Abstract
Motivation: Single-particle cryo electron microscopy (cryoEM) typically produces density maps of macromolecular assemblies at intermediate to low resolution (∼5–30 Å). By fitting high-resolution structures of assembly components into these maps, pseudo-atomic models can be obtained. Optimizing the quality-of-fit of all components simultaneously is challenging due to the large search space that makes the exhaustive search over all possible component configurations computationally unfeasible.
Results: We developed an efficient mathematical programming algorithm that simultaneously fits all component structures into an assembly density map. The fitting is formulated as a point set matching problem involving several point sets that represent component and assembly densities at a reduced complexity level. In contrast to other point matching algorithms, our algorithm is able to match multiple point sets simultaneously and not only based on their geometrical equivalence, but also based on the similarity of the density in the immediate point neighborhood. In addition, we present an efficient refinement method based on the Iterative Closest Point registration algorithm. The integer quadratic programming method generates an assembly configuration in a few seconds. This efficiency allows the generation of an ensemble of candidate solutions that can be assessed by an independent scoring function. We benchmarked the method using simulated density maps of 11 protein assemblies at 20 Å, and an experimental cryoEM map at 23.5 Å resolution. Our method was able to generate assembly structures with root-mean-square errors <6.5 Å, which have been further reduced to <1.8 Å by the local refinement procedure.
Availability: The program is available upon request as a Matlab code package.
Contact: alber@usc.edu and m.topf@cryst.bbk.ac.uk
Supplementary information: Supplementary data are available at Bioinformatics Online.
1 INTRODUCTION
To understand biological mechanisms of cellular processes, high-resolution structures of macromolecular assemblies are needed (Alber et al., 2008; Robinson et al. 2007). Single-particle cryo electron microscopy (cryoEM) and image processing typically produces 3D density maps of large assemblies at intermediate to low levels of resolution (∼5–30 Å). Although the number of maps at subnanometer resolution is increasing significantly in recent years, allowing the identification of secondary structure elements and even the tracing of the backbone (Jiang et al., 2008; Lindert et al., 2009; Yu et al., 2008), for most cryoEM maps the level of resolution is still not sufficient to directly determine the structure at atomic detail. However, a pseudo-atomic picture of the entire macromolecule can be determined by integrating information about the atomic structures of the individual components with the density map of the assembly (Baumeister and Steven, 2000; Fabiola and Chapman, 2005; Wriggers and Chacon, 2001). This integration is done via a process called density fitting, where structures are fitted into the density maps by optimizing a quality-of-fit measure between the cryoEM map and the density of the probe structure at a corresponding level of resolution (Ceulemans and Russell, 2004; Chacón and Wriggers, 2002; Dror et al., 2007; Garzon et al., 2007; Jiang et al., 2001; Kovacs et al., 2003; Navaza et al., 2002; Rath et al., 2003; Roseman, 2000; Rossmann, 2000; Rossmann et al., 2001; Topf et al., 2005; Velazquez-Muriel et al., 2006; Volkmann and Hanein, 2003).
A considerable challenge is the fitting of multiple components into the density map of an assembly if no a priori knowledge about the location of the components is available. Sequential fitting of components often fails when they cannot be unambiguously placed in the density map as is often the case for maps of ∼10–30 Å resolution and for assemblies with a large number of components. In such cases, all components must be fitted simultaneously into the map to identify the global optimum of the quality-of-fit measure. The simultaneous fitting of components is difficult as the large search space makes an exhaustive search protocol that uniformly samples over all degrees of freedom computationally unfeasible.
To overcome this problem, Lasker et al. (2009) uses discrete sampling in combination with an inference optimizer and expands the quality-of-fit measure by additional information such as shape complementarity between interacting components. Other fitting strategies simplify the search problem by reducing the complexity of the 3D volumetric density and structures. In one method, the initial density distribution of assembly and components are approximated by a small set of Gaussian functions to efficiently use gradient-based optimization methods for the structural optimization of component orientations (GMFIT) (Kawabata, 2008). Other methods reduce the complexity of density maps to a small set of feature points (so-called codebook vectors) that are meant to best reproduce the density map's gross features, such as its shape and mass distribution (Birmanns and Wriggers, 2007). The optimal positions of feature points can be determined by the vector quantization (VQ) technique (Martinetz et al., 1993; Wriggers et al., 1998). The fitting problem then effectively reduces into a common point set matching problem. This matching has been achieved by an exhaustive search method (Wriggers et al., 1999) for single-molecule fitting and a heuristic anchor-point registration method for component fitting into an assembly map (Birmanns and Wriggers, 2007). The latter method uses a hierarchical alignment of the point sets and reduces the search-space complexity by an integrated tree pruning technique. Although the method is very fast, it does not allow the simultaneous fitting of all components.
Here, we present an efficient mathematical programming algorithm to fit multiple component structures simultaneously into density maps of assemblies. Integer quadratic programming (IQP) is used for matching two point sets that represent the reduced density distribution of components and assembly. Numerical computation and theoretical analysis show that IQP can be relaxed into the corresponding quadratic programming (QP) scheme, which almost always ensures an integer solution. Therefore, a QP algorithm can be adopted to efficiently solve IQP problems without any approximation. In contrast to other point set matching approaches, we are able to match multiple component point sets simultaneously while considering both information about the geometric architecture of the point distributions, as well as the consistency of the density in the immediate neighborhood of the points. Other major advantages of this method are the ability to fit all the components of a given assembly simultaneously and its ability to allow straightforward integration of additional information about the assembly into the method.
Reducing the complexity of the density information to point sets is accompanied by an inevitable loss in accuracy in the fitting process. To overcome this challenge, we perform a large number of independent point set matches and generate an ensemble of candidate solutions. This ensemble is then assessed using an independent scoring function that measures the quality-of-fit between the components structures and the assembly map using the cross-correlation function (CCF). Finally, the best scoring structures are refined to locally optimize the fit between assembly and component density maps. To this end, we also present an efficient refinement procedure based on the weighted Iterative Closest Point (wICP) registration algorithm for refining the coarse fitting. The wICP procedure detects the optimal registration between weighted points that represent the voxels of two density grids. The method has a large radius of convergence and is able to refine the coarse component positions in the assembly that were generated by the IQP method.
Both theoretical and numerical results demonstrate that the proposed method is effective and general. We tested the method on a benchmark of 11 protein assemblies. The component structures are fitted simultaneously and refined in the context of their native assembly density maps simulated at 20 Å resolution. In addition, we tested the method also on an experimental cryoEM density map at 23.5 Å resolution. We have implemented our method as a MATLAB software package, which is available from the authors upon request.
2 MATERIALS AND METHODS
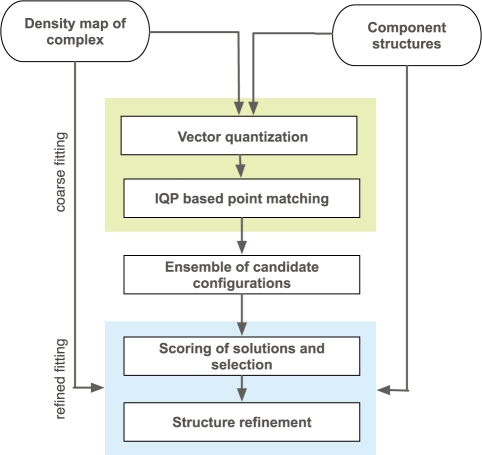
In the following section, we describe our protocol for simultaneous fitting of component structures into density maps (Fig. 1). In the first stage, the density of assembly and components are represented by sets of feature points, whose optimal location are determined by VQ. At the location of each feature point, a density value is calculated such that it captures the characteristic properties of the density distribution in the proximity of the feature point. The simultaneous fitting of components into the assembly map is then achieved by solving a point set matching problem that considers both information about the geometric architecture of the point distributions and the consistency of their density values. The point set matching problem between component and assembly point sets is solved by IQP. By varying starting parameters in individual fitting processes an ensemble of candidate configurations of the assembly structure is generated. All the resulting candidate configurations are then assessed by a quality-of-fit measure between the component structures and the assembly density map. Finally, the best scoring candidate structures are refined by performing a local optimization of the fit of the component structures with respect to the assembly density map.
Fig. 1.
Our protocol for simultaneous fitting of component structures into density maps is divided into two stages. First, approximate positions of all components are determined at a coarse information level by our IQP point matching approach (upper grey shading). By varying the initial parameter settings, an ensemble of solutions is generated. At a second stage, all candidate structures are assessed and structurally refined using the initial density map and the density of the component structures simulated at the same resolution (lower grey shading).
2.1 VQ
To extract feature points from a density map we follow a procedure by Wriggers et al. (1999) and adopt a fast VQ technique based on the neural gas clustering technique. Feature points are defined as the centers of density clusters, which as a whole capture the characteristic features of the density distribution. As the interior details of a density map take key roles in the reduced point matching problem (Birmanns and Wriggers, 2007), we apply the Laplacian edge enhancement filter to the density maps, which boosts the contrast of the map and enhances the contour as well as the interior detail. The Laplacian density map is normalized and only the more robust interior map information is used in the VQ procedure by considering only voxels with a Laplacian density value above a given threshold. As the optimal value for this threshold is unknown beforehand we perform independent VQ by varying the Laplacian density cutoff. Moreover, due to numerical instabilities, independent VQ runs with identical starting conditions can produce slightly different point configurations. To account for the variability of feature point configurations, 10 independent VQ runs are performed for each density map, with five different Laplacian density cutoffs for each run, resulting in 50-point configurations. These configurations are used as input for the IQP-based fitting and are henceforth named the ‘point configuration ensemble’. The variance for a given point position in the point configuration ensemble can be up to 3 Å (observed for a component in 2REC).
2.2 Multiple component matching by IQP
Our method for efficient rigid-body matching relies on the distance matrix representation of 3D point sets, which is defined as the square matrix of distances between all pairs of points in the system. The distance matrix representation has been applied to many structural problems, including protein structure alignment (Caprara et al., 2004; Holm and Sander, 1993). The distance matrix as well as the related contact map overlap problem (Caprara et al., 2004) are both NP-hard. Solutions for the distance matrix problem were approximated by using Monte Carlo optimizations (Dali) (Holm and Sander, 1993) and an algorithm involving heuristic cutoffs on pairwise distance scores [Combinatorial Extension, (CE)] (Bourne and Shindyalov, 1998), whereas the contact map overlap problem has been addressed by using integer programming and Lagrangian relaxation (Caprara et al., 2004). The point matching problem is also related with the weighted maximum common subgraph problem (Jain and Lappe, 2007). In all these methods, the matching between two configurations is based purely on the geometrical equivalence and is applied only to two matching point sets. Here, we introduce a method, that can not only consider simultaneous matching of multiple components, but can also consider additional feature point properties in the matching process. In the case of density fitting, each feature point can be assigned a rotation invariant density measure that captures the local density distribution in the immediate neighborhood of the point. Two corresponding feature points should have roughly equivalent density measures in addition to their geometric matching. In the following section, we formulate the weighted points matching problem based on distance matrix as well as density information.
Formally, we are given two point sets V1={v11,···, vm1} and V2={v12,···, vn2}, with corresponding density values U1={u11,···, um1} and U2={u12,···, un2}. Each density value ui is defined as an average over all voxels in the neighborhood (within 5 units of distance) of the corresponding point vi. The distance matrices of the point configurations V1 and V2 are A=(aij)m×m and B=(bij)n×n, respectively, where aij=‖vi1−vj2‖ is the Euclidean distance between points i and j. The distances bij are defined in the same way.
In our approach, the matching between vi1∈V1 and vj2∈V2 is represented by a binary variable xij,
xij therefore indicates the relationship between one pair of points and X={xij} represents a complete point-by-point matching. The ‘optimal’ score associated with X is defined by two objective functions, namely the point density score and the point-to-point distance score.
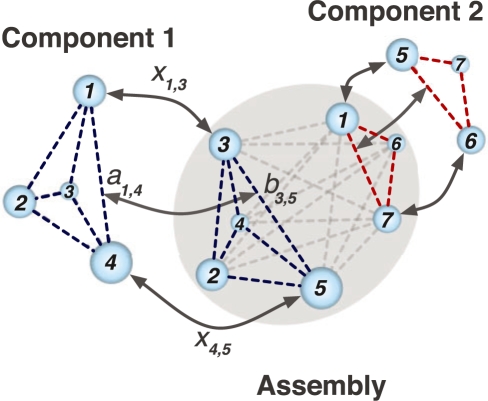
The matching problem is to maximize the similarity score F(U1, U2, V1, V2) between point sets V1 and V2 with density values U1 and U2 among all feasible combinations X (Fig. 2). A solution can be found using IQP:
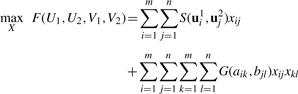 |
(1) |
where the functions S and G represent the contributions of density matching and geometric matching, respectively. They are defined as follows:
| (2) |
The objective function is subject to three constraints. First, each point in V1 can match at most one point in V2. Second, each point in V2 can match at most one point in V1. Third, the variable xij is binary.
Fig. 2.
An illustration of the feature point matching procedure. The goal is to match simultaneously the point sets of Component 1 and Component 2 with the Assembly point set. All point sets are shown as spheres, where the size of a sphere represents the averaged density value in a defined volume of the density map, which is within five grid voxels of the corresponding feature point. The dashed lines between the spheres represent all possible distances within each component and within the assembly. aik and bjl are the distances between points i and k in Component 1 and points j and l in the Assembly, respectively. The value of the binary variable xij is set to 1 if point i in Component 1 matches with point j in the Assembly and xij is set to zero otherwise. Correspondingly, the product xijxkl is 1 if distance aik in Component 1 matches with distance bjl in the Assembly and is 0 otherwise. The aim of IQP is to find the best matching xij with maximized IQP score F(U1, U2, V1, V2) (see Section 2).
Since we are simultaneously matching several structural components to the overall assembly map, we can combine the individual distance matrices of the components into a single composite matrix, that is then compared to the assembly matrix. The composite matrix A is defined as follows:
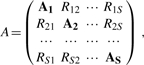 |
(3) |
where As is the distance matrix of component s, s=1,···, S (number of components) and each submatrix R of appropriate size describes the possible distance values between feature points of two components. If no information about the interaction between two components is available, all elements of the corresponding submatrix R are set to zero. Moreover, the function G(a, b) is set to zero for these elements, so that the corresponding pairs of elements do not contribute to the objective function. In this article, we assume that no additional knowledge about component interactions is available. The aforementioned IQP constraints prevent any structural overlap between components in the resulting assembly, because each feature point can be matched to at most one component point. Therefore, the IQP constraints enable simultaneous fitting of all components into an assembly map.
The a priori knowledge of the protein interactions can be incorporated by defining specific values in the corresponding inter-component matrices (R). In such a case, the estimated distance between the corresponding feature points in the two interacting components can be added to the R matrix. Any positive value in A ensures that the corresponding pairwise distance will be considered in the objective function, while setting an element to 0 excludes the distance between two feature points from the optimization process. Therefore inter-component matrices are an efficient tool to integrate additional information such as chemical cross-linking or yeast two-hybrid experiments into the IQP framework (see Supplementary Figure S1 for an illustrative example for this).
It can be shown that IQP belongs to the class of NP-hard problems. However, it can easily be demonstrated that in some cases, the types of IQP constraints used here make such problems unimodular. This property implies that the system of equations can be reduced to a QP problem with an integral solution. Furthermore, even if unimodularity does not apply, the corresponding QP still has an optimal integer solution in most cases (Li et al., 2007). For a non-integer solution, a rounding strategy can be adopted to determine an approximate integer solution of the QP procedure. The relaxed QP problem has been solved and implemented in Matlab based on an efficient interior algorithm (Ye, 1992). Although IQP is a local optimization method, it is still expected to determine the global optimum because the number of feature points is relatively small.
2.3 Ensemble of candidate structures
IQP is a local method, therefore, we must sample several IQP runs with the identical feature point sets to ensure that the global minimum in the scoring function is found. To test the scope of the necessary sampling, we analyzed the variability of the outcome of multiple IQP runs. When for each feature point set 10 independent IQP runs are performed, we can observe on average only around three different configurations (Supplementary Figure S2). Therefore, in each of our test cases, 10 IQP runs are sufficient to determine the global minimum for IQP point matching. The IQP generally converges to a stable solution within a small number of iterative steps (<20). As described earlier, the density map of an assembly is represented by 50 VQ point configurations. For each of these point configurations IQP point matching is repeated 10 times with random initial values for the variables {xij} set between 0 and 1. As a result, a total of 500 structures is generated from independent IQP and VQ runs, some of which may not be unique. These 500 structures are henceforth refereed to as the ‘ensemble’.
2.4 Scoring of candidate structures
An independent criteria for the fitting quality is performed by a scoring scheme that was not used in generating the structures. To pick the best set of results from the IQP fitting, each arrangement of assembly components is scored within the density map using a normalized CCF. For this calculation, each component structure needs to be converted into a probe density with the same size and sampling as the target density. First, the atomic coordinates are mapped out on to a grid sampled at 1 Å/pixel. Then it is convoluted in Fourier space with a Gaussian function of σ = 0.356 • resolution (so that the Gaussian width at 1/e maximum height equals the resolution). The resultant grid is then resampled to match the spacing in the target density map.
The CCF between the target and probe densities is given by:
| (4) |
where ρt(i) is the density value at position i in the target map, ρp(i) is the density value at position i in the probe density, < ρt > and < ρp > are the mean values of the target density and the probe density, respectively. σt and σp are the standard deviation of the target and probe densities respectively. The IQP results are additionally scored using the CCF Equation (4) on the Laplacian-filtered probe (Laplacian-CCF) and target densities, as described in Chacón and Wriggers (2002). All of the above was implemented in Python with components from the Scipy package (http://www.scipy.org/).
2.5 Refinement with weighted ICP algorithm
After having established the position and orientations of all the components, we refine their positions using the density maps at their initial resolution. We define a variant of the Iterative Closest Point (ICP) algorithm, originally introduced by (Besl and McKay, 1992 and Rusinkiewicz and Levoy, 2001) and commonly used in computer vision and pattern recognition. Here, we introduce a weighted registration formalism (wICP) where the density contribution of the maps is considered in the registration process. Each grid voxel in the density maps is treated as a weighted point with a position and density value, and the task of registration is to determine the optimal transformation that minimizes the deviation of position and density values between two point sets. Given the initial orientation of two 3D rigid point sets X and Y, the wICP algorithm in its simplest form iterates two steps repeatedly. First, the correspondence between points in the two configurations is identified based on the proximity between them. A point in one configuration corresponds to the closest point in the second configuration. Based on the correspondence, a transformation matrix is then calculated by singular value decompositions (SVD) and applied to X to determine its new point positions, which in effect leads to a new correspondence relationship between the points. Iterations of these two steps progressively reduces a given error metric. To balance the density consistency and the 3D rigid matching, we introduce the following weighted root-mean-square (RMS) error metric,
where X and Y are the coordinates of two voxel point sets and ϕ(i) is the index function representing the corresponding points in the two sets. In order to incorporate the density information into the registration procedure, we use a Gaussian weighting parameter. The weight is defined as
where ρ(xi) and ρ(yj) are the density values for voxel points xi and yj; c = 0.2 is a scaling factor whose optimal value was determined by test calculations. Varying c between 0.1 and 0.3 does not affect the outcome of our calculations. In this context, the fitting of two density maps is equivalent to finding an optimal correspondence index ϕ : i → j and a transformation that minimizes the wRMS (Supplementary Material for a detailed flowchart of the method). The wICP algorithm that has been proven to converge monotonically to a local minimum, allows the optimization of the component position and orientation without the need to calculate a gradient of an objective function.
The computational complexity of the above algorithm is of O(CMN), where C is the number of iterated steps. The complexity of the SVD-based least square fitting is of the order O(N). So, the most computationally expensive part of both wICP and ICP is the exhaustive search for the point correspondence with a time of the order O(MN). The complexity of this search can be reduced to O(NlogM) by employing a k dimensional binary search tree (k-D tree) (Akca and Gruen, 2005; Besl and McKay, 1992). Other types of acceleration strategies include reducing the number of iterations and the number of employed points (Akca and Gruen, 2005).
2.6 Assessment of structural solutions
Two criteria were used to assess the accuracy of the IQP-fitted conformations. First, the Cα RMS error (referred to here as the RMS error) between the corresponding Cα atoms in the fitted and correct structures is calculated. However, since RMS error is highly dependent on the size and shape of each component, a second assessment score is used—the component placement score (CPS) is also used for assessment (Lasker et al., 2009; Topf et al., 2008). The CPS calculates the difference between the orientation and position of equivalent components in the fitted and native structure. This gives two values for each assembly component, i.e. the shift and rotation angle needed to superpose the fitted component onto the native one.
We define a correctly predicted structure as one with a RMS error <7 Å to its native structure, and a CPS shift score and angle score <6 Å and 25○, respectively.
3 RESULTS AND DISCUSSIONS
We tested our approach on a set of 11 protein assemblies that are diverse in assembly size, number of components (between two and seven components, including symmetrical and unsymmetrical assemblies), and global shape of the assemblies (Table 1). The density maps for each assembly were simulated at 20 Å resolution using the PDB2VOL program of the Situs 2.0 package with voxel size of 3 Å.
Table 1.
Summary of benchmark results
| Assembly | Comp. | Sym. | Feat. Points | Time (s) (Total time in min) | Lowest RMSD structure |
Best CCF ranking structure |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CCF (Lapl.-CCF) | CPS (Å, ○) | RMSD | RMSD* | CPS (Å, ○) | RMSD | RMSD* | |||||
| 1DOR | 2 | Y | 10 | 0.16 (1.33) | 2 (1) | (1.1, 6.8) | 2.1 | 1.1 | (0.6, 9.5) | 2.5 | 1.2 |
| 1AFW | 2 | Y | 10 | 0.15 (1.25) | 2 (1) | (2.3, 14.4) | 4.8 | 0.9 | (2.5, 15.0) | 4.9 | 0.9 |
| 1PC8 | 2 | N | 10 | 0.10 (0.83) | 6 (10) | (1.1, 3.1) | 1.3 | 0.5 | (0.8, 6.4) | 1.6 | 0.5 |
| 1TX4 | 2 | N | 11 | 0.14 (1.17) | 8 (6) | (1.2, 2.8) | 2.6 | 0.4 | (0.7, 2.9) | 3.0 | 0.4 |
| 1NIC | 3 | Y | 15 | 0.65 (5.42) | 1 (1) | (5.6, 5.1) | 5.9 | 1.1 | (5.6, 5.1) | 5.9 | 1.1 |
| 1CS4 | 3 | N | 11 | 0.16 (1.33) | 8 (7) | (2.4, 24.0) | 6.5 | 1.8 | (2.3, 55.5) | 12.8 | 11.7 |
| 2DQJ | 3 | N | 12 | 0.20 (1.67) | 34(11) | (2.0, 21.1) | 4.5 | 1.7 | (1.4, 62.1) | 9.5 | 7.8 |
| 1F1X | 4 | Y | 12 | 0.42 (3.50) | 2 (18) | (2.4, 14.6) | 4.6 | 0.9 | (2.3, 168.4) | 28.2 | 26.1 |
| 2BO9 | 4 | N | 18 | 0.75 (6.25) | 1 (1) | (1.1, 4.6) | 1.7 | 1.1 | (1.1, 4.6) | 1.7 | 1.1 |
| 2REC | 6 | Y | 30 | 2.56 (21.33) | 1 (1) | (1.3, 4.2) | 1.7 | 1.0 | (1.3, 4.2) | 1.7 | 1.0 |
| 1J2P | 7 | Y | 28 | 2.48 (20.67) | 1 (3) | (1.6, 16.2) | 4.4 | 1.5 | (1.6, 16.2) | 4.4 | 1.5 |
The individual columns are: Assembly the PDB ID (Bernstein et al., 1977) of the assembly structure being used; Comp., the number of components of the assembly; Sym., indicates if the assembly structure is symmetric (Y) or non symmetric (N); Feat. Points, the number of feature points being used; Time, the average running time for an IQP run, and the total time of 500 IQP runs is shown in brackets; CCF (Lapl. -CCF), the rank of the structure with the lowest RMS error based on the CCF and Laplacian CCF, respectively; CPS, Component placement score composed of two elements (the shift and orientation). The average component placement score for all components is shown. RMSD, the root-mean-square error (RMS error) between the corresponding Cα atoms in the fitted and the native structures. RMSD*, the RMS error of the assembly after wICP refinement.
3.1 IQP-based fitting and generation of ensemble structures
To test if the IQP procedure can sample accurate results, we examined if the correctly predicted structures are present in the ensemble. A correctly predicted structure for each assembly is a structure for which all the components are occupying their correct positions and are oriented correctly (Section 2). Here, this is typically the case if the RMS error of a structure with respect to the native structure is <7 Å.
We now discuss the accuracy of the structures in the ensemble with the lowest RMS error with respect to the native structures (Table 1). The RMS errors for all of these structures range between 0.4 Å and 6.5 Å, which corresponds to an average shift of a component from its native position by ≤5.6 Å and an average difference of the orientation of each component by ≤24.0○ (see Section 2). At such values, our local refinement method was able to produce improved structures with RMS errors between 0.4 and 1.8 Å (see below). This result indicates that for all the test cases correct solutions can be identified in the ensemble of 500 structures, and the proposed IQP matching procedure can efficiently produce accurate fitting results. For example, the best fitted structure of the four-component assembly 2BO9 has an RMS error of 1.7 Å. This structure can be further optimized by the local wICP refinement protocol to an RMS error of only 1.1 Å (Table 1). Our results indicate, therefore, that the IQP matching procedure is able to efficiently produce accurate fitting results.
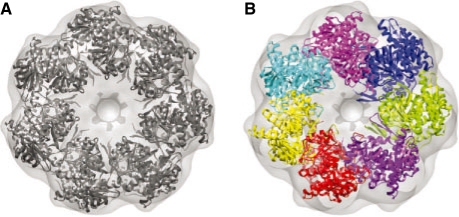
We also tested our approach on a 23.5 Å resolution experimental cryoEM map of Apo-GroEL. Specifically, we used the density map of Escherichia coli GroES-ADP7-GroEL-APT7 EMD id: 1046;(Ranson et al., 2001) and generated the apo-GroEL ring by manual segmentation. For the fitting, we used seven identical copies of one of the apo-GroEL components taken from an x-ray crystal structure of GroEL-GroES (PDB id: 1AON). After simultaneous IQP fitting, the lowest RMS error for the fitted structure is 8.6 Å with respect to the crystal structure, and only slightly larger than our results observed for simulated maps. The average shift of a component with respect to the crystal structure is 5.7 Å and the average difference in the orientation of each component is 17.7○. This result is not surprising because experimental maps are a greater challenge due to the inherent noise levels.
We now analyze the structural variability among the structures in the ensemble. Indeed, not all the structures in each ensemble are correct solutions. For instance, in the case of 2REC the range of observed RMS errors for structures in the ensemble ranges between 1.7 Å and 35.4 Å. This observation indicates the need for an independent scoring system to identify the correct solutions in the ensemble. Such a scoring system should preferably use the initial density maps as input information instead of the reduced feature point representation.
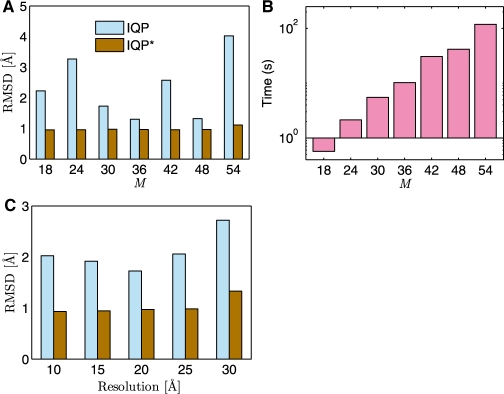
Next, we analyze how the number of feature points and the resolution of the density map affect the outcome of the IQP calculations. For example, the assembly 2REC has been fitted using IQP point matching with various numbers of feature points per component. All these IQP runs produce results with RMS errors <4.2 Å (Fig. 4A), indicating that the correct structure can be predicted. This result shows the robustness of the method and indicates that accurate results can already be generated with a relatively small number of feature points. In addition, for 2REC density maps at resolutions between 10 Å and 30 Å produces very similar IQP fitting results. This observation is not surprising considering that the VQ process produces a reduced representation of the density maps, and consequently, the accuracy of the IQP fitting is less dependent on the initial resolution of the map.
Fig. 3.
Simultaneous fitting of all seven components of the APO-GroEL into the experimental density map of GroEL-GroES at 23.5 Å resolution. (A) Experimental map and fitted atomic model, as provided by Ranson et al. (2001). (B) Fitted component structures with an RMS error of 8.6 Å with respect to the native structure in (A).
Fig. 4.
Simultaneous fitting of six components into the symmetric hexamer 2REC. (A) The dependency of fitting accuracy, measured by the RMS error (RMSD) with respect to the number of feature points M of the assembly. (B) The dependency of running time (in seconds) on the number of feature points M of the assembly. (A) and (B) are calculated using density maps at 20 Å resolution. (C) The fitting accuracy at different resolutions of the initial density map. The calculations for (C) are performed with 30 feature points per assembly. Results are shown after IQP fitting (IQP) and after additional refinement with wICP (IQP*).
IQP-based matching of multicomponent assemblies is very fast and the run time for each of the test cases is <3 s allowing for multiple fitting runs and the generation of an ensemble of structural solutions. Although, the running time increases exponentially with the number of points, IQP is sufficiently fast to deal with large number of points (∼54) (Fig. 4B), enabling the fitting of protein assemblies with a large number of components.
3.2 Density-based scoring of ensemble structures
To identify the correctly predicted solutions in the ensemble, we used two density-based scoring systems: the CCF that measures a quality-of-fit between the density maps of the assembly and its components, and the corresponding CCF for the Laplacian of the density maps (Laplacian-CCF) (see Section 2). We assessed the different scores by calculating the rank of the most accurate structure in the ensemble, which is the solution with the lowest RMS error from the native structure (Table 1). For all cases, except 2DQJ (see discussion below), this structure lies within the top 10 ranked solutions (out of 500) for both scores (Table 1), indicating their equally good performance.
Next, we analyze the accuracy of the best scoring structures in the ensemble. In eight out of the 11 test cases, the best scoring structure correctly predicts the positions and orientations of all its components with RMS errors from the native structures ranging between 1.6 Å and 5.9 Å and corresponding average component placement score ranging from 0.6 Å to 5.6 Å for the shifts and 2.9○ to 16.2○ for the angles of the components relative to the native positions (Table 1). When the wICP refinement is applied to these structures (see below) the RMS errors are further reduced and range only between 0.4 Å and 1.5 Å (Table 1). Thus, our method allows us to predict the correct assembly structure with relatively high accuracy.
For two cases (2DQJ and 1CS4), the best scoring solution predicts the positions of all components correctly, but the orientation of one of the component proteins differs significantly from the native structure. For example, for the three component assembly 2DQJ, the best CCF-scoring structure correctly predicts two components in position and orientation, with an RMS error of only 3.4 and 2.2 Å and component placement scores of {0.6 Å, 20.6○} and {2.1 Å, 8.6○}, respectively. The third component shows a component placement score of {1.6 Å, 158.1○}, indicating that the position of the center of mass is close to the native structure. However, the component is wrongly oriented, which leads to an RMS error of 15.4 Å with respect to the native structure. For the homotetramer 1F1X, the best CCF-ranked structure positions the component's center of mass within 2.5 Å from the native structure. However, all the components show an incorrect orientation that leads to a large RMS error of ∼28 Å.
For all these three cases, the correctly predicted structures are not top-ranked by the CCF score. Here, we discuss in more detail the possible cause of these scoring problems. The densities of all the components that were incorrectly ranked share specific self-symmetric characteristics. In 1CS4, the misaligned component protein is formed by two domains of the same fold family (adenylyl and guanylyl cyclase catalytic domain). This peculiarity may cause difficulties in CCF-based scoring systems when low resolution density maps are used, because a rotation along the pseudo-symmetry axis could in principle lead to similar CCF-scoring results. We observe a similar situation for the structure 1F1X, where all four components are formed by two domains of the same fold family (extradiol dioxygenases scope family). The misaligned protein component in 2DQJ is formed by the immunoglobulin-like fold, which is composed of two similar β-strands that are stacked in such a way that a rotation along the central pseudo-symmetric axis of the protein may produce similar cross-correlation scores at low resolution densities. In summary, the three problematic proteins include pseudo-symmetric components for which the correct orientation could not be identified by CCF-based scoring in low-resolution density maps.
In conclusion, the IQP method in combination with CCF-based scoring is an efficient tool to simultaneously fit components into assembly density maps. To further improve the accuracy of the fitting, the IQP point matching and scoring must be combined with a refinement strategy that uses the initial density maps as input information. To this end, we have developed a refinement method to improve the placement of components. In the following section, we describe the results of our wICP refinement method.
3.3 wICP refinement
Structures determined by IQP are refined by the wICP method using the density maps of components and assembly as input information. wICP is an iterative optimization procedure, which does not depend on the gradient of the objective function. The wICP refinement significantly reduces the RMS error after initial IQP-based fitting from ∼6 Å to very low values (between 0.4 Å and 1.8 Å) (Table 1). For instance, for the seven component assembly 1J2P, wICP refinement reduces the RMS error from 4.4 Å to 1.5 Å (Table 1).
To test the efficiency of the wICP refinement, we systematically explored a number of random starting configurations that can be refined to assembly structures with an RMS error <1 Å. A suite of random rotation matrices is generated, which increasingly deviate from the native orientation. For all test cases, the result of wICP refinement converges to the native orientation even if the starting orientation differs by an angle of ∼50○ (Supplementary Figure S3). wICP is therefore an efficient tool to refine the IQP-derived structures. As the wICP refinement relies on the density maps of assemblies and components, the method is sensitive to the resolution of the maps (Fig. 4C). The RMS error increases slightly as the resolution is reduced (Fig. 4C); however, even for low resolutions (∼30 Å) assembly structures can be fitted with high accuracy (RMS error ∼1 Å); (Fig. 4C).
3.4 Comparison with other methods
The present work aims to develop a tool for fitting components into the density map of an assembly. To achieve this goal, we have proposed an efficient point matching method that uses sets of feature points obtained by VQ. Compared with the initial fitting method (QDOCK program) that introduced the usage of VQ (Birmanns and Wriggers, 2007; Wriggers et al., 1999) for density fitting, the proposed IQP procedure is competitively fast [within a few seconds even for large complexes with six components (Table 2)]. In contrast to QDOCK, the IQP framework can simultaneously fit all the components into the assembly, while QDOCK can only fit one component at a time.
Table 2.
Comparison of the performance between IQP and GMFIT fitting
| Assembly | Comp. | GMFIT |
IQP |
|||
|---|---|---|---|---|---|---|
| Time (s) | RMSD | Time (s) | RMSD | RMSD* | ||
| 1AFW | 2 | 7.1 | 1.0 | 0.15 | 4.8 | 0.9 |
| 1NIC | 3 | 16.0 | 1.8 | 0.65 | 5.9 | 1.1 |
| 2REC | 6 | 110.9 | 2.3 | 2.56 | 1.7 | 1.0 |
RMS errors between the fitted and native structures are shown for three different assemblies. GMFIT used 16 GDFs (Gaussian distribution functions) to represent the atomic structures of each component and 12 GDFs to represent the density map of the assembly (Kawabata, 2008). The IQP model used 5 feature points per component for each assembly. The individual columns are: Assembly, the PDB ID (Bernstein et al., 1977) of the assembly structure being used; Comp., the number of components of the assembly; RMSD, RMS error between the corresponding Cα atoms in the fitted and assembly structures; RMSD*, the RMS error after refinement.
Most of the existing fitting tools are not able to perform simultaneous fitting of assembly components. An exception is the program GMFIT, that was recently developed based on the Gaussian Mixture Model (Kawabata, 2008). Here, we have used three homo-oligomers to directly compare the performance of our method to GMFIT. IQP-based fitting is significantly faster than GMFIT. Although IQP produces structures with slightly larger RMS error values in comparison to those generated by GMFIT, the resulting structures still capture the correct orientation and position of all components. However, the combination of IQP with wICP refinement produces more accurate results than GMFIT (Table 2). These results demonstrate that the combination of IQP point matching with wICP refinement is an efficient simultaneous fitting procedure.
4 CONCLUSIONS
The VQ technique provides an efficient way to transform low-resolution density maps of a macromolecular structure into a set of feature points. The fitting of structures into density maps can then be formulated as a point matching problem, which has been solved by exhaustive or heuristic search methods (Birmanns and Wriggers, 2007; Wriggers et al., 1999). Here, we describe a more flexible framework for solving this problem based on an efficient mathematical programming procedure (IQP) that considers information about the geometry of the point configurations, as well as the consistency of the density distribution in the neighborhood of feature points. The proposed IQP procedure enables a fast and reliable fitting of atomic structures into density maps (or maps into maps) within few seconds even for very large assemblies. More importantly, in contrast to previous methods, the proposed IQP procedure can tackle the simultaneous fitting of multiple components into an assembly map without adding computational complexity. Moreover, it is possible to incorporate existing knowledge about protein interactions and protein binding interfaces in the point matching procedure, providing an ideal framework for comprehensive data integration. These advantages could help increase the applicability of the method to large complexes, which most existing fitting methods cannot handle. It could also be used to generate an ensemble of solutions that would be further refined and assessed by flexible fitting methods.
The applicability of the IQP procedure depends on the intrinsic limitations and robustness of the VQ technique for feature point determination. For some proteins, the positions of feature points in the isolated components can vary greatly in comparison to the corresponding positions observed for the same component in the environment of the assembly. This problem occurs when components differ largely in size or when the binding interface between the proteins occupies a large fraction of the protein surfaces. These problems explain the observed errors in our IQP calculations (∼1.3–6.5 Å RMS errors), which are due to inaccurate positioning of feature points. Moreover, the VQ algorithm is based on stochastic gradient descent optimizations. Therefore, it is possible that for different runs slightly different feature point positions are generated. For each VQ solution, an optimal matching can then be determined by our IQP procedure and the structure with the best density-based CCF score among all solutions is selected. To further improve the results, we also introduced an efficient wICP algorithm for the refinement of position and orientation of assembly components based on the initial density maps. wICP shows a large radius of convergence and therefore it can serve as an efficient refinement procedure (Supplementary Figure S3).
Our fitting of components into density maps of assemblies were performed with component structures in their bound conformational state. It is possible that in some cases large conformational differences between bound and unbound states could reduce the accuracy of the presented fitting process. In our future work, we will address these cases by including flexible fitting approaches (Topf et al., 2008) in our framework.
In summary, we have proposed a fast mathematical programming method and an efficient refinement procedure for determining the accurate positions and orientations of atomic structures of components in 3D density maps of their assembly. The present method is time efficient, can be applied to simultaneous fitting of multiple components and allows an effective way to incorporate additional experimental information about an assembly. Future directions will include the improvement of the scoring as well as the extension with flexible fitting.
Supplementary Material
ACKNOWLEDGEMENT
We would like to thank Prof. Xianghong Jasmine Zhou for her contributions and valuable suggestions.
Funding: Human Frontier Science Program (RGY0079/2009-C to F.A. and M.T.), Alfred P. Sloan Research foundation (to F.A.); F.A. is a Pew Scholar in Biomedical Sciences, supported by the Pew Charitable Trusts; BBSRC (to D.V.), MRC Career Development Award (G0600084) (to M.T.) and the ‘Special Presidential Prize and Excellent PhD Thesis Award - Scientific Research Foundation of the Chinese Academy of Sciences (to S.Z.).
Conflict of Interest: none declared.
REFERENCES
- Akca D, Gruen A. Fast correspondence search for 3D surface matching. ISPRS Workshop Laser scanning, Enschede, the Netherlands, International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences. 2005;XXXVI:186–191. part 3/W19. [Google Scholar]
- Alber F, et al. Integrating diverse data for structure determination of macromolecular assemblies. Ann. Rev. Biochem. 2008;77:443–477. doi: 10.1146/annurev.biochem.77.060407.135530. [DOI] [PubMed] [Google Scholar]
- Baumeister W, Steven AC. Macromolecular electron microscopy in the era of structural genomics. Trends Biochem. Sci. 2000;25:624–631. doi: 10.1016/s0968-0004(00)01720-5. [DOI] [PubMed] [Google Scholar]
- Bernstein FC, et al. The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Besl P, McKay N. A method for registration of 3-D shapes. IEEE Trans. Pattern Anal. Mach. Intell. 1992;14:239–256. [Google Scholar]
- Birmanns S, Wriggers W. Multi-resolution anchor-point registration of biomolecular assemblies and their components. J. Struct. Biol. 2007;157:271–280. doi: 10.1016/j.jsb.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Bourne PE, Shindyalov IN. Protein structure alignment by incremental combinatorial extension of optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- Caprara A, et al. 1001 Optimal PDB structure alignments: integer programming methods for finding the maximum contact map overlap. J. Comp. Bio. 2004;11:27–52. doi: 10.1089/106652704773416876. [DOI] [PubMed] [Google Scholar]
- Ceulemans H, Russell RB. Fast fitting of atomic structures to lowresolution electron density maps by surface overlap maximization. J. Mol. Biol. 2004;338:783–793. doi: 10.1016/j.jmb.2004.02.066. [DOI] [PubMed] [Google Scholar]
- Chacón P, Wriggers W. Multi-resolution contour-based fitting of macromolecular structures. J. Mol. Biol. 2002;317:375–384. doi: 10.1006/jmbi.2002.5438. [DOI] [PubMed] [Google Scholar]
- Dror O, et al. EMatch: an efficient method for aligning atomic resolution subunits into intermediate-resolution cryo-EM maps of large. Acta Crystallogr. D. Biol. Crystallogr. 2007;63:42–49. doi: 10.1107/S0907444906041059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabiola F, Chapman MS. Fitting of high-resolution structures into electron microscopy reconstruction images. Structure. 2005;13:389–400. doi: 10.1016/j.str.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Garzon JI, et al. ADP_EM: fast exhaustive multi-resolution docking for high-throughput coverage. Bioinformatics. 2007;23:427–433. doi: 10.1093/bioinformatics/btl625. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Jain BJ, Lappe M. the 1st International Confernce on Bioinformatics Research and Development (BIRD07) Berlin / Heidelberg: Springer; 2007. Joining soft assign and dynamic programming for the contact map overlap problem; pp. 410–423. [Google Scholar]
- Jiang W, et al. Bridging the information gap: computational tools for intermediate resolution structure interpretation. J. Mol. Biol. 2001;308:1033–1044. doi: 10.1006/jmbi.2001.4633. [DOI] [PubMed] [Google Scholar]
- Jiang W, et al. Backbone structure of the infectious e15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–1134. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- Kawabata T. Multiple subunit fitting into a low-resolution density map of a macromolecular complex using a gaussian mixture model. Biophys. J. 2008;95:4643–4658. doi: 10.1529/biophysj.108.137125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs JA, et al. Fast rotational matching of rigid bodies by fast Fourier transform acceleration of five degrees of freedom. Acta Crystallogr. D. Biol. Crystallogr. 2003;59:1371–1376. doi: 10.1107/s0907444903011247. [DOI] [PubMed] [Google Scholar]
- Lasker K, et al. Inferential optimization for simultaneous fitting of multiple components into a cryoEM map of their assembly. J. Mol. Biol. 2009;388:180–194. doi: 10.1016/j.jmb.2009.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, et al. Alignment of molecular networks by integer quadratic programming. Bioinformatics. 2007;23:1631–1639. doi: 10.1093/bioinformatics/btm156. [DOI] [PubMed] [Google Scholar]
- Lindert S, et al. Hybrid approaches: applying computational methodsin cryo-electronmicroscopy. Curr. Opin. Struc. Biol. 2009;19:218–225. doi: 10.1016/j.sbi.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinetz TM, et al. ‘Neural-gas’ network for vector quantization and its application to time-series prediction. IEEE Trans. Neural Netw. 1993;4:558–569. doi: 10.1109/72.238311. [DOI] [PubMed] [Google Scholar]
- Navaza J, et al. On the fitting of model electron densities into EM reconstructions: a reciprocal-space formulation. Acta Crystallogr. D. Biol. Crystallogr. 2002;58:1820–1825. doi: 10.1107/s0907444902013707. [DOI] [PubMed] [Google Scholar]
- Ranson NA, et al. ATP-bound states of GroEL captured by cryo-electron microscopy. Cell. 2001;107:869–879. doi: 10.1016/s0092-8674(01)00617-1. [DOI] [PubMed] [Google Scholar]
- Rath BK, et al. Fast 3D motif search of EM density maps using a locally normalized cross-correlation function. J. Struct. Biol. 2003;144:95–103. doi: 10.1016/j.jsb.2003.09.029. [DOI] [PubMed] [Google Scholar]
- Robinson CV, et al. The molecular sociology of the cell. Nature. 2007;450:973–982. doi: 10.1038/nature06523. [DOI] [PubMed] [Google Scholar]
- Roseman AM. Docking structures of domains into maps from cryo-electron microscopy using local correlation. Acta Crystallogr. D. 2000;56:1332–1340. doi: 10.1107/s0907444900010908. [DOI] [PubMed] [Google Scholar]
- Rossmann MG. Fitting atomic models into electron-microscopy maps. Acta Crystallogr. D. 2000;56:1341–1349. doi: 10.1107/s0907444900009562. [DOI] [PubMed] [Google Scholar]
- Rossmann MG, et al. Combining electron microscopic with X-ray crystallographic structures. J. Struct. Biol. 2001;136:190–200. doi: 10.1006/jsbi.2002.4435. [DOI] [PubMed] [Google Scholar]
- Rusinkiewicz S, Levoy M. Internal Conference on 3-D Digital Imaging and Modeling. Quebec City, Canada: IEEE Computer Society; 2001. Efficient variants of the ICP algorithm; pp. 145–152. [Google Scholar]
- Topf M, et al. Structural characterization of components of protein assemblies by comparative modeling and electron cryo-microscopy. J. Struct. Biol. 2005;149:191–203. doi: 10.1016/j.jsb.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Topf M, et al. Fitting and refinement of atomic structures guided by cryoEM density structure. Structure. 2008;16:295–307. doi: 10.1016/j.str.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Muriel J.A, et al. Flexible fitting in 3D-EM guided by the structural variability of protein superfamilies. Structure. 2006;14:1115–1126. doi: 10.1016/j.str.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Volkmann N, Hanein D. Docking of atomic models into reconstructions from electron microscopy. Meth. Enzymol. 2003;374:204–225. doi: 10.1016/S0076-6879(03)74010-5. [DOI] [PubMed] [Google Scholar]
- Wriggers W, et al. Selforganizing neural networks bridge the biomolecular resolution gap. J. Mol. Biol. 1998;284:1247–1254. doi: 10.1006/jmbi.1998.2232. [DOI] [PubMed] [Google Scholar]
- Wriggers W, Chacon P. Modeling tricks and fitting techniques for multiresolution structures. Structure. 2001;9:779–788. doi: 10.1016/s0969-2126(01)00648-7. [DOI] [PubMed] [Google Scholar]
- Wriggers W, et al. Situs: a package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 1999;125:185–195. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- Ye Y. On affine-scaling algorithm for nonconvex quadratic programming. Math. Program. 1992;56:285–300. [Google Scholar]
- Yu X, et al. 3.88 Å structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.