Abstract
Motivation: Recent years have seen the development of a wide range of biomedical ontologies. Notable among these is Sequence Ontology (SO) which offers a rich hierarchy of terms and relationships that can be used to annotate genomic data. Well-designed formal ontologies allow data to be reasoned upon in a consistent and logically sound way and can lead to the discovery of new relationships. The Semantic Web Rules Language (SWRL) augments the capabilities of a reasoner by allowing the creation of conditional rules. To date, however, formal reasoning, especially the use of SWRL rules, has not been widely used in biomedicine.
Results: We have built a knowledge base of human pseudogenes, extending the existing SO framework to incorporate additional attributes. In particular, we have defined the relationships between pseudogenes and segmental duplications. We then created a series of logical rules using SWRL to answer research questions and to annotate our pseudogenes appropriately. Finally, we were left with a knowledge base which could be queried to discover information about human pseudogene evolution.
Availability: The fully populated knowledge base described in this document is available for download from http://ontology.pseudogene.org. A SPARQL endpoint from which to query the dataset is also available at this location.
Contact: matthew.holford@yale.edu; mark.gerstein@yale.edu
1 INTRODUCTION
In recent years, formal ontologies have been suggested as a solution to the problem of describing complicated realms of biomedical knowledge (Rubin et al., 1997). Well-designed ontologies possess a number of positive aspects including, (i) the ability to define controlled vocabularies of terms, (ii) the ability to inherit and extend existing terms, (iii) the ability to declare relationships between existing terms and (iv) the ability to infer new relationships by reasoning over existing terms. Through the technologies known collectively as the Semantic Web, most especially the Web Ontology Language (OWL) (OWL2, 2009), researchers are able to share and extend ontologies throughout the scientific community. Although biomedical ontologies have existed for a number of years, scientists are far from realizing the full benefits of their use. There is still room for considerable advancement in this area, especially in the application of formal reasoning.
A unique strength of formal ontologies in the area of knowledge representation is their ability to be reasoned upon in a logically provable way. This reasoning is performed using Description Logic (DL) , a form of logic developed to reason on objects, both individual objects and classes of objects. Software called reasoners [examples include Pellet (Sirin et al., 2007), Fact++ (Tsarkov and Horrocks, 2006) and KAON (KAON, 2010)] use the rules of DL to perform particular operations on knowledge bases. The most important of these are: (i) consistency checking: the adherence of the ontological model to the rules of DL; (ii) satisfiability checking: the ability for classes described to be realized by actual individuals; and (iii) classification: the expansion of relationships between objects inferred from explicitly stated relationships. The DL version of the OWL language assures that DL reasoners can perform these services in a computationally tractable manner. While the first two services are imperative for ensuring the integrity of data, the third, the ability to infer new relationships, is especially appealing to scientists as it hints at the possibility of new discovery. Moreover, the relationships discovered are instantly provable, given the reasoner's adherence to the rules of formal logic. The OWL language offers a rich set of properties for inference, including class subsumption, property subsumption, transitivity of properties and inverse properties. We consider a simple example. We define a class Father as a subclass of a class Ancestor and a class Son as a subclass of a class Descendent. We also define a property has_father as a sub-property of has_ancestor with an inverse property has_son that is a sub-property of has_descendent. Now, if say that Matt has_father Ted, the reasoner can automatically infer that Ted has_descendant Matt. Taken in isolation, such examples seem trivial and obvious but they can be very helpful when sifting through huge amounts of data.
Despite the richness of OWL's set of relational properties, it does not cover the full range of expressive possibilities for object relationships that we might like. For example, it is often useful to declare data relationships in terms of conditional statements or production rules. For this purpose, a specialized rule language is useful. The Semantic Web Rule Language (SWRL) incorporates an existing rule language (RuleML) with OWL (SWRL, 2005). Rules are defined in two parts: antecedents and consequents. If all statements in the antecedent clause are determined to be true, then all statements in the consequent clause are applied. In this way, new properties can be assigned to individuals in an ontology based upon the current state of the knowledge base. A popular example is the ‘Uncle Rule’, which states that if a person's father has a brother, that brother is the person's uncle. So, if Matt has_father Ted and Ted has_brother Doug, the reasoner can infer that Matt has_uncle Doug. SWRL also specifies a library of built-in functions which can be applied to individuals. These include numerical comparison, simple arithmetic and string manipulation. At present, SWRL is the most widely used rule language in the Semantic Web community. The popular ontology development environment Protégé includes a SWRLTab plugin for creating and processing SWRL rules (SWRLTab, 2010). SWRL is supported by the Pellet reasoner up to the point where rules can be determined to be ‘DL-safe’, i.e. they may be realized in a computationally tractable fashion. Firing of SWRL rules is performed by Pellet as part of the classification process and new entailments thus generated can be added to an existing ontology.
We wish to exploit these reasoning capabilities in our research on pseudogenes. For this purpose, we can build upon Sequence Ontology (SO)(Eilbeck et al., 2005), among the most notable of ontologies created in the biomedical community. SO aims to provide the full set of terms and relationships necessary to perform sequence annotation. Despite its central importance to molecular biology, sequence annotation has historically been more difficult than required due to a divergence in naming standards. This makes the sharing of data a challenge. SO provides a controlled hierarchy of terms that describe elements that may be found upon a sequence, referred to as sequence_features. These may represent anything from genes and pseudogenes to smaller units such as individual bases. A class sequence_variant describes variable elements on the sequence such as alleles and copy_number_variations. Annotative tags which may be attached to elements on the sequence are subclasses of the sequence_attribute class. Examples include conserved, retrotransposed and transgenic. In addition to this structured set of terms, SO defines relationships for how sequence elements are inter-related. In particular, the authors rigorously define part-whole relationships employing formalisms from the philosophical discipline of mereology (Winston et al., 1987).
The most common usage of SO is to label sequence annotations with appropriate SO terms. Typically this is done by attaching terms to an annotation in a separate format, such as a flat file or a database which describes instances of sequence data (Eilbeck and Lewis, 2004). The developers have created a modular relational database schema called CHADO for this purpose (Mungall et al., 2007). Here, notably, sequence elements are not hard-coded with their location on a particular sequence but are linked to featureloc elements which contain individual location information. CHADO and its companion mark-up format CHADO-XML offer a robust approach to annotating sequences in a formal and logically coherent manner. Its strengths are particularly evident in the handling of large volumes of data. We wished to explore an alternative approach. We decided to create a full knowledge base by populating the SO ontology with individual instances of sequence features. We would then perform reasoning using relationships defined as part of SO, extensions to these relationships and SWRL rules based upon these relationships. From this. we hoped to discover new relationships between sequence features and thereby strengthen our pseudogene annotation.
2 APPROACH
Pseudogenes form an almost ideal subdomain for ontology development in that they are connected to normal genomic features while maintaining a large enough number of unique aspects to form an area for independent description. Pseudogenes represent bits of genomic sequence that were once functional but have become inactive. They are often the result of various genomic copying processes, principally duplication and retro-transposition. There are two basic types of pseudogenes—processed and duplicated. The former arise through the process of retro-transposition, the latter through duplication events. As artifacts of a history of copying, pseudogenes offer us a glimpse of evolutionary history, both of individual genes and of the genome as a whole. An understanding of how and when particular pseudogenes were derived in relation to other genomic features is significant to our comprehension of both genomics and evolutionary biology (Zhang and Gerstein, 2003). To this end, we have been involved in annotating pseudogenes in collaboration with researchers at the Sanger Institute and at UCSC, including researchers who were involved in the development of SO.
As SO already defines a number of classes related to pseudogenes, we were able use these terms to fill our knowledge base with individuals. For those terms not present, we were able to extend existing SO classes. SO currently defines a class pseudogene with several subclasses including processed_pseudogene. We added additional subclasses duplicated_pseudogene and unitary_pseudogene for instances of these types of pseudogene. Pseudogenes whose specific type was ambiguous were left as instances of the base class pseudogene. All pseudogenes are defined as non-functional copies of parent genes. These genes are generally identified using the transcribed protein. For this reason, even though pseudogenes may derive from any type of gene including RNA genes, in our ontology all parent genes were instantiated as instances of the SO's protein_coding_gene class. We created a sub-property of SO's property non_functional_homolog_of to describe the link between a pseudogene and the parent from which it derived. This sub-property, has_parent_gene restricts the range of values to instances of protein_coding_gene and restricts the maximum cardinality to a single instance. We used identifiers from the existing pseudogene ontology (PGO), which was created as part of the Pseudofam project (Lam et al., 2009). We also incorporated information, where available, about the location of particular exons and introns within the pseudogenes, noting of course that these no longer have the same meaning in a non-functional context. Here, we were able to use existing SO classes, pseudogenic_exon and intron. To express containment of these features within a pseudogene, we adopted SO's recommended usage of the part_of property, which is defined as a core relationship in the OBO Relationship Ontology (RO) (Smith et al., 2007). To simplify querying later, we created sub-properties contains_pseudogenic_exon and contains_pseudogenic_intron. These constrain the domain of the property to members of the pseudogene class and the range of the property to pseudogenic_exons and introns, respectively. We also defined inverse properties pseudogenic_exon_in_pseudogene and pseudogenic_intron_in_pseudogene. These constrain the RO's has_part property which is the inverse of part_of. These inverse properties can, of course, be automatically inferred by the reasoner.
One focus of our research is the relation of pseudogenes to segmental duplications (SDs). SDs are defined as continuous stretches of DNA that map to multiple locations on the genome. A common ground rule is that they are 1000 or more base pairs in length and have sequence similarities of 90% or more. Like pseudogenes, they are residual artifacts of a history of copying. Apprehending their origins is similarly important to understanding the evolutionary history of the genome (Bailey and Eichler, 2006). To include SDs in our annotation, we defined a class sd_segment as a subclass of the relatively low-level SO term biological_region. To keep track of the one or more duplicate segments each sd_segment has, we defined a property is_sd_pair_of. The sd_segment class acts as both domain and range of this property which is a sub-property of the SO term similar_to. We also wanted to determine what if any pseudogenes or genes were located within a segment in an SD pair. We used the same technique of constraining the has_part property that we did for contains_pseudogenic_exons. Properties called contains_pseudogene and contains_gene were created, as were the inverse properties pseudogene_in_segment and gene_in_segment. Finally, we created a property to indicate the number of pseudogenes (has_pseudogene_count) and the number of genes (has_gene_count) within a segment. These were necessary for certain SWRL rules we created later. On the surface it appears a deficiency that we must specify the size of the lists of genes and pseudogenes as a separate property. Indeed, the SWRL built-in library provides an operator to determine the length of a list. However, this is not considered DL-safe as it would violate the open-world assumption which is a central tenet of DL and the Semantic Web. Essentially, though we list certain genes within a segment, it is not automatically guaranteed that these are the only genes unless we explicitly say so. Knowledge that is unstated is not presumed to be false; it is merely presumed to be missing.
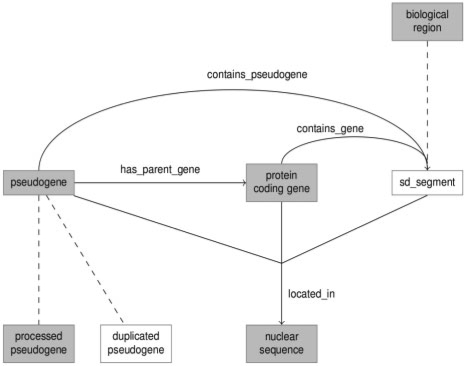
Attaching elements of the genome to a particular location is problematic in sequence annotation, as exact coordinate locations will always vary between individual members of a species. For this reason, model sequences have been developed for a number of organisms. Even where such sequences do exist, however, there will always be incompatibility between builds or versions of the model. Previously, sequence annotations using SO terms have handled the description of individual sequence features outside of the ontology. CHADO, for example, mitigates the issue of exact coordinates by abstracting location away from features through the use of a featureloc object. For our purposes, however, because we perform reasoning on the locations of features, we must incorporate coordinates from an individual build into our knowledge base. Thus, we needed to create a few relationships to specify exact feature location. We stored genome loci using the SO class nuclear_sequence. This class served as the domain for five new properties which help to spell out an exact location: in_build, on_chromosome, has_start_point, has_end_point and on_strand. The RO property located_in is then used to link a sequence feature to its location. The in_build property specifies which version of the model sequence we are using. We declared custom datatypes to limit the legal values for chromosomes (1–22, X, Y) and for strands (positive, negative). Our instances of the classes pseudogene (and its subclasses), protein_coding_gene and sd_segment all make use of these properties to specify their precise locations on the genome. An outline view of the basic classes and relationships in our ontology is provided in Figure 1.
Fig. 1.
Diagram showing the relationships between some of the base classes of the ontology. Dashed lines are used to indicate subclass relationships. Regular lines indicate property relationships. Classes in SO are highlighted in gray, while those which were added to our ontology have a white background.
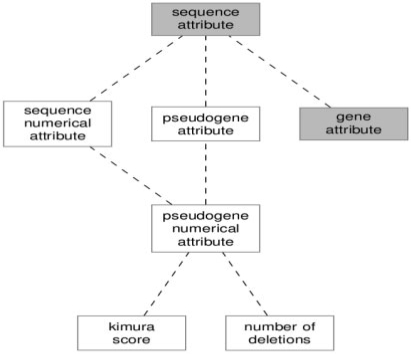
Using SO, one annotates a feature by assigning it an instance of one of the subclasses of sequence_attribute. This is accomplished using the has_quality property. Sequence attributes currently defined in SO are for labeling conditions that are either present or absent. We found that many of the attributes we wanted to use took numerical values, be they counts, ratios or scores. We decided to create a subclass of sequence_attribute for these types of attributes called sequence_numerical_attribute. This new class serves as the domain for a has_numerical_value property which is used to represent the data value. To help organize our attributes, we created a pseudogene_attribute class akin to the SO class gene_attribute. We further subclassed this with a pseudogene_numerical_attribute class. Specific classes of attribute were created using this hierarchy for the counts (number_of_insertions, number_of_deletions, number_of_stops, number_of_shifts, disablements and polyA) and scores (log_kimura_score, fraction, evalue and identity) we wanted to track. Figure 2 illustrates the hierarchy of pseudogene annotation attributes.
Fig. 2.
Diagram showing the hierarchy of annotation attributes for our pseudogene ontology. The dashed lines denote subclass relationships. Classes from SO are highlighted in gray while classes add by our ontology have a white background.
With the base relationships of our ontology in place, we created a set of SWRL rules to infer new relationships based upon them. Their design was guided by the goals of our research and they will be discussed in detail in Section 5.
Having defined the terms and relationships of our ontology, we now populated it with instances of pseudogenes, parent genes and SDs. We then performed consistency and satisfiability checking using the reasoner and classified the data. At this point, we were able to query the resulting knowledge base to answer biological questions. Here, we were presented with two options. We could leave the data in the reasoner and query against it programmatically. Or we could export an XML document containing our fully entailed data and import it into a triple store, a piece of software that functions like a database for relational data. In both cases, we would be querying the data using a variant of the SPARQL language (SPARQL, 2008), and RDF query language comparable with SQL for relational data. Generally, triple stores do not support the degree of DL reasoning that actual reasoners do. In fact, the plain SPARQL language does not expect data to be reasoned upon. Reasoners typically use a reasoning-enhanced version of SPARQL such as Pellet's SPARQL-DL. The tradeoff however is speed; because they do not need to perform reasoning, triple stores can generally retrieve results much more quickly.
3 METHODS
Data about pseudogenes were obtained from the pseudogene.org (Karro et al., 2007) website. These data reference proteins by Ensembl identifiers in defining parent proteins for the pseudogenes. We obtained the links between these proteins and the genes that code for them using the Ensembl (Hubbard et al., 2002) website. To assign an exact location to a parent gene, we used the lowest start value and the highest stop value for all the transcripts of the gene. Information on SDs was obtained from the web resources of the Eichler lab (Duplication, 2010). The data were parsed from flat files using a custom program written in Java. This program employed the OWLAPI library (Bechhofer and Philip Lord, 2003) to build an OWL 2 compatible ontology which included the SWRL rules that we used. The program used the Pellet reasoner to check the ontology for consistency and satisfiability and then to classify it. The full set of entailments generated by the reasoner were serialized into an OWL document. This document was then loaded into the open-source version of Virtuoso, a universal database which includes a triple store (Virtuoso, 2010). Because the full set of inferences was pre-created, we were able to take advantage of Virtuoso's fast performance without losing the advantages of reasoning. Virtuoso provides an HTTP interface to a SPARQL endpoint, which we were able to query through either a web interface or programmatically.
A key focus of our research is to explore the relationship between pseudogenes and SDs and determine what this can teach us about the evolution of the genome. We were particularly interested in finding examples of two scenarios. In the first (Case 1), we sought to find situations that would allow us to directly compare the evolution rate of a pseudogene and its parent gene. To do this, we would need to find cases where a pseudogene and its parent were located on the separate segments of an SD pair. Additionally, we needed to verify that no other pseudogenes or genes were on the same segment as that containing the parent gene. We could then examine the relative substitution rates between the pseudogene and its surrounding area and the parent gene and its surrounding area. We did this using the Kimura score metric. If the log of this value fell within a specific range of values, we could argue that the pseudogene was evolving more rapidly, less rapidly or at an equal rate as its parent. In the second scenario (Case 2), we tried to find pseudogenes that arose not from duplication of a parent gene but from duplication of another pseudogene. To find these, we again needed to locate cases where a pseudogene and its parent were on the separate segments of an SD pair. In this case, however, we wanted other pseudogenes to be present on the segment containing the parent gene. We then looked at whether the original pseudogene was aligned with its parent or with another pseudogene. In the latter scenario, we were able to conclude that the pseudogene arose from duplication of another pseudogene and suggested that it formed a new category, the duplicated-processed pseudogene. It is worth noting the possibility of other scenarios, for example, the orginating pseudogene may be located on a third duplicate segment distinct from that of the duplicated pseudogene and the parent gene. More complex possibilities such as these are discussed in detail in Khurana et al. (manuscript in preparation).
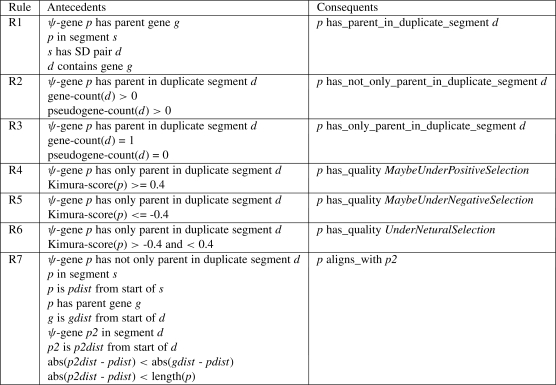
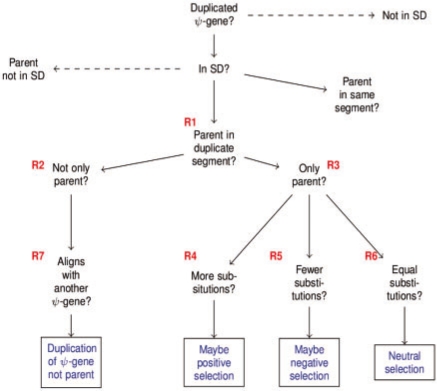
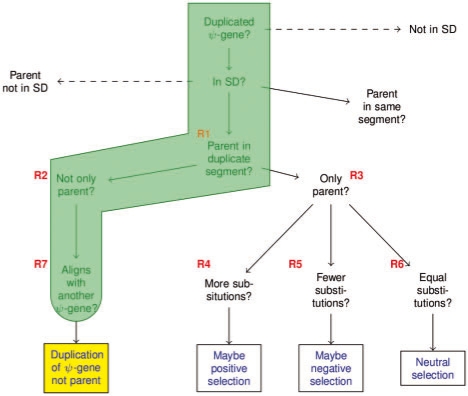
These cases suggest a sort of flowchart which can be traversed by a series of rules which build upon each other. We list these rules in Figure 3. The flowchart can be seen in Figure 4. Further discussion of this decisions tree, including the strategies employed and their biological rational can be found in the forth-coming paper by Khurana et al. We created a total of seven rules to reach our goal. Rule 1 is a foundational rule, necessary for all that follow it. Its goal is to mark all pseudogenes that are in the segment of an SD pair whose parent gene is located in the other segment. These pseudogenes are assigned the property has_parent_in_duplicate_segment whose value is the segment of the parent. Rule 2 uses Rule 1 to find parent segments and then checks the has_gene_count and has_pseudogene_count values to determine if other genes or pseudogenes are present in the segment. If these other features are present, the pseudogene is given the property has_not_only_parent_in_duplicate_segment. Rule 3 functions similarly except that it expects the has_pseudogene_count value to be 0 and the has_gene count to be 1 (the parent gene). Matching pseudogenes are given the property has_only_parent_in_duplicate_segment. At this point, we can move to directly answer the questions raised by Case 1. Rule 4 uses Rule 3 to find segments containing only the parent gene. It then retrieves the log of the Kimura score of the pseudogene. If its value is above a high cutoff, it determines the pseudogene is under positive selection and assigns it the sequence attribute maybe_positively_selected. Rule 5 behaves as Rule 4, except that for log Kimura scores below a low cutoff, it assigns the pseudogene the maybe_negatively_selected attribute. The path taken by Rule 5 is illustrated in Figure 5. Rule 6 closes out Case 1 by assigning maybe_neutrally_selected to eligible pseudogenes with log Kimura scores between the high and low cutoff values. We chose 0.4 and −0.4 as high and low cutoffs, as these correspond with the distribution of scores for all pseudogenes (Khurana et al., manuscript in preparation). Rule 7 handles Case 2 by comparing the alignment of the pseudogene and its parent gene with the alignment of the pseudogene and the other pseudogenes on the duplicate segment. It uses Rule 2 to find pseudogenes on one segment of an SD pair whose parent gene is on the other segment along with other pseudogenes. It then uses the SWRL built-in arithmetic capabilities to measure the distance from the start of pseudogene (p1) to the start of its segment (p1dist). It then looks at the distance from start of the features on the duplicate segment. If the distance from start for one of the other pseudogenes (p2dist) is closer to p1dist than the distance from start of the parent gene is and if p2dist is within close enough range of p1dist (within the length of p1), it is determined that p1 is aligned with the pseudogene on the duplicate pair. This allows us to spot potential duplicated-processed pseudogenes which can be given the property aligned_to_pseudogene. Figure 6 indicates the path traversed by Rule 7.
Fig. 3.
Informal pseudocode description of the rules implemented in SWRL to traverse the flowchart.
Fig. 4.
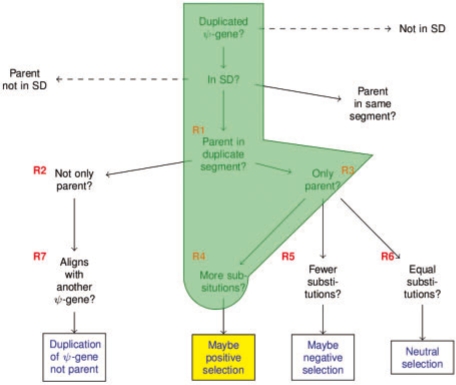
The decision tree to be traversed by SWRL rules. Dashed lines indicate a ‘No’ answer; solid lines indicate a ‘Yes’ answer. The same convention is used in Figures 5 and 6.
Fig. 5.
The path traversed by Rule 5 on the decision tree. This path follows Case 1 in looking to examples of pseudogenes which evolve at a less rapid pace than their parent genes.
Fig. 6.
The path traversed by Rule 7 on the decision tree. This path follows Case 2 in looking for pseudogenes which have arisen from the duplication of another pseudogene rather than their parent gene.
4 RESULTS
With a fully entailed ontology loaded into a triple store, we were able to issue SPARQL queries to find pseudogenes matching the criteria specified by Cases 1 and 2. In both cases, the SPARQL queries are quite simple and direct. Recall that in Case 1, we are trying to compare the evolution rate of a pseudogene with that of its parent. Using Rules 1–6, we isolated pseudogenes and parent genes which occur on the segments of an SD pair, making sure that no other genes or pseudogenes were present on the segment containing the parent gene. By analyzing the substitution rate we attached a quality to the pseudogene indicating whether it might be positively, negatively or neutrally selected. We can now find pseudogenes of interest by naming those that possess this quality. For example, to find quickly evolving pseudogenes one might issue the following:
SELECT ?p
WHERE
?p #has_quality #maybe_positively_selected
For the sake of brevity, we are skipping the necessary import statements. In Case 2, we used Rule 7 to locate pseudogenes which were derived from the duplication of another pseudogene rather than a parent gene. These were found by testing the alignment of a pseudogene to other pseudogenes present on the same segment of the SD pair as the parent gene. A property relationship was created between these aligned pseudogenes. To find examples of these duplicated-processed pseudogenes, we ask for pseudogenes fulfilling this relationship:
SELECT ?p
WHERE
?p #aligned_to_pseudogene ?p2
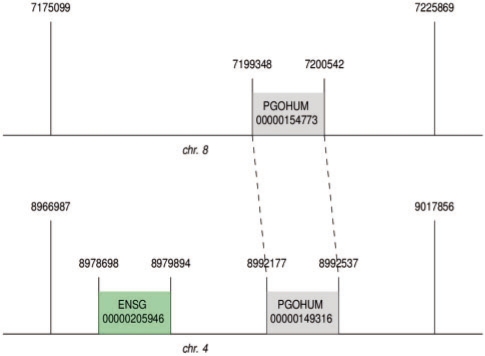
As a result of this query, we discovered that PGOHUM00000154773 is potentially a duplicated-processed pseudogene, as it is more closely aligned with the pseudogene PGOHUM00000154773 than to its parent gene, ENSG00000205946. This relationship is illustrated in Figure 7.
Fig. 7.
A potential duplicated-processed pseudogene found by aligning one pseudogene with another on the same segment as the parent gene. The pseudogene, PGOHUM00000154773, is located on chromosome 8 of the reference sequence between bases 7199348 and 7200542. Its parent gene, ENSG00000205946 (USP17L6P), is found on chromosome 4 between bases 8978698 and 8979894. PGOHUM00000154773 is found on an SD segment located between 7199348 and 7200542 on chromosome 8. The parent gene is on the duplicate segment located between 8966987 and 9017856 on chromosome 4. The duplicate segment also contains another pseudogene, PGOHUM00000149316 between bases 8992177 and 8992537. Because this other pseudogene is a similar distance from the start of the segment as PGOHUM00000154773 is to the start of its segment (25 190 bp versus 24 249 bp) and the parent gene is in a different portion of the segment (11 711 bp from the start), the deduction that PGOHUM00000154773 is aligned to PGOHUM00000149316 rather than ENSG00000205946 makes sense. This was found by applying SWRL Rule 7.
5 DISCUSSION
Because of their status as genomic fossils, pseudogenes are of interest not only for how they currently appear but how they arose and developed. Much like examining and dating bones to a paleontologist, the issue of ascertainment is central to the student of pseudogenes. In this, a certain amount of uncertainty is inherent. For a number of pseudogenes, we can precisely describe their origin and place in time; for others we are less certain. We can see an example of this in Case 1 above, where a higher substitution rate suggests that a pseudogene may be positively selected it also raises the possibility that the surrounding region is negatively selected. We cannot say with full certainty which possibility is the case. The handling of uncertainty is a problematic issue when formally describing pseudogenes. OWL and most other mainstream ontology languages do not deal with the concept of probability with respect to knowledge. This is largely because DL itself only deals with data that is certain. Other branches of logic exist to handle situations of uncertainty, such as fuzzy or probabilistic logic and extensions to OWL have been proposed to build knowledge bases using these logics (Ding and Peng, 2004). At present, however, these are confined to the more experimental reaches of knowledge representation studies. The alternative would be to define terms using a conventional ontology to represent different levels of certainty with regards to ascertainment. As our knowledge base grows, we hope to explore this area more fully.
Performance presents another challenge to builders of biomedical ontologies. Although OWL-DL guarantees computational tractability, it does not promise that classification can be completed using an amount of time and memory that we may find acceptable. It is also an unfortunate truth that computational expense increases as an ontology becomes more expressive. These problems are particularly acute for genomics researchers, where vast amounts of data can quickly bog down a DL reasoner even on a well apportioned machine. For example, we initially ran out of memory while trying to classify our ontology using the full set of pseudogenes. This occurred even when running the reasoner on a 32 GB server. After some experimentation, we were able to get the ontology to classify by performing two steps. First, we removed all properties that were not used for the creation of entailments for production rules, generating the full set of inferences and then re-inserting the non-essential properties. Second, we changed individual instances of protein_coding_gene to a custom class SimpleGene which extends SO's biological_region class. This freed the reasoner from applying the restrictions defined by SO for protein coding genes and saved considerable amounts of memory. We felt this workaround was acceptable because our production rules do not make use of these restrictions. After the reasoner had finished generating entailments, we added an assertion declaring SimpleGene a subclass of protein_coding_gene, thus allowing future inferences to be drawn upon the genes using SO. After applying these techniques, classification of the full set of pseudogenes took around 19 min using a 28 GB heap size. A diminished set containing one of every 10 pseudogenes took around 11 min to classify and the set containing one of every 4 took around 13 min. It is abundantly clear that at present our approach could not be used for large-scale annotations, such as that of an entire genome. For large amounts of data, the integration of SO terms with relational database technology through the CHADO schema and CHADO-XML offer a far quicker solution. It is promising that RDF data can be queried quickly using triple stores, but the process of creating the full set of entailments by classifying the data through the reasoner is still a significant performance bottleneck. We can only hope that the future will continue to bring performance improvements in this area, both through more efficient algorithms and faster technology.
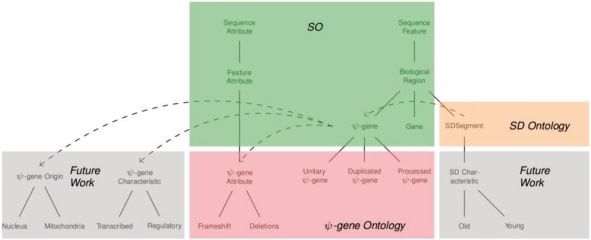
The ontology we have presented here is an extension of SO that joins additions related to pseudogenes with additions related to SDs. It forms a useful prototype for describing pseudogenes and provides a useful framework for reasoning and drawing biological inferences. It stops short, however, of providing a canonical ontology of the domain of pseudogenes. As part of our future research, we intend to build upon the structure presented here to form a more complete ontology. For example, it would be useful to add classes and relationships to describe pseudogene characteristics such as regulatory and transcribed. These terms could be incorporated from previous work by Lam et al. (2009). We also wish to incorporate the notion of derivation of a pseudogene, whether from the nucleus or mitochondria. We hope to enlist the support of other pseudogene researchers in this endeavor. Finally, we see the potential for further development leading to an ontology of SDs. Figure 8 illustrates the present coverage of our ontology and areas we hope to include in the future.
Fig. 8.
Informal depiction of the coverage provided by the current ontology, including portions derived from SO, as well as areas to be covered in future work. In the diagram, plain lines indicate class hierarchy (is-a) relationships, while dashed lines indicate property (has-a) relationships.
6 CONCLUSION
We used the SO to build a knowledge base of pseudogenes, extending SO terms where necessary to describe our data and borrowing identifiers from the PGO ontology. We created a series of custom SWRL rules to find situations of interest involving our research on the relation between pseudogenes and SDs. Using these rules and the inherent capabilities of DL reasoners, we were able to infer new relationships about our existing data. We moved this fully entailed knowledge base into a triple store with a SPARQL endpoint to allow us to query it for biologically relevant information.
Funding: The National Institutes of Health and AL Williams Professorship funds; National Institute of Health grants P01 DC04732 and R01 DA021253 (to K.C.).
Conflict of Interest: none declared.
REFERENCES
- Bailey JA, Eichler EE. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat. Rev. Genet. 2006;7:552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- Bechhofer S, Philip Lord RV. ISWC 2003. Berlin: Springer; 2003. Cooking the semantic web with the OWL API; pp. 659–675. [Google Scholar]
- Ding Z, Peng Y. Proceedings of the 37th Hawaii International Conference On System Sciences (HICSS-37) Big Island: IEEE.; 2004. A probabilistic extension to ontology language owl. [Google Scholar]
- Duplication,S. 2010. [last accessed date January 8, 2010]. Available at http://humanparalogy.gs.washington.edu/build36/build36.htm. [Google Scholar]
- Eilbeck K, Lewis SE. Sequence ontology annotation guide. Comp. Funct. Genomics. 2004;5:642–647. doi: 10.1002/cfg.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilbeck K, et al. The sequence ontology: a tool for the unification of genome annotations. Genome Biol. 2005;6:1–12. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T, et al. The ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAON. 2010. Available at http://kaon2.semanticweb.org. [Google Scholar]
- Karro JE, et al. Pseudogene.org: a comprehensive database and comparison platform for pseudogene annotation. Nucleic Acids Res. 2007;35:D55–D60. doi: 10.1093/nar/gkl851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H.YK, et al. Pseudofam: the pseudogene families database. Nucleic Acids Res. 2009;37:D738–D743. doi: 10.1093/nar/gkn758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall CJ, et al. A chado case study: an ontology-based modular schema for representing genome-associated biological information. Bioinformatics. 2007;23:i337–i346. doi: 10.1093/bioinformatics/btm189. [DOI] [PubMed] [Google Scholar]
- OWL2. 2009. [last accessed date February 1, 2010]. Available at http://www.w3.org/TR/owl2-profiles/ [Google Scholar]
- Rubin DL, et al. Biomedical ontologies: a functional perspective. Brief. Bioinformatics. 1997;9:75–90. doi: 10.1093/bib/bbm059. [DOI] [PubMed] [Google Scholar]
- Sirin E, et al. Pellet: A practical owl-dl reasoner. Web Semant. 2007;5:51–53. [Google Scholar]
- Smith B, et al. The obo foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. biotechnol. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPARQL. 2008. [last accessed date January 23, 2010]. Available at http://www.w3.org/tr/rdf-sparql-query. [Google Scholar]
- SWRL. 2005. [last accessed date January 8, 2010]. Available at http://www.w3.org/submission/swrl. [Google Scholar]
- SWRLTab. 2010. [last accessed date December 23, 2009]. Available at http://protege.cim3.net/cgi-bin/wiki.pl?swrltab. [Google Scholar]
- Tsarkov D, Horrocks I. Automated Reasoning. Berlin: Springer; 2006. Fact++ description logic reasoner: system description; pp. 292–297. [Google Scholar]
- Virtuoso. 2010. [last accessed date January 8, 2010]. Available at virtuoso-openlinksw.com. [Google Scholar]
- Winston ME, et al. A taxonomy of part-whole relations. Cogn. Sci. 1987;11:417–444. [Google Scholar]
- Zhang Z, Gerstein M. Patterns of nucleotide subsitution, insertion and deltion in the human genome inferred from pseudogenes. Nucleic Acids Res. 2003;31:5338–5348. doi: 10.1093/nar/gkg745. [DOI] [PMC free article] [PubMed] [Google Scholar]