Abstract
Respiratory deficiency increases the sensitivity of the pathogenic fungi Candida albicans and C. glabrata to oxidative stress induced by photodynamic therapy (PDT) sensitized by the cationic porphyrin meso-tetra (N-methyl-4-pyridyl) porphine tetra tosylate (TMP-1363). Since disruption of electron transport chain (ETC) function increases intracellular levels of reactive oxygen species in yeast, we determined whether interference with ETC assembly or function increased sensitivity to TMP-1363-PDT in C. albicans, C. glabrata and the non-pathogenic yeast Saccharomyces cerevisiae. Metabolic inhibitor antimycin A and defined genetic mutants were used to identify ETC components that contribute to the sensitivity to PDT. Inhibition of cytochrome bc1 (Complex III) with antimycin A increases mitochondrial levels of reactive oxygen species. PDT performed following pretreatment with antimycin A reduced colony forming units (CFU) of C. albicans and C. glabrata by approximately two orders of magnitude relative to PDT alone. A S. cerevisiae mitochondrial glutaredoxin grx5 mutant, defective in assembly of Fe-S clusters critical for Complex III function, displayed increased sensitivity to PDT. Furthermore, C. glabrata and S. cerevisiae mutants in cytochrome c oxidase (Complex IV) synthesis and assembly were also significantly more sensitive to PDT. These included suv3, encoding an ATP-dependent RNA helicase critical for maturation of cytochrome c oxidase subunit transcripts, and pet117, encoding an essential cytochrome c oxidase assembly factor. Following PDT, the reduction in CFU of these mutants was one to two orders of magnitude greater than in their respective parental strains. The data demonstrate that selective inhibition of ETC Complexes III and IV significantly increases the sensitivity of C. albicans, C. glabrata and S. cerevisiae to PDT sensitized with TMP-1363.
Keywords: photodynamic therapy, Candida, oxidative stress, respiration
1. Introduction
Candida species commonly colonize the epithelial surfaces of the body in the majority of the human population. However, few healthy carriers develop clinical signs of candidiasis until the body's physical and immunological defenses are compromised in some way. Patients with diseases such as cancer, HIV/AIDS, or diabetes, as well as premature infants and patients requiring intensive care, are among the groups at risk of developing infection from Candida. As a result of its widespread colonization of mucosal and cutaneous surfaces, the bulk of Candida infections are located at these sites [1].
While C. albicans accounts for nearly half of Candida infections, C. glabrata is the second most prevalent pathogenic Candida species [2]. Infections with non-albicans species such as C. glabrata often emerge after treatment for an initial C. albicans infection, or during prophylaxis for C. albicans infection, by virtue of their inherent resistance to commonly used azole antifungals [3]. For example, fluconazole-resistant Candida species colonize approximately 81% of AIDS patients receiving therapy for oral candidiasis [4]. In oral candidiasis, C. glabrata is seen as either a co-infecting agent with C. albicans or as the sole etiologic agent [5]. C. glabrata is also emerging as a prominent cause of vaginal infections [2]. Finally, catheters and prosthetic devices serve as substrates for Candida biofilms [6], providing a reservoir of drug-resistant cells that can initiate life-threatening disseminated infection [7]. These trends underscore the importance of developing novel strategies for treatment of Candida infection, as the microbiology and resistance patterns of clinical isolates evolve in response to selective pressures of current antifungal therapy.
The application of photodynamic therapy (PDT) to microbial infections, also referred to as photodynamic antimicrobial chemotherapy or PACT [8, 9], is gaining attention as an alternative treatment against organisms resistant to conventional chemotherapy [10]. However, the mechanisms by which PDT exerts antimicrobial activity are just beginning to be examined. Mammalian cells require mitochondrial respiration for survival and normal growth. However, certain fungi, such the model eukaryotic yeast Saccharomyces cerevisiae [11] and its pathogenic relatives C. albicans [12] and C. glabrata [13], can grow without functional mitochondria if provided with a fermentable carbon source such as glucose. Hence, respiratory-deficient mutant yeasts are useful tools for the study of mitochondrial function. Respiratory-deficient Candida display pleiotropic resistance to a number of toxic stresses, including antifungal drugs [13-16].
In contrast, we recently made the striking observation that respiratory-deficient mutants of Candida [17], characterized by the deletion of major segments or total absence of mitochondrial DNA [18], are significantly more sensitive than their parental counterparts to oxidative stress induced by PDT mediated by the cationic photosensitizer meso-tetra (N-methyl-4-pyridyl) porphine tetra tosylate (TMP-1363). This indicates that there are inherent mechanisms in Candida linked to respiratory function that help the pathogen directly counter the oxidative stress and/or repair the damage induced by PDT. Exploiting our understanding of these mechanisms may lead to therapeutic strategies to increase the sensitivity of Candida to PDT.
Disruption of the respiratory pathway at selected points can increase intracellular levels of reactive oxygen species in yeast [19, 20]. Using a metabolic inhibitor of respiration and defined genetic mutants deficient in respiratory pathway function, we determined whether interference with electron transport chain assembly or function increased sensitivity to PDT in C. albicans, C. glabrata and the non-pathogenic yeast S. cerevisiae.
2. Materials and methods
2.1 Organisms and culture conditions
C. albicans, C. glabrata and S. cerevisiae strains used in this study are described in Table 1. Organisms were grown overnight at 30°C in yeast extract-peptone-dextrose (YPDextrose) broth (Difco, Detroit, MI). Organisms were washed twice with sterile dH2O and adjusted to 107 cells/ml in sterile dH2O prior to PDT. We have used conventional yeast genetics nomenclature throughout the paper. For example, PET117 refers to the wild type gene, pet117 refers to a mutant in that allele and Pet117 refers to the protein encoded by PET117.
Table 1.
Strains used in this study.
| Name | Description | Reference |
|---|---|---|
| C. albicans | ||
| SC5314 | Parental | 17 |
| C. glabrata | ||
| MRO-084-R | Parental | 17 |
| 66032 | Parental | 32 |
| BG2 | Parental (RCa) | 23 |
| Δpet117 | ZeoR insertion mutant in BG2; cytochrome c oxidase assembly (RDb) | This study |
| BG462 | URA+ transformant of ura3 BG2 (RC) | 23 |
| BG663 | suv3::Tn7; ATP-dependent RNA helicase (RD) | 23 |
| BG769 | she9::Tn7; mt inner memb. protein (RC) | 23 |
| BG824 | FLCRc derivative of she9 (RD) | 23 |
| BG643 | mrpl4::Tn7; mt large ribosomal subunit protein (RC) | 23 |
| BG820 | FLCR derivative of mrpl4 (RD) | 23 |
| S. cerevisiae | ||
| S288C | Parental (RC) | 29 |
| Δpet117 | S288C deletion mutant; cytochrome c oxidase assembly (RD) | 29 |
| Δgrx5 | Deletion mutant; mt glutaredoxin (RD) | 29 |
RC Respiratory competent.
RD Respiratory deficient.
FLCR Selected on YPD plates supplemented with 256 μg/mL fluconazole. 23
2.2 Recombinant DNA manipulations
Plasmid DNA was isolated using the QIAprep spin kit (QIAGEN, Valencia, CA). Restriction endonucleases, calf intestinal phosphatase, and T4 DNA ligase were used as specified by the supplier (New England Biolabs, Beverley, MA). DNA fragments were isolated from 0.7% Tris-borate-EDTA agarose gels using the Freeze and Squeeze Kit (Bio-Rad, Hercules, CA). PCR was performed using the PuReTaq Ready-To-Go PCR beads (GE Healthcare Bio-Sciences Corp., Piscataway, NJ) in a MJ Mini Gradient Thermal Cycler (Bio-Rad). Oligonucleotide primers were obtained from Sigma Genosys (The Woodlands, TX). Candida transformation was performed using the Frozen EZ Yeast Transformation II kit (Zymo Research, Orange, CA).
2.3 Photodynamic treatment
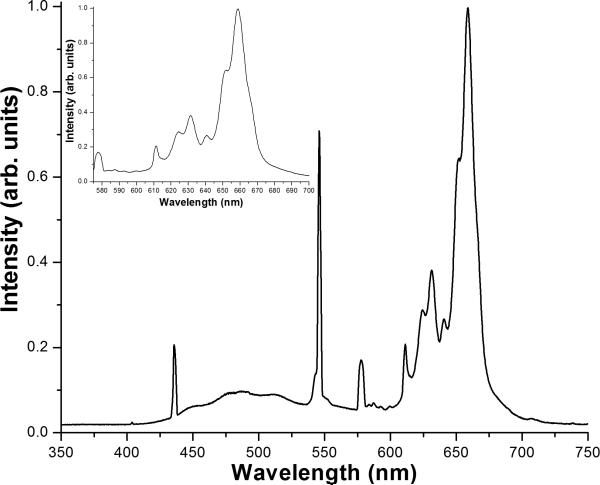
For studies on the metabolic inhibition of the electron transport chain, Candida species were grown overnight in YPDextrose at 30°C with vigorous aeration, in either the presence or absence of the inhibitor of respiratory Complex III inhibitor antimycin A (10 μM AA) (Sigma-Aldrich, St. Louis, MO) [21]. Following this treatment, organisms were treated with 10 μg/ml of the cationic photosensitizer meso-tetra (N-methyl-4-pyridyl) porphine tetra tosylate (TMP-1363; Frontier Scientific, Logan, UT) for 10 min. The organism suspension (2 ml) was added to six well plates for the irradiation step and irradiated at 2.4 J/cm2 with visible light from a 48 cm × 48 cm light box equipped with a bank of fluorescent lamps (Sylvania GRO-LUX, 15 W, part no. F15T8/GRO). The irradiance at the surface of the light box was 4.0 mW/cm2, and the spectrum of the light, shown in Figure 1, was such that approximately 67% of the power was emitted within the range of 575–700 nm. The fluence of 2.4 J/cm2 corresponded to a 10 min. irradiation time with this light source. Previous work [17] has shown that these parameters give a significant PDT response against parental strains but with enough range in the assay to observe enhanced sensitivity of the respiratory-deficient mutants. Untreated organisms and organisms treated with TMP-1363 but shielded from light were used as controls. For subsequent studies, organisms were incubated for 10 min. with different concentrations of TMP-1363 as indicated in the figures, and PDT was carried out as described.
Figure 1. Spectrum of visible radiation from the light box used for PDT.
The inset is an expanded view of the region of the spectrum in the wavelength range of 575 - 700 nm that accounts for ~ 67% of the total power emitted from the light source.
2.4 Colony forming unit (CFU) phototoxicity assay
Following PDT, organism suspensions were diluted in dH2O. For screening a range of TMP-1363 concentrations, dilutions of the different experimental groups were spotted (2 μl/spot) on YPDextrose plates and grown overnight at 30°C to assay phototoxicity. To assay phototoxicity quantitatively following PDT, organisms were diluted in water, 200 μl was plated on YPDextrose agar and plates were incubated 24–48 h at 30°C to allow colony formation. Data were expressed as CFU/ml and represented as a log10 reduction compared to untreated organisms.
2.5 C. glabrata genomic DNA preparation
Organisms were grown overnight at 30°C in YPDextrose broth, cells from 3 ml of culture were washed twice with dH2O and resuspended in 200 μl of breaking buffer (2% Triton X-100, 1% SDS, 0.1 M NaCl, 1 mM EDTA) containing 0.5 mm Zirconia/Silica beads (BioSpec Products, Bartlesville, OK), followed by the addition of 200 μL of phenol:chloroform:isopropanol (25:24:1) in 10 mM Tris (pH 7.6):1 mM EDTA. Organisms were vortexed for 1.5 min, and placed on ice for the same amount of time. This cycle was repeated 4 times. Following these steps, 200 μL of TE was added the suspension and the contents were centrifuged for 3 min in a microfuge. The DNA was then concentrated from the supernatant by precipitation, and resuspended in 100 μl water.
2.6 Construction of the C. glabrata PET117 disruption mutant
Genetic recombination with a homologous linear DNA fragment was used to generate an insertion mutant in C. glabrata PET117 [22]. Genomic DNA from C. glabrata BG2 [23] was used as a template for PCR to amplify fragments from the 5’ and 3’ ends of PET117 and its flanking regions of 399 bp and 424 bp, respectively. PCR primers with unique restriction sites were used to facilitate cloning into pTEF1/Zeo (Invitrogen Corp., Carlsbad CA) containing a Zeocin resistance (ZeoR) cassette. The restriction sites are underlined in the primer sequences, and the respective restriction enzyme is listed after the primer sequence. The 5’ fragment amplified using primers PET117 5’F (5’ ATGACAGATCTAACTGCAAGGACGTGAGCTTC 3’; BglII) and PET117 5’R (5’ ATGACGCGGCCGCTCTTACTGGCTCGGCTCATTG 3’; NotI) was cloned into the 5’ polylinker upstream of the ZeoR cassette, and the 3’ fragment amplified using primers PET117 3’F (5’ ATGACGAATTCCACTATGTGCAGGAGCTGGAGC 3’; EcoRI) and PET117 3’R (5’ TACGGAAAGCTTACGCCAATGATTCCATTCTC 3’; HindIII) was cloned into the 3’ polylinker downstream of the ZeoR cassette, resulting in the ZeoR cassette flanked by the 5’ and 3’ PET117 fragments.
The entire insert was released by restriction digestion, and 100 μg was used to transform both C. glabrata BG2 and C. glabrata 66032 using the Frozen EZ Yeast Transformation II kit (Zymo Research, Orange, CA). Disruption of native PET117 was achieved by homologous recombination into the chromosome by the fragment containing the ZeoR cassette. To allow expression of Zeocin resistance, organisms were incubated in YPDextrose broth for 24 h prior to plating in selective media containing Zeocin concentrations ranging from 200-400 μg/mL. Recombinant clones were further screened for inability to grow on YP + 2% glycerol (YPGlycerol) to confirm their respiratory-deficient status.
Disruption of PET117 by the ZeoR cassette was confirmed by PCR using C. glabrata-specific primers flanking those employed to generate the recombinogenic fragment and primers internal to the Zeocin cassette. Primers PET117 A-F (5’ AACTGCAAGGACGTGAGCTTC 3’) and ZeocinR (5’ TCAACACCGCCCCTTAGATT 3’) generated an amplicon of approximately 500 bp from the region upstream of the Zeocin cassette; primers ZeocinF (5’ GGTCGTGTCCACGAACTTCC 3’) and PET117 B-R (5’ ACGCCAATGATTCCATTCTC 3’) generated an amplicon of approximately 900 bp from the region downstream of the Zeocin cassette. The amplicons were characterized by nucleotide sequencing, and specific disruption of PET117 was confirmed by comparison to sequences in the C. glabrata genome database (http://cbi.labri.fr/Genolevures/cagl.php#).
2.7 Spectrophotometric analysis of photosensitizer association with yeast
C. glabrata parental strain BG2 and ATP-dependent RNA helicase mutant suv3, C. glabrata BG2 and cytochrome c oxidase assembly mutant pet117, and S. cerevisiae parental strain S288C and glutaredoxin mutant grx5 were incubated with the same concentration of TMP-1363 (10 μg/mL) for approximately 30 minutes, washed with dH2O to remove unbound photosensitizer and resuspended in dH2O. Steady state fluorescence spectra from these samples were obtained using optimal TMP-1363 excitation wavelengths (near 425 nm) determined experimentally from excitation spectra. In a separate series of experiments, we performed fluorescence excitation and emission spectroscopy on these organisms in the absence of TMP-1363 incubation in order to evaluate a possible role for endogenous porphyrins in conferring enhanced sensitivity to PDT. Excitation and emission spectra were acquired using a Varian Cary Eclipse fluorometer (Varian Instruments, Walnut Creek, CA). Sample volumes were 3 mL and contained approximately 2-4 × 108 organisms/mL.
2.8 Statistical analysis
To obtain an adequate sample number for statistical analysis, each experimental condition was assayed in triplicate, and each experiment was performed at least three times. Data points for a given condition from each experiment were combined for subsequent data analysis and analyzed as the mean ± standard deviation. Comparisons of samples in the same group, and between-group comparisons, were made using the Student's t test. Comparisons among strains or treatment groups were made by one-way analysis of variance (ANOVA) to permit comparison of multiple groups or treatment conditions in the same analysis. In each case, p values ≤0.05 were considered significant.
3. Results
3.1 Metabolic inhibition of respiration enhances sensitivity of Candida to oxidative stress induced by PDT
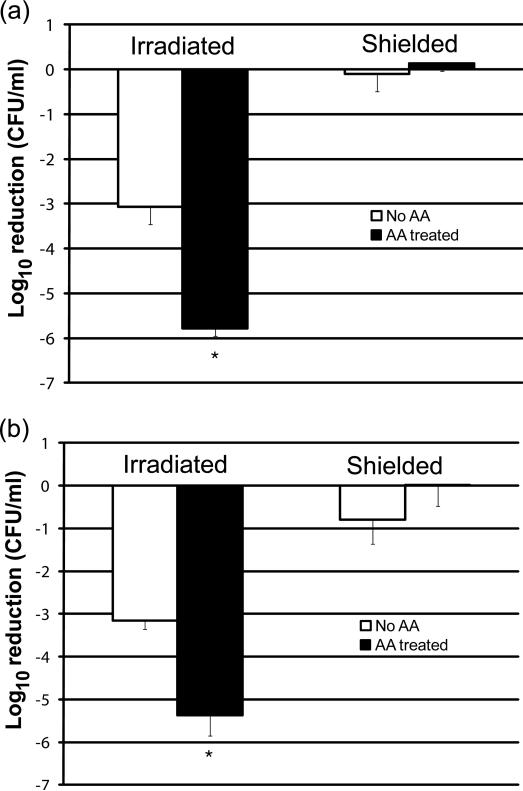
C. glabrata possesses only the conventional, cyanide-sensitive cytochrome c respiratory pathway[13]. C. albicans also possesses the cyanide-insensitive alternative oxidase (AOX) respiratory pathway [24]. Antimycin A (AA) specifically inhibits the Rieske iron-sulfur (Fe-S) protein in the bc1 complex (Complex III) of the conventional respiratory pathway [25] by a mechanism that increases oxidative stress in yeast [26]. However, AA does not inhibit the AOX pathway of C. albicans [21]. To determine whether AA pre-treatment would increase the sensitivity of C. albicans and C. glabrata to PDT, organisms were grown overnight in either the presence or absence of AA, and subjected to PDT [17, 27]. Pre-treatment with AA significantly enhanced (p<0.001) the sensitivity of both C. albicans and C. glabrata to PDT compared to untreated organisms (Figure 2), resulting in an additional 2-log10 reduction in CFU compared to cells grown in the absence of AA. AA treatment did not appear to exert toxicity to either species in organisms shielded from irradiation.
Figure 2. Metabolic inhibition of respiratory Complex III by antimycin A increases the sensitivity of C. albicans and C. glabrata to PDT.
C. albicans (panel a) and C. glabrata (panel b) were grown overnight to early stationary phase in YPDextrose at 37°C with vigorous aeration, in either the presence (closed bars) or absence (open bars) of the inhibitor of respiratory complex III inhibitor antimycin A (10 μM AA). Organisms were adjusted to 107 cells/mL, incubated with 10 μg/mL of TMP-1363 for 10 min and irradiated at 2.4 J/cm2 with broadband visible light. Untreated organisms and organisms treated with TMP-1363 but shielded from light were used as controls. Organism killing was determined by the colony forming unit (CFU) assay and represented as a log10 reduction compared to untreated organisms. Data represent the mean ± S.D. of three experiments performed in duplicate. Pre-treatment with AA significantly (*) enhanced (p<0.001) the sensitivity of both C. albicans and C. glabrata to PDT compared to untreated organisms.
3.2 Specific respiratory-deficient mutants display increased sensitivity to PDT
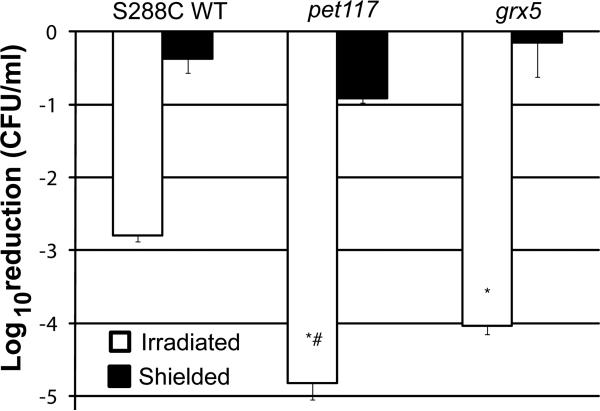
The observations in Figure 2 and our previous studies [17] led us to investigate the role of specific gene products in conferring sensitivity to singlet oxygen-mediated stress induced by PDT. Using S. cerevisiae as a model, previous studies by Herrero and colleagues [28] compared gene expression in respiratory-deficient mutants pet117 and grx5 following oxidative stress, and observed grx5 to be hypersensitive to oxidative stress. We used a S. cerevisiae non-essential gene, haploid deletion library [29] available to us (courtesy of Dr. Damian J. Krysan, University of Rochester, Rochester, NY) to compare pet117 and grx5 to parental strain S288C for their sensitivity to PDT (Figure 3). Pet117 participates in the assembly of cytochrome c oxidase [30]. Grx5 is a glutaredoxin found in the mitochondrial matrix, where it participates in Fe-S cluster formation in proteins, including cytochromes [31]. Both S. cerevisiae respiratory-deficient mutants pet117 and grx5 displayed significantly increased sensitivity to oxidative stress mediated by PDT (p<0.001) compared to parental strain S288C. The pet117 mutant was also significantly more sensitive to PDT (p=0.003) compared to grx5. These data indicate that cytochrome assembly and maturation are likely important in determining sensitivity to PDT.
Figure 3. Genetic evidence that Pet117 and Grx5 participate in sensitivity to PDT in S.
Early stationary phase yeast of S. cerevisiae S288C, pet117 and grx5 were incubated with 10 μg/ml TMP-1363 for 10 min and irradiated with a fluence of 2.4 J/cm2 broadband visible light (open bars). Untreated organisms and organisms treated with TMP-1363 but shielded from light (closed bars) were used as controls. Organism killing was determined by the CFU assay and represented as a log10 reduction compared to untreated organisms. Data represent the mean ± S.D. of three experiments performed in triplicate. Both S. cerevisiae respiratory-deficient mutants pet117 and grx5 displayed significantly (*) increased sensitivity to oxidative stress mediated by PDT (p<0.001) compared to parental strain S288C. The pet117 mutant was also significantly (#) more sensitive to PDT (p=0.003) compared to grx5.
3.3 C. glabrata pet117 insertion mutants are respiratory deficient and display increased sensitivity to PDT
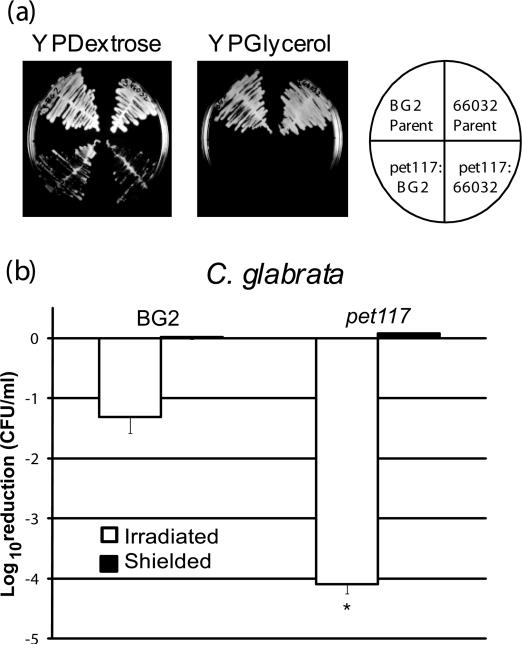
To compare mutants in S. cerevisiae demonstrating increased sensitivity to PDT with mutants in orthologous genes in C. glabrata, we utilized the method of Alderton, et al. [22] for generating targeted mutants in C. glabrata by homologous recombination. Briefly, linear DNA is recombinagenic in yeasts such as S. cerevisiae and C. glabrata. Transformation of contiguous genomic DNA separated by a selectable marker will undergo homologous recombination at high frequency, and selection is achieved by plating on the appropriate medium. We initiated our studies by targeting C. glabrata PET117. To corroborate phenotypes observed in C. glabrata parental strain BG2 [23], we also transformed a second strain, C. glabrata 66032 [32] (American Type Culture Collection, Manassas, VA). Since we predicted C. glabrata pet117 would be respiratory-deficient, we screened transformants by colony transfer to YPDextrose and YPGlycerol agar plates to identify clones that grew on YPDextrose but not YPGlycerol, since glycerol is a non-fermentable carbon source. We identified numerous Zeocin-resistant transformants in each strain background that were respiratory-deficient based on their inability to grow on YPGlycerol. Representative clones are shown in Figure 4A. In addition, we confirmed the genomic location of the Zeocin resistance cassette in the PET117 locus by PCR amplification, nucleotide sequencing and BLAST analysis of the amplicons (data not shown). As seen in Figure 4B, C. glabrata pet117 exhibited significantly enhanced sensitivity to PDT compared to parental strain BG2 (p<0.001), with a reduction of more than 4-log10 CFU in the irradiated samples. In contrast, wild type BG2 had approximately 1.5-log10 reduction of CFU. There was no dark toxicity observed in the shielded samples of either the parental strain or pet117.
Figure 4. C. glabrata pet117 insertion mutants are respiratory deficient and display increased sensitivity to PDT.
(a) Representative clones in both the C. glabrata BG2 and C. glabrata 66032 background incubated at 30oC for 48 h are shown. The diagram on the rightshows the organization of the isolates on the YPDextrose and YPGlycerol agar plates. Both transformants show the “petite” phenotype of smaller colony size and reduced growth on YPDextrose, and the putative pet117 insertion mutants are unable to grow on YPGlycerol, confirming their respiratory deficiency (lower quadrants). The parental strains BG2 and 66032 grew well on both media (upper quadrants).
(b) Early stationary phase yeast of C. glabrata BG2 and pet117 were treated with 0.5 μg/ml TMP-1363 for 10 min and irradiated at a fluence of 2.4 J/cm2 with broadband visible light (open bars). Untreated organisms and organisms treated with TMP-1363 but shielded from light (closed bars) were used as controls. Sensitivity to PDT was assessed by the CFU assay. Data are expressed as a log10 reduction in CFU compared to untreated organisms, and represent the mean + S.D. of three experiments performed in triplicate. C. glabrata pet117 exhibited significantly (*) enhanced sensitivity to PDT compared to parental strain BG2 (p<0.001).
3.4 Selected pathways leading to respiratory deficiency in C. glabrata contribute to increased susceptibility to PDT
We proceeded to examine Tn7 insertion mutants of C. glabrata with disruptions in genes known to contribute to mitochondrial function for increased sensitivity to PDT. The mutants have been described in detail [23] and were generously provided by Dr. Brendan P. Cormack (Johns Hopkins University, Baltimore, MD). We addressed the question of whether disruption of all pathways that led to respiratory deficiency resulted in increased sensitivity to PDT. Examination of a limited set of mitochondrial Tn7 insertion mutant strains [23] showed that this was not the case, and illustrates that selected pathways contribute more significantly to sensitivity to PDT.
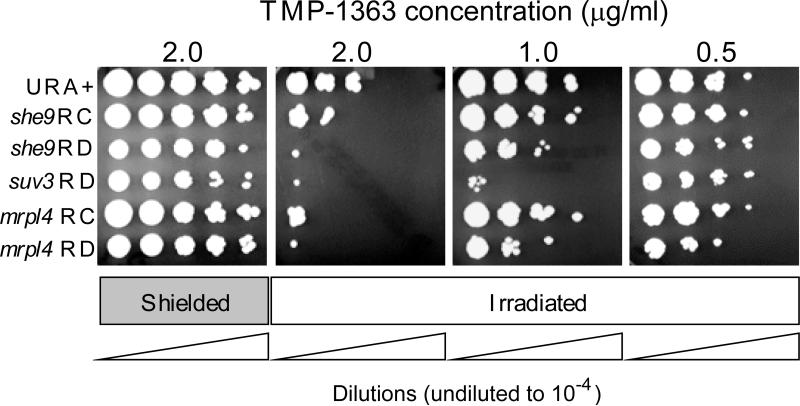
MRPL4 encodes a structural protein in the large subunit of the mitochondrial ribosome. The mrpl4 mutant is respiratory competent and fluconazole sensitive. However, when mrpl4 was challenged by plating on YPDextrose agar supplemented with fluconazole [23], clones were selected that were respiratory-deficient, based on an inability to grow using the non-fermentable substrate glycerol as a carbon source (mrpl4 RD). SHE9 encodes a mitochondrial inner membrane protein of unknown function. Similar to the mrpl4 strains, the original she9 mutant was respiratory-competent, but following challenge with fluconazole, respiratory-deficient clones were again isolated. It is unknown why the survivors of fluconazole treatment became respiratory-deficient. SUV3 encodes an ATP-dependent mitochondrial RNA helicase [33, 34]. In contrast to mrpl4 and she9, the initial suv3 mutant was respiratory deficient in C. glabrata. We compared the sensitivity of the mutant strains to PDT with a URA+ transformant that is used as a control strain for the C. glabrata mutants. The URA+ strain has equivalent sensitivity to PDT to the parental BG2 strain from which the mutants were derived (data not shown). Representative results are shown in Figure 5. PDT using 1.0 μg/ml TMP-1363 showed the greatest contrast among the strains. While the she9 and mrpl4 respiratory-deficient strains were somewhat more sensitive to PDT than their respiratory-competent counterparts, suv3 was at least an order of magnitude more sensitive than the other RD mutants, and over 2 orders of magnitude more sensitive than the URA+ strain. Taken together, the data in Figure 5 suggest not all mitochondrial mutations result in sensitivity to oxidative stress mediated by PDT, and some respiratory deficient mutants are more sensitive to PDT than others.
Figure 5. Selected pathways leading to respiratory deficiency in C. glabrata contribute to increased susceptibility to PDT.
To better reveal differences in sensitivity to PDT among the strains, organisms were adjusted to 107 cells/ml, incubated with a range of concentrations (0.5 – 2.0 μg/ml) of TMP-1363 for 10 min and irradiated with a fluence of 2.4 J/cm2 broadband visible light (Irradiated). Organisms treated with TMP-1363 but shielded from light were used as controls (Shielded). Organism killing was determined by spotting 2 μl of ten-fold serial dilutions of treated cell suspensions on YPDextrose plates, followed by incubation at 37°C for 24 h. Representative results are shown. Abbreviations: RC (respiratory-competent); RD (respiratory-deficient).
3.5 Hypersensitivity of respiratory-deficient mutants to PDT is not a result of increased accumulation of TMP-1363
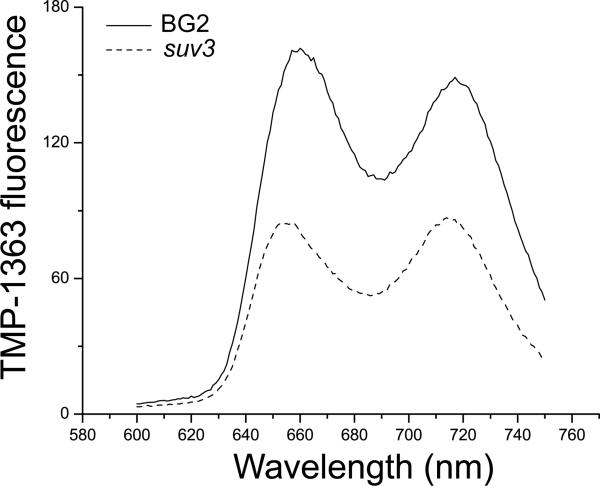
We investigated the possibility that the hypersensitivity to PDT of the respiratory-deficient mutant strains resulted from differences in uptake of the photosensitizer by comparing three parental/respiratory-deficient mutant pairs. C. glabrata parental strain BG2 and ATP-dependent RNA helicase mutant suv3, C. glabrata BG2 and cytochrome c oxidase assembly mutant pet117, and S. cerevisiae parental strain S288C and glutaredoxin mutant grx5 were incubated with the same concentration of TMP-1363 (10 μg/mL) for approximately 30 minutes, washed with dH2O to remove unbound photosensitizer and resuspended in dH2O. Steady state fluorescence spectra from these samples were obtained using optimal TMP-1363 excitation wavelengths (near 420 nm) determined experimentally from excitation spectra. Fluorescence characteristic of TMP-1363 was the overwhelmingly dominant feature in all emission spectra. Peak TMP-1363 emission from wild type strains was consistently higher than that from the mutants, as indicated in the representative pair of spectra for the C. glabrata BG2/suv3 pair shown in Figure 6. In each case, TMP-1363 emission was approximately 2-fold to 8-fold higher in the respective parental strains vs. their mutant counterparts, thus enabling a clear rejection of the hypothesis that increased sensitizer accumulation contributed to the hypersensitivity to PDT of these specific mutant strains.
Figure 6. Comparison of cell-associated photosensitizer levels in C. glabrata parental and respiratory-deficient strains.
TMP-1363 steady state fluorescence from wild type C. glabrata BG2 (solid line) and respiratory-deficient mutant suv3 (dashed line) strains. The excitation wavelength was 425 nm. The shapes of the TMP-1363 excitation spectra (not shown) from these two strains were indistinguishable, thus the differences in amplitude reflect significant differences in sensitizer concentration. In this case, the level of TMP-1363 associated with respiratory-deficient mutant C. glabrata suv3 was less than its parent BG2.
3.6 Hypersensitivity of respiratory-deficient mutants to PDT is not a result of increased accumulation of endogenous porphyrins
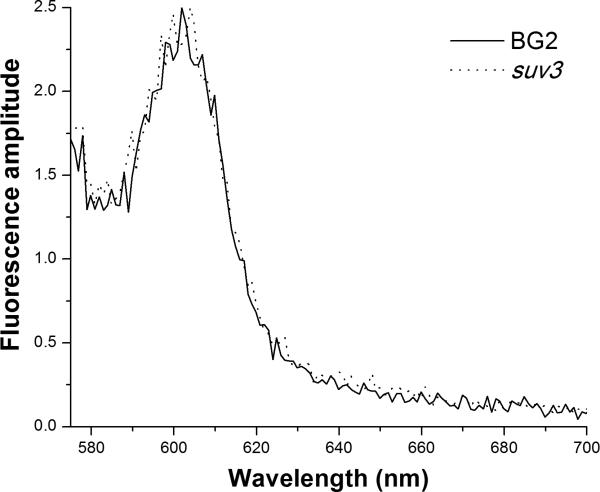
We also considered the possibility that mitochondrial mutants could accumulate abundant endogenous porphyrins as a result of defects in the heme synthesis pathway. Indeed, it has been suggested [35] that endogenous porphyrins contribute to photosensitivity of yeast. We investigated this possible mechanism in two ways. First, we performed steady state excitation and fluorescence emission spectroscopy on the 3 pairs of wild type and corresponding mutant organisms described above. No detectable emission was observed when cells were excited at 425 nm, the optimal wavelength for TMP-1363 emission spectra (Figure 6). Excitation spectroscopy revealed a peak near 500 nm. Using this excitation wavelength, we did not observe any fluorescence spectral features or amplitudes that distinguished the wild type C. glabrata cells from their mutant counterpart, as shown in Figure 7. Similar results were observed with the other 2 pairs of wild type and mutant organisms.
Figure 7. Comparison of endogenous fluorescence levels in C. glabrata parental and respiratory-deficient mutant strains.
Endogenous fluorescence from wild type C. glabrata BG2 (solid line) and respiratory-deficient mutant suv3 (dashed line) was excited at 500 nm. The spectral shape and amplitude of the emission from these two strains were indistinguishable. The displayed amplitudes were scaled to illustrate the 50-100-fold lower emission intensities of the endogenous fluorescence relative to that of TMP-1363 (Fig. 6).
Further, the amplitudes of the endogenous fluorescence signatures were 50-100 fold lower than those of TMP-1363, making it extremely unlikely that these fluorophores contribute to the PDT response of wild type or mutant cells. Finally, the fluorescence spectra were not consistent with that of protoporphyrin IX or with any of the fluorescent precursors in the heme synthesis pathway. Second and more directly, we have performed light-only (no TMP-1363) control experiments in all of the PDT-sensitive respiratory-deficient mutant strains and have observed no phototoxicity, even at fluences 3-fold above those used in PDT experiments (data not shown).
4. Discussion
In the model yeast S. cerevisiae, mitochondrial function is required for innate resistance to oxidative stress caused by reactive oxygen species (ROS) produced during oxidative phosphorylation occurring in the mitochondrial inner membrane, independent of the adaptive response to oxidative stress [36]. Therefore, intact mitochondrial function is likely to provide a basal level of innate anti-oxidant defense against PDT-induced phototoxicity in both S. cerevisiae and Candida. In an earlier study [17], we found that respiratory-deficient mutants of C. albicans and C. glabrata were significantly more susceptible to killing by PDT using TMP-1363 compared to their respective parental strains.
In the present report, we sought to identify specific pathways affecting mitochondrial function that contributed to this increase in sensitivity to PDT. While multiple pathways lead to impairment of respiratory function in fungi [37], our results suggest that selected, not all, pathways leading to respiratory deficiency contribute to increased susceptibility to oxidative stress induced by PDT. The pathways we identified that increase sensitivity to PDT involve the assembly and function of the electron transport chain (ETC). Based on extensive studies in S. cerevisiae (reviewed in [25, 31, 38]) and recent work on Candida [20], we propose that impairment of ETC function at selected sites adds to oxidative stress in the cell, thereby increasing its sensitivity to the added oxidative stress induced by PDT.
The increased sensitivity of Candida to PDT following pre-treatment with cytochrome bc1 (Complex III) inhibitor antimycin A supports this concept. During respiration, Complex III embedded in the mitochondrial inner membrane accepts two electrons from ubiquinol near the outer surface of the inner membrane. Superoxide anion is produced during each of the two electron transfers via the leakage of electrons to molecular oxygen present within the inner membrane. Binding of antimycin A to the cytochrome b subunit blocks electron flux by maintaining the b cytochromes in a reduced state. As a consequence, electron flow to the Rieske iron-sulfur protein is also blocked [38]. In S. cerevisiae, this results in a significant increase in both mitochondrial superoxide anion [26] and hydrogen peroxide production [39].
Antimycin A binds stoichiometrically with comparable affinity to cytochrome bc1 complexes from S. cerevisiae and bovine heart mitochondria [40]. The Complex III inhibitor ilicicolin H binds to the same (Qi) site on cytochrome bc1 as antimycin A, but less tightly to the bovine enzyme by nearly two orders of magnitude compared to the yeast enzyme [40]. This observation would indicate that structural differences between fungal and mammalian cytochrome b subunits could be exploited to advantage by increasing fungal sensitivity to PDT while reducing potential toxicity of the Complex III inhibitor to host tissue.
A second potential target within respiratory Complex III is illustrated by the increased sensitivity of S. cerevisiae grx5 to PDT compared to its parental strain. We initially chose respiratory-deficient S. cerevisiae grx5 for analysis because of its increased sensitivity to other oxidants [28]. Grx5 is located in the mitochondrial matrix where its primary biological role is participation in the late stages of Fe-S cluster incorporation into proteins, including the Rieske Fe-S protein of Complex III of the respiratory chain [41]. S. cerevisiae grx5 displays inactive Fe-S enzymes and accumulates intracellular iron [31, 41]. The excess iron can replace the manganese cofactor of S. cerevisiae mitochondrial superoxide dismutase Sod2, leading to enzyme inactivation [42]. Following PDT, the excess iron may also participate in the formation of additional reactive oxygen radicals via the Fenton reaction, thereby increasing damage to macromolecules [43]. This possibility is supported by the observation that constitutive carbonylation of proteins, an indicator of protein oxidation, is observed in S. cerevisiae grx5 even without the application of external oxidative stress, suggesting Grx5 protects against oxidative damage of proteins [44]. Together, these events potentially lead to increased oxidative stress in grx5 [28] that likely increases its sensitivity to PDT compared to its parental strain.
Initial studies in S. cerevisiae examining gene expression in grx5 following oxidative stress [28] utilized respiratory-deficient mutant pet117 [30] as a comparator to control for the effects of the petite [18] phenotype on the transcriptome. Pet117 is an assembly factor required for cytochrome c oxidase (Complex IV) and, as indicated by the pet designation, is essential for respiratory function. Interestingly, when we compared the sensitivity to PDT of S. cerevisiae pet117 and grx5 (Figure 3), pet117 was significantly more sensitive to PDT than both parental strain S288C and grx5, indicating Complex IV of the respiratory pathway is also important for sensitivity to PDT.
Given the close orthologous relationship of Pet117 in S. cerevisiae and C. glabrata (Saccharomyces Genome Database; www.yeastgenome.org), we generated a pet117 insertion mutant in two different C. glabrata parental strains. In both backgrounds, C. glabrata pet117 was more sensitive to PDT than its corresponding parental strain, suggesting the increased sensitivity of pet117 was not strain specific. While respiratory chain components are highly conserved in structure, several assembly factors for Complex IV found in yeast are absent in mammals including Pet100, Cox14, Cox17, and Coa1 (Saccharomyces Genome Database; www.yeastgenome.org), and thus constitute potential therapeutic targets.
Taking advantage of a set of C. glabrata transposon insertion mutants related to mitochondrial function [23], we demonstrated that suv3 displayed the highest degree of sensitivity to PDT. In S. cerevisiae, the ATP-dependent RNA helicase Suv3 and exonuclease Dss1 form a tight functional complex termed the “degradosome” [45] that participates in the processing of 3’ ends of the eight multi-genic C. glabrata mitochondrial transcriptional units [46] to within two bases of a conserved dodecamer protected by a specific, phosphorylated binding protein [33]. Suv3 is also predicted to participate in degradation of three spliced introns [34] from the cytochrome c oxidase subunit 1 transcript in C. glabrata [46].
In yeast, a notable characteristic of the mitochondrial gene expression machinery is the close physical association of its components [47]. Suv3 and Dss1 demonstrate a tight functional interdependence in S. cerevisiae, and co-purify as a complex when expressed in recombinant form. Furthermore, Suv3 is not functional in the absence of Dss1, and Dss1 is only minimally active in the absence of Suv3 [45]. It is reasonable to suggest that the same physical interaction will be true for C. glabrata, since no other candidates for the mitochondrial degradosome have been identified in this pathogen. Since neither Suv3 nor Dss1 has a predicted transmembrane domain, it is also likely that these two proteins interact in the mitochondrial matrix at or near the site of translation, the mitochondrial ribosome, in both S. cerevisiae [48] and C. glabrata. A potential strategy to increase the sensitivity of C. glabrata to PDT is to disrupt the interaction between these two components of the mitochondrial “degradosome” to induce respiratory deficiency [48].
The tetra-cationic photosensitizer TMP-1363 has been highly effective in our previous in vitro studies of PDT against both C. albicans and C. glabrata [17, 27]. TMP-1363 is also effective in antibacterial PDT in vitro in the presence of serum [8]. Preliminary studies indicate that efficacy of PDT against Candida is reduced using photosensitizers with fewer cationic substitutions compared to TMP-1363. Similar trends are observed in both parental and respiratory-deficient strains. At present, it is unknown whether respiratory deficiency enhances sensitivity to PDT in other pathogenic microorganisms. Our data suggest a possible two-step therapeutic strategy, where fungal respiration is first selectively inhibited, increasing susceptibility to oxidative stress induced by PDT and leading to organism death. Characterizing specific metabolic pathways and gene products that can be targeted to increase the sensitivity of Candida to PDT brings us closer to the identification of specific inhibitors that may be used to initiate such a two-step strategy. This may permit the use of lower PDT doses to eliminate fungal infection, while increasing the therapeutic index.
Acknowledgements
The authors thank Dr. Brendan P. Cormack for providing C. glabrata strains and Dr. Damian J. Krysan for providing S. cerevisiae strains. We thank Dr. David Kessel for providing us with the light box used for PDT irradiation.
Funding
This work was supported by grants from the National Institutes of Health (DE016537 to C.G.H.; CA68409 to T.H.F.). Y.C.R. was supported in part from the National Institutes of Health by a training grant (T32AI07362). F.J.D.A. was supported by a Post-Baccalaureate Research Program for Minority Students (PREP) from the National Institutes of Health (R25 GM064133-06). There was no participation from any of the funding sources in the work described or the preparation of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
None to declare.
References
- 1.Calderone RA. Candida and Candidiasis. ASM Press; Washington, D.C.: 2002. [Google Scholar]
- 2.Fidel PL, Jr., Vazquez JA, Sobel JD. Candida glabrata: review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin. Microbiol. Rev. 1999;12:80–96. doi: 10.1128/cmr.12.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfaller MA, Messer SA, Hollis RJ, Jones RN, Doern GV, Brandt ME, Hajjeh RA. Trends in species distribution and susceptibility to fluconazole among blood stream isolates of Candida species in the United States. Diag. Microbiol. Infect. Dis. 1999;33:217–222. doi: 10.1016/s0732-8893(98)00160-6. [DOI] [PubMed] [Google Scholar]
- 4.Johnson EM, Warnock DW, Luker J, Porter SR, Scully C. Emergence of azole drug resistance in Candida species from HIV-infected patients receiving prolonged fluconazole therapy for oral candidosis. J. Antimicrob. Chemother. 1995;35:103–114. doi: 10.1093/jac/35.1.103. [DOI] [PubMed] [Google Scholar]
- 5.Li L, Redding S, Dongari-Bagtzoglou A. Candida glabrata: an emerging oral opportunistic pathogen. J. Dent. Res. 2007;86:204–215. doi: 10.1177/154405910708600304. [DOI] [PubMed] [Google Scholar]
- 6.Kojic EM, Darouiche RO. Candida infections of medical devices. Clin. Microbiol. Rev. 2004;17:255–267. doi: 10.1128/CMR.17.2.255-267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramage G, Martinez JP, Lopez-Ribot JL. Candida biofilms on implanted biomaterials: a clinically significant problem. FEMS Yeast Research. 2006;6:979–986. doi: 10.1111/j.1567-1364.2006.00117.x. [DOI] [PubMed] [Google Scholar]
- 8.Donnelly RF, Cassidy CM, Loughlin RG, Brown A, Tunney MM, Jenkins MG, McCarron PA. Delivery of Methylene Blue and meso-tetra (N-methyl-4-pyridyl) porphine tetra tosylate from cross-linked poly(vinyl alcohol) hydrogels: a potential means of photodynamic therapy of infected wounds. J. Photochem. Photobiol. B. 2009;96:223–231. doi: 10.1016/j.jphotobiol.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 9.Wainwright M. Photodynamic antimicrobial chemotherapy (PACT). J. Antimicrob. Chemother. 1998;42:13–28. doi: 10.1093/jac/42.1.13. [DOI] [PubMed] [Google Scholar]
- 10.Jori G, Brown SB. Photosensitized inactivation of microorganisms. Photochem. Photobiol. Sci. 2004;3:403–405. doi: 10.1039/b311904c. [DOI] [PubMed] [Google Scholar]
- 11.Traven A, Wong JM, Xu D, Sopta M, Ingles CJ. Interorganellar communication. Altered nuclear gene expression profiles in a yeast mitochondrial dna mutant. J. Biol. Chem. 2001;276:4020–4027. doi: 10.1074/jbc.M006807200. [DOI] [PubMed] [Google Scholar]
- 12.Cheng S, Clancy CJ, Zhang Z, Hao B, Wang W, Iczkowski KA, Pfaller MA, Nguyen MH. Uncoupling of oxidative phosphorylation enables Candida albicans to resist killing by phagocytes and persist in tissue. Cell. Microbiol. 2007;9:492–501. doi: 10.1111/j.1462-5822.2006.00805.x. [DOI] [PubMed] [Google Scholar]
- 13.Brun S, Aubry C, Lima O, Filmon R, Berges T, Chabasse D, Bouchara JP. Relationships between respiration and susceptibility to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 2003;47:847–853. doi: 10.1128/AAC.47.3.847-853.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gyurko C, Lendenmann U, Troxler RF, Oppenheim FG. Candida albicans mutants deficient in respiration are resistant to the small cationic salivary antimicrobial peptide histatin 5. Antimicrob. Agents Chemother. 2000;44:348–354. doi: 10.1128/aac.44.2.348-354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng S, Clancy CJ, Nguyen KT, Clapp W, Nguyen MH. A Candida albicans petite mutant strain with uncoupled oxidative phosphorylation overexpresses MDR1 and has diminished susceptibility to fluconazole and voriconazole. Antimicrob. Agents Chemother. 2007;51:1855–1858. doi: 10.1128/AAC.00182-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanglard D, Ischer F, Bille J. Role of ATP-binding-cassette transporter genes in high-frequency acquisition of resistance to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 2001;45:1174–1183. doi: 10.1128/AAC.45.4.1174-1183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chabrier-Rosello Y, Foster TH, Mitra S, Haidaris CG. Respiratory deficiency enhances the sensitivity of the pathogenic fungus Candida to photodynamic treatment. Photochem. Photobiol. 2008;84:1141–1148. doi: 10.1111/j.1751-1097.2007.00280.x. [DOI] [PubMed] [Google Scholar]
- 18.Slonimski PP, Perrodin G, Croft JH. Ethidium bromide induced mutation of yeast mitochondria: complete transformation of cells into respiratory deficient non-chromosomal “petites”. Biochem. Biophys. Res. Comm. 1968;30:232–239. doi: 10.1016/0006-291x(68)90440-3. [DOI] [PubMed] [Google Scholar]
- 19.Barros MH, Netto LE, Kowaltowski AJ. H(2)O(2) generation in Saccharomyces cerevisiae respiratory pet mutants: effect of cytochrome c. Free Rad. Biol. Med. 2003;35:179–188. doi: 10.1016/s0891-5849(03)00307-1. [DOI] [PubMed] [Google Scholar]
- 20.Ruy F, Vercesi AE, Kowaltowski AJ. Inhibition of specific electron transport pathways leads to oxidative stress and decreased Candida albicans proliferation. J. Bioenerget. Biomem. 2006;38:129–135. doi: 10.1007/s10863-006-9012-7. [DOI] [PubMed] [Google Scholar]
- 21.Helmerhorst EJ, Murphy MP, Troxler RF, Oppenheim FG. Characterization of the mitochondrial respiratory pathways in Candida albicans. Biochim. Biophys. Acta. 2002;1556:73–80. doi: 10.1016/s0005-2728(02)00308-0. [DOI] [PubMed] [Google Scholar]
- 22.Alderton AJ, Burr I, Muhlschlegel FA, Tuite MF. Zeocin resistance as a dominant selective marker for transformation and targeted gene deletions in Candida glabrata. Mycoses. 2006;49:445–451. doi: 10.1111/j.1439-0507.2006.01271.x. [DOI] [PubMed] [Google Scholar]
- 23.Kaur R, Castano I, Cormack BP. Functional genomic analysis of fluconazole susceptibility in the pathogenic yeast Candida glabrata: roles of calcium signaling and mitochondria. Antimicrob. Agents Chemother. 2004;48:1600–1613. doi: 10.1128/AAC.48.5.1600-1613.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shepherd MG, Chin CM, Sullivan PA. The alternate respiratory pathway of Candida albicans. Arch. Microbiol. 1978;116:61–67. doi: 10.1007/BF00408734. [DOI] [PubMed] [Google Scholar]
- 25.Turrens JF. Mitochondrial formation of reactive oxygen species. J. Physiol. 2003;552:335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turrens JF, Alexandre A, Lehninger AL. Ubisemiquinone is the electron donor for superoxide formation by complex III of heart mitochondria. Arch. Biochem. Biophys. 1985;237:408–414. doi: 10.1016/0003-9861(85)90293-0. [DOI] [PubMed] [Google Scholar]
- 27.Chabrier-Rosello Y, Foster TH, Perez-Nazario N, Mitra S, Haidaris CG. Sensitivity of Candida albicans germ tubes and biofilms to photofrin-mediated phototoxicity. Antimicrob. Agents Chemother. 2005;49:4288–4295. doi: 10.1128/AAC.49.10.4288-4295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belli G, Molina MM, Garcia-Martinez J, Perez-Ortin JE, Herrero E. Saccharomyces cerevisiae glutaredoxin 5-deficient cells subjected to continuous oxidizing conditions are affected in the expression of specific sets of genes. J. Biol. Chem. 2004;279:12386–12395. doi: 10.1074/jbc.M311879200. [DOI] [PubMed] [Google Scholar]
- 29.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M'Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 30.McEwen JE, Hong KH, Park S, Preciado GT. Sequence and chromosomal localization of two PET genes required for cytochrome c oxidase assembly in Saccharomyces cerevisiae. Curr. Genet. 1993;23:9–14. doi: 10.1007/BF00336742. [DOI] [PubMed] [Google Scholar]
- 31.Herrero E, Ros J, Belli G, Cabiscol E. Redox control and oxidative stress in yeast cells. Biochim. Biophys. Acta. 2008;1780:1217–1235. doi: 10.1016/j.bbagen.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 32.Vermitsky JP, Earhart KD, Smith WL, Homayouni R, Edlind TD, Rogers PD. Pdr1 regulates multidrug resistance in Candida glabrata: gene disruption and genome-wide expression studies. Mol. Microbiol. 2006;61:704–722. doi: 10.1111/j.1365-2958.2006.05235.x. [DOI] [PubMed] [Google Scholar]
- 33.Dziembowski A, Stepien PP. Genetic and biochemical approaches for analysis of mitochondrial degradosome from Saccharomyces cerevisiae. Meth. Enzymol. 2001;342:367–378. doi: 10.1016/s0076-6879(01)42559-6. [DOI] [PubMed] [Google Scholar]
- 34.Margossian SP, Li H, Zassenhaus HP, Butow RA. The DExH box protein Suv3p is a component of a yeast mitochondrial 3'-to-5' exoribonuclease that suppresses group I intron toxicity. Cell. 1996;84:199–209. doi: 10.1016/s0092-8674(00)80975-7. [DOI] [PubMed] [Google Scholar]
- 35.Strakhovskaya MG, Shumarina AO, Fraikin G, Rubin AB. Endogenous porphyrin accumulation and photosensitization in the yeast Saccharomyces cerevisiae in the presence of 2,2'-dipyridyl. J. Photochem. Photobiol. B - Biology. 1999;49:18–22. doi: 10.1016/s1011-1344(98)00215-2. [DOI] [PubMed] [Google Scholar]
- 36.Grant CM, MacIver FH, Dawes IW. Mitochondrial function is required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. FEBS Lett. 1997;410:219–222. doi: 10.1016/s0014-5793(97)00592-9. [DOI] [PubMed] [Google Scholar]
- 37.Dimmer KS, Fritz S, Fuchs F, Messerschmitt M, Weinbach N, Neupert W, Westermann B. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:847–853. doi: 10.1091/mbc.01-12-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guzy RD, Schumacker PT. Oxygen sensing by mitochondria at complex III: the paradox of increased reactive oxygen species during hypoxia. Exp. Physiol. 2006;91:807–819. doi: 10.1113/expphysiol.2006.033506. [DOI] [PubMed] [Google Scholar]
- 39.Fang J, Beattie DS. External alternative NADH dehydrogenase of Saccharomyces cerevisiae: a potential source of superoxide. Free Rad. Biol. Med. 2003;34:478–488. doi: 10.1016/s0891-5849(02)01328-x. [DOI] [PubMed] [Google Scholar]
- 40.Rotsaert FA, Ding MG, Trumpower BL. Differential efficacy of inhibition of mitochondrial and bacterial cytochrome bc1 complexes by center N inhibitors antimycin, ilicicolin H and funiculosin. Biochim. Biophys. Acta. 2008;1777:211–219. doi: 10.1016/j.bbabio.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodriguez-Manzaneque MT, Tamarit J, Belli G, Ros J, Herrero E. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol. Biol. Cell. 2002;13:1109–1121. doi: 10.1091/mbc.01-10-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang M, Cobine PA, Molik S, Naranuntarat A, Lill R, Winge DR, Culotta VC. The effects of mitochondrial iron homeostasis on cofactor specificity of superoxide dismutase 2. EMBO J. 2006;25:1775–1783. doi: 10.1038/sj.emboj.7601064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Redmond RW, Kochevar IE. Spatially resolved cellular responses to singlet oxygen. Photochem. Photobiol. 2006;82:1178–1186. doi: 10.1562/2006-04-14-IR-874. [DOI] [PubMed] [Google Scholar]
- 44.Rodriguez-Manzaneque MT, Ros J, Cabiscol E, Sorribas A, Herrero E. Grx5 glutaredoxin plays a central role in protection against protein oxidative damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:8180–8190. doi: 10.1128/mcb.19.12.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malecki M, Jedrzejczak R, Stepien PP, Golik P. In vitro reconstitution and characterization of the yeast mitochondrial degradosome complex unravels tight functional interdependence. J. Mol. Biol. 2007;372:23–36. doi: 10.1016/j.jmb.2007.06.074. [DOI] [PubMed] [Google Scholar]
- 46.Koszul R, Malpertuy A, Frangeul L, Bouchier C, Wincker P, Thierry A, Duthoy S, Ferris S, Hennequin C, Dujon B. The complete mitochondrial genome sequence of the pathogenic yeast Candida (Torulopsis) glabrata. FEBS Lett. 2003;534:39–48. doi: 10.1016/s0014-5793(02)03749-3. [DOI] [PubMed] [Google Scholar]
- 47.Krause K, Lopes de Souza R, Roberts DG, Dieckmann CL. The mitochondrial message-specific mRNA protectors Cbp1 and Pet309 are associated in a high-molecular weight complex. Mol. Biol. Cell. 2004;15:2674–2683. doi: 10.1091/mbc.E04-02-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dziembowski A, Piwowarski J, Hoser R, Minczuk M, Dmochowska A, Siep M, van der Spek H, Grivell L, Stepien PP. The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J. Biol. Chem. 2003;278:1603–1611. doi: 10.1074/jbc.M208287200. [DOI] [PubMed] [Google Scholar]