Abstract
Staphylococcus aureus possesses an impressive arsenal of complement evasion proteins that help the bacterium escape attack of the immune system. The staphylococcal complement inhibitor (SCIN) protein exhibits a particularly high potency and was previously shown to block complement by acting at the level of the C3 convertases. However, many details about the exact binding and inhibitory mechanism remained unclear. In this study, we demonstrate that SCIN directly binds with nanomolar affinity to a functionally important area of C3b that lies near the C terminus of its β-chain. Direct competition of SCIN with factor B for C3b slightly decreased the formation of surface-bound convertase. However, the main inhibitory effect can be attributed to an entrapment of the assembled convertase in an inactive state. Whereas native C3 is still able to bind to the blocked convertase, no generation and deposition of C3b could be detected in the presence of SCIN. Furthermore, SCIN strongly competes with the binding of factor H to C3b and influences its regulatory activities: the SCIN-stabilized convertase was essentially insensitive to decay acceleration by factor H and the factor I- and H-mediated conversion of surface-bound C3b to iC3b was significantly reduced. By targeting a key area on C3b, SCIN is able to block several essential functions within the alternative pathway, which explains the high potency of the inhibitor. Our findings provide an important insight into complement evasion strategies by S. aureus and may act as a base for further functional studies.
The bacterium Staphylococcus aureus is a widely disseminated and persistent human pathogen that has a longstanding and increasingly negative impact on human health. It is a primary etiologic agent of numerous disorders that range from the seemingly mild, such as skin and soft tissue infections, to several potentially fatal conditions, including bacteremia, necrotizing pneumonia, and endocarditis, as well as infections associated with implanted medical devices, such as intravascular catheters, pacemakers, and delivery pumps (1). Treatment options for staphylococcal disease have become more limited in recent years in large part because of the proliferation of antibiotic-resistant strains of the organism. Thus, it has become clear that if effective therapies and preventative measures against S. aureus infection are to be provided in the future, a more complete understanding of this complex organism and how it interacts with various human physiological systems is required today.
Perhaps one of the most striking features of S. aureus biology is its ability to colonize and survive in a number of unique microenvironments within its host. This ability contributes to its propensity to form long-lasting and repetitive infections (1), even in the presence of a robust immune response. Over the last several years, studies from a number of different investigators have revealed that S. aureus achieves this ability in part by producing a variety of immunomodulators that affect both arms of the host immune system (2, 3). Because the complement system is an essential first line of defense against invading microbial pathogens and is a central component of immunity, it is a prime target of staphylococcal immune evasion strategies (4). In fact, S. aureus secretes at least 10 unique proteins that disrupt processes essential to either complement initiation or amplification, or downstream chemotactic events that depend upon the anaphylatoxins generated during complement activation (reviewed in Ref. 4–6). Very recently, it was discovered that S. aureus may even disrupt the activation of adaptive immune responses by inhibiting the interaction of C3d with complement receptor (CR)4 type 2 (7).
Among this suite of immune evasion factors, the staphylococcal complement inhibitor (SCIN) family is comprised of four small (~9.8 kDa), secreted proteins that adopt a three-helix bundle fold (8, 9). SCIN proteins are structurally homologous to the IgG-binding modules of staphylococcal protein A (4, 9), and are related more distantly to the other complement inhibitors extracellular fibrinogen-binding protein (Efb) (2, 10), Efb homologous protein (Ehp) (11), and staphylococcal Ig-binding protein (Sbi) (12) from the same organism (4). Within the SCIN family, three of the members (SCIN-A, SCIN-B, and SCIN-C) inhibit complement activation, whereas a fourth form, open reading frame (ORF)-D, lacks this activity (9). This observation, coupled with the higher levels of sequence identity among SCIN-A, SCIN-B, and SCIN-C, was used by Rooijakkers et al. (9) to map the active site of the functional SCIN family members to an 18 residue segment comprised of Leu31-Gly48. According to the crystal structure of SCIN-A, this active region spans the extreme C-terminal portion of helix α1 through approximately the N-terminal half of helix α2.
The effectiveness of the complement-mediated immune response relies on the rapid recognition of microbial cell surface patterns by the classical, lectin, or alternative pathway (of complement activation). All pathways lead to the cleavage of complement component C3 into its active fragments C3a and C3b by bimolecular C3 convertase complexes. Whereas C3a acts as a potent immunomodulator, C3b becomes covalently attached to foreign surfaces. Deposited C3b is not only responsible for the self-amplification of the complement response via the alternative pathway, but it also induces a variety of downstream immune responses. The SCIN proteins are of particular interest for complement evasion because they are capable of potent inhibition of all three complement initiation pathways (8, 13). This feature stands in contrast with the other complement inhibitors Efb, Ehp, and Sbi, which exert their effects primarily through disruption of the alternative pathway (10, 11, 13, 14). Furthermore, the inhibitory mechanism of SCIN is also unique in that they serve to lock the structure of the C3 convertases in a more stable yet inactive state, whereas the native regulators of complement activation factor H (fH), decay acceleration factor, and CR1 all promote accelerated decay rates of the transiently stable convertase assemblies (15). Though the molecular aspects of convertase “stabilization” are not yet elucidated, recent studies suggest that binding of SCIN may restrict the conformational flexibility of the convertase (16). In this regard, it is surprising that SCIN proteins achieve this strong inhibitory effect on the convertase even though they do not appear to bind directly to any complement components other than the fully assembled convertases (8). This is very unusual because all known inhibitory proteins that affect the structure/function of C3 convertases under either normal or pathological states must also bind directly to one or more of the proteins (or their precursors) that are present in the convertase assembly to exert their activities.
This prompted us to investigate the nature of the protein-protein interactions that lie at the heart of the SCIN mechanism. Surprisingly, we observed that active members of the SCIN family bind directly to C3b with nanomolar affinity. Further competition binding studies revealed that the SCIN binding site overlaps with that of fH and factor B (fB) on C3b, a result which suggests that SCIN locks the C3 convertase complex in an inactive state and prevents its accelerated decay by blocking fH binding to the convertase-bound C3b substituent. These findings support and extend our knowledge about the inhibitory mechanism of SCIN and may offer new opportunities for studying S. aureus virulence and for developing therapeutic interventions.
Materials and Methods
Proteins
All complement proteins (C3, C3b, iC3b, fH, fB, factor D (fD), and factor I (fI)) were prepared using established protocols. C3b, iC3b, and C3d were site-specifically biotinylated at the thiol group of their thioester moiety as previously described (17). fH(1–4) was expressed and purified as described recently (18), and fH(19–20) and CR2(1–15) were expressed as reported previously (19, 20).
DNA fragments encoding the predicted mature forms of SCIN (SCIN-A, SCIN-B, or ORF-D) were amplified from S. aureus genomic DNA (strain Mu50) and subcloned into the prokaryotic expression vector pT7HMT (21). Following sequence verification, expression vectors were transformed into Escherichia coli strain BL21(DE3). Each protein was overexpressed, purified by immobilized metal ion affinity chromatography under denaturing conditions, refolded by rapid dilution, and concentrated according to previously described protocols (21). Following removal of the affinity tag by site-specific proteolysis, the proteins were again purified by immobilized metal ion affinity chromatography; in this case, it was the unbound fraction that contained the recombinant SCIN proteins. The unbound fractions were exchanged into 20 mM ethanolamine buffer (pH 9.0) and the proteins contained therein were separated by Resource S cation exchange chromatography. The final purity of each protein was assessed by SDS-PAGE or MALDI-TOF mass spectrometry before experimentation.
Direct binding studies between SCIN and C3 fragments
All surface plasmon resonance (SPR) studies were performed on a Biacore 2000 instrument (GE Healthcare) at 25°C. Scrubber (version 2; BioLogic Software) was used for data processing and evaluation. For an initial screening of C3 fragment binding to the immobilized inhibitor, SCIN (1 µM in 10 mM sodium acetate (pH 5.5)) was attached to a CM5 chip via amine coupling using activation and deactivation contact times of 5 min. Solutions of 1 µM C3b, fB, and fD alone as well as mixtures of C3b plus fB, C3b plus fD, fB plus fD, and C3b plus fB plus fD (1 µM each) were prepared in running buffer (10 mM HEPES (pH 7.4), 150 mM NaCl, 1 mM NiCl2, 0.005% Tween 20) and injected for 3 min at a flow rate of 10 µl/min. After 5 min of undisturbed dissociation, the surface was regenerated for 1 min with 3 mM EDTA. The signals were referenced using the responses from a plain sensor surface and an ensemble of buffer blank injections was subtracted from the processed data (double referencing (22)).
For a more physiological evaluation of the direct binding of SCIN to C3 fragments, thioester-specifically biotinylated forms of C3b, iC3b, and C3d were captured on separate flow cells of a streptavidin-covered sensor chip. A 2-fold dilution series of SCIN (1 nM-10 µM) was injected for 1 min at 20 µl/min in PBS-T (10 mM sodium phosphate, 150 mM NaCl, 0.005% Tween 20, pH 7.4) and the dissociation was observed for 2 min. No regeneration was needed because all signal readily returned to baseline within the dissociation phase window. For comparing the specificity toward the three C3 fragments, the observed signals at 1 µM SCIN were normalized by both the molecular weight and surface density of the corresponding fragment.
Solution phase binding between SCIN and C3b was evaluated by isothermal titration calorimetry (ITC) using an iTC200 instrument (MicroCal). Both C3b and SCIN were dialyzed against sample buffer (PBS without Tween 20). SCIN (125 µM in the syringe) was injected into C3b (12 µM in the cell) at a cell temperature of 25°C using 19 injections of 2.5 µl(5 s), injection intervals of 180 s, a reference power of 8 µcal/s, and a stirring speed of 1000 rpm. Thermograms were evaluated using Origin (version 7; OriginLab), and the data sets were fitted to either a “one set of sites” or “two sets of sites” model.
Structural analysis of C3b-SCIN complexes by small angle X-ray scattering (SAXS)
All protein samples were prepared in a buffer of 20 mM Tris (pH 8.0), 200 mM NaCl, 5% (v/v) glycerol, and passed through a 0.22-µm syringe filter before data collection. SAXS data were collected at the Advanced Light Source (beamline 12.3.1; Lawrence Berkeley National Laboratory) (23). In particular, the wavelength, λ, was set to 1.033 Å and the sample-to-detector distance was fixed at 1.5 m; this resulted in scattering vectors, q, that ranged from 0.008 Å−1 to 0.2 Å−1. The scattering vector is defined as 4π sinθ/λ, where 2θ is the scattering angle. All experiments were performed at 20°C and data was processed as described previously (23). Data acquired at both short and long time exposures (0.5 and 5 s) were merged for calculations that required the entire scattering spectrum. Experimental SAXS data for protein concentrations of 1 and 2 mg/ml were analyzed for potential aggregation using Guinier plots (24). The radius of gyration, Rg, was derived by the Guinier approximation formula I(q) = I0exp(−q2Rg2/3), where the experimental limit of qRg < 1.6 was used. SAXS curves were measured for two C3b and C3b-SCIN concentrations (1 and 2 mg/ml), and displayed no concentration-dependence using this criterion. The program GNOM (25) was used to compute the pair-distance distribution function, P(r). This approach also provided the maximum particle dimension of the macromolecule, Dmax. The molecular weights of C3b and the C3b-SCIN complexes were determined by extrapolating each sample intensity at the zero angle and comparing the value to a standard curve derived from four reference proteins (26). Theoretical SAXS curves for atomic models were calculated by CRYSOL and compared with the experimental curve as originally described by Svergun et al. (27). To further dissect the nature of the C3b-SCIN interaction, scattering data from this complex were analyzed with the program OLIGOMER (28). This program fit the experimental scattering curve to weighted distribution of the theoretical scattering curves for free and dimeric C3b, using a crystal structure of C3b (Brookhaven Protein Data Bank (PDB) code 2I07 (29)) as a model. A plausible structure for a C3b dimer was prepared by rotation of the C3b molecule along the crystallographic 2-fold symmetry axis (c = 147.03 Å); this orientation was chosen because it is the only solution that describes the physiological scenario in which two C3b molecules are immobilized on a continuous surface via their thioester-containing domain (TED). Best-fit solutions obtained by deconvolution of these components suggest that the sample exists as a mixture of the components. ORF-D was used as a negative control in SAXS experiments (9, 13).
Competition assays with fH and fB
The ability of SCIN to compete with fB for binding to C3b was investigated using SPR. For this purpose, a constant concentration of fB (500 nM) in a 5-fold dilution series of SCIN (from 4 nM to 12.5 µM; i.e., molar ratio of fB to SCIN from 125:1 to 1:25) was injected for 1 min at 20 µl/min in running buffer (PBS-T). After 3 min of dissociation, the surface was regenerated by consecutive injections of 2 M NaCl and 0.2 M sodium carbonate (pH 9.0) for 30 s each. The signal intensity immediately before injection end was evaluated, and the responses of the pure SCIN series were subtracted from the corresponding competition signal. These corrected signals were divided by the responses of fB alone (500 nM; i.e., 100% binding) to determine the residual binding. The same experiment was repeated with fH (500 nM) and the competitive strength of SCIN toward both complement factors was compared.
Cofactor activity assay
Because SCIN may interfere with the functionality of fH to act as a co-factor for fI in the degradation of C3b, this possibility was assessed both on the surface and in solution. In an SPR assay, a mixture of fI (0.4 µM) and fH (0.4 µM) in presence or absence of SCIN (1 µM) was injected for 10 min at 2 µl/min and 25°C to immobilized C3b. To monitor the conversion of C3b to iC3b, solutions of fH(1–4), fH(19–20), and CR2 (1 µM each) were injected before and after enzyme treatment and their relative signals were evaluated. In addition, the inhibition of cofactor activity in solution was investigated using a SDS-PAGE-based cofactor assay as described (30).
C3 convertase formation, stabilization, and function
SPR was used to dissect the molecular effect of SCIN on the C3 convertase. In an on-chip convertase formation assay, a 1:1 mixture of fB and fD (100 nM each) was injected onto thioester-immobilized C3b on a streptavidin-coated chip for 2 min at 10 µl/min to establish C3 convertase formation. The regular convertase decay was observed for 1 min before either SCIN (1 µM) or a buffer control was injected for 1 min. Finally, fH(1–4) (1 µM) was injected to accelerate convertase decay, and the postinjection baseline was compared.
To assess the effect of SCIN competition with fB during convertase formation, the convertase was formed by injecting fB plus fD in presence or absence of SCIN as premixed solutions. The association and decay phases of the respective C3 convertases were evaluated and compared.
Finally, the ability of the SCIN-stabilized convertase to bind and cleave native C3 was compared with the unmodified convertase. For this purpose, C3b was immobilized to a CM5 sensor chip via amine coupling and the convertase was formed on-chip as earlier described. Native C3 was injected for 1 min onto the established convertase, and both the slope and postinjection increase in baseline were monitored. SCIN (1 µM) or a buffer control was injected for 1 min, and the injection of native C3 was repeated.
Results
SCIN binds directly to C3b
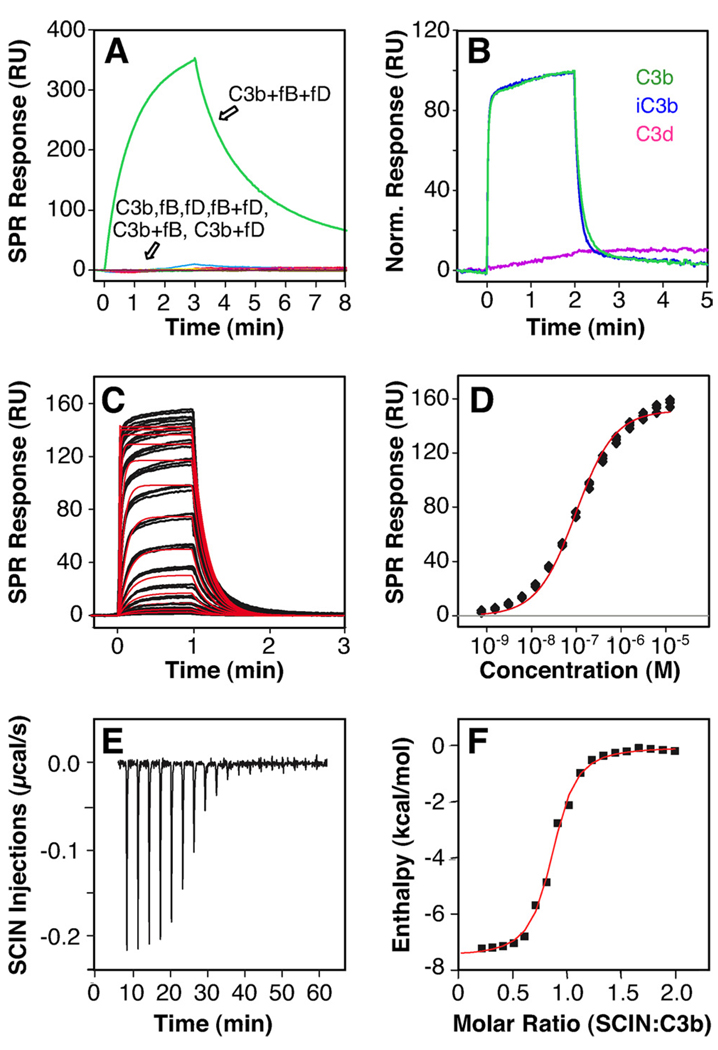
SCIN was previously described to bind to the assembled alternative pathway C3 convertase (i.e., C3bBb) but not to any of its individual components (8, 13). Indeed, when SCIN-A was immobilized on a sensor chip surface via random primary amine groups, no significant binding was detected for C3b, fB, fD, or two-component mixtures thereof. In contrast, an SPR signal was observed when C3b was mixed with fB and fD in a nickel-containing buffer (Fig. 1A). Although a similar response was noted in buffer containing magnesium, no signal was detected in the absence of these divalent metal ions (data not shown). Together, these results strongly suggested binding to the assembled convertase and confirmed the lack of binding for the individual components under these assay conditions.
FIGURE 1.
SCIN binds to both surface-bound and solution-based C3b with nanomolar affinity. A, When SCIN was immobilized on a SPR chip surface, only the convertase mix (C3b plus fB plus fD) showed significant affinity. B, In contrast, solution-based SCIN (1 µM) bound with similar activity to C3b (green) and iC3b (blue) that have been immobilized via their thioester moiety to the SPR sensor chip. No significant binding was observed for C3d (magenta). C, The binding mode and affinity of SCIN was further elucidated using SPR by injecting a dilution series of SCIN (0.8 nM–12.5 µM) to site-specifically immobilized C3b. Despite some heterogeneity at higher concentrations, the interaction featured reproducible triplicate injections (black SPR signals) and fitted reasonably well to a Langmuir 1:1 kinetic model (red fitting curves). D, In agreement with the kinetic analysis, the steady state signals (black filled symbols) could be fitted to a single-binding site model and showed clear signs of saturation (red fitting curve). A direct interaction between SCIN and C3b was also detected in solution using ITC. E, Injection of SCIN (120 µM) into C3b (12 µM) produced a dose-dependent, exothermic response. F, The integrated data (filled black symbols) could be fitted to a single set of sites (red fitting curve). RU, Resonance units. A schematic representation of the assays is available (see supplemental Fig. S6, A–C).5
We likewise used a more physiological assay format, wherein C3b was site-specifically captured on a sensor chip via its thioester group. Remarkably, soluble SCIN-A showed substantial binding to this surface (Fig. 1B, green signal). Furthermore, although the signal intensity was nearly identical for the inactive form of C3b (i.e., iC3b), no significant binding was observed for the C3d fragment (Fig. 1B, blue and magenta signals). These data strongly suggested that SCIN interacts with the C3c part of C3b, and also that the TED is not involved in formation of this complex. Consistent with previous functional data, no interaction was detected for the inactive member of the SCIN family, ORF-D, whereas SCIN-A and SCIN-B bound with apparently similar activity (see supplemental Fig. S1; SCIN-A is referred to as SCIN in subsequent experiments).5
To further characterize the interaction profile, a 2-fold serial-dilution series of SCIN (0.8 nM-12.5 µM) was injected on immobilized C3b. SCIN showed dose-dependent binding with rather fast kinetic on- and off-rates (Fig. 1C). Although clear signs of saturation were seen in the selected concentration range, both the signal intensity and shape deviated slightly from a simple 1:1 model, especially at higher concentrations. To estimate the affinity of the interaction, the “steady-state” phases were evaluated; this analysis revealed a reasonable fit to a single-binding site model with an apparent KD of 177 nM (Fig. 1, D and Table I). Although the quality of fit for the experimental data was improved significantly when a two-binding site model was applied (see supplemental Fig. S2),5 no quantitative assessment of the two affinity constants was performed due to the lack of concentration coverage. Similarly, kinetic analysis of these data sets revealed deviations from a pure Langmuir 1:1 binding model, particularly in the association phase (data not shown). Nevertheless, a kinetic off-rate of 0.09 s−1 could be extracted, which translates to a rather short dissociation half-life of nearly 10 s. When compared with the 60-min half-life for the C3b-Efb-C complex (10), the kinetic stability of the C3b-SCIN complex is much lower. As might be predicted by the highly electropositive nature of SCIN, the interaction with C3b was dependent on the ionicity of the buffer system. This suggests that electrostatic steering effects contribute substantially to the interaction (see supplemental Fig. S3),5 as has also been reported for the C3d-Efb-C complex (31). To examine separately the binding event between C3b and SCIN in solution and to characterize its thermodynamic profile, we also performed an ITC study. Injection of SCIN into C3b generated clear exothermic binding signals that resulted in saturation (Fig. 1E), and therefore demonstrated that SCIN binds to C3b in solution. Even though the data set could be fit to a single set of sites with a KD of 264 nM and an enthalpy of ΔH = −7 kcal/mol, the stoichiometry value of N = 0.75 again indicated a deviation from a pure 1:1 model (Fig. 1F and Table I).
Table I.
Thermodynamic and kinetic profile of the direct interaction between SCIN and C3ba
| kon (105/M s) | koff (10−2/s) | KD,KIN (nM)b | KD,SS (nM)c | KD,ITC (nM)d | ΔHITC (kcal/mol)d | |
|---|---|---|---|---|---|---|
| SCIN-A | 5.5 ± 0.9 | 9.1 ± 1.7 | 169 ± 34 | 177 ± 34 | 264 ± 6 | −7.1 ± 0.5 |
Profile measured using SPR and ITC. Data are mean affinity ± SD.
Calculated as KD = koff/kon; concentration range from 0.8 nM to 12.5 µM; Langmuir 1:1 binding model.
Determined from steady state plot; concentration range from 0.8 nM to 12.5 µM; single-binding site model.
Fitted to a “one set of sites” model with stoichiometry; N = 0.75 ± 0.09 and entropy; ΔS = 6.3 ± 1.7 cal/mol.
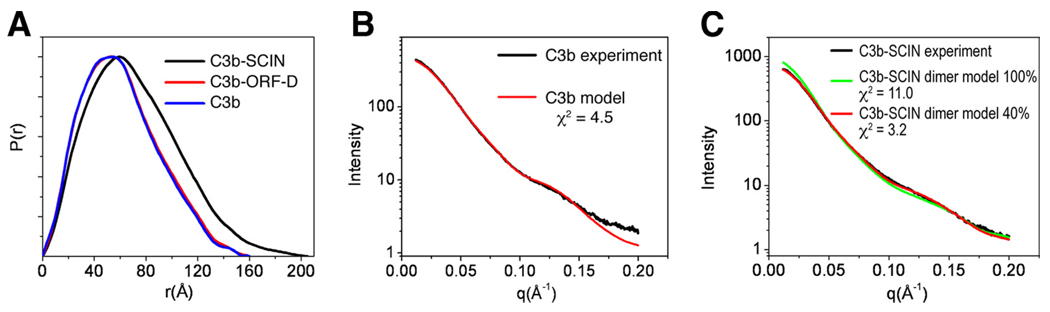
The observed deviations from a 1:1 binding model were further investigated using SAXS, which has evolved into a powerful tool to achieve structural information on protein-protein interactions in solution. This technique is sensitive to changes in protein conformations and can also be used to better understand defined monomer-to-oligomer transitions that occur upon binding. To examine the interactions between C3b and SCIN, we collected SAXS data on C3b alone and in the presence of either SCIN or ORF-D (Fig. 2 and Table II). The pair-distance distribution functions, P(r), derived from the scattering curves for C3b alone or in the presence of equimolar (but nonbinding) ORF-D were indistinguishable. In contrast, the pair-distance distribution function for C3b-SCIN was consistent with substantial increases in the particle size of this complex, as judged by changes in both the radius of gyration (Rg) and the long-tailing nature of the pair-distance distribution function (Fig. 2A). Estimation of the observed molecular weight for each sample by calculating the normalized scattering intensity at zero angle (q = 0; I0) relative to four standard, control proteins (Table II) (see supplemental Fig. S4),5 revealed a nearly 40% increase in the apparent mass of C3b-SCIN relative to the ORF-D control or C3b alone. This change was surprising because SCIN binding to C3b was expected to increase the theoretical molecular mass of the complex by less than 5% relative to C3b alone.
FIGURE 2.
SCIN and C3b form a mixture of monomeric and dimeric complexes when analyzed by SAXS. A, Pair-distance distribution function (P(r)) calculated for scattering data collected on solutions of C3b-SCIN (black), C3b-ORF-D (red), and C3b alone (blue). Note the shift in r(Å) value, where pair-distance distribution function is maximal and the long-tailing pair-distance distribution function curve for C3b-SCIN relative to the control samples. B, Experimental scattering curve of unliganded C3b observed in solution (black) compared with the theoretical scattering curve for an unliganded C3b crystal structure (PDB code 2I07 (29)). C, Guinier analysis of the C3b-SCIN experimental data as compared with theoretical scattering curves for a sample comprised entirely of C3b dimer (green) or 40/60 percentage C3b dimer to C3b monomer (red), as determined by the program OLIGOMER (28). The value for the quality of fit parameter (χ2) is shown in B and C.
Table II.
Dimensional constants for C3b and SCIN complexes derived from SAXS data
| Sample | Rg (Å)a | Dmax (Å)b | Molecular Mass (kDa)c |
|---|---|---|---|
| C3b | 46.9 | 160 | 185 |
| C3b-ORF-D | 47.6 | 160 | 188 |
| C3b-SCIN | 54.2 | 215 | 265 |
Radius of gyration (Rg) given by the Guinier approximation (45).
Dmax is maximum protein distance estimated from pair-distance distribution (P(r)) function.
Calculated from molecular mass versus I0/concentration relationship (see supplemental Fig. S4).5
The results from direct binding studies presented above suggested that although equimolar, the C3b-SCIN complex might exist as equilibrium between 1:1 and 2:2 species. Given the unexpectedly large molecular weight of C3b-SCIN derived from the scattering curve (Table II), we examined whether this complex might be composed of a defined mixture between heterodimeric and heterotetrameric species. The program OLIGOMER was used to deconvolute the experimental C3b-SCIN scattering curve into a weighted distribution of the theoretical scattering curves for free and dimeric C3b (Fig. 2, B and C). Although this analysis for C3b alone confirmed that the scattering curve was described well by the crystal structure of C3b (χ2 = 4.5), we found that the C3b-SCIN complex existed most likely as a percentage 40:60 distribution of tetrameric to dimeric species (χ2 = 3.2). It has to be noted that the small deviation of calculated and experimental scattering observed at high scattering vector (q > 0.18Å−1) may be due to intrinsic flexibility of C3b in solution relative to its crystal structure. This ability may lead to additional variability in the conformations of C3b and probably resulted in the observed deviation in the high scattering vector range. Nevertheless, these SAXS data, in conjunction with the binding studies described, demonstrated that SCIN binds directly to purified C3b, and strongly suggested that the complex exists as an equilibrium mixture of 1:1 and 2:2 species.
SCIN binds to an area near the C terminus of the C3b β-chain
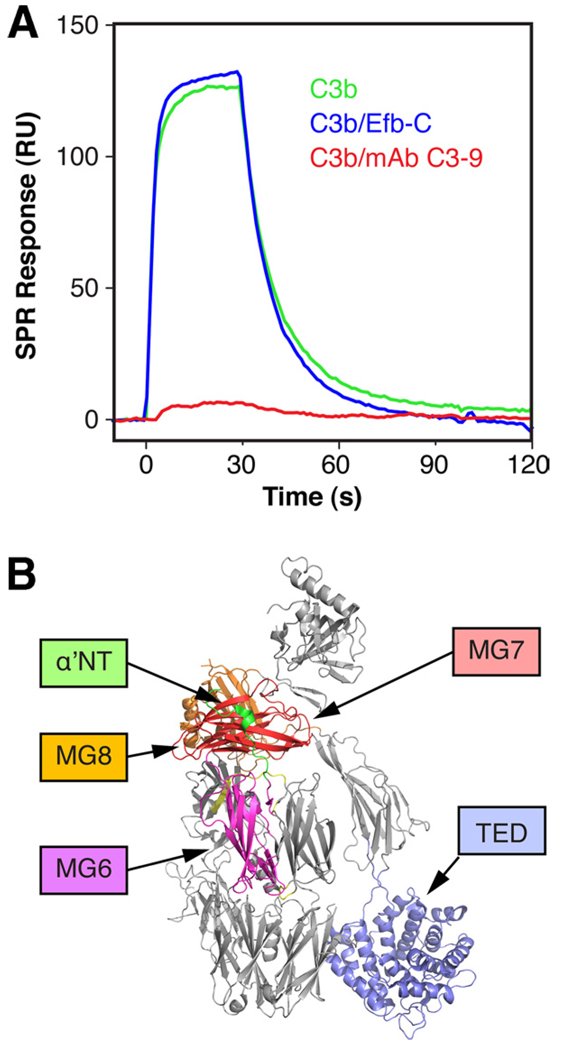
The ability of SCIN to bind C3b and iC3b but not C3d indicated that the SCIN binding site is located within the C3c fragment and not the C3d/TED of C3. In agreement with this observation, formation of a stable complex between C3b and Efb-C, which is known to bind to the TED, did not significantly influence the interaction with SCIN (Fig. 3, blue). In contrast, complex formation with the monoclonal anti-C3c Ab C3–9 almost completely eliminated binding of SCIN to C3b, which confirms the localization on the C3c segment (Fig. 3; red). As negative-stained EM imaging has previously defined the binding area for mAb C3–9 to the vicinity of the β-chain C terminus (i.e., macroglobulin (MG)7/8 domains) (32), a SCIN binding site at or in the proximity of a similar region of C3b seems likely.
FIGURE 3.
The SCIN binding site can be localized at a functionally important area on C3b. A, The binding of SCIN to immobilized C3b was compared in the presence or absence of mAb C3–9 (anti-C3c) and the bacterial complement inhibitor Efb-C. Whereas SCIN bound similarly well to the C3b-Efb-C complex (blue) compared with free C3b (green), mAb C3–9 led to a significant decrease in binding activity (red). B, The domains involved in binding of mAb C3–9 (MG7, MG8 (32)), Efb-C (TED (10)), and the fragment fH(1–2), and Ba (α′NT, MG6, MG7 (18)) have been highlighted on the crystal structure of C3b (PBD code 2I07 (29)). A schematic representation of the assays is available (see supplemental Fig. S6D).5
SCIN competes with complement fB and fH for C3b binding
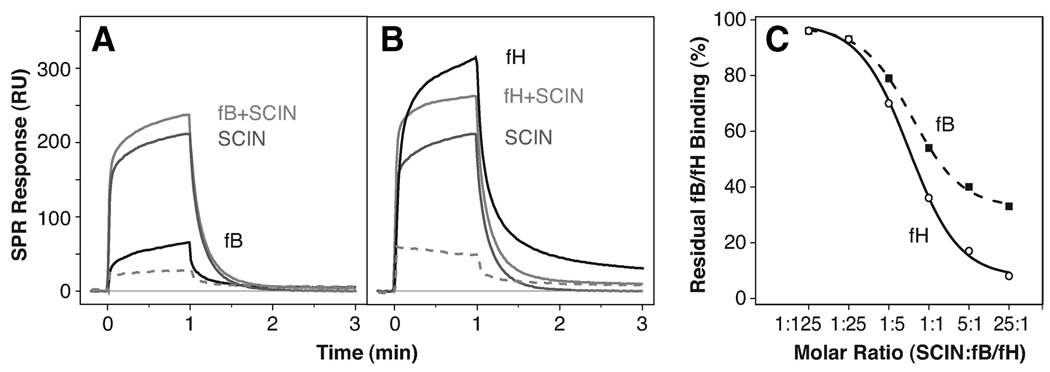
Previous observations suggested that SCIN binds the C3bBb convertase complex and locks it in an inactive state (8). It is therefore reasonable that SCIN may bind to an area on C3b that is adjacent to the contact region for fB (or its Bb fragment). In contrast, the complement regulator fH is known to accelerate decay of the convertase by competing with Bb for C3b. As a consequence, a shared binding region for fB, fH, and SCIN on C3b seemed highly likely. We therefore performed a series of solution competition experiments with SCIN and both fB and fH for binding to C3b (Fig. 4). For this purpose, a constant concentration of the respective complement factor was mixed with increasing concentrations of SCIN and injected onto a biosensor surface on which C3b had been uniformly immobilized via its thioester group. If SCIN and the complement factor bound independently, the resulting SPR signal should be additive. In the case of both fB (Fig. 4A) and fH (Fig. 4B), subtraction of the pure SCIN response revealed a significantly lower binding response in the presence of the inhibitor. The most straightforward interpretation of these data is that SCIN, fB, and fH all compete for an overlapping binding site on C3b. In contrast to fH, which was almost completely inhibited at a 25-fold molar excess of SCIN, the binding of fB to C3b was only partially restricted at the same ratio (Fig. 4C).
FIGURE 4.
SCIN competes with fB and fH for binding to C3b. In an SPR experiment, constant concentrations (500 nM) of either fB (A) or fH (B) in HBS-Mg buffer were injected to immobilized C3b either alone (black line) or as mixtures with increasing concentrations (20 nM–12.5 µM) of SCIN (light gray line). The same dilution series of SCIN alone (dark gray line) was injected and subtracted from the corresponding signal of the fB plus SCIN mixture (dashed curve). For reasons of simplicity, the plots show only one concentration of SCIN (2.5 µM) and fB or fH (500 nM). C, The residual signals for both competition series were divided by the uninhibited fB or fH response and plotted against the molar ratio between SCIN and fH or fB concentrations. Although both complement factors compete with SCIN for binding to C3b, the effect is significantly more pronounced for fH than fB. A schematic representation of the assays is available (see supplemental Fig. S6, E and F).5
SCIN inhibits the degradation of surface-bound C3b by fl and fH
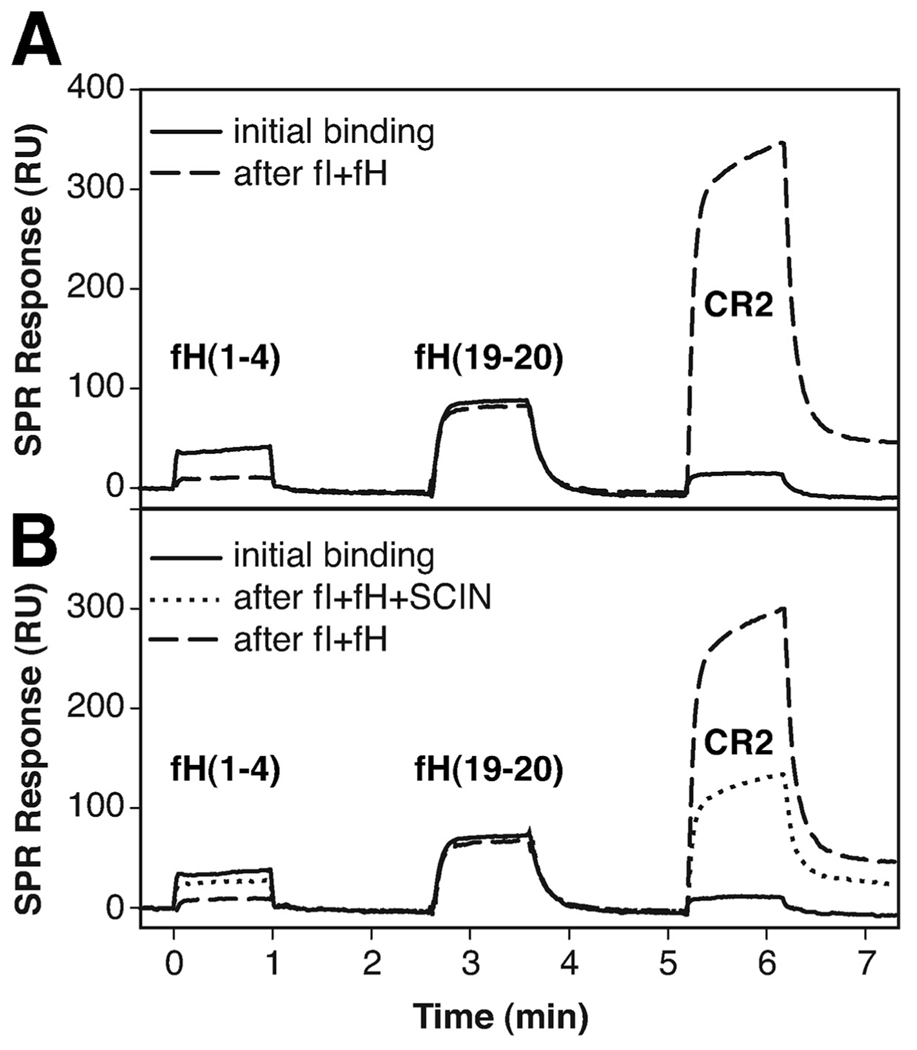
The strong competition with fH for binding to C3b suggested that SCIN might interfere with the activities of this complement factor. As the major regulator of the alternative pathway of complement activation, fH suppresses complement activation on host cells by both destabilizing the convertase (decay acceleration activity) and acting as a cofactor for fI-catalyzed degradation of C3b to iC3b (cofactor activity). We therefore tested the influence of SCIN on the cleavage of surface-bound C3b by fI plus fH using an SPR assay. Differential ligand binding was used to monitor the progress of degradation; although the N-terminal fragment of fH (i.e., fH(1–4)) is known to lose activity after the conversion of C3b to iC3b, CR2 shows a clear selectivity for iC3b over C3b. The C terminus of fH (i.e., fH(19–20)) was not expected to change activity at all and was included as a negative control. Indeed, when the three ligands were injected onto immobilized C3b, only the two fH fragments showed significant binding. After premixed fI and fH were injected for 5 min, the binding pattern changed completely between fH(1–4) and CR2 (Fig. 5A). When SCIN was added to the fI plus fH mixture, the extent of degradation of C3b was significantly lower (Fig. 5B, dotted line). Subsequent injection of the uninhibited enzyme mixture again led to the full extent of cleavage (Fig. 5B, dashed line). Although the removal of the C3f fragment during the conversion of C3b to iC3b was also visible as a drop in the mass of the surface-bound protein, the rate and extent of this mass decrease was much lower in the presence of SCIN (see supplemental Fig. S5).5 An SDS-PAGE-based assay system was used to detect a corresponding effect on cofactor activity of fH in solution. However, the inhibitory effect of SCIN was very low even at a 50-fold molar excess of SCIN over fH (data not shown). This discrepancy in cofactor inhibition may be either based on the sensitivity of the assay system or a result of differential activity for SCIN toward surface-bound and solution-based C3b, as has been observed for Efb-C (13).
FIGURE 5.
SCIN inhibits the cofactor activity of fH. A, The fI-mediated degradation of surface-bound C3b by fI was observed by using SPR with fH(1–4) as a probe for intact C3b and CR2 for degraded C3b (i.e., iC3b). fH(19–20) was used as a control that is not affected by the cleavage. After sequential injection of the three probes over intact C3b (solid curve), a degradation mixture of fI plus fH was injected, and the probes were assessed again (dashed curve): degradation of C3b to iC3b was manifested in an impairment of fH(1–4) binding with a simultaneous increase in CR2 activity. B, The SPR experiment in A was repeated with the same degradation mixture in presence of SCIN. Injection of this mixture led only to a partial degradation with significantly lower effects on fH(1–4) and CR2 (dotted curve). When the degradation mixture was injected in absence of SCIN, full degradation could be achieved again (dashed curve). A schematic representation of the assays is available (see supplemental Fig. S6G).5
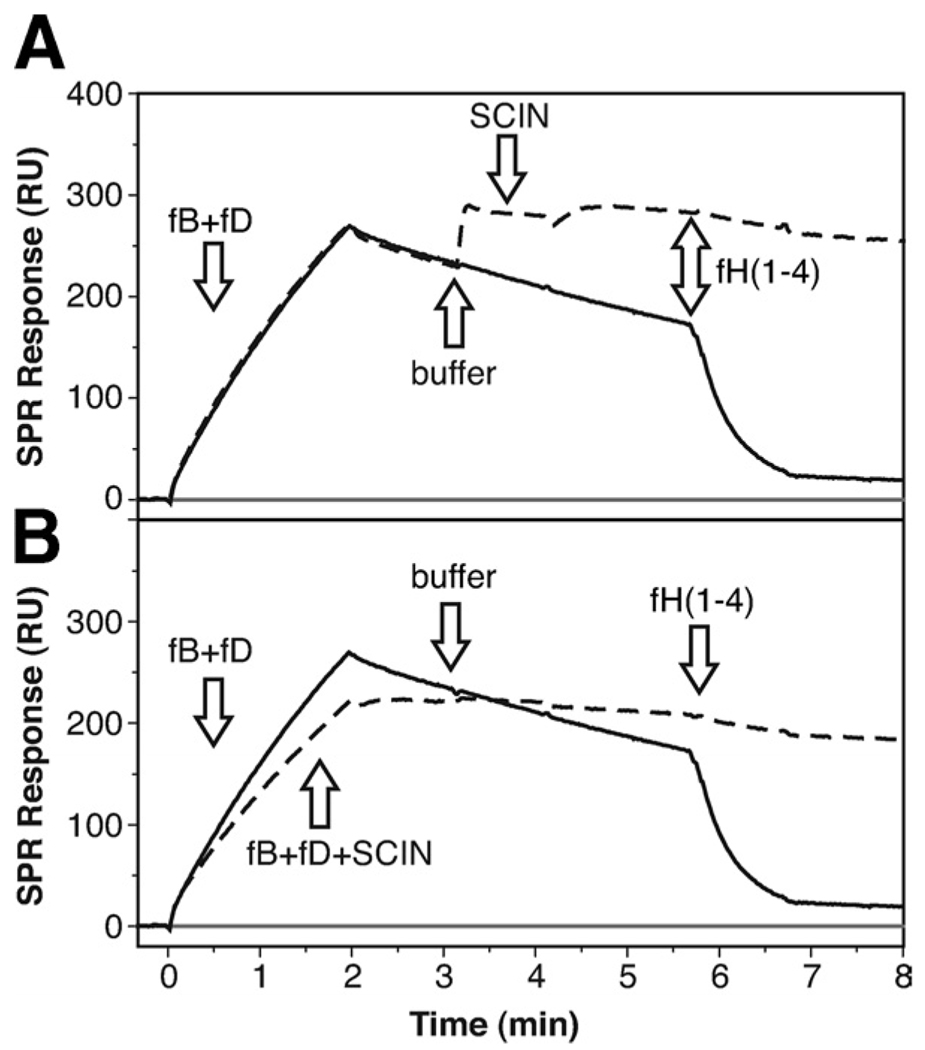
SCIN binds to and prevents decay of the assembled convertase
Due to its competition with both fB and fH, SCIN is expected to have direct consequences on the formation and stability of the C3 convertase. As previously demonstrated, the assembly and decay of the convertase can be monitored directly on a SPR sensor chip when a mixture of complement fB and fD is injected over immobilized C3b (18, 33, 34). In the analyses shown in Fig. 1A, the C3 convertase readily formed when the two factors were injected on thioester-specifically immobilized C3b and showed a decay half-life of 5–6 min in magnesium-containing buffer at 25°C. Injection of fH(1–4) induced a large acceleration of this decay as indicated by a drop of the postinjection signal to near-baseline levels (Fig. 6A, solid line). When SCIN was injected after a short stretch of regular decay, the inhibitor rapidly bound to the assembled convertase and significantly slowed down the decay phase (Fig. 6A, dashed line). Even more, the subsequent injection of fH(1–4) had nearly no effect on the signal (Fig. 6A, dashed line). This indicated that SCIN not only slows down the regular decay of the convertase but also prevents accelerated decay by fH.
FIGURE 6.
SCIN influences the formation, decay, and stability of the alternative pathway C3 convertase. A, The on-chip formation and decay of the C3 convertase was established by injecting a 1:1 mixture of fB and fD (100 nM each) onto immobilized C3b using SPR (solid curve). After injection end (2 min), a moderate decay of the convertase could be observed, which was greatly accelerated upon injection of fH(1–4) (1 µM). When SCIN (1 µM) was injected after formation of the convertase (dashed curve), the inhibitor bound to the complex, slowed the decay rate, and impaired the decay accelerating action of fH(1–4). B, When SCIN (1 µM) was added to the fB plus fD mixture, it partially reduced the formation of the C3bBb complex, and showed the same effect on the stability and fH(1–4) resistance as in A. A schematic representation of the assays is available (see supplemental Fig. S6H).5
Separately from its stabilizing effect on the assembled C3 convertase, the direct competition of SCIN with fB previously observed (Fig. 4) may also affect formation of the C3 convertase. We therefore preincubated SCIN with fB plus fD and monitored formation of the convertase in the presence of SCIN. Indeed, the rate of complex formation was slightly but significantly reduced in presence of the inhibitor (Fig. 6B, dashed line). The convertase formed in this manner again exhibited a much slower decay phase when compared with the same experiment in absence of SCIN (Fig. 6B, solid line). In summary, these results indicate that binding of SCIN to a site in proximity to the binding areas for fH and fB affects both the formation and decay of the surface-assembled alternative pathway C3 convertase.
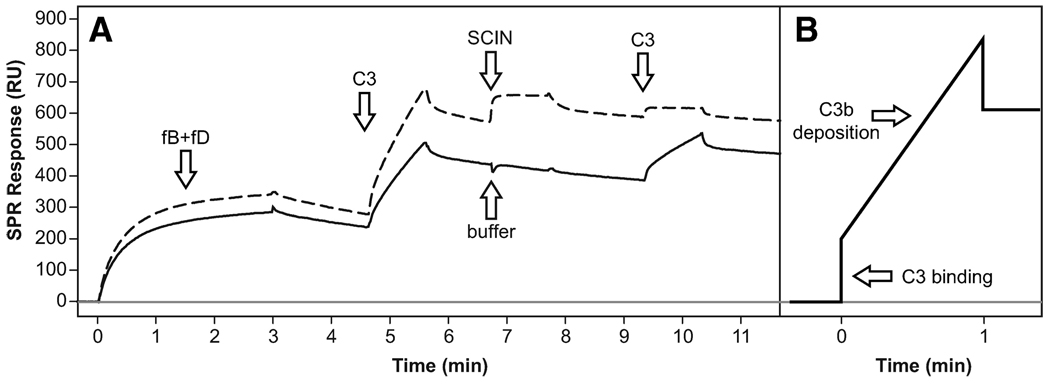
SCIN prevents the cleavage by but not the binding of C3 to the convertase
To exhibit any inhibitory effect on C3 activation, binding of SCIN to the convertase must result in a complex that is incapable of generating C3b. Cleavage of C3 has been shown to generate a very short-lived acyl-imidazole species at the thioester moiety that binds covalently to amine- and hydroxyl-rich patches on cellular surfaces. It has previously been shown that the carboxymethyl dextran-based matrix of SPR sensor chips is accepted as a target surface and that C3b deposition can be observed when C3 is injected onto the assembled convertase (34). We therefore formed the C3 convertase as described and injected native C3 during the decay phase (Fig. 7). After a short and rapid signal increase, a slower but steady increase followed that indicated deposition of C3b. After the injection cycle ceased, the baseline remained at a significantly higher level due to the covalently bound C3b (Fig. 7A, solid line). Because of the ongoing decay of the convertase, a second injection with C3 generated a similar yet smaller increase. After regeneration of the convertase by fH(1–4), the complex was freshly formed by another injection of fB plus fD. This time, the formation rate was slightly higher due to the additional C3b on the surface. Again, a first injection of native C3 led to deposition of C3b. However, when SCIN was injected to the assembled convertase, the subsequent injection of C3 only generated a small binding response but not deposition of C3b (Fig. 7A, dashed line). These findings suggested that SCIN does not prevent the initial binding of C3 to the convertase, but instead inhibits its cleavage to active C3b.
FIGURE 7.
SCIN prevents C3 cleavage and C3b deposition by the C3 convertase. A, In a first cycle (solid curve), the C3 convertase was formed by injecting fB plus fD over immobilized C3b. When native C3 was injected, an initial sharp signal increase corresponding to C3 binding to the convertase and a slower binding slope representing deposition of cleaved C3b on the sensor chip surface could be observed (see schematic representation in panel B). After a buffer injection, C3 was injected again and a similar yet smaller slope could be detected (due to the regular decay of the convertase). In a subsequent cycle (dashed curve), a higher convertase formation rate was observed due to the previously deposited C3b. Again, injection of C3 led to binding of C3 and deposition of cleaved C3b. However, after SCIN was injected to the convertase, only C3 binding but no deposition of cleaved C3b was detected, therefore indicating that SCIN prevented the activation of C3 to C3b by the convertase. A schematic representation of the assays is available (see supplemental Fig. S6I).5
Discussion
SCIN is one of the most potent inhibitors of complement activation identified to date and may be an important determinant in the virulence of S. aureus. Previous studies have identified the C3 convertases of the alternative and classical complement pathways (C3bBb and C4b2a, respectively) as primary targets for this inhibitor (8, 13), and demonstrated that SCIN inhibited the activation of all three complement pathways, opsonization by C3b, phagocytosis, bacterial killing by neutrophils, and generation of C5a (8, 13). Despite this wealth of information about the functional implications of SCIN, its binding mode and molecular mechanism remained elusive. In this study, we focused our attention on molecular effects of SCIN on the alternative pathway, as this pathway is responsible for a majority of the usually observed complement response through its amplification loop (35). In addition, more structural and functional data are available for this pathway, which facilitates a more informative interpretation and correlation of our experimental data. While dissecting the molecular targets of SCIN, initial binding studies indicated that SCIN only binds to the assembled convertase complex (e.g., C3bBb) but not to individual proteins (i.e., C3b and fB) or to the proenzyme complex C3bB (8). Considering the high activity and specificity of the inhibitor, as well as its structural similarity to C3b-binding proteins from S. aureus (4), this selectivity for the convertase was rather surprising. When using a similar assay setup with immobilized SCIN on a sensor chip and complement fragments in solution, we could indeed reproduce this selectivity in our study experiments (Fig. 1A). However, this model only poorly describes the in vivo situation in which C3b and C4b are firmly attached on cell surfaces and form the respective convertases, and in which the rather small and highly charged SCIN protein might interact more freely with these surface structures. In an attempt to mimic these conditions, we specifically immobilized C3b via its thioester to a sensor chip and injected SCIN in solution. Using this reversed experimental format, SCIN indeed showed concentration-dependent binding to C3b with nanomolar affinity.
Although the different results obtained from these inverted assay formats are surprising, they are not without explanation. First, surface-immobilized SCIN may suffer from a partial inactivation during the random amine immobilization procedure. Examination of the SCIN sequence indeed reveals the presence of two lysine residues (Lys41 and Lys45) within its reported active site (9). Modification of these locations may well have altered the affinity and selectivity of the protein. Secondly, surface attraction effects may also contribute to this discrepancy. Much like the cell wall of S. aureus (36), the carboxymethyl dextran matrix of the sensor chip exhibits a negative net charge and is rich in hydroxyl groups (37). Previous studies have shown that such SPR surfaces are readily recognized as targets for C3b opsonization (34, 38). The highly electropositive SCIN (pI ≈ 9) may therefore be attracted to and enriched at the surface, thus leading to a stronger apparent affinity toward C3b. In its biological environment, such a behavior makes sense because soluble SCIN, although secreted, is supposed to inhibit complement convertase activity at the surface of S. aureus. A similar effect may also be responsible for the observation that Efb-C and Ehp, which are similarly electropositive, primarily act on surface-bound rather than fluid phase convertases (13). Here again, these inhibitors may be enriched at the surface of S. aureus or on zymosan, which were both used in the study.
Although the effect of surface attraction may also be reflected in the small, but significant difference in affinity between SCIN binding to surface-bound C3b (i.e., SPR) and soluble C3b (i.e., ITC), our ITC results clearly demonstrate that surface attraction is not a requirement for this interaction and that SCIN alone possesses a strong binding affinity for C3b. The observed KD values in the range of 170–265 nM are weaker than those of the structurally similar Efb-C protein (i.e., 1–2 nM) (10) but are clearly stronger than for domains III-IV of Sbi (14), which also belongs to this structural family (i.e., 1.4 µM for C3dg and weaker still for C3b). In both binding assays, the C3b-SCIN interaction slightly but significantly deviated from a 1:1 binding model. In the case of SPR, this effect was more pronounced at the higher SCIN concentrations we used. As heterogeneity effects could be largely excluded (high purity of the proteins, oriented immobilization or solvent state of all analytes), the availability of two separate binding sites with different affinity is more likely. Extrapolation of the SPR fit to a two-binding site model (see supplemental Fig. S2)5 suggests a KD of greater than 200 µM for the low-affinity site. However, neither the SPR nor the ITC assays are capable of covering such a high concentration range, which makes an exact determination of this value unfeasible. Nevertheless, this additional binding site is likely to explain the deviations from the 1:1 model, and cooperative binding of two SCIN and two C3b molecules might lead to the formation of rather stable dimeric complexes in solution. This hypothesis was further supported by the solution scattering data, which clearly indicated that SCIN and C3b are forming both 1:1 and 2:2 complexes. In addition, the recent description of low-resolution crystal data for the C3b-SCIN complex also revealed a rather large unit cell that was consistent with the presence of either two 1:1 complexes or a single 2:2 complex in the asymmetric unit (39). Most importantly, our findings are in agreement with structural data from the inhibited alternative pathway C3 convertase, in which SCIN is in contact with two C3b molecules and forms highly stable SCIN2C3b2Bb2 complexes (16).
The fact that SCIN interacts with both C3b and iC3b (with similar activity) but not with C3d clearly indicates that the binding site for the inhibitor must be located at the C3c domain. The nearly complete blockage of the SCIN-C3b interaction by the anti-C3c mAb C3–9 not only confirmed this observation but also allowed us to further resolve the primary binding site. Electron microscopy studies with the F(ab)2 fragment of mAb C3–9 have shown that this Ab binds to an area around MG7 and MG8 domains on C3b, which undergoes a large conformational transformation upon activation of C3 to C3b (32). Indeed, formation of this neo-epitope allows mAb C3–9 to distinguish between native C3 and active C3b (40). The direct competition with this mAb for C3b binding strongly suggests that SCIN binds to a similar area at the C terminus of the β-chain of C3b. The functional importance of this region has been demonstrated earlier, as both mAb C3–9 and a novel therapeutic anti-C3b mAb (S77), which primarily binds to MG7, block the binding of all fH, fB, and CR1 to C3b (41, 42). In the case of fH, we have recently shown that mAb C3–9 only blocks the binding of the N-terminal four domains (fH(1–4)) but the not the C-terminal domains (fH(19–20)) (18). These four domains are sufficient for both regulatory properties of fH, i.e., the acceleration of convertase decay and the cofactor function for fI (43).
We therefore investigated whether the potential binding of SCIN to a similar region would allow the inhibitor the activity of fB and fH and found that both factors are substantially inhibited by SCIN for their binding to C3b. Interestingly, the degree of this competition effect was distinct for the two complement proteins. Although fH was nearly completely blocked at a 25-fold molar excess of SCIN, the binding of fB was only partially competed and leveled off at around 30% residual binding. This observation is in agreement with the known mechanism of SCIN to lock the convertase in an inactive state, which requires that the Bb domain is still associated with C3b and not replaced by SCIN. It is therefore more likely that SCIN partially restricts the initial binding of full-length fB to C3b via its Ba domain. Even though this competition with fB did slightly influence the formation rate of the convertase complex on the chip (Fig. 6B), the effect on the stability and function of the assembled convertase is more striking. When added to the C3bBb complex, SCIN bound rapidly and decreased the decay rate of the convertase (Fig. 6A). Even more importantly, the SCIN-stabilized convertase was unable to cleave C3, as no C3b deposition to the chip surface could be detected in the presence of SCIN (Fig. 7). As a consequence, we can now confirm earlier hypotheses that SCIN locks the convertase in an inactive state (13) at a molecular level.
Interestingly, C3 still seems to be able to bind the convertase (as indicated by a block-shaped signal upon injection; Fig. 7B), which suggests that only contact and cleavage through the enzymatic entity (i.e., Bb) but not the initial binding to the convertase is inhibited by SCIN. When combining this information with recent structural studies about the C3 convertase and the complement inhibitor compstatin (16, 44), it emerges that the area around MG4/5 is more important for this initial binding, whereas the primary SCIN binding site around MG6–8 is essential for the functional activity of C3b or the convertase. Binding of SCIN at this area forms a trimeric complex with C3b and Bb (16), which explains the much slower convertase decay observed in our study. Although it was speculated that SCIN might inhibit the convertase by either preventing substrate binding or hindering essential domain movements that allow cleave of C3 (16), our data indicate that binding of C3 is still possible and are therefore in favor of the conformational hypothesis. The observation that the SCIN-stabilized convertase appears to exclusively form a dimeric complex (SCIN2C3b2Bb2) in solution (16) opened the question of whether this dimerization is required for blocking the convertase. In this regard, ligand density and uniform immobilization of C3b molecules in our SPR assay makes it highly unlikely that any dimeric convertase assemblies could form. We therefore conclude that SCIN is able to inhibit the surface-bound convertase in its monomeric form.
In the case of fH, we observed that binding of SCIN to C3b or to the C3 convertase affects both key regulatory functions of the factor. First, the ability of fH to act as a cofactor for fI in the cleavage of surface-bound C3b to iC3b was investigated by monitoring the change in binding specificity for fH(1–4) and CR2, which have opposite selectivity for the active and inactive form of C3b (45, 46). Addition of SCIN to the enzyme/cofactor mix partially inhibited the conversion to iC3b as visible both in a lower mass loss caused by removal of the C3f fragment (see supplemental Fig. S5)5 and in a less pronounced change in binding selectivity toward CR2 (Fig. 5). The functional relevance of this inhibitory effect is difficult to judge because it may require rather high local SCIN concentrations and S. aureus may actually benefit from the degradation of C3b. However, iC3b and its downstream fragment C3d are both able to participate in signaling pathways and keeping C3b locked up in the convertase may be advantageous. Perhaps more important for the functional properties of SCIN is that this bacterial protein effectively eliminates the ability of fH(1–4) to accelerate the decay of the convertase (Fig. 6). Again, this observation is in good agreement with the direct and complete competition between SCIN and fH for C3b binding as described. As a consequence, SCIN not only decelerates the decay of the inactive C3bBb complex but also makes it insensitive for an intercalation by fH to remove the Bb fragment (Fig. 8). In solution, binding of SCIN to C3b will likely induce dimeric convertase complexes and therefore impair complement activation in circulation (16). It has to be further investigated, though, whether other SCIN-mediated effects such as impairing cofactor activity of fH are relevant in solution, especially because they are expected to require rather high concentration to be effective (as e.g., seen in the case of the solution-based cofactor assay using SDS-PAGE).
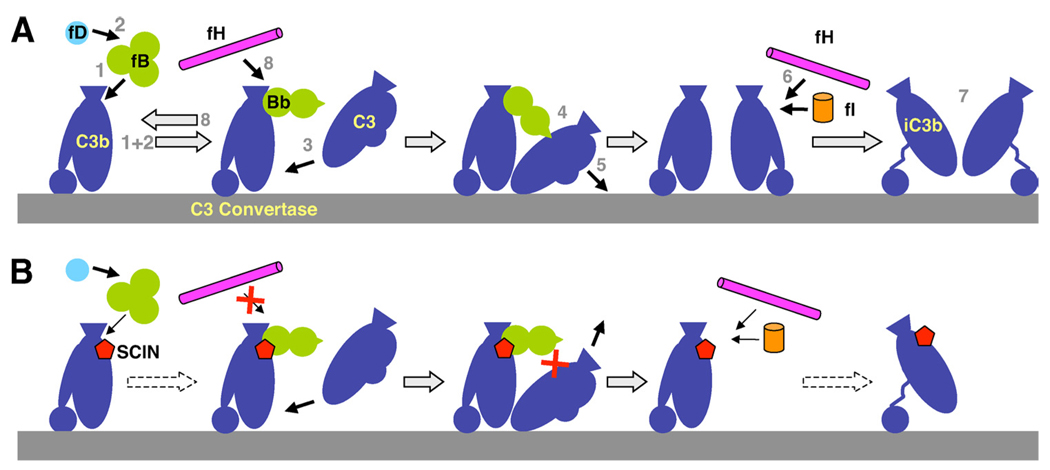
FIGURE 8.
Potential effects of SCIN on complement activation and regulation. A, During normal complement activation, fB binds to C3b (1) and is converted to the C3 convertase (C3bBb) by fD (2). Native C3 binds to the convertase complex (3), becomes cleaved by Bb (4), and the resulting C3b is deposited on the surface (5). Surface-bound C3b can again participate in convertase formation (self-amplification) or can be degraded by fI and fH (6) to iC3b, which does not bind fB but is still involved in signaling (7). Finally, fH also regulates complement activity by accelerating the decay of the C3 convertase (8). B, By binding at a focal point on C3b, SCIN seems to interfere with this response in various ways: it slows down convertase formation but kinetically stabilizes the convertase complex in a state in which C3 can still bind but does not get cleaved. It also impairs the actions of fH by preventing decay acceleration and potentially decelerating the degradation to iC3b.
In summary, our data clearly demonstrate that SCIN directly interacts with C3b at a functional “hot spot” on this central complement component and thereby exerts a variety of its functional and inhibitory activities. Of these, entrapment of the C3 convertase complex in an inactive state that does not allow cleavage of C3 has to be considered the most important and effective immune evasion strategy. Though “stabilization” of the central active entity in the cascade (C3 convertase) and “inhibition” of a potent down-regulator (fH) appears highly contradictory for a microorganism, S. aureus has developed a malicious yet ingenious strategy to turn these actions to its advantage (Fig. 8). In fact, the bacterium profits much more from a locked, dysfunctional convertase rather than an intact decay acceleration activity because the former case not only leads to a direct inhibition of the C3b self-amplification loop but may also block C3b from participation in any proinflammatory downstream activities of the cascade. Still, SCIN may also influence additional complement functions by competing with fH and fB (and possibly other regulators) for binding to C3b, some of which may benefit complement evasion (e.g., slowing the rate of convertase formation) and some of which may be less obvious or mere “side effects” of its successful inhibition (e.g., inhibition of cofactor activity of fH) (Fig. 8). Our data also suggest that electrostatic effects may contribute to an increased activity of SCIN around negatively charged surfaces (such as the S. aureus cell wall). The versatility and high activity of its inhibitory effects not only put SCIN into the spotlight for elucidating its contribution on the virulence of S. aureus but also renders it into an interesting molecular template for the design of novel therapeutics against complement-mediated diseases (4, 15). In this regard, more molecular details about the binding interface, the exact mechanism of blocking C3 cleavage and its effects on the classical pathway convertase or the C5 convertases should be addressed in future studies.
Supplementary Material
Acknowledgments
We thank the Lawrence Berkeley Lab Advanced Light Source and SIBLYS staff at beamline 12.3.1 for providing generous access to X-ray scattering facilities, and Kasra X. Ramyar for critically reading the manuscript.
Footnotes
This work was supported by Grants AI071028, AI068730, AI30040, AI072106, and CA53615 from the National Institutes of Health, and in part by the Office of Science, Office of Biological and Environmental Research, U.S. Department of Energy, under Contract DE-AC02-05CH11231 for SIBLYS beamline efforts
Abbreviations used in this paper: CR, complement receptor; SCIN, staphylococcal complement inhibitor; SPR, surface plasmon resonance; ORF, open reading frame; Efb, extracellular fibrinogen-binding protein; Ehp, Efb homologous protein; fB, factor B; fD, factor D; fH, factor H; fI, factor I; ITC, isothermal titration calorimetry; MG, macroglobulin domain; SAXS, small angle X-ray scattering; Sbi, staphylococcal Ig-binding protein; TED, thioester-containing domain.
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Lowy FD. Staphylococcus aureus infections. N. Engl. J. Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Lee LY, Hook M, Haviland D, Wetsel RA, Yonter EO, Syribeys P, Vernachio J, Brown EL. Inhibition of complement activation by a secreted Staphylococcus aureus protein. J. Infect. Dis. 2004;190:571–579. doi: 10.1086/422259. [DOI] [PubMed] [Google Scholar]
- 3.Lee LY, Miyamoto YJ, McIntyre BW, Hook M, McCrea KW, McDevitt D, Brown EL. The Staphylococcus aureus map protein is an immunomodulator that interferes with T cell-mediated responses. J. Clin. Invest. 2002;110:1461–1471. doi: 10.1172/JCI16318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambris JD, Ricklin D, Geisbrecht BV. Complement evasion by human pathogens. Nat. Rev. Microbiol. 2008;6:132–142. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chavakis T, Preissner KT, Herrmann M. The anti-inflammatory activities of Staphylococcus aureus. Trends Immunol. 2007;28:408–418. doi: 10.1016/j.it.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Foster TJ. Immune evasion by staphylococci. Nat. Rev. Microbiol. 2005;3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 7.Ricklin D, Ricklin-Lichtsteiner SK, Markiewski MM, Geisbrecht BV, Lambris JD. Cutting edge: members of the Staphylococcus aureus extracellular fibrinogen-binding protein family inhibit the interaction of C3d with complement receptor 2. J. Immunol. 2008;181:7463–7467. doi: 10.4049/jimmunol.181.11.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rooijakkers SH, Ruyken M, Roos A, Daha MR, Presanis JS, Sim RB, van Wamel WJ, van Kessel KP, van Strijp JA. Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat. Immunol. 2005;6:920–927. doi: 10.1038/ni1235. [DOI] [PubMed] [Google Scholar]
- 9.Rooijakkers SH, Milder FJ, Bardoel BW, Ruyken M, van Strijp JA, Gros P. Staphylococcal complement inhibitor: structure and active sites. J. Immunol. 2007;179:2989–2998. doi: 10.4049/jimmunol.179.5.2989. [DOI] [PubMed] [Google Scholar]
- 10.Hammel M, Sfyroera G, Ricklin D, Magotti P, Lambris JD, Geisbrecht BV. A structural basis for complement inhibition by Staphylococcus aureus. Nat. Immunol. 2007;8:430–437. doi: 10.1038/ni1450. [DOI] [PubMed] [Google Scholar]
- 11.Hammel M, Sfyroera G, Pyrpassopoulos S, Ricklin D, Ramyar KX, Pop M, Jin Z, Lambris JD, Geisbrecht BV. Characterization of Ehp, a secreted complement inhibitory protein from Staphylococcus aureus. J. Biol. Chem. 2007;282:30051–30061. doi: 10.1074/jbc.M704247200. [DOI] [PubMed] [Google Scholar]
- 12.Upadhyay A, Burman JD, Clark EA, Leung E, Isenman DE, van den Elsen JM, Bagby S. Structure-function analysis of the C3 binding region of Staphylococcus aureus immune subversion protein Sbi. J. Biol. Chem. 2008;283:22113–22120. doi: 10.1074/jbc.M802636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jongerius I, Kohl J, Pandey MK, Ruyken M, van Kessel KP, van Strijp JA, Rooijakkers SH. Staphylococcal complement evasion by various convertase-blocking molecules. J. Exp. Med. 2007;204:2461–2471. doi: 10.1084/jem.20070818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burman JD, Leung E, Atkins KL, O’Seaghdha MN, Lango L, Bernado P, Bagby S, Svergun DI, Foster TJ, Isenman DE, van den Elsen JMH. Interaction of human complement with SBI, a staphylococcal immunoglobulin-binding protein: indications of a novel mechanism of complement evasion by Staphylococcus aureus. J. Biol. Chem. 2008;283:17579–17593. doi: 10.1074/jbc.M800265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ricklin D, Lambris JD. Complement-targeted therapeutics. Nat. Biotechnol. 2007;25:1265–1275. doi: 10.1038/nbt1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rooijakkers SH, Wu J, Ruyken M, van Domselaar R, Planken KL, Janssen BJC, Ricklin D, Tzekou A, Lambris JD, van Strijp JA, Gros P. Structural and functional implications of the alternative complement pathway C3 convertase stabilized by a staphylococcal inhibitor. Nat. Immunol. 2009;10:721–727. doi: 10.1038/ni.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarrias MR, Franchini S, Canziani G, Argyropoulos E, Moore WT, Sahu A, Lambris JD. Kinetic analysis of the interactions of complement receptor 2 (CR2, CD21) with its ligands C3d, iC3b, and the EBV glycoprotein gp350/220. J. Immunol. 2001;167:1490–1499. doi: 10.4049/jimmunol.167.3.1490. [DOI] [PubMed] [Google Scholar]
- 18.Wu J, Wu Y-Q, Ricklin D, Janssen B, Lambris JD, Gros P. Structure of complement fragment C3b–factor H and implications for host protection by complement regulators. Nat. Immunol. 2009;10:728–733. doi: 10.1038/ni.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herbert AP, Uhrin D, Lyon M, Pangburn MK, Barlow PN. Disease-associated sequence variations congregate in a polyanion recognition patch on human factor H revealed in three-dimensional structure. J. Biol. Chem. 2006;281:16512–16520. doi: 10.1074/jbc.M513611200. [DOI] [PubMed] [Google Scholar]
- 20.Asokan R, Hua J, Young KA, Gould HJ, Hannan JP, Kraus DM, Szakonyi G, Grundy GJ, Chen XS, Crow MK, Holers VM. Characterization of human complement receptor type 2 (CR2/CD21) as a receptor for IFN-α: a potential role in systemic lupus erythematosus. J. Immunol. 2006;177:383–394. doi: 10.4049/jimmunol.177.1.383. [DOI] [PubMed] [Google Scholar]
- 21.Geisbrecht BV, Bouyain S, Pop M. An optimized system for expression and purification of secreted bacterial proteins. Protein Expr. Purif. 2006;46:23–32. doi: 10.1016/j.pep.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Myszka DG. Improving biosensor analysis. J. Mol. Recognit. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 23.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 24.Guinier A, Fournet F. Small Angle Scattering of X-rays. New York: Wiley Interscience; 1955. [Google Scholar]
- 25.Svergun D. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 1992;25:495–503. [Google Scholar]
- 26.Mylonas F, Svergun DI. Accuracy of molecular mass determination of proteins in solution by small-angle X-ray scattering. J. Appl. Cryst. 2007;40:s245–s249. [Google Scholar]
- 27.Svergun D, Baraberato C, Koch MH. CRYSOL: a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 1995;28:768–773. [Google Scholar]
- 28.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 2003;36:1277–1282. [Google Scholar]
- 29.Janssen BJ, Christodoulidou A, McCarthy A, Lambris JD, Gros P. Structure of C3b reveals conformational changes that underlie complement activity. Nature. 2006;444:213–216. doi: 10.1038/nature05172. [DOI] [PubMed] [Google Scholar]
- 30.Alsenz J, Lambris JD, Schulz TF, Dierich MP. Localization of the complement-component-C3b-binding site and the cofactor activity for factor I in the 38kDa tryptic fragment of factor H. Biochem. J. 1984;224:389–398. doi: 10.1042/bj2240389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haspel N, Ricklin D, Geisbrecht BV, Kavraki LE, Lambris JD. Electrostatic contributions drive the interaction between Staphylococcus aureus protein Efb-C and its complement target C3d. Protein Sci. 2008;17:1894–1906. doi: 10.1110/ps.036624.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishida N, Walz T, Springer TA. Structural transitions of complement component C3 and its activation products. Proc. Natl. Acad. Sci. USA. 2006;103:19737–19742. doi: 10.1073/pnas.0609791104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris CL, Abbott RJ, Smith RA, Morgan BP, Lea SM. Molecular dissection of interactions between components of the alternative pathway of complement and decay accelerating factor (CD55) J. Biol. Chem. 2005;280:2569–2578. doi: 10.1074/jbc.M410179200. [DOI] [PubMed] [Google Scholar]
- 34.Nilsson B, Larsson R, Hong J, Elgue G, Ekdahl KN, Sahu A, Lambris JD. Compstatin inhibits complement and cellular activation in whole blood in two models of extracorporeal circulation. Blood. 1998;92:1661–1667. [PubMed] [Google Scholar]
- 35.Harboe M, Ulvund G, Vien L, Fung M, Mollnes TE. The quantitative role of alternative pathway amplification in classical pathway induced terminal complement activation. Clin. Exp. Immunol. 2004;138:439–446. doi: 10.1111/j.1365-2249.2004.02627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neuhaus FC, Baddiley J. A continuum of anionic charge: structures and functions of D-alanyl-teichoic acids in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2003;67:686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Löfås S, Johnsson B. A novel hydrogel matrix on gold surfaces in surface plasmon resonance sensors for fast and efficient covalent immobilization of ligands. J. Chem. Soc. Chem. Commun. 1990:1526–1528. [Google Scholar]
- 38.Ricklin D, Lambris JD. Exploring the complement interaction network using surface plasmon resonance. Adv. Exp. Med. Biol. 2007;598:260–278. doi: 10.1007/978-0-387-71767-8_19. [DOI] [PubMed] [Google Scholar]
- 39.Garcia BL, Tzekou A, Ramyar KX, McWorther WJ, Ricklin D, Lambris JD, Geisbrecht BV. Crystallization of human complement component C3b in the presence of a staphylococcal complement-inhibitor protein (SCIN) Acta Crystallogr. Sec. F Struct. Biol. Cryst. Commun. 2009;65:482–485. doi: 10.1107/S174430910901207X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hack CE, Paardekooper J, Smeenk RJ, Abbink J, Eerenberg AJ, Nuijens JH. Disruption of the internal thioester bond in the third component of complement (C3) results in the exposure of neodeterminants also present on activation products of C3: an analysis with monoclonal antibodies. J. Immunol. 1988;141:1602–1609. [PubMed] [Google Scholar]
- 41.Becherer JD, Alsenz J, Esparza I, Hack CE, Lambris JD. Segment spanning residues 727–768 of the complement C3 sequence contains a neoantigenic site and accommodates the binding of CR1, factor H, and factor B. Biochemistry. 1992;31:1787–1794. doi: 10.1021/bi00121a029. [DOI] [PubMed] [Google Scholar]
- 42.Katschke KJ, Jr, Stawicki S, Yin J, Steffek M, Xi H, Sturgeon L, Hass PE, Loyet K, Deforge L, Wu Y, et al. Structural and functional analysis of a C3b-specific antibody that selectively inhibits the alternative pathway of complement. J. Biol. Chem. 2009;284:10473–10479. doi: 10.1074/jbc.M809106200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gordon DL, Kaufman RM, Blackmore TK, Kwong J, Lublin DM. Identification of complement regulatory domains in human factor H. J. Immunol. 1995;155:348–356. [PubMed] [Google Scholar]
- 44.Janssen BJ, Halff EF, Lambris JD, Gros P. Structure of compstatin in complex with complement component C3c reveals a new mechanism of complement inhibition. J. Biol. Chem. 2007;282:29241–29247. doi: 10.1074/jbc.M704587200. [DOI] [PubMed] [Google Scholar]
- 45.Weis JJ, Tedder TF, Fearon DT. Identification of a 145,000 Mr membrane protein as the C3d receptor (CR2) of human B lymphocytes. Proc. Natl. Acad. Sci. USA. 1984;81:881–885. doi: 10.1073/pnas.81.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DiScipio RG. The binding of human complement proteins C5, factor B, β1H and properdin to complement fragment C3b on zymosan. Biochem J. 1981;199:485–496. doi: 10.1042/bj1990485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.