Abstract
Motivation: The growth of sequence data has been accompanied by an increasing need to analyze data on distributed computer clusters. The use of these systems for routine analysis requires scalable and robust software for data management of large datasets. Software is also needed to simplify data management and make large-scale bioinformatics analysis accessible and reproducible to a wide class of target users.
Results: We have developed a workflow management system named Ergatis that enables users to build, execute and monitor pipelines for computational analysis of genomics data. Ergatis contains preconfigured components and template pipelines for a number of common bioinformatics tasks such as prokaryotic genome annotation and genome comparisons. Outputs from many of these components can be loaded into a Chado relational database. Ergatis was designed to be accessible to a broad class of users and provides a user friendly, web-based interface. Ergatis supports high-throughput batch processing on distributed compute clusters and has been used for data management in a number of genome annotation and comparative genomics projects.
Availability: Ergatis is an open-source project and is freely available at http://ergatis.sourceforge.net
Contact: jorvis@users.sourceforge.net
1 INTRODUCTION
Workflow management systems (WMS) include software systems that execute and manage computational pipelines. These have become important tools in bioinformatics because they enable researchers to analyze the massive quantities of data generated by modern laboratory equipment. There are a number of WMS targeted to bioinformatics that differ in scope and approach for construction and execution of workflows (Romano, 2008; Tiwari and Sekhar 2007). One class of WMS enables biologists to manipulate data in a manner that would normally require some level of scripting ability or the use of a collection of local tools and web forms. The scope of this class usually includes querying preexisting datasets and transforming results. Operations include retrieving sequences from public collections, extraction of subsequences, converting among file formats and performing set operations on collections of results. Applications in this class include ISYS, a local application and development framework (Siepel et al., 2001), and Galaxy, a comprehensive web-based interface designed for tool and database integration (Giardine et al., 2005). Galaxy, in particular, excels at enabling users to gather data from diverse sources and execute a set of queries. Pipelines are represented as a history of user actions within the system that can be reused.
BioMOBY (Wilkinson and Links, 2002) and Taverna (Oinn et al., 2004) are notable instances of a class of WMS that organize and integrate a disparate collection of web service providers. In this model, data are exchanged using a common format and protocol, usually XML and SOAP, respectively, between the host and any number of providers during the course of a pipeline execution. Users of these systems do not need extensive local hardware resources since all computes are performed at each provider's site. Indeed, the number of providers available for common and overlapping services has grown so large that new strategies have been developed to assist in managing and ranking them (DiBernardo et al., 2008). For data-intensive pipelines, web service approaches can be limited by network performance, service availability and I/O compatibility between providers.
Other WMS typically assume direct access to component executables and manage the execution of pipelines either locally or on a compute cluster. Wildfire (Tang et al., 2005) and Pegasys (Shah et al., 2004) aim to enable construction of components into a pipeline using local graphical user interfaces, representing these pipelines in a form that can be executed on distributed resources.
Ergatis is a WMS that fits into this last class and is targeted toward the analysis of genome sequence data. Ergatis is designed to be accessible to bioinformaticians and biologists alike. Using an intuitive web-based interface, biologists can use the suite of integrated components to construct new analysis pipelines or reuse existing pipeline templates. These pipelines can be executed on a single desktop or distributed across large compute clusters. Their output can be converted into common formats or loaded into a relational database. The underlying workflow system provides structured pipeline representation, monitoring, audit capability and task execution on managed compute resources such as a grid or compute cluster. Our system allows construction of pipelines from common bioinformatics analysis tools as well as providing an architecture within which these pipelines can be reused and applied to new datasets. Ergatis is the only WMS among these to provide these features and contain prepackaged pipelines for whole-genome annotation, comparative genomics and pan-genome diversity studies.
2 ARCHITECTURE AND METHODS
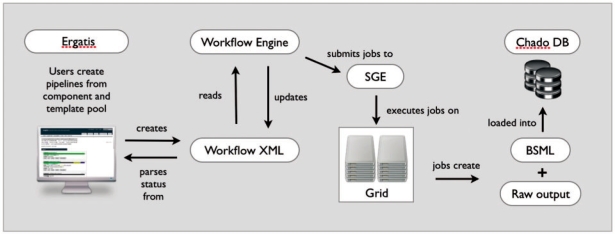
The architecture of Ergatis is shown in Figure 1.
Fig. 1.
Architecture diagram showing process and data flow from pipeline creation in Ergatis, processing of wXML by Workflow Engine, job scheduling on a computational grid by SGE and finally optional data loading into a Chado relational database instance.
Ergatis consists of a web interface for building workflows, a processing engine for execution and reusable components and templates that incorporate a suite of bioinformatics tools and tasks. It has a web interface that allows users to create a pipeline description in XML in a simple point and click manner. The steps of a pipeline are described in XML that is read by the processing engine. The processing engine uses Sun Grid Engine (SGE) to schedule and manage jobs on a compute cluster.
2.1 Pipeline components and templates
Ergatis allows users to build pipelines out of modular analysis components. A component is a series of steps where each step is a script or binary executable. Most components consist of a bioinformatics package, such as a sequence alignment or gene prediction program along with pre- and post-processing Perl scripts. A component is described by an XML definition file and a configuration file that, together, define the steps of the component and its configurable parameters.
A set of components constitutes a pipeline. Components can be combined in series or in parallel to form any desired order of execution. Pipelines can be executed locally or distributed across a compute grid. They can also be saved as an XML file for future execution or sharing with other users.
2.2 Inputs and outputs
Ergatis was designed to support batch processing of sequences in a high-throughput environment using compute grids. Parallelism is achieved by splitting inputs into groups and distributing each group to a node of the compute cluster. Support for generating groups is included in each component. The automatic input grouping described above helps to alleviate load on job schedulers and avoid file system limitations using an output directory.
Each component in Ergatis has a defined set of required input files. Input files include traditional FASTA formatted or Bioinformatic Sequence Markup Language (BSML) files (LabBook). BSML is an XML format that provides a simple encoding for sequences, annotation and search results. Our current Ergatis release includes scripts for parsing and generating BSML from common input formats such as GFF3 and the GenBank flat file format and most components in Ergatis include scripts to also transform the native tool's output to BSML. Using BSML as a common output format has the added advantage of regularizing data by using terms from controlled vocabularies such as the Sequence Ontology (Eilbeck et al., 2005) and Gene Ontology (Ashburner et al., 2000).
Ergatis also includes support for the Chado relational database schema. Chado is a community-supported schema for biological data that relies heavily on the use of ontologies for typing data (Mungall and Emmert, 2007). Ergatis includes a component (initdb) that can initialize a Chado database with a set of ontologies described in OBO file format (Day-Richter). Genome sequences, annotation and search evidence that are encoded in BSML can be written to and read from a Chado database instance using the bsml2chado and chado2bsml Ergatis components. The Ergatis database components support multiple database vendors, including PostgreSQL, MySQL and Sybase.
2.3 Workflow processing and grid support
A scientific workflow can be imagined as a directed acyclic graph (DAG) where the nodes of the graphs are scientific processes and the edges represent the path for data and process flow. Ergatis uses a processing engine called Workflow that has a simple XML format for describing steps in a pipeline called wXML. The wXML represents the procedural specification of such scientific workflows in a machine-readable language that describes the nodes of the DAG as Command or CommandSet elements. Command elements represent a single atomic process while CommandSet elements represent a collection of Command and CommandSet elements, allowing the capability to nest such elements and construct complex, hierarchical workflows. Additionally, the CommandSet construct permits the ability to iterate sequentially or concurrently over a subset of the elements of a workflow encapsulated in a CommandSet.
The workflow engine is written in Java and is multithreaded to support multiprocessors on a local machine or computational grid. Workflow Engine supports SGE and Condor through use of the Distributed Resource Management Application API. The supporting hardware architecture includes a distributed compute cluster and a shared file system, such as Network File System.
The workflow engine executes the wXML, distributing jobs on a local server or a compute cluster. The engine tracks the execution of the workflow steps and maintains detailed audit information for each command in the wXML. Workflow engine has the ability to recover from errors and resume execution from the last point of failure or roll back execution to a user-defined arbitrary location in the workflow and resume execution from that point.
2.4 Pipeline build/monitor interface
Ergatis provides a web interface to build a wXML description of a pipeline. The pipeline wXML is built from reusable modular components that come prepackaged with Ergatis. The pipeline wXML contains a complete description of the pipeline including start and end times, error messages, return values, execution host, logging and execution strings of every command in the pipeline. The Ergatis web interface reads the pipeline wXML to provide status information to the user.
The Ergatis interface allows users to build pipelines by easily adding components in a pipeline and dynamically arranging them to execute either serially, in parallel or in any nested combination of these. A subset of Ergatis components is listed in Table 1.
Table 1.
Selected Ergatis components by classification
| Component type | Count | Examples |
|---|---|---|
| Gene prediction | 14 | fgenesh, glimmer3, genscan, RNAmmer |
| HMM alignment | 4 | hmmpfam, panther |
| Sequence masking | 2 | repeatmasker, seg |
| Functional prediction | 12 | SignalP, tmhmm, pFunc |
| Phylogeny/binning | 3 | RDP, stap |
| Multiple alignment | 3 | clustalw, MUSCLE |
| Pairwise alignment | 14 | NCBI blast suite, WU-BLAST, BER |
Ergatis release v2.r12 currently contains 162 components that can be used to form complex bioinformatics analysis pipelines.
While monitoring a pipeline the interface periodically updates the status of each component, with progress bars color coded by status. Command counts and a label of the current execution step are displayed. Not limited to this pipeline-level abstract view, users can click through to see details on each component and down to the level of each individual command, allowing any individual command line string to be copied and run within a terminal.
2.5 Collaborative pipeline development
Important features of a WMS include pipeline reuse and exportability. Pipelines built with Ergatis can be saved as a ‘pipeline template’ and reused within the system or exported to other users. The template contains the pipeline layout and each component's configuration options, allowing the pipeline to be exactly reproduced. Importing a pipeline requires that it be placed inside of directory in the Ergatis installation. The templates are also small enough to be sent via e-mail—the 47-component prokaryotic annotation pipeline described below is only 28 kb. We encourage developers in the bioinformatics community to utilize the public Subversion repository, located on the Ergatis SourceForge project site, to build and contribute new components.
3 PIPELINES
Ergatis has served as a data management tool in bioinformatics cores with compute grids of up to 600 CPU cores at both the J. Craig Venter Institute and the Institute for Genome Sciences, University of Maryland School of Medicine. It has been used to build and run analysis pipelines that have been incorporated into numerous published individual genome (Nene et al., 2007; Ouyang et al., 2007; Carlton et al., 2007) and comparative genomic studies (Hotopp et al., 2006; El-Sayed et al., 2005). Ergatis contains several components for gene/RNA prediction, repeat masking, BLAST, HMM searching, subcellular localization prediction and more (Table 1). Ergatis also includes a number of multicomponent analysis pipelines. These include pipelines for bacterial genome annotation, an orthology identification pipeline and a pan-genome analysis pipeline.
3.1 Genome annotation
3.1.1 Bacterial genome annotation
Ergatis includes a pipeline template for automated prokaryotic genome annotation that is composed of 36 analysis components. These include gene structure prediction and evaluation, RNA analysis, homology searches, frameshift analysis and functional prediction. Predicted transcripts are assigned functional names, Enzyme Commission numbers, gene symbols and GO terms where possible. The final output is a BSML file that can be loaded into a Chado database instance. Execution of this pipeline on a Pyrobaculum species, with a genome size of 2.2 megabases, yielded 2863 predicted genes. The pipeline had 202 704 commands overall and ran in 216 CPU hours, or 3.8 actual hours when distributed on our compute grid. Ergatis and this pipeline template are used to support the IGS annotation engine (Giglio).
3.1.2 Eukaryotic genome annotation
The complexity of eukaryotic gene structures makes purely automated prediction with a single pipeline difficult. Ergatis contains components for over a dozen different gene prediction programs, such as GeneWise (Birney et al., 2004), GeneMark (Besemer and Borodovsky, 2005) and FGENESH (SoftBerry), as well as RNA prediction. Gene models can be used as inputs to other components for functional annotation using sequence searches, signal sequence prediction using SignalP and protein subcellular localization with components such as TargetP and TMHMM (Emanuelsson et al., 2007). Ergatis has been the primary data management tool for several eukaryotic genome projects, including Aedes aegypti (Nene et al., 2007) and Oryza sativa (Ouyang et al., 2007).
3.2 Comparative genomics
The decreasing cost of genome sequencing has provided data for the comparative analysis of related whole genomes. Ergatis provides a pipeline template to identify putative paralogs and orthologs within a collection of organisms. The pipeline is based on all-vs-all BLASTP searches and a reciprocal best BLAST clustering of proteins. Putative paralogs are flagged from BLAST hits that span at least 80% of sequence length at least 80% identity (Crabtree et al., 2007). The pipeline template consists of eight components and provides default cutoffs for the BLAST and clustering steps. The output is set of gene clusters encoded in a BSML file that can be loaded into Chado and visualized using Sybil (Crabtree et al., 2007).
This comparative pipeline was used to build gene clusters for the Pathema resource center (Brinkac et al., 2010). The analysis contained an all-vs-all protein BLAST of the 131 008 polypeptide sequences in this organism set. The BLAST search ran in 172 CPU hours and the complete pipeline ran in 381 compute hours, which included output validation and format conversion steps to a tab-delimited format and BSML. Distributed across a 60-node compute cluster, this component took just 8.2 h to complete.
The Ergatis comparative pipeline was also used to build gene clusters for the ‘Strepneumo’ website (Tettelin) with a group of 32 Streptococcal strains to identify putative ortholog and paralog gene clusters. This collection of organisms contained 72 101 coding genes, which were predicted to form 1543 paralogous clusters and 3342 orthologous clusters. This pipeline contained 79 106 commands that were executed in 87 CPU hours which, when distributed across a 100-node computation cluster, had a wall-clock runtime of 1 h and 53 min.
Ergatis also includes a pipeline template to summarize genomic diversity in a pan-genome (Tettelin et al., 2008). The pipeline classifies genes in the pan-genome as core (conserved across all input genomes), shared (conserved across a subset of input genomes) and unique (present in a single genome) used the method described in Tettelin et al. (2008). The results are plotted and a regression is fit to determine whether the data suggest more sequenced genomes would uncover more new genes (an open pan-genome) or not (a closed pan-genome). A related plot estimates size of the entire gene repertoire (or pan-genome) for the species being sampled. The Ergatis pan-genome pipeline was used on the analysis of 14 Yersinia pestis genomes in Eppinger et al. (2010). The input of 4844 unique protein-coding genes ran for 16 h when distributed across 150 compute nodes of our cluster consuming 519 compute hours.
4 DISCUSSION
The Ergatis uses a modular, scalable and extensible approach to pipeline creation and management on local or distributed compute resources. It provides a wide array of analysis components and pre-configured pipeline templates with which users can build their own custom pipelines. While flexible and extensible, this modular approach is not necessarily the most efficient way to execute some pipelines. Each modular component accepts a set of input types and creates one or more output types. Because of this, Ergatis pipelines may involve a series of format conversion steps that can incur extra computational overhead. We believe that the benefits of the component abstraction layer, such as modular construction and pipeline reuse, outweigh drawbacks such as this. We believe this approach is preferable over webservice-based systems for institutions who possess adequate computational hardware and who wish to ensure maximal resource availability and customization. Pipelines can be executed in Ergatis without concern for service availability or data transfer limitations to remote service sites.
Though initially intended as a local pipeline management tool, Ergatis has been extended and employed by some users to drive publicly available computational web resources. One example of this is the Integrative Services for Genomics Analysis, a web-based prokaryotic annotation server that uses Ergatis as its back-end (Hemmerich et al., 2010). Ergatis has also been extended to serve as the pipeline framework for CloVR, a virtual appliance that integrates genomics tools on cloud computing platforms for viral, prokaryotic, metagenomic and eukaryotic sequencing projects (Fricke).
Current and future work includes improvement to the web interface, training documentation and addition of new components and pipeline templates to Ergatis for analysis of metagenomics and transcriptomics data. This work will also enable Ergatis to serve as one of the access portals for the Data Intensive Academic Grid, a publicly available 1000+ core computational infrastructure currently under development. The Ergatis software is open source and freely available at http://ergatis.sf.net.
ACKNOWLEDGEMENTS
We would like to thank both the developers on the project and the user community who continue to suggest new features and improve the project.
Funding: National Institute of Allergy and Infectious Diseases Microbial Sequencing Contract (NIH-N01-AI-30071); National Institutes of Health BRC contract (NIH-N01-AI-30071) in part.
Conflict of Interest: none declared.
REFERENCES
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J, Borodovsky M. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005;33:W451–W454. doi: 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, et al. GeneWise and genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkac LM, et al. Pathema: a clade-specific bioinformatics resource center for pathogen research. Nucleic Acids Res. 2010;38(Database issue):D408–D414. doi: 10.1093/nar/gkp850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JM, et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree J, et al. Sybil: methods and software for multiple genome comparison and visualization. Methods Mol. Biol. 2007;408:93–108. doi: 10.1007/978-1-59745-547-3_6. [DOI] [PubMed] [Google Scholar]
- Day-Richter J. “OBO Flat File Format Specification, version 1.2.”. Available at http://www.geneontology.org/GO.format.obo-1_2.shtml(last accessed date April 23, 2010) [Google Scholar]
- DiBernardo M, et al. Semi-automatic web service composition for the life sciences using the BioMoby semantic web framework. J. Biomed. Inform. 2008;41:837–847. doi: 10.1016/j.jbi.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Eilbeck K, et al. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed NM, et al. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309:404–409. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, et al. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Eppinger M, et al. Genome sequence of the deep-rooted Yersinia pestis strain Angola reveals new insights into the evolution and pangenome of the plague bacterium. J. Bacteriol. 2010 doi: 10.1128/JB.01518-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke WF. [(last accessed date April 23, 2010)];CloVR: A Genomics Tool for Automated and Portable Sequence Analysis Using Virtual Machines and Cloud Computing. Available at http://clovr.igs.umaryland.edu/ [Google Scholar]
- Giardine B, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglio M. [(last accessed date April 23, 2010)];“Institute for Genome Sciences – Annotation Engine.”. Available at http://ae.igs.umaryland.edu. [Google Scholar]
- Hemmerich C, et al. An Ergatis-based prokaryotic genome annotation web server. Bioinformatics. 2010;26:1122–1124. doi: 10.1093/bioinformatics/btq090. [DOI] [PubMed] [Google Scholar]
- Hotopp JC, et al. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2006;2:e21. doi: 10.1371/journal.pgen.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LabBook I. [(last accessed date April 23, 2010)];“Bioinformatic Sequence Markup Language (BSML).”. Available at http://www.xml.com/pub/r/1311. [Google Scholar]
- Mungall CJ, Emmert DB. A Chado case study: an ontology-based modular schema for representing genome-associated biological information. Bioinformatics. 2007;23:i337–i346. doi: 10.1093/bioinformatics/btm189. [DOI] [PubMed] [Google Scholar]
- Nene V, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oinn T, et al. Taverna: a tool for the composition and enactment of bioinformatics workflows. Bioinformatics. 2004;20:3045–3054. doi: 10.1093/bioinformatics/bth361. [DOI] [PubMed] [Google Scholar]
- Ouyang S, et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano P. Automation of in-silico data analysis processes through workflow management systems. Brief Bioinform. 2008;9:57–68. doi: 10.1093/bib/bbm056. [DOI] [PubMed] [Google Scholar]
- Shah SP, et al. Pegasys: software for executing and integrating analyses of biological sequences. BMC Bioinformatics. 2004;5:40. doi: 10.1186/1471-2105-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, et al. ISYS: a decentralized, component-based approach to the integration of heterogeneous bioinformatics resources. Bioinformatics. 2001;17:83–94. doi: 10.1093/bioinformatics/17.1.83. [DOI] [PubMed] [Google Scholar]
- SoftBerry. [(last accessed date April 23, 2010)];“Gene finding in Eukaryota.”. Available at http://www.softberry.com. [Google Scholar]
- Tang F, et al. Wildfire: distributed, grid-enabled workflow construction and execution. BMC Bioinformatics. 2005;6:69. doi: 10.1186/1471-2105-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H. [(last accessed date April 23, 2010)];Sybil: strepneumo: home. Available at http://strepneumosybil.igs.umaryland.edu. [Google Scholar]
- Tettelin H, et al. Comparative genomics: the bacterial pan-genome. Curr. Opin. Microbiol. 2008;11:472–477. doi: 10.1016/j.mib.2008.09.006. Available at http://strepneumosybil.igs.umaryland.edu(last accessed date April 23, 2010) [DOI] [PubMed] [Google Scholar]
- Tiwari A, Sekhar AK. Workflow based framework for life science informatics. Comput. Biol. Chem. 2007;31:305–319. doi: 10.1016/j.compbiolchem.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Wilkinson MD, Links M. BioMOBY: an open source biological web services proposal. Brief Bioinform. 2002;3:331–341. doi: 10.1093/bib/3.4.331. [DOI] [PubMed] [Google Scholar]