Abstract
Motivation: Phylogenetic tree-building methods use molecular data to represent the evolutionary history of genes and taxa. A recurrent problem is to reconcile the various phylogenies built from different genomic sequences into a single one. This task is generally conducted by a two-step approach whereby a binary representation of the initial trees is first inferred and then a maximum parsimony (MP) analysis is performed on it. This binary representation uses a decomposition of all source trees that is usually based on clades, but that can also be based on triplets or quartets. The relative performances of these representations have been discussed but are difficult to assess since both are limited to relatively small datasets.
Results: This article focuses on the triplet-based representation of source trees. We first recall how, using this representation, the parsimony analysis is related to the median tree notion. We then introduce SuperTriplets, a new algorithm that is specially designed to optimize this alternative formulation of the MP criterion. The method avoids several practical limitations of the triplet-based binary matrix representation, making it useful to deal with large datasets. When the correct resolution of every triplet appears more often than the incorrect ones in source trees, SuperTriplets warrants to reconstruct the correct phylogeny. Both simulations and a case study on mammalian phylogenomics confirm the advantages of this approach. In both cases, SuperTriplets tends to propose less resolved but more reliable supertrees than those inferred using Matrix Representation with Parsimony.
Availability: Online and JAVA standalone versions of SuperTriplets are available at http://www.supertriplets.univ-montp2.fr/
Contact: vincent.ranwez@univ-montp2.fr
1 INTRODUCTION
Representing the evolutionary history of a set of taxa is usually performed by using a phylogenetic tree. External nodes (leaves) correspond to operational units, and the root corresponds to its origin. The whole branching order from the root to each leaf provides a representation of the evolutionary relationships among the operational units studied. However, one could readily observe variations among different trees inferred for a given phylum, depending on stochastic errors due to site and taxon sampling, systematic errors induced by model misspecifications during probabilistic inferences, and biological errors due to hidden paralogy, lateral gene transfer and incomplete lineage sorting of ancestral polymorphism (see Swofford et al., 1996, for a discussion on the causes of incongruence). Summarizing various trees obtained from independent inferences into a single tree is therefore a recurrent issue in molecular systematics and phylogenomics. Having a single tree at hand is not just a simplification facilitating the management of topological information; it is in most cases a necessity to enable further analyses (see Jeffroy et al., 2006).
Pooling a collection of phylogenetic trees when all of them are defined on the same leaf set can be done with a consensus tree (Adams, 1972), such as the widely used strict or majority one (see Bryant, 2003, for an overview). Actually, the consensus tree is a particular case of the more general problem of combining trees defined on partially overlapping leaf sets. The latter case is known as the supertree problem (Gordon, 1986), whereby the aim is to build a tree (the supertree) summarizing the topological information induced by a collection of source (input) trees as well as possible. It has long been proposed that the median tree, i.e. the tree minimizing the sum of distances to the source trees, could be considered as an adequate supertree candidate (Bryant, 1997; Cotton and Wilkinson, 2007; Wilkinson et al., 2001). Defining the supertree as a median turns out to be not only an intuitive definition but also a guarantee of the consistency of so-defined supertree methods (Steel and Rodrigo, 2008). Even though this ensures the consistency of any median supertree, these authors stressed the importance of choosing appropriate metrics with regards to the biological setting and expected computing times. The triplet distance, based on rooted, binary, three-leaf topologies, has the advantage of being fine-grained and easy to compute in polynomial time (see Methods section for more formal definitions). We thus worked on designing a supertree method that seeks the triplet median supertree (TMS).
Among the numerous supertree techniques that have been proposed over the past 20 years, Matrix Representation with Parsimony (MRP; Baum, 1992; Ragan, 1992; see also Doyle, 1992) is the most widely used. In brief, MRP first encodes the whole topological signal induced by the source trees into a matrix of binary pseudocharacters (i.e. the Matrix Representation of the source trees, here noted MR), and secondly considers the optimal supertree(s) as the most parsimonious (MP) tree(s) reconstructed from the MR. If there is more than one MP tree, the standard approach is to consider the supertree as the strict consensus of these MP trees (e.g. Wilkinson et al., 2005a; see also Thorley, 2000 for some combinatorial properties). Although alternative consensus methods are available to merge the topological information of all optimal MP trees into a supertree (Wilkinson et al., 2007), considering their strict consensus is a standard way for inferring a non-ambiguous supertree from a collection of source trees. For these reasons, all MRP-based supertrees considered in this manuscript are strict consensus of MP trees. To build the MR, several binary encodings have been proposed. In the standard encoding, denoted here b-MR, each pseudocharacter represents one bipartition (b) of the leaf set of the corresponding source tree (Baum, 1992; Doyle, 1992; Farris et al., 1970; Ragan, 1992). Alternatively, in the triplet-based encoding (Nelson and Ladiges, 1994; Wilkinson et al., 2004; Williams and Humphries, 2003), denoted here t-MR, each pseudocharacter represents a triplet (t) of the leaf set of the corresponding source tree (Fig. 1).
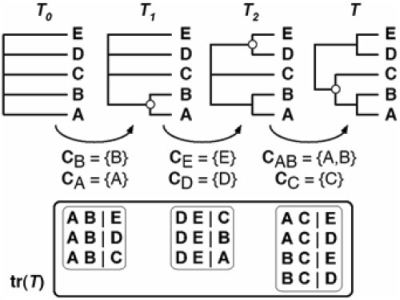
Fig. 1.
Matrix representation based on triplets (t-MR). Topology T is decomposed into its set of triplets. The closer phylogenetic affinities of A and B relative to C, or D or E are indicated by the AB|C, AB|D and AB|E triplets. This correspondence is also indicated by black circles. Rooting is ensured by an artificial all-zero taxon.
In this article, we first recall that, using t-MR, the MP tree(s) could be viewed as a kind of triplet-based median of the source trees. After describing some limits of the corresponding supertree method, i.e. triplet-based MRP (t-MRP), we then propose a new, simple and fast supertree method, named SuperTriplets, that constructs an initial tree using a polynomial algorithm close to the Neighbor Joining scheme (NJ; Saitou and Nei, 1987) followed by iterative improvement based on nearest neighbor interchange (NNI) moves. SuperTriplets is dedicated to triplets and explicitly searches for a triplet-based median supertree. Unlike the TH(SPR/TBR) triplet supertree heuristics recently proposed by Lin et al. (2009), SuperTriplets uses an asymmetric dissimilarity measure and is able to propose supertrees that are not fully resolved. After describing the SuperTriplets method, we compare its properties to those of b- and t-MRP, and those of another triplet-based supertree method, named TILI (Mosses, 2005), which is the triplet adaptation of the QILI supertree algorithm (Piaggio-Talice et al., 2004). Then, to compare these methods, we present a simulation protocol close to those used in previous supertree studies (Criscuolo et al., 2006; Eulenstein et al., 2004). There are several other triplet-based supertree building heuristics, such as MaxCut (Moran et al., 2005) and TH(SPR/TBR) (Lin et al., 2009), but since there is currently no publicly available implementation of these approaches, we could not incorporate them in our comparative analysis. The computer simulation protocol based on larger taxon sets and larger collections of source trees enables both comparison of the relative accuracy and running time performances of SuperTriplets, t-MRP, b-MRP and TILI, and validation of their previously discussed properties. In particular, we show that SuperTriplets infers supertrees with much fewer wrong triplets (triplets absent from the initial model tree), and thus represents a pertinent alternative to the b- and t-MRP approaches. Lastly, the utility of SuperTriplets is illustrated by a phylogenomics inference on 33 mammalian taxa from ∼13 000 source trees collected in the OrthoMaM database (Ranwez et al., 2007).
2 METHODS
A phylogenetic tree on a set L of leaves is a rooted tree whose leaves are each bijectively labeled by an element of L (see Semple and Steel, 2003; for simplicity sake, we use in the following the term tree without ambiguity). Given a forest F (i.e. a collection of trees), L(F) denotes its set of leaves; similarly L(T) denotes the leaf set of a tree T. In the following, k denotes the number of tree(s) inside a forest F (i.e. k=|F|), and n is the total number of leaves in F (i.e. n=|L(F)|). A clade within a phylogenetic tree is the set of all leaves that descend from a given node. For example in Figure 1, the clades of tree T are {A,B}, {A,B,C}, {D,E}, and the trivial ones composed by each of its leaves {A}, {B}, {C}, {D}, {E} and the root-clade L(T). A triplet is a three-leaf tree. Given three leaves X, Y, Z, a triplet is denoted XY|Z when its only non-trivial clade is {X,Y}. With these three leaves, the only possible triplets are then XY|Z, XZ|Y, YZ|X, and the unresolved one, denoted XYZ. Given a tree T, any subset L′ ⊆L(T) induces another tree on leaf set L′ denoted as T|L′. Informally, this restriction of T is the subtree of T that connects the taxa of L′. In particular, T|{X,Y,Z} is a triplet said to be displayed by T (see Fig. 1). A tree T can be completely described by all of its non-trivial clades. It can also be described by the set tr(T) of all the triplets it induces (Bryant, 1997; Grunewald et al., 2007). The triplet distance d3(T1, T2) between two trees T1 and T2 having the same leaf set is the cardinality of the symmetric difference between tr(T1) and tr(T2) (Critchlow et al., 1996; Dobson, 1975). This definition is similar to that of the bipartition distance (Bourque, 1978; Robinson and Foulds, 1981), except that the respective sets of triplets of T1 and T2 are considered instead of the sets of leaf bipartitions induced by each of their internal branch. When two trees have the same leaf set, the triplet distance is thus the number of resolved triplets present in only one of these two trees. It directly follows from its definition that d3 is a dissimilarity, but it is also a distance since it satisfies the triangle inequality (Bansal et al., 2008).
2.1 Triplet asymmetric median supertree
Several distances have been proposed to compare two trees with the same leaf set (e.g. Bourque, 1978; Critchlow et al., 1996; Dobson, 1975; Robinson and Foulds, 1981; see also Steel and Penny, 1993). These distances cannot be directly used to compare a supertree T with a source tree Ti since both do not necessarily have the same leaf set. The similarity between Ti and T is thus generally evaluated through the distance between Ti and the supertree T restricted to L(Ti) (e.g. Cotton and Wilkinson, 2007; Creevey and McInerney, 2005). The resulting measure between T and Ti does not satisfy distance axioms (e.g. trees on different leaf sets can have a distance of 0). Yet in what follows we will, for simplicity sake, refer to this measure as the distance between the supertree T and a source tree Ti. Since a supertree is supposed to summarize a forest, a natural candidate is the median tree minimizing the sum of distances to the source trees (Bryant, 1997; Wilkinson et al., 2001; Creevey and McInerney, 2005). Several authors have underlined the equivalence between the majority rule consensus and the median tree based on bipartition metrics, and Cotton and Wilkinson (2007) generalize this approach to the supertree setting. Defining supertree as a median turns out to be not only an intuitive definition but also a guarantee of fundamental theoretical properties. Using a model of phylogenetic error, Steel and Rodrigo (2008) consider supertree in a maximum likelihood (ML) framework, with the so-defined ML-supertree being simply a median tree (with respect to tree metrics). This ML framework allows the authors to demonstrate the statistical consistency of the median supertree approach (i.e. converging to the true tree as more source trees are added). Even though this result ensures the consistency of any median supertree definition, the authors stress the importance of choosing appropriate metrics. The triplet distance has the advantage of being easily computed (unlike the SPR distance; Bordewich and Semple 2004; Hickey et al., 2008; but see Lin et al., 2009) and more fine-grained than the bipartition distance (e.g. Steel and Penny, 1993). As stated by Williams (2004), it seems that ‘three item data are more sensitive to how information accumulates to produce overall summary solutions’.
Considering a source tree Ti and the supertree T, the set of triplets over L(T) can be partitioned into five sets (Page, 2002): S(Ti, T) and D(Ti, T), the triplets resolved in Ti and T|L(Ti) that have same and different topologies, respectively; R1(Ti, T), the triplets resolved in Ti and not resolved in T|L(Ti); R2(Ti, T), the triplets not resolved in Ti and resolved in T|L(Ti); X(Ti, T), the triplets unresolved in both Ti and T|L(Ti). Using this notation, a triplet distance between a source tree and a supertree can be formulated as dt(Ti, T)=2|D(Ti, T)|+|R1(Ti, T)|+|R2(Ti, T)|. Minimizing this distance is closely related to maximizing the similarity |S(Ti, T)| as done by Lin et al. (2009). Our distance formulation has the advantage of describing the way unresolved triplets are handled. Indeed, this distance is increased by the presence of triplet(s) resolved in the supertree T but not in some source trees Ti, i.e. the set R2(Ti, T). Supertrees resolving triplets that are not resolved in some source trees are thus penalized. The use of the (symmetric) distance dt results in considering irresolution in source trees as a signal to take into account (hard polytomies; Maddison, 1989) rather than as an absence of signal (soft polytomies; Bansal et al., 2008). Since polytomies in source trees are generally considered as soft ones, it seems more suitable to consider the asymmetric triplet distance: δt(Ti, T)=2|D(Ti, T)|+|R1(Ti, T)|. Note that δt is no longer a distance between Ti and T|L(Ti) (due to its asymmetry, δt is quite a divergence). The corresponding median supertree definition is therefore no longer guaranteed to be consistent overall. However, the consistency still holds in the special case where source trees are all fully resolved, since in this case there is no difference between dt and δt. Using the asymmetric function δt weakens the theoretical properties of the corresponding supertree approach but seems to be more in line with the biological acceptation of polytomies. Moreover, the advantages of an asymmetric approach have already been pointed out and exploited in the consensus setting (Phillips and Warnow, 1996).
Given a forest F={T1, T2 … Tk}, the two closely related computational problems of finding a TMS (i.e. according to dt) and finding a triplet asymmetric median supertree (TAMS, i.e. according to δt) can be stated as finding a supertree T with L(T)=∪i L(Ti) that minimizes dt(F, T)=∑i dt(Ti, T) and δt(F, T)=∑i δt(Ti, T), respectively. As conjectured by Bansal et al. (2008), the TMS problem is NP-hard due to the NP-completeness of the maximum triplet compatibility problem (Semple and Steel, 2003). Since both TMS and TAMS problems are equivalent when source trees are fully resolved, we conjecture that the TAMS problem is also NP-hard (nothing indicates that instances of the TMS problem where source trees are fully resolved are easier than others). The TAMS optimization problem can be turned into a classical MP optimization problem by using a t-MR of the source trees (Wilkinson et al., 2001, 2005a, 2007). As unresolved triplets of sources trees are not encoded in the matrix, the MP optimization does, indeed, tackle the TAMS problem and not the TMS problem. This approach benefits from long-term efforts to optimize MP local search programs (e.g. Goloboff et al., 2008) but is hampered by some limitations due mostly to its inefficient representation of triplet information. We therefore designed SuperTriplets, a heuristic dedicated to the TAMS optimization problem.
2.2 Agglomerative construction of a TAMS
The first step of SuperTriplets aims at converting in O(kn3) the (rooted) source trees into weighted triplets, where the weight of each triplet is the number of source trees containing it. Then the agglomerative scheme is used to construct an initial supertree. For our triplet-based strategy, this algorithmic scheme is well adapted, since when an agglomeration is performed, all resolved triplets that this agglomeration generates in the final supertree are completely determined (see Fig. 2).
Fig. 2.
Detail of the agglomeration process. Each taxon agglomeration step is denoted by an open circle, and creates some new triplets. Their union is tr(T), the set of triplets of the final binary tree.
Considering the agglomerative scheme as an iterative improvement of the current supertree T, the aim is to select, during each iteration, the best agglomeration with respect to the TAMS criterion to minimize. When modifying a tree T into T′ by proposing a new clade resulting from the joining of two existing clades CA and CB, SuperTriplets creates new triplets AB|X for any set of three taxa {A,B,X} such that A ∈CA, B ∈CB and X ∉ CA∪CB (Fig. 2). The difference between δt(F, T) and δt(F, T′) depends only on such newly resolved triplets. Consequently, after agglomerating CA and CB, the criterion value δt(F, T′) can be expressed from δt(F, T) as δt(F, T′)=δt(F, T)−N+(CA, CB)+N−(CA, CB), where N+(CA, CB) is the number of times the new triplets AB|X appear in F, and N−(CA, CB) the number of times an alternative resolution (i.e. AX|B or BX|A) appears in F. Obviously, δt(F, T′)<δt(F, T) if N+(CA, CB)>N−(CA, CB). A natural approach is then to agglomerate the two clades such that N+(CA, CB)−N−(CA, CB) is maximal. Note, however, that all agglomerations do not resolve the same number of triplets, and that the importance of a given triplet is related to the number of times it appears in F, i.e. its weight. During this agglomeration process, SuperTriplets thus does not fully take triplet weights into account, but only uses weights to determine, for each group of three taxa, the most frequent resolution in source trees (i.e. the triplet with the highest weight). If a triplet AB|X is more frequent in source trees than its alternative resolutions (i.e. AX|B and BX|A), it is called a most frequent triplet, denoted mft. At each step, SuperTriplets selects the agglomeration leading to the highest proportion of mft. This criterion ensures that SuperTriplets will recover the correct phylogeny for every dataset such that, for any three-taxon set {A,B,C}, the mft is the correct one. Indeed, in such cases, every creation of a correct clade leads to 100% of mft, while this proportion is smaller for incorrect clades. Moreover, the tree obtained during this agglomeration step will contain every mft and is therefore the one minimizing the TAMS criterion. More formally, the selected agglomeration is randomly chosen among those maximizing Nmft+(CA, CB)/[Nmft+(CA, CB)+Nmft−(CA, CB)], where Nmft+(CA, CB) is the number of newly resolved triplets that are mft, and Nmft−(CA, CB) the number that conflict with mft. It follows that to evaluate the agglomeration of any pair CA and CB, SuperTriplets only needs to have at hand the corresponding values for Nmft+(CA, CB) and Nmft−(CA, CB).
During the first iteration of the agglomerative process, clades are reduced to a single taxon, so that for any of the O(n2) pairs of such trivial connected component CA and CB, the values Nmft+(CA, CB) and Nmft−(CA, CB) correspond to the creation of new triplets AB|X, where A and B are fixed taxa and where X can take n − 2 label values. All the Nmft+(CA, CB) and Nmft−(CA, CB) values are therefore initialized in O(n3). When two clades CA1 and CA2 are merged in a new clade CA, this does not change the value of Nmft+(CX, CB) as long as both CX and CB differ from CA1, CA2 and CA. Moreover, the new value Nmft+(CA, CB) can easily be obtained from Nmft+(CA1, CB) and Nmft−(CA2, CB). Indeed, any triplet AB|X that will be newly resolved by an agglomeration between CA and CB is a triplet that would have been newly resolved by agglomerating CA1 and CB, or CA2 and CB. This is simply due to the fact that if A is inside CA, it was either in CA1 or in CA2. On the other hand, the triplets A1B|X that would have been newly resolved by the agglomeration of CA1 and CB will also be newly resolved by the agglomeration of CA and CB, except if X is within CA2. It follows that Nmft+(CA, CB) is simply the sum of Nmft+(CA1, CB) and Nmft+(CA2, CB) minus a correction. This correction is equal to twice the number of mft A1A2|B (with A1∈CA1, A2∈CA2 and B ∈CB) since these triplets are counted in both Nmft+(CA1, CB) and Nmft+(CA2, CB), and should not be counted in Nmft+(CA, CB). The same remark is easily transposed to Nmft−(CA, CB).
Once the initial supertree is obtained thanks to the above agglomerative heuristics, SuperTriplets checks whether any of its neighbor trees is closer to the set of source trees F. If so, this particular neighbor is used as the new current supertree to recheck its tree neighbors. This is repeated until a supertree is reached such that none of its neighbor trees is better fitted than itself. The neighborhood used is that defined by the NNI, where a tree T′ is a tree neighbor of T if T′ can be obtained from T by swapping two subtrees of T, both of which are rooted on the two extremities of a given edge e. Each NNI is therefore related to a specific internal branch. This heuristics is often used in fast phylogenetic reconstruction algorithms (e.g. PhyML; Guindon and Gascuel, 2003). Given an NNI related to an edge e of T, the resolution of a triplet AB|X will be changed after the NNI if and only if e is the sole edge separating a clade containing A and B but not X from the first clade containing the three taxa. To evaluate the whole set of possible NNIs, SuperTriplets therefore twice considers the occurrences of such triplets as well as the occurrence of their alternative resolutions, so this can be done in O(n3). Note that this is already faster than consulting all triplets since most of them are not considered at all during this process (no current NNI can change their resolution). Furthermore, for each NNI around an edge e, one can store the value NNI(e) that represents the difference between the actual criterion and the best value for the two trees that differs from T by an NNI around e. Storing those values makes NNI selection very fast, since once an NNI is actually done around an edge e, the value NNI(e′) only changes for a handful of edges e′ close to e. Since NNI exploration is really fast, SuperTriplets uses this advantage to better explore the topology space using random NNI modifications that are allowed to temporarily decrease the quality of the current tree.
2.3 Reliability estimation of supertree edges
Collapsing an edge e transforms the supertree T into a less resolved supertree T′. Indeed, a set of resolved triplets AB|X is dependent upon the presence of the edge e and will become unresolved inside T′. The difference between δt(F, T) and δt(F, T′) depends only upon such newly unresolved triplets. Following the notation used to describe the agglomerative process, we have δt(F, T′)=δt(F, T)−N−(e)+N+(e), where N+(e) represents the number of times the triplets AB|X induced by e appear in F, and N−(e) the number of appearances of their alternative resolutions (i.e. AX|B or BX|A). A measure of the reliability of an edge e in a tree T can thus be obtained by considering the modification of T by collapsing e. SuperTriplets estimates the reliability of e as N+(e)/[N+(e)+N−(e)]. This criterion value ranges from 0 to 1, and represents the percentage of triplets that supports the edge e. Note that this measure only takes a small subset of the triplets included in the current tree into account. Indeed, some triplets require the collapse of several edges of T to become unresolved. Using this measure, a supertree can contradict some input triplets while having all its edges with maximal support. This can be avoided by considering a global measure involving all triplets. Each edge defines a clade that can be incompatible with some triplets present in the source trees. A global measure can thus be obtained by considering triplets of source trees that disagree (or agree) with this clade. From this standpoint, a triplet AB|X of the source trees will influence the support of every branch of the supertree that defines a clade containing A and B but not X (i.e. those that should be collapsed to obtain an irresolution on A, B and X). The number of such clades varies among triplets. In order to obtain a measure such that all triplets have the same importance, the contribution of each triplet is equally divided among these clades, as suggested by Wilkinson et al. (2005b). Hence, all triplets have the same importance, but while some of them focus on a single edge, others influence several edge supports. This provides a global measure of agreement, i.e. Ng+(e), and contradiction, i.e. Ng−(e), between source trees and a given edge e. These measures are then used to define a global support of edge e defined by Ng+(e)/[Ng+(e)+Ng−(e)]. Though each triplet influences all the edges within a path in the supertree and not a single one, the support of each edge can be estimated in O(n3). SuperTriplets uses these measures to detect edges that must be collapsed. The modification of the supertree, which involves collapsing edges with local support smaller than 0.5, always reduces the value of δt(F, T). Since these supports are interdependent, SuperTriplets collapses all such edges simultaneously to avoid arbitrary choice between edges. Then all remaining edges with global support <0.5 are also simultaneously collapsed. Note that these edges may have a local support >0.5, for instance if the taxon sampling in source trees implies that this local support is estimated based on very little information. Collapsing a single one of these branches will not reduce the divergence δt(F, T) between the source trees and the supertree, but collapsing several of them possibly will. Finally, the local supports of the remaining edges are evaluated and displayed in the final supertree as a reliability indication. Using this conservative procedure, the supertree returned by SuperTriplets is more reliable by displaying strongly supported clades, but is also less resolved as the topological information contained in the source trees is ambiguous.
3 RESULTS
3.1 Simulation Results
Simulations were conducted to compare the behavior of SuperTriplets to that of the well-known b-MRP and the alternative t-MRP methods when analyzing collections of source trees of varying size and taxon overlap. We also compare these three methods with the more recently implemented TILI algorithm (Mosses, 2005), which is the triplet variant of the quartet-based supertree algorithm QILI (Piaggio-Talice et al., 2004).
An ultrametric model topology Tmodel with n=100 ingroup taxa was generated by the r8s software (Sanderson, 2003) according to a Yule-Harding branching process (Harding, 1971; Yule, 1925). One outgroup taxon was a posteriori added, with a root-to-tip branch length of 1.1 substitution per site. To highlight the substitution rate heterogeneity among taxa and genes, Tmodel was duplicated 50 times, and a global deformation of these identical clocklike trees was achieved by individual branch length deviation followed by 50 independent total tree length rescaling according to Criscuolo et al. (2006). We thus obtained 50 rooted 101-taxon trees with the same topology as Tmodel but with different branch lengths and different induced global evolutionary rates. Each of these 50 non-clocklike phylogenetic trees was used by Seq-Gen (Rambaut and Grassly, 1997) to generate 50 nucleotide alignments of 101 taxa under a Kimura (1980) model with transition/transversion ratio of 2.0. The number of aligned sites was uniformly drawn from 200 to 1000 bp. For each alignment, ingroup taxa were randomly deleted with a probability d=25, 50 and 75% following the procedure first suggested by Eulenstein et al. (2004).
For each of the three taxon deletion rates (d=25, 50, 75%), 50 ML trees were then inferred from the 50 partially deleted alignments with PhyML (Guindon and Gascuel, 2003) under the Kimura (1980) model with the transition/transversion ratio left as free parameter. The whole procedure of topology generation, phylogram deformation, sequence generation, taxon deletion with three conditions and phylogeny reconstruction by PhyML was repeated 100 times. Each tree returned by PhyML was then rooted by the outgroup taxon. One thus obtains 100 sets of 50 trees for each of the three values of d.
For each taxon deletion rate d and each of the 100 corresponding sets of trees, the collections of the first k=10, 20, 30, 40, 50 source trees were submitted to SuperTriplets and TILI. The same forests were analyzed with both b- and t-MRP. Hence, the b-MR and the weighted site t-MR were first built and, secondly, the parsimony analyses were performed by the TNT software (Goloboff et al., 2008) using 10 random addition sequences and TBR branch swapping. The ratchet option was used for b-MRP, but not for t-MRP since this approach uses t-MRs of extremely large size (e.g. currently more than 500 000 pseudocharacters with d=25% and k=50, despite the site weighting procedure during the t-MR computing step) which require huge running times to be analyzed (e.g. at most 20 h, whereas several days are necessary for one supertree inference with the ratchet option).
To evaluate the degree to which the inferred supertrees agree with their corresponding model tree Tmodel, we used bipartition- and triplet-based distances between trees. Given the five previously described triplet partitions S, D, R1, R2, X (see Section 2.1), and the five corresponding triplet rates s, d, r1, r2, x, respectively, one can define two error types. The proportion of triplets that are in the supertree but not in the model tree is called the type I error and corresponds to erroneous information displayed in the supertree (sometimes called false positive). Reciprocally, the proportion of triplets that are not in the supertree but in the model tree is called the type II error, and corresponds to topological information that is missing in the supertree (sometimes called false negative). The type I error rate between Tmodel and its related supertree T is then defined as etI=(d+r2)/(d+s+r2); reciprocally, the type II error rate is defined as etII=(d+r1)/(d+s+r1). It should be stressed that the type II error rate is closely related to the triplet-fit similarity from Page (2002), which is defined as 1-etII.
Since b-MRP optimizes a bipartition-based criterion, not a triplet-based criterion, both type I and II errors were also computed at a bipartition level. The bipartition-based error rate etI (etII, respectively) is defined as the proportion of bipartitions of L(T) induced by each of the internal branches of Tmodel|L(T) (of T, respectively) that are not present in T (in Tmodel|L(T), respectively).
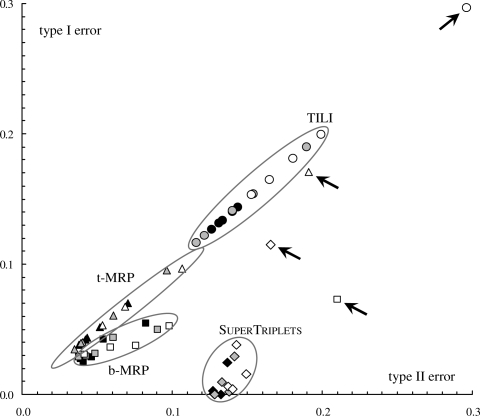
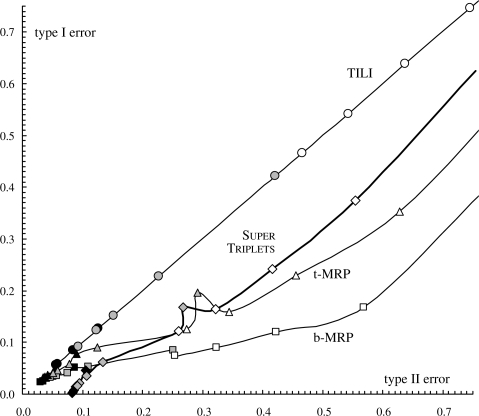
A low type I error rate means that the supertree T displays few leaf bipartitions (or triplets) that are not present in Tmodel. Respectively, a high type II error means that the supertree T does not represent all the phylogenetic information in Tmodel. For each of the four supertree methods studied (i.e. b- and t-MRP, SuperTriplets and TILI), each taxon deletion rate (i.e. d=25, 50, 75%) and each forest size (i.e. k=10–50), triplet- and bipartition-based etI and etII values were computed and averaged over the 100 replicates. These results are graphically represented in Figure 3 for the triplet-based errors and in Figure 4 for the bipartition-based errors.
Fig. 3.
Type I error rate of supertree methods as a function of type II error rate at the triplet level. Supertrees were inferred from forests by four methods under three different simulation conditions. Points represent the average over 100 replicates of the supertree error rates. Symbols correspond to the methods: b-MRP (open squares), t-MRP (open triangles), SuperTriplets (open diamonds) and TILI (open circles). For clarity, symbols corresponding to each of the four methods are encircled. Colors correspond to the three taxon deletion rates: d=25% (black), 50% (grey) and 75% (white). Arrows point to the difficult case d=75% and k=10 (see Section 3.1 for more details).
Fig. 4.
Type I error rate as a function of type II error rate at the bipartition level. Symbols and colors have the same meaning as in Figure 3. For clarity, symbols that correspond to the same method are connected by a line. The difficult case d=75% and k=10 is not represented.
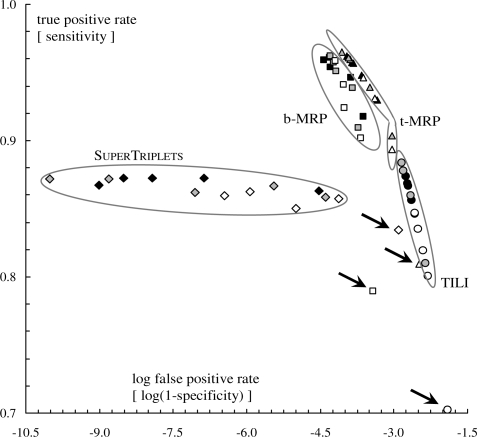
The simulation results can be alternatively represented by focusing on the sensitivity (i.e. the true positive rate) and specificity (i.e. the complementary of the false positive rate) of the methods. This representation, unlike the previous one, takes the size of the triplet (or bipartition) space into account. Using notations introduced in Section 2.1, the number of false positive triplets is FP=|D|+|R2|, the number of false negative triplets is FN=|D|+|R1|, and the number of true positive triplets is TP=|S|. Finally, since the total number of resolved triplets on n taxa is n(n−1)(n−2)/2, the number of true negative triplets is TN =n(n−1)(n−2)/2− (TP + FP + FN). Using these notations, the true positive and false negative rates are defined as TP/(TP + FN) and FP/(FP + TN), respectively. Figure 5 depicts the plot of these two rates (i.e. the so-called ROC graph; e.g. Fawcett, 2004) for the various methods and simulation conditions. Based on the fact that the total number of non-trivial bipartition on n taxa is 2n−1−n−1, a similar approach can be used to obtain the bipartition-based ROC graph displayed in Figure 6. It should be stressed that in ROC graphs (Figs 5 and 6) the best results are displayed close to top left corner (i.e. the method is both sensitive and specific; Fawcett, 2005) whereas in the type I/type II plot (Figs 3 and 4) the best results are close to the bottom left corner (i.e. reduced type I and II errors.)
Fig. 5.
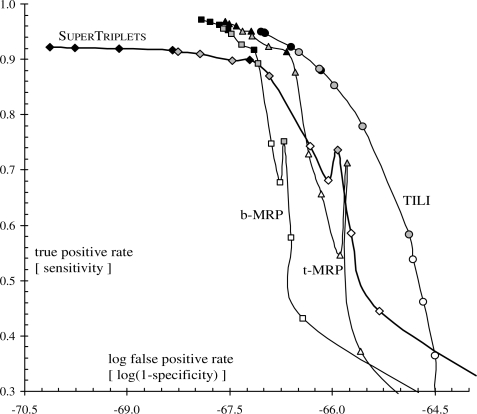
ROC graph displaying true positive rates as a function of false positive rates at the triplet level. Symbols, colors and arrows have the same meaning as in Figure 3. Note that the x-axis is on a logarithmic scale.
Fig. 6.
ROC graph displaying true positive rates as a function of false positive rates at the bipartition level. Symbols that correspond to the same method are connected by a line. Symbols and colors have the same meaning as in Figure 4. Note that the x-axis is on a logarithmic scale.
SuperTriplets displays the lowest triplet-based type I errors (Fig. 3) and false positive rates (Fig. 5) in all cases but the very difficult one (k=10 and d=75%, i.e. few source trees with low taxon overlap). Indeed, its triplet-based type I errors and false positive rates are often close to 0, and are significantly lower than those obtained for the three other methods (as assessed by a sign test; e.g. Dixon and Mood, 1946). This shows that a supertree inferred by SuperTriplets displays very few triplets that are not present in the corresponding model tree (i.e. so-called false positive triplets). In addition, SuperTriplets bipartition-based type I errors (Fig. 4) and false positive rates (Fig. 6) are among the lowest. However, it should be stressed that b-MRP supertrees show better minimization of bipartition-based false positive rates for difficult cases, i.e. when the taxon overlap among source trees is minor (e.g. d=50% and k≤20, or d=75%). This latter result is still expected since b-MRP optimizes a bipartition-based encoding of the source topologies. Conversely, SuperTriplets displays higher triplet-based type II errors and lower true positive rates than both b- and t-MRP (Figs 3 and 5) whatever the number of source trees (except for k=10 and d=75%). Note also that, although based on triplet decomposition, TILI has the worst type I and II errors even when considering triplet error rates (Fig. 3). For this reason, we will not further discuss results obtained with the TILI supertree method.
Overall, SuperTriplets has a much lower type I error whereas b- and t-MRP have a much lower type II error. Our simulations therefore suggest that SuperTriplets infers less-resolved but more reliable conservative supertrees, whereas b- and t-MRP infers more resolved but less reliable liberal ones. It is not clear, however, whether or not both error types have the same importance. Suppose that starting from a given supertree T, it is possible to modify it into T′ to resolve 10 additional triplets—everything apart from these 10 triplets would be the same in T and T′. Among these 10 triplets, suppose that four of them are wrong whereas the six others are correct. It is thus hard to determine which of these two supertrees is the most informative summary of the source trees. This can be formulated as ‘how many true triplets do we require to accept the introduction of a wrong one?’. The answers certainly vary depending upon the end-users and the analyses they plan to conduct with the supertree. As described previously, b-MRP, t-MRP and SuperTriplets answer this question in different ways.
3.2 Triplet-based supertree in a phylogenomics context
To stress the benefits of SuperTriplets for inferring a species tree, we collected the 12 958 trees available in the fifth release of the OrthoMaM database (Ranwez et al., 2007). These ML trees have been inferred from alignments of orthologous gene coding sequences, as identified by EnsEMBL v. 54. The alignment sizes range from 0.1 to 42 kb, and the number of taxa from 6 to 33 mammals. A supertree was obtained by using SuperTriplets on these 12 958 equally-weighted trees after having rooted them via two outgroup taxa, i.e. one marsupial (Monodelphis) and one monotreme (Ornithorhynchus). The running time was very fast (30 s on a MacPro computer with 2.66 GHz dual-core Intel Xeon processor).
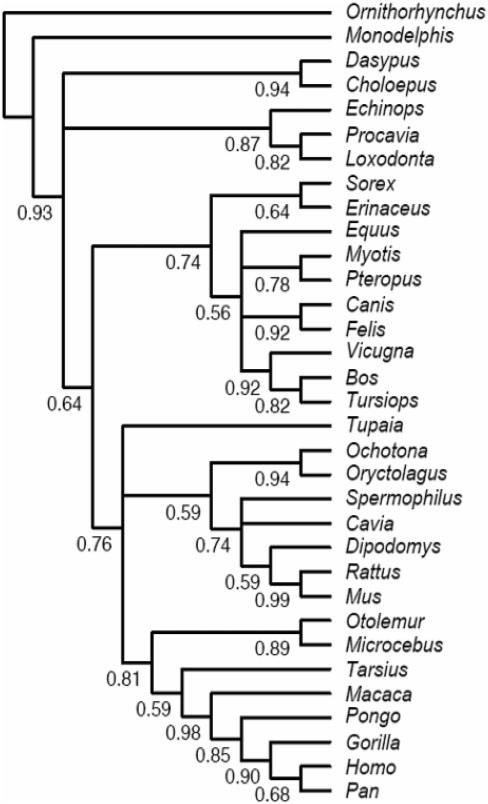
The topology inferred (see Fig. 7) is in line with current knowledge about mammalian phylogenetics (e.g. Prasad et al., 2008). SuperTriplets branch support values range from high to moderate for defining several supra-ordinal clades: Cetartiodactyla (0.92), Afrotheria (0.87), Paenungulata (0.82), Euarchontoglires (0.76), Laurasiatheria (0.74), Boreoeutheria (0.64), Glires (0.59) and Scrotifera (0.56). This branch support hierarchy (showing the level of congruence among source trees) is compatible with those reported by Beck et al. (2006). Several multifurcations in the supertree correspond to areas of documented phylogenetic uncertainty: placental tree root (Afrotheria + Xenarthra, versus either Afrotheria or Xenarthra first; Churakov et al., 2009), relationships among Laurasiatheria (Prasad et al., 2008), Tupaia affinities (Janecka et al., 2007), and relationships among the three major Rodentia clades (Blanga-Kanfi et al., 2009). Rodents and primates are monophyletic (respective supports: 0.74 and 0.81), and the following within-order clades are supported: Dipodomys + Rattus + Mus (0.59), Strepsirrhini (0.89) and Haplorrhini (0.59), Catarrhini (0.98) and Hominoidea (0.85).
Fig. 7.
Phylogenomic case study: reconstruction by SuperTriplets of the supertree of 33 mammals for which complete genome data are available. The SuperTriplets supertree has been inferred from the 12 958 source trees of OrthoMaM v. 5, i.e. ML trees estimated from DNA orthologous gene coding sequences (CDS). SuperTriplets node supports are given.
Given the huge number of independent multiple sequence alignments that can be harvested from comparative genomics projects, there is a need for efficient tools to quickly summarize the whole phylogenetic signal of the numerous trees inferred from these various loci. In such phylogenomic cases, when using a supermatrix approach (obtained by concatenating all alignments), very large matrices must be dealt with that can exceed the memory capacity of most today computers. There would also be many missing data, which could interfere with the optimization process (Criscuolo et al., 2006). Supertree approaches offer a more tractable alternative in this setting. SuperTriplets is especially well tailored to applications under these conditions since the number of input trees has little impact on the computer resources it uses. Here, for instance, assembling the 12 958 CDSs into a supermatrix would have resulted in a huge matrix made of ∼27-million of sites containing 28% missing character states (some taxa with low-coverage genomes such as Myotis or Otolemur are not represented for many CDSs). This matrix is so huge that its phylogenetic analysis would be problematic due to memory limitations.
Supertree methods are also used for summarizing previously published phylogenies into a single one that may contain hundreds to thousands of taxa (e.g. Beck et al., 2006; Bininda-Emonds et al., 2007). We thus compared the computation time of b-MRP, t-MRP and SuperTriplets on the OrthoMaM CDS dataset, and on the mammalian family-level dataset of Beck et al. (2006) that contains a total of 115 taxa and 725 weighted source trees (i.e. 6724 trees to account for the differential weighting of individual source topologies). The computation times were measured on a 3-GHz Intel® Core™2 Duo PC with 1.9 Gb RAM. For t-MRP, the compressed t-MR was obtained in few minutes using the homemade MRtools software (available on the SuperTriplets webpage). Since no substantial efforts were made to optimize this step, we did not include it when measuring the overall computation time of b-MRP and t-MRP methods. We thus slightly underestimate their running times, especially for t-MRP, since the building of the t-MR is an O(kn3) step. For the three methods, we used the same parameters as those used for assessing their accuracy (e.g. ratchet for b-MRP but not for t-MRP, see Section 3.1). On the OrthoMaM dataset, b-MRP runs in ∼18 min while t-MRP and SuperTriplets run in a couple of seconds. On the mammalian family dataset, b-MRP runs in ∼21 min, t-MRP in ∼7 min, and SuperTriplets in ∼10 s. Moreover, the supertrees obtained by b-MRP and SuperTriplets are in agreement with current knowledge on the mammalian phylogeny. SuperTriplets thus seems to be a reliable and fast alternative to the widespread b-MRP method.
4 DISCUSSION
Summarizing various trees obtained from different inferences into a single one is a frequent task in phylogenetic analysis of multigene datasets. Supertree methods are able to achieve this task even when the source trees only have partially overlapping taxon sets. A natural candidate supertree is the tree that is the closest to the source trees in terms of a given dissimilarity measure. SuperTriplets is a method specially designed to find the asymmetric median supertree according to triplet dissimilarity δt, and that has several advantages over the closely related t-MRP method.
In the first step of the t-MRP approach, the t-MR of the source trees depicts them as a set of binary sequences (one sequence per taxon, plus an extra one consisting only of ‘0’ for an artificial outgroup leaf corresponding to the root of all source trees). Given a fully resolved tree T, the parsimony value for a site encoding XY|Z is one if T entails this triplet resolution and two if T entails an alternative resolution (i.e. XZ|Y or YZ|X). It follows that the parsimony value of a binary supertree T for the t-MR of a forest F is simply the number of pseudocharacters (i.e. the total t-MR length) plus δt(F, T)/2 (Moran et al., 2005; Wilkinson, 1994). A t-MRP supertree T is thus one of the binary trees that minimizes the triplet dissimilarity δt(F, T) (Wilkinson et al., 2001, 2005a, 2007). Nevertheless, there are some practical problems that hamper standard MP analysis performed to optimize this triplet-based criterion.
First, each pseudocharacter of the t-MR is one of the O(n3) triplets induced by one of the k source trees. For comparison, with the b-MR, one have O(n) pseudocharacters. The number of source trees does not impact the number of pseudocharacters since it is common practice to pre-process them in order to keep only one (weighted) representative for the many identical pseudocharacters.
Second, for each pseudocharacter of the t-MR, there are only four informative character states (Fig. 1). It follows that, for instance, when inferring a supertree on 99 taxa, for each pseudocharacter the t-MR uses 100 states (99 taxa+root), 96 of which are missing character states. The t-MR is then made of 96% uninformative character states and this proportion is constant regardless of whether or not identical sites are compressed using a weighting procedure. If the supertree is defined on 999 taxa, the rate of uninformative character states grows to 99.6%. Each of the O(n3) triplets, i.e. stored in O(1) memory space by SuperTriplets, is stored in O(n) by the t-MR. It follows that, when dealing with t-MRP, both the MR size and the proportion of missing character states rapidly grows. Even with weighted pseudocharacters, the size of the t-MR remains large and requires huge memory to compute the t-MRP supertree (see Section 3.1). Moreover, a high proportion of missing character states is known to often slow down any MP tree inference method, since it may increase both the size and number of local parsimony optima. By directly considering all weighted triplets (instead of a t-MR) and by combining them through a polynomial time algorithm, SuperTriplets is better suited for achieving this task than t-MRP.
Third, the parsimony criterion does not allow comparison of a fully resolved tree T with a tree T′ obtained by collapsing edges of T. Indeed, T′ will always have a parsimony value greater or equal to that of T. By definition, the supertree returned by t-MRP is a summary (using strict consensus) of all MP (fully) resolved trees. This supertree is not the best according to the MP criterion; it is just a practical summary of the MP trees. On the other hand, the criterion optimized by SuperTriplets (i.e. asymmetric triplet dissimilarity δt) naturally allows comparison of trees with different degrees of resolution.
5 CONCLUSION
We propose SuperTriplets, a method that first constructs a supertree using a polynomial agglomerative scheme, and, secondly, performs fast local search to find the asymmetric median supertree according to a triplet dissimilarity. We point out that, by using triplet representation (i.e. t-MR), this median supertree is one of the MP binary trees. Computer simulations and a biological case study indicate that SuperTriplets tends to propose more reliable but less resolved supertrees than MRP methods.
We think that this work provides new prospects for designing supertree methods. On the one hand, SuperTriplets could certainly be significantly improved. Heuristics searching for the MP tree(s) have strongly benefited from more than 20 years of improvements, and have a considerable advantage with respect to method tuning and implementation tricks (e.g. Goloboff et al., 2008) as compared to recently proposed approaches. On the other hand, the confidence values displayed by SuperTriplets at the supertree edges are well-founded, and having an indication of the supertree branch support is an important feature. Several related confidence values dedicated to supertree-based comparative studies have already been proposed (Bininda-Emonds, 2003; Burleigh et al., 2006; Cotton et al., 2006; Moore et al., 2006; Wilkinson et al., 2005b). Comparing their relevance and accuracy is far from easy, as pointed out by Cotton et al. (2006), who concluded: ‘Our example at least shows that there is no single correct view of support for supertrees…’. This remark is relevant for supertree approaches but also reflect the intrinsic difficulty of comparing any clade support value in the supertree or supermatrix context (Burleigh et al., 2006; Douady et al., 2003).
ACKNOWLEDGEMENTS
We thank the three referees for their suggestions on the manuscript. We also thank Guillaume Dugas for the SuperTriplets webpage and Khalid Belkhir for helpful advices.
Funding: This work was supported by the Research Networks Program in Bioinformatics of the High Council for Scientific and Technological Cooperation between France and Israel; the bioinformatics cluster of ISE-M; and the French Agence Nationale de la Recherche ‘Domaines Emergents’ [ANR-08-EMER-011 ‘PhylAriane’]. This publication is contribution No. 2010-021 of the Institut des Sciences de l'Evolution de Montpellier (UMR 5554 - CNRS), France.
Conflict of Interest: none declared.
REFERENCES
- Adams EN. Consensus techniques and the comparison of taxonomic trees. Syst. Zool. 1972;21:390–397. [Google Scholar]
- Bansal MS, et al. Comparing and aggregating partially resolved trees. Lect. Notes Comput. Sci. 2008;4957:72–83. [Google Scholar]
- Baum BR. Combining trees as a way of combining data sets for phylogenetic inference, and the desirability of combining gene trees. Taxon. 1992;41:3–10. [Google Scholar]
- Beck R.MD, et al. A higher-level MRP supertree of placental mammals. BMC Evol. Biol. 2006;6:93. doi: 10.1186/1471-2148-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bininda-Emonds O.RP. Novel versus unsupported clades: assessing the qualitative support for clades in MRP supertrees. Syst. Biol. 2003;52:839–848. [PubMed] [Google Scholar]
- Bininda-Emonds O.RP, et al. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- Blanga-Kanfi S, et al. Rodent phylogeny revised: analysis of six nuclear genes from all major rodent clades. BMC Evol. Biol. 2009;9:71. doi: 10.1186/1471-2148-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordewich M, Semple C. On the computational complexity of the rooted subtree prune and regraft distance. Ann. Combinat. 2004;8:409–423. [Google Scholar]
- Bourque M. PhD Thesis. Montréal, Canada: University of Montréal; 1978. Arbres de Steiner et réseaux dont varie l'emplagement de certains sommets. (In French) [Google Scholar]
- Bryant D. PhD Thesis. Canterbury, New Zealand: University of Canterbury; 1997. Building trees, hunting for trees and comparing trees. [Google Scholar]
- Bryant D. A classification of consensus methods for phylogenies. In: Janowitz M, et al., editors. Bioconsensus. Providence, RI: DIMACS, AMS; 2003. pp. 163–184. [Google Scholar]
- Burleigh JG, et al. Supertree bootstrapping methods for assessing phylogenetic variation among genes in genome scale data. Syst. Biol. 2006;55:426–440. doi: 10.1080/10635150500541722. [DOI] [PubMed] [Google Scholar]
- Churakov G, et al. Mosaic retroposon insertion patterns in placental mammals. Genome Res. 2009;19:868–875. doi: 10.1101/gr.090647.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton JA, Wilkinson M. Majority-rule supertrees. Syst. Biol. 2007;56:445–452. doi: 10.1080/10635150701416682. [DOI] [PubMed] [Google Scholar]
- Cotton JA, et al. Discriminating supported and unsupported relationships in supertrees using triplets. Syst. Biol. 2006;55:345–350. doi: 10.1080/10635150500481556. [DOI] [PubMed] [Google Scholar]
- Creevey CJ, McInerney JO. CLANN: investigating phylogenetic information through supertree analyses. Bioinformatics. 2005;21:390–392. doi: 10.1093/bioinformatics/bti020. [DOI] [PubMed] [Google Scholar]
- Criscuolo A, et al. SDM: a fast distance-based approach for (super)tree building in phylogenomics. Syst. Biol. 2006;55:740–755. doi: 10.1080/10635150600969872. [DOI] [PubMed] [Google Scholar]
- Critchlow DE, et al. The triples distance for rooted bifurcating phylogenetic trees. Syst. Biol. 1996;45:323–334. [Google Scholar]
- Dixon WJ, Mood AM. The statistical sign test. J. Am. Statist. Assoc. 1946;41:557–566. doi: 10.1080/01621459.1946.10501898. [DOI] [PubMed] [Google Scholar]
- Dobson AJ. Comparing the shapes of trees. In: Street AP, Wallis WD, editors. Combinatorial Mathematics III, LNCS. Vol. 452. New York: Springer; 1975. pp. 95–100. [Google Scholar]
- Douady CJ, et al. Comparison of Bayesian and maximum likelihood bootstrap measures of phylogenetic reliability. Mol. Biol. Evol. 2003;20:248–254. doi: 10.1093/molbev/msg042. [DOI] [PubMed] [Google Scholar]
- Doyle JJ. Gene trees and species trees: molecular systematics as one-character taxonomy. Syst. Bot. 1992;17:144–163. [Google Scholar]
- Eulenstein O, et al. Performance of flip supertree construction with a heuristic algorithm. Syst. Biol. 2004;53:299–308. doi: 10.1080/10635150490423719. [DOI] [PubMed] [Google Scholar]
- Farris JS, et al. A numerical approach to phylogenetic systematics. Syst. Zool. 1970;19:172–191. [Google Scholar]
- Fawcett T. Technical Report HPL-2003-4. Palo Alto, CA: HP Labs; 2004. ROC graphs: notes and practical considerations for researchers. [Google Scholar]
- Fawcett T. An introduction to ROC analysis. Pattern Recogn. Lett. 2005;27:86–874. [Google Scholar]
- Goloboff PA, et al. TNT, a free program for phylogenetic analysis. Cladistics. 2008;24:774–786. [Google Scholar]
- Gordon AD. Consensus supertrees: the synthesis of rooted trees containing overlapping sets of labeled leaves. J. Classif. 1986;3:335–348. [Google Scholar]
- Grunewald S, et al. Closure operations in phylogenetics. Math. Biosci. 2007;208:521–537. doi: 10.1016/j.mbs.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Harding E. The probabilities of rooted tree-shapes generated by random bifurcation. Adv. Appl. Probab. 1971;3:44–77. [Google Scholar]
- Hickey G, et al. SPR distance computation of unrooted trees. Evol. Bioinform. Online. 2008;4:17–27. doi: 10.4137/ebo.s419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janecka JE, et al. Molecular and genomic data identify the closest living relative of primates. Science. 2007;318:792–794. doi: 10.1126/science.1147555. [DOI] [PubMed] [Google Scholar]
- Jeffroy O, et al. Phylogenomics: the beginning of incongruence? Trends Genet. 2006;22:225–231. doi: 10.1016/j.tig.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;2:87–90. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Lin HT, et al. Triplet supertree heuristics for the tree of life. BMC Bioinformatics. 2009;10(Suppl. 1):S8. doi: 10.1186/1471-2105-10-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP. Reconstructing character evolution on polytomous cladograms. Cladistics. 1989;5:365–377. doi: 10.1111/j.1096-0031.1989.tb00569.x. [DOI] [PubMed] [Google Scholar]
- Moore BR, et al. Increasing data transparency and estimating phylogenetic uncertainty in supertrees: approaches using nonparametric bootstrapping. Syst. Biol. 2006;55:662–676. doi: 10.1080/10635150600920693. [DOI] [PubMed] [Google Scholar]
- Moran S, et al. Using semi-definite programming to enhance supertree resolvability. In: Istrail S, et al., editors. Algorithms in Bioinformatics, Proceedings of WABI 2005. Vol. 3692. Berlin, ALLEMAGNE: Springer; 2005. pp. 89–103. of LNCS. [Google Scholar]
- Mosses C. PhD Thesis. Aarhus, Denmark: University of Aarhus; 2005. Triplet supertrees. [Google Scholar]
- Nelson G, Ladiges PY. Three-item consensus: empirical test of fractional weighting. In: Scotland RW, et al., editors. Models in Phylogeny Reconstruction. Clarendon, Oxford: 1994. pp. 193–209. [Google Scholar]
- Page R.DM. Modified MinCut supertrees. In: Guigó R, Gusfield D, editors. of LNCS. Vol. 2452. London, UK: Springer; 2002. pp. 537–551. [Google Scholar]
- Phillips C, Warnow TJ. The asymmetric median tree–a new model for building consensus trees. Discr. Appl. Math. 1996;71:311–355. [Google Scholar]
- Piaggio-Talice R, et al. Quartet supertrees. In: Bininda-Emonds O.RP, editor. Phylogenetic Supertrees: Combining Information to Reveal the Tree of Life. Dordrecht: Kluwer Academic Publishers; 2004. pp. 173–191. [Google Scholar]
- Prasad AB, et al. Confirming the phylogeny of mammals by use of large comparative sequence data sets. Mol. Biol. Evol. 2008;25:1795–1808. doi: 10.1093/molbev/msn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragan MA. Phylogenetic inference based on matrix representation of trees. Mol. Phyl. Evol. 1992;1:53–58. doi: 10.1016/1055-7903(92)90035-f. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Grassly NC. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput. Appl. Biosci. 1997;13:235–238. doi: 10.1093/bioinformatics/13.3.235. [DOI] [PubMed] [Google Scholar]
- Ranwez V, et al. OrthoMaM: a database of orthologous genomic markers for placental mammal phylogenetics. BMC Evol. Biol. 2007;7:241. doi: 10.1186/1471-2148-7-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DF, Foulds LR. Comparison of phylogenetic trees. Math. Biosci. 1981;53:131–147. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- Semple C, Steel M. Phylogenetics. Oxford: Oxford University Press; 2003. [Google Scholar]
- Steel MA, Penny D. Distribution of tree comparison metrices - some new results. Syst. Biol. 1993;42:126–141. [Google Scholar]
- Steel MA, Rodrigo A. Maximum likelihood supertree. Syst. Biol. 2008;57:243–250. doi: 10.1080/10635150802033014. [DOI] [PubMed] [Google Scholar]
- Swofford DL, et al. Phylogenetic inference. In: Hillis DM, et al., editors. Molecular Systematics. Massachussets: Sinauer Associates; 1996. pp. 407–514. [Google Scholar]
- Thorley JL. PhD Thesis. Bristol, UK: University of Bristol; 2000. Cladistic information, leaf stability and supertree construction. [Google Scholar]
- Wilkinson M. Three-taxon statements: when is a parsimony analysis also a clique analysis? Cladistics. 1994;10:221–223. [Google Scholar]
- Wilkinson M, et al. Towards a phylogenetic supertree for platyhelminthes? In: Littlewood DT, Bray RA, editors. Interrelationships of the Platyhelminthes. London: Taylor and Francis, Chapman Hall; 2001. pp. 292–301. [Google Scholar]
- Wilkinson M, et al. The information content of trees and their matrix representations. Syst. Biol. 2004;53:989–1001. doi: 10.1080/10635150490522737. [DOI] [PubMed] [Google Scholar]
- Wilkinson M, et al. The shape of supertrees to come: tree shape related properties of fourteen supertree methods. Syst. Biol. 2005a;54:419–431. doi: 10.1080/10635150590949832. [DOI] [PubMed] [Google Scholar]
- Wilkinson M, et al. Measuring support and finding unsupported relationships in supertrees. Syst. Biol. 2005b;54:823–831. doi: 10.1080/10635150590950362. [DOI] [PubMed] [Google Scholar]
- Wilkinson M, et al. Properties of supertree methods in the consensus setting. Syst. Biol. 2007;56:330–337. doi: 10.1080/10635150701245370. [DOI] [PubMed] [Google Scholar]
- Williams DM. Supertrees, components and three-item data. In: Bininda-Emonds O.RP, editor. Phylogenetic Supertrees: Combining Information to Reveal the Tree of Life. Dordrecht: Kluwer Academic Publishers; 2004. pp. 389–408. [Google Scholar]
- Williams DM, Humphries CJ. Component coding, three-item coding, and consensus methods. Syst. Biol. 2003;52:255–259. doi: 10.1080/10635150390192753. [DOI] [PubMed] [Google Scholar]
- Yule GU. A mathematical theory of evolution, based on the conclusions of Dr J.C. Willis. Philos. Trans. Roy. Soc. B. 1925;213:21–87. [Google Scholar]