Google Scholar]
- 6.Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, et al. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- 7.Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 8.Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, et al. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science. 2008;320:942–945. doi: 10.1126/science.1153795. [DOI] [PubMed] [Google Scholar]
- 9.Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA. 2008;105:803–808. doi: 10.1073/pnas.0709559105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yadav RK, Girke T, Pasala S, Xie M, Reddy GV. Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc Natl Acad Sci USA. 2009;106:4941–4946. doi: 10.1073/pnas.0900843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mustroph A, Zanetti ME, Jang CJ, Holtan HE, Repetti PP, Galbraith DW, et al. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:18843–18848. doi: 10.1073/pnas.0906131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawaguchi R, Bailey-Serres J. mRNA sequence features that contribute to translational regulation in Arabidopsis. Nucleic Acids Res. 2005;33:955–965. doi: 10.1093/nar/gki240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Branco-Price C, Kaiser KA, Jang CJ, Larive CK, Bailey-Serres J. Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J. 2008;56:743–755. doi: 10.1111/j.1365-313X.2008.03642.x. [DOI] [PubMed] [Google Scholar]
- 14.Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An “electronic fluorescent pictograph” browser for exploring and analyzing largescale biological data sets. PloS One. 2007;2:718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franke R, Schreiber L. Suberin—a biopolyester forming apoplastic plant interfaces. Curr Opin Plant Biol. 2007;10:252–259. doi: 10.1016/j.pbi.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Pollard M, Beisson F, Li Y, Ohlrogge JB. Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci. 2008;13:236–246. doi: 10.1016/j.tplants.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Franke R, Briesen I, Wojciechowski T, Faust A, Yephremov A, Nawrath C, et al. Apoplastic polyesters in Arabidopsis surface tissues—a typical suberin and a particular cutin. Phytochemistry. 2005;66:2643–2658. doi: 10.1016/j.phytochem.2005.09.027. [DOI] [PubMed] [Google Scholar]
- 18.Todd J, Post-Beittenmiller D, Jaworski JG. KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J. 1999;17:119–130. doi: 10.1046/j.1365-313x.1999.00352.x. [DOI] [PubMed] [Google Scholar]
- 19.Franke R, Höfer R, Briesen I, Emsermann M, Efremova N, Yephremov A, et al. The DAISY gene from Arabidopsis encodes a fatty acid elongase condensing enzyme involved in the biosynthesis of aliphatic suberin in roots and the chalaza-micropyle region of seeds. Plant J. 2009;57:80–95. doi: 10.1111/j.1365-313X.2008.03674.x. [DOI] [PubMed] [Google Scholar]
- 20.Lee SB, Jung SJ, Go YS, Kim HU, Kim JK, Cho HJ, et al. Two Arabidopsis 3-ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J. 2009;60:462–475. doi: 10.1111/j.1365-313X.2009.03973.x. [DOI] [PubMed] [Google Scholar]
- 21.Millar AA, Clemens S, Zachgo S, Giblin EM, Taylor DC, Kunst L. CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell. 1999;11:825–838. doi: 10.1105/tpc.11.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lolle SJ, Berlyn GP, Engstrom EM, Krolikowski KA, Reiter WD, Pruitt RE. Developmental regulation of cell interactions in the Arabidopsis fiddlehead-1 mutant: a role for the epidermal cell wall and cuticle. Dev Biol. 1997;189:311–321. doi: 10.1006/dbio.1997.8671. [DOI] [PubMed] [Google Scholar]
- 23.Rossak M, Smith M, Kunst L. Expression of the FAE1 gene and FAE1 promoter activity in developing seeds of Arabidopsis thaliana. Plant Mol Biol. 2001;46:717–725. doi: 10.1023/a:1011603923889. [DOI] [PubMed] [Google Scholar]
- 24.Beaudoin F, Wu X, Li F, Haslam RP, Markham JE, Zheng H, et al. Functional characterization of the Arabidopsis beta-ketoacyl-coenzyme A reductase candidates of the fatty acid elongase. Plant Physiol. 2009;150:1174–1191. doi: 10.1104/pp.109.137497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Beisson F, Koo AJ, Molina I, Pollard M, Ohlrogge J. Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proc Natl Acad Sci USA. 2007;104:18339–18344. doi: 10.1073/pnas.0706984104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Höfer R, Briesen I, Beck M, Pinot F, Schreiber L, Franke R. The Arabidopsis cytochrome P450 CYP86A1 encodes a fatty acid omega-hydroxylase involved in suberin monomer biosynthesis. J Exp Bot. 2008;59:2347–2360. doi: 10.1093/jxb/ern101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao F, Goodwin SM, Xiao Y, Sun Z, Baker D, Tang X, et al. Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO J. 2004;23:2903–2913. doi: 10.1038/sj.emboj.7600290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li-Beisson Y, Pollard M, Sauveplane V, Pinot F, Ohlrogge J, Beisson F. Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proc Natl Acad Sci USA. 2009;106:22008–22013. doi: 10.1073/pnas.0909090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wellesen K, Durst F, Pinot F, Benveniste I, Nettesheim K, Wisman E, et al. Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid omegahydroxylation in development. Proc Natl Acad Sci USA. 2001;98:9694–9699. doi: 10.1073/pnas.171285998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Compagnon V, Diehl P, Benveniste I, Meyer D, Schaller H, Schreiber L, et al. CYP86B1 is required for very long chain omega-hydroxyacid and alpha, omega-dicarboxylic acid synthesis in root and seed suberin polyester. Plant Physiol. 2009;150:1831–1843. doi: 10.1104/pp.109.141408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molina I, Li-Beisson Y, Beisson F, Ohlrogge JB, Pollard M. Identification of an Arabidopsis feruloylcoenzyme a transferase required for suberin synthesis. Plant Physiol. 2009;151:1317–1328. doi: 10.1104/pp.109.144907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurdyukov S, Faust A, Trenkamp S, Bär S, Franke R, Efremova N, et al. Genetic and biochemical evidence for involvement of HOTHEAD in the biosynthesis of long-chain alpha-, omega-dicarboxylic fatty acids and formation of extracellular matrix. Planta. 2006;224:315–329. doi: 10.1007/s00425-005-0215-7. [DOI] [PubMed] [Google Scholar]
- 33.Beisson F, Li Y, Bonaventure G, Pollard M, Ohlrogge JB. The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell. 2007;19:351–368. doi: 10.1105/tpc.106.048033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gou JY, Yu XH, Liu CJ. A hydroxycinnamoyltransferase responsible for synthesizing suberin aromatics in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:18855–18860. doi: 10.1073/pnas.0905555106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lü S, Song T, Kosma DK, Parsons EP, Rowland O, Jenks MA. Arabidopsis CER8 encodes LONGCHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J. 2009;59:553–564. doi: 10.1111/j.1365-313X.2009.03892.x. [DOI] [PubMed] [Google Scholar]
- 36.Schnurr J, Shockey J, Browse J. The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell. 2004;16:629–642. doi: 10.1105/tpc.017608. [DOI] [PMC free article] [PubMed] [Google Scholar]