Abstract
Motivation: Tiled serial section Transmission Electron Microscopy (ssTEM) is increasingly used to describe high-resolution anatomy of large biological specimens. In particular in neurobiology, TEM is indispensable for analysis of synaptic connectivity in the brain. Registration of ssTEM image mosaics has to recover the 3D continuity and geometrical properties of the specimen in presence of various distortions that are applied to the tissue during sectioning, staining and imaging. These include staining artifacts, mechanical deformation, missing sections and the fact that structures may appear dissimilar in consecutive sections.
Results: We developed a fully automatic, non-rigid but as-rigid-as-possible registration method for large tiled serial section microscopy stacks. We use the Scale Invariant Feature Transform (SIFT) to identify corresponding landmarks within and across sections and globally optimize the pose of all tiles in terms of least square displacement of these landmark correspondences. We evaluate the precision of the approach using an artificially generated dataset designed to mimic the properties of TEM data. We demonstrate the performance of our method by registering an ssTEM dataset of the first instar larval brain of Drosophila melanogaster consisting of 6885 images.
Availability: This method is implemented as part of the open source software TrakEM2 (http://www.ini.uzh.ch/∼acardona/trakem2.html) and distributed through the Fiji project (http://pacific.mpi-cbg.de).
Contact: tomancak@mpi-cbg.de
1 INTRODUCTION
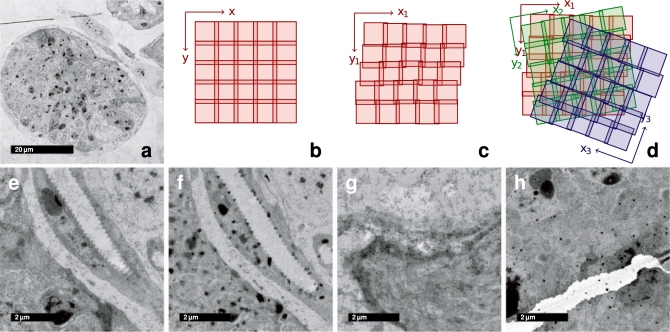
Transmission electron microscopy (TEM) offers currently the highest available resolution to investigate the structure of biological samples. The superior resolution of TEM is classically used to reveal subcellular structures such as organelles and some of the largest macromolecular assemblies inside cells. For anatomical studies on the tissue level, TEM finds most applications in neurobiology to reveal neuronal connectivity maps that underlie the function of the nervous system. The reconstruction of the complete neuronal connectivity map in Caenorhabditis elegans remains the pinnacle of these studies (White et al., 1986). On the nanometer scales, brains are gigantic structures and many overlapping images must be assembled as a mosaic to cover a substantial portion of the tissue. Moreover, serial sectioning is required to capture the volume of the specimen (Fig. 1a–d).
Fig. 1.
Properties of ssTEM image mosaics. (a) Low magnification overview of a single section mosaic consisting of 9 × 9 registered tiles. (b) Each section is imaged as a sequence of overlapping tiles that is assumed to be a regular grid. (c) Each tile's true pose in the local section coordinate frame is affected by odometry errors of the microscope, that are propagated over consecutive tiles. (d) The relative pose of two consecutive sections is arbitrarily shifted and rotated. Clean sections are relatively rare (e); typically sections contain artifacts such as dirt (f), staining precipitate (g) and folds or cracks (h).
Digital imaging and high-precision beam or specimen shifting equipment (Suloway et al., 2005) simplifies the reconstruction of mosaics within one section by providing a good initial estimate of the tile configuration (Fig. 1b). However, due to the minute distances involved the precision of the instruments is insufficient for seamless stitching of the tiles (Fig. 1c). Adjacent sections are rotated and translated arbitrarily with respect to each other (Fig. 1d). Physical sectioning, the subsequent manipulation of the sections by contrast gaining staining procedures and imaging with the electron beam introduce significant distortions and artifacts into the captured images (Fig. 1e–h). The goal in reconstruction of serial section TEM (ssTEM) data is to identify the configuration of all tiles that best preserves both the continuity and geometry of 3D structures in the specimen making it amenable for subsequent segmentation and analysis. Particularly in neurobiology, it is important to preserve the continuity of axons across large distances and the ability to distinguish and localize synaptic connections between neurons.
Anderson et al. (2009) propose a registration pipeline for large ssTEM datasets based on correlation of image intensities within and across sections. They are able to identify overlapping tiles in unordered sets automatically, mosaic them by propagating pairwise translation and refine this initial layout by warping each tile to its preceding partners. The resulting section mosaics are registered relative to each other by searching a rigid transformation initially and subsequently warping one section to the other. This solution guarantees continuity of 3D structures by maximizing the overlap of similar image content in adjacent sections. On the other hand, it cannot preserve geometrical properties because the unavoidable registration error is accumulated with each consecutive registration step.
Instead of using all image intensities, it is possible to register two or more images using corresponding salient points as a statistic about the image content as proposed by Brown and Lowe (2003) for panorama stitching. Reducing the problem from full resolution image content to a relatively small number of corresponding points simplifies the estimation of the globally optimal configuration for a large number of tiles.
State of the art techniques (Bay et al., 2008; Lowe, 2004; Matas et al., 2002; Mikolajczyk and Schmid, 2004; Tuytelaars and Van Gool, 2004) allow both automatic detection of interest points and extraction of affine invariant local descriptors for these points. Affine invariant matching becomes nearest neighbor search in a local feature descriptor space. It was shown by Mikolajczyk and Schmid (2003) that the local descriptor as used in the Scale Invariant Feature Transform (SIFT; Lowe, 2004) outperforms competing techniques in both distinctiveness and robustness with respect to significant image deformation and other disturbing artifacts.
In this article, we present a fully automatic registration method for large ssTEM image mosaics that globally minimizes the registration error. We identify overlapping images by corresponding image content within and across sections using SIFT features and a geometric consistency constraint. We approximate the non-linear transformation between adjacent sections by an independent rigid-body transformation (rotation and translation) for each image tile of the section mosaics. In a TEM section series, none of the sections can be considered non-deformed and thus, no section can serve as a template for registration. Therefore, we define the registration problem to be template free and, instead, explicitly minimize the non-rigid deformation applied to all sections. This is achieved by globally minimizing the square displacement of all corresponding SIFT landmarks within and across sections. By this means, the resulting tile configuration is non-rigid but as-rigid-as-possible. In order to evaluate the performance of this approach, we developed a synthetic ground truth dataset that is designed to mimic the properties of ssTEM data. We were able to register 6885 images from 85 serial sections through the Drosophila first instar larval brain; each section consisting of 9 × 9 tiles of 2048 × 2048 px. The registered dataset serves as a starting point for characterizing the fine architecture of this large brain at unprecedented resolution.
2 METHODS
2.1 Definition of the task
Specimens prepared for serial sectioning are typically embedded in a block of rigid medium such as resin, plastic or ice. Using a microtome, ultrathin consecutive slices are cut from the rigidly mounted block. The slices float on the surface of a liquid in the ‘knife boat’ and are manually picked up onto a TEM grid. While the section thickness may vary slightly within and across sections, in this work, we consider sections to be planar and of constant thickness. In reality, the cross-section variation can be minimized by careful operation of the microtome, and, as both the specimen itself and the knife of the microtome are rigidly mounted, intrasection variation is compensated throughout the section series by complementary variation in other sections.
Each section is imaged as a set of overlapping tiles. For each tile t, the microscope registers a 2D translation vector tt=(u, v)T in a relative section reference frame ℝ2s, whereas the section index s is given by the human operator. These coordinates are inaccurate due to measurement errors θt=(u, v)T (Fig. 1b and c). A section's pose relative to its adjacent sections is an unknown rigid transformation Rs (Fig. 1d). Moreover, sectioning, preparation after sectioning and even the imaging process itself induce noticeable deformations of both sections as a whole (Ds) as well as on the scale of single tiles (Dt) (Fig. 1a and e–h).
Therefore, the task is to reconstruct a 3D image volume from serial sections that have to be reconstructed from single tiles while compensating for noticeable deformation within and across sections. Particularly, the deformation–compensation poses the question how to invert the largely unknown transforming process that mapped a 3D coordinate (x, y, z)T in scene space ℝ3S into a 2D coordinate (u, v)T in tile space ℝ2t. Assuming planar sections of constant thickness as mentioned above, we attach scene and section spaces by a permutation Pzs between the scene dimension z and section index s. Now this mapping shows as
| (1) |
with RsDtDs and (x, y, z)T being unknown. Without precise knowledge about the deforming process DtDs, the appropriate solution is that with minimal non-rigid deformation for all sections, a model being as-rigid-as-possible. This coincides with the researcher's request not to introduce unmotivated artificial deformation to the image data that would adulterate his observations. Declaring a tile being the unit of rigidity, we approximate RsDtDs+tt by a rigid-per-tile transformation Rt such that
| (2) |
with ωt being the transformation error introduced by the approximation. Maximal consistency relative to the scene is guaranteed by minimizing the remaining error ϵt=ωt+θt.
Without knowing the scene, it is impossible to estimate ϵt directly. However, consistency with respect to the scene implies consistency relative to overlapping tiles. Let T be the set of all tiles t∈T and R be the set of all rigid transformations Rt∈R, then the best configuration R is that minimizing the sum of all relative transfer errors of pairwise overlapping tiles:
| (3) |
Using landmark correspondences as a statistic about corresponding image content, ϵto is the sum of all square correspondence point displacements. Let Cto be a set of landmark correspondences (x, y) between two tiles t∈T and o∈T∖{t} with x∈t and y∈o, then the best configuration R is that minimizing the sum of all square correspondence displacements:
| (4) |
with f∈T being a fixed tile that defines the scene reference frame. All tiles must represent one single interconnected graph being pairwise connected by sets of landmark correspondences.
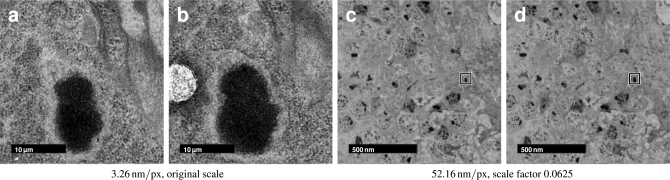
This optimization task requires a reliable landmark correspondence detector. Within a section, the appropriate interest point detector needs to be covariant to translation and robust against small amounts of non-rigid deformation as the overlapping parts of two consecutive tiles show the projection of the same tissue slightly deformed by heat during imaging. For cross-section matching, the appropriate interest point detector needs covariance to rotation as well as an automatism to detect only structures that appear similar, since two adjacent tiles show different portions of the tissue and are related by a rigid transformation and some plastic deformation. In ssTEM data, depending on section thickness, corresponding locations can be identified by local appearance of large enough structures sectioned perpendicularly. Such structures appear on different scales (for instance, mitochondria 500 nm, nuclei 2 μm and neuropile compartments 40 μm) and thus detecting structures with large (x, y)T-scale raises the chance to detect the same structures in adjacent sections (Fig. 2).
Fig. 2.
Similarity of serial sections depends on the scale of observation. Two consecutive 60 nm sections from the registered Drosophila first instar larval brain dataset showing a part of the cortex and the neuropile. The same region is shown with a resolution of 3.26 nm/px (a,b) and 52.16 nm/px (c,d). It is clearly visible, that the latter, having approximately isotropic resolution, shows significant similarity across sections while it is hard to identify corresponding pixels in the higher resolution example.
For its scale invariant nature and robustness, we decided to use SIFT (Lowe, 2004) for both intra- and intersection landmark correspondence detection.
2.2 SIFT
SIFT (Lowe, 2004) detects blobs of arbitrary size as interest points using the Difference of Gaussian detector, estimates one or more dominant orientations for each detection and extracts a scale and orientation invariant local descriptor for each. Such blob-like structures are common in most textured images including TEM data. Typically, we detect several hundreds to thousands of interest points in a TEM micrograph of 2000 × 2000 px image size.
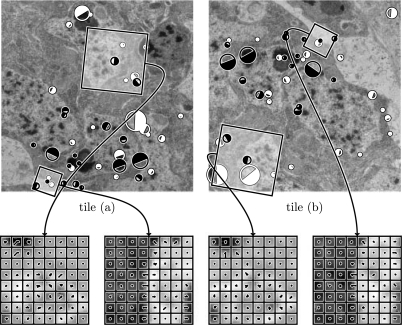
The detection's size, orientation and 2D location define a local coordinate frame for extraction of an invariant descriptor. We found empirically, that a larger descriptor built from 8 × 8 local histograms with eight bins each (512 dimensions) provides substantially better matching results for our data than the originally proposed 4 × 4 × 8 descriptor. The smaller descriptors were proposed for natural images where viewpoint change and occlusion play a significant role. In TEM data, where two regions are related only by a rigid transformation with some deformation, a larger descriptor region necessarily increases the distinctiveness (Fig. 3).
Fig. 3.
SIFT-feature correspondences in two overlapping tiles from adjacent sections. 1003 feature candidates were extracted in tile (a), 971 in tile (b). 41 correspondence pairs were identified by local feature descriptor matching, 19 of them are true matches consistent to common transformation model. False matches are displayed in white, true matches in black. The size of the circles is proportional to the features scale, the filled part visualizes a feature's orientation. Two true correspondence pairs are selected as an example for the local SIFT-descriptor. The values of the local histogram bins are shown as combs on top of the local region the descriptor is extracted from.
Interest point correspondence candidates are identified by nearest neighbor matching in the local descriptor space. As suggested by Lowe (2004), good candidates are those that are significantly better than the next nearest neighbor. Significantly better means that the ratio between the Euclidean distances to the nearest and the next nearest neighbor is smaller than a given threshold. Empirically, we identified a threshold of 0.92 for TEM images to yield a reasonable number of correspondence candidates. Since real-time performance is not required, we identify the exact nearest neighbor by exhaustive search.
Local descriptor matching results in a significant number of false correspondences. We use the Random Sample Consensus (RANSAC) (Fischler and Bolles, 1981) to separate true correspondences that behave consistently with respect to a rigid transformation up to a maximal correspondence displacement ϵmax (see Algorithm 1). For estimation of the optimal rigid transformation by means of least square correspondence displacement, we use the closed form solution described by Schaefer et al. (2006). Significant deformation requires ϵmax to be relatively tolerant thus accepting some false correspondences. We filter these with a robust regression scheme that iteratively removes correspondences with a displacement larger than 3 × the median displacement. For details, see Saalfeld and Tomančák (2008).

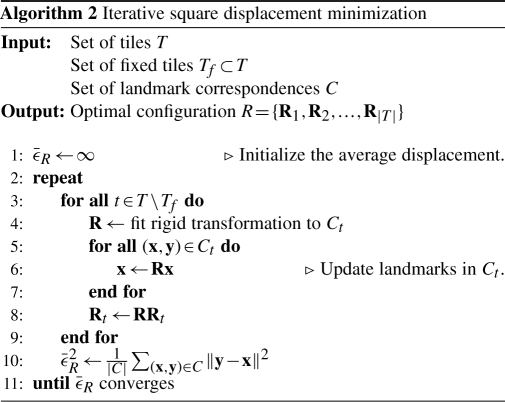
2.3 Global optimization of the tile configuration
Having the set of true landmark correspondences identified successfully, the optimal configuration R of all tiles has to be estimated. As shown in Equation (4), this is the configuration with minimal square correspondence displacements.
We minimize this term using an iterative optimization scheme (see Algorithm 2). In case that prior knowledge about the gross configuration of all tiles is available, the minimization is initialized with this configuration. Otherwise all tiles start from Rt=I. In each iteration i, the optimal Rt for each single tile t∈T∖{f} relative to the current configuration is estimated using the closed form least square solution by Schaefer et al. (2006) and applied to all landmark coordinates in this tile. The scheme terminates on convergence of the average correspondence displacement 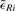 that is estimated after each iteration. As convergence criteria, we require
that is estimated after each iteration. As convergence criteria, we require 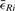 to have a low absolute slope
to have a low absolute slope 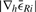 and an absolute value below some threshold given by the user. It is crucial to prevent being trapped in local minima or at long plateaus. We address this effectively by requiring
and an absolute value below some threshold given by the user. It is crucial to prevent being trapped in local minima or at long plateaus. We address this effectively by requiring 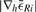 to be very low for all h from a set of decreasing lengths. The maximal length h of a plateau or local valley, the maximal total number of iterations and the maximal accepted remaining error are user-defined parameters with empirically estimated defaults suggested.
to be very low for all h from a set of decreasing lengths. The maximal length h of a plateau or local valley, the maximal total number of iterations and the maximal accepted remaining error are user-defined parameters with empirically estimated defaults suggested.
2.4 Notes on implementation
The described registration algorithm including SIFT, RANSAC, closed form least square error solutions for a variety of transformation models, the robust regression scheme and the iterative optimizer are implemented using the ? programming language and provided as an Open Source library through the ImageJ distribution Fiji (Schindelin, 2008). The registration procedure is included in the software toolkit TrakEM2 (Cardona, 2006) for management, registration and analysis of massive serial section microscopy datasets and, by that, in daily use in a large number of laboratories allover the world.
3 RESULTS
3.1 Overview of the registration algorithm
Principally, the registration procedure consists of extracting landmarks from all tiles, identifying landmark correspondences between tile pairs and estimating the tile configuration that minimizes the sum of all square correspondence displacements as shown in Algorithm 3.
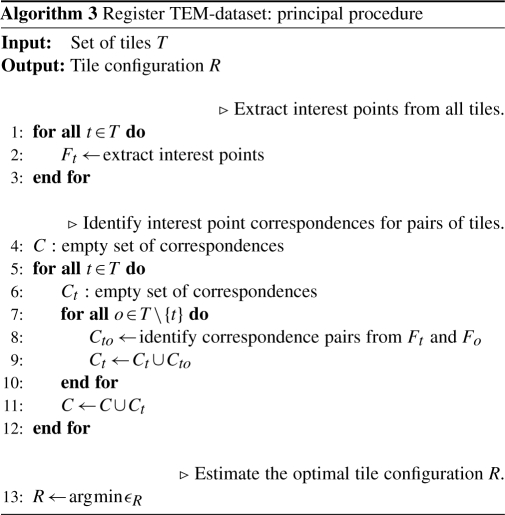
Practically, since adjacent sections are related by a rigid transformation Rs plus some non-rigid deformation Ds that is to be compensated, we would like to use the transformation Rs as initialization for the sought after configuration R and as a hint which tiles to exclude from pairwise feature comparison. Unfortunately, single sections do not always represent a single landmark-interconnected graph of tiles, while the whole volume eventually does. This is a typical scenario when imaging tree-like structures and the operator following only the branches ignoring the rest of the volume. That is, instead of the section as a whole, all graphs of a section have to be tested against all graphs present in the previous section. Two graphs that can be registered to each other are then joined into a single one by matching and filtering the features of overlapping tiles. Eventually, this will result in one single graph representing the whole volume. After joining two graphs, the configuration of the resulting graph is optimized thus sequentially building up the optimal global configuration from optimal intermediate configurations.
We applied this registration approach to test sets of TEM data of the Drosophila first instar larval brain sectioned into 60 nm sections imaged at 3.26 nm/px resolution. We developed a visualization of the global optimization progress where for each iteration the corresponding landmarks and the residual transfer error between them are highlighted by green dots and red lines, respectively. Movie 1 available as supplementary on-line material shows the progress of the optimization for a single section dataset consisting of 6 × 6 tiles.1 It highlights the ability of the method to correctly place even tiles that contain mostly background (lower left corner tile) and gain very little SIFT feature detections, as long as these are attached to the graph. If a tile lacks SIFT correspondences altogether, it represents an independent graph on its own. The user can choose to drop or hide all but the largest graph from the dataset. Movie 2 shows the progress of the optimization on three serial sections imaged as 4 × 4 tile mosaics demonstrating that the procedure for multiple section is essentially the same as for a single section.2
3.2 Quantitative assessment of registration results on synthetic evaluation data
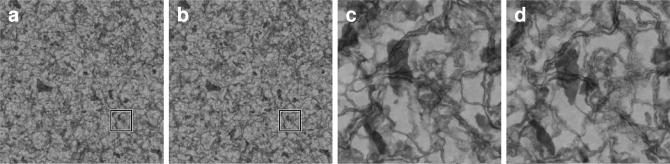
We created an artificial dataset with the ray-tracing program POV-ray (Cason et al., 2007) that allows to define the interior of a volume as a density pattern based on an arbitrary volumetric function, where high density scatters more light than low density (Fig. 4).3 This simulates the imaging process in the Transmission Electron Microscope where structures with a higher density of heavy-metal atoms scatter more electrons than structures with lower density and appear darker at the screen. The interior of the evaluation dataset contains filamentous and ‘blob’-like structures, both over three scale octaves with multiscale turbulence. In this way, it mimics typical structures in the stained tissue, like membranes, nucleoli and mitochondria.
Fig. 4.
Artificially generated evaluation dataset. The dataset simulates thin transilluminated volumes of 20 px thickness with membrane- and blob-like structures at various scales. Structures are defined by volumetric density functions. Locations with higher density scatter more ‘light’ than those with lower density and appear darker in the shadow projection. Two adjacent sections are shown to visualize the cross-section change of visible structures. In (a,b), the full dataset (4096 × 4096 px) is shown at low magnification, an area of 512 × 512 px is marked and shown in (c,d) at high magnification.
To emulate the ssTEM data we generated 16 projections of 4096 × 4096 px each through consecutive volumes of 20 px thickness, which is roughly equivalent to the ratio of x, y versus z resolution in typical TEM data. We split the synthetic ground truth data into 1024 × 1024 px tiles with 102 px overlap resulting in 25 images per section. We ran our registration program on the resulting 400 images as if there was nothing known about the tiles poses other than the section index. The registration results in a stack that is anchored to a randomly selected tile in the world reference frame. Thus, missing comparable world coordinates, we evaluate the quality of the registration by estimating the coordinate translation that gives the minimal average displacement when applied to the registered stack. This identifies the world reference frame of the registered stack and gives an average registration error of a pixel relative to the original scene.
We select a random sample of 1000 locations in each tile and transfer all locations into the world reference frame using the true and the registered transformation model of the tile. The translation between registration and ground truth domain is that between the centroids of both point clouds. The average residual transfer error serves as a quantitative measure of the registration process. We estimated an average displacement  =4.14 px, SD σϵ=3.63 px and maximal displacement ϵmax=15.71 px for the registration result. These results demonstrate that, with the proposed method, we can both identify overlap in an unknown configuration of images and register them.
=4.14 px, SD σϵ=3.63 px and maximal displacement ϵmax=15.71 px for the registration result. These results demonstrate that, with the proposed method, we can both identify overlap in an unknown configuration of images and register them.
3.3 Registration of the Drosophila first instar larval brain ssTEM dataset
As an example of real biological data, we registered the Drosophila first instar larval brain ssTEM dataset consisting of 85 sections of 60 nm thickness covering lateral neuronal layers and part of the neuropile of the left hemisphere. Each section was imaged with TEM using a moving stage operated by the Leginon software as 9 × 9 tiles overlapping by ∼6%. Tiles have a size of 2048 × 2048 px and a resolution of 4.68 nm/px. All 6885 tiles were registered fully automatically. Intrasection configurations were initialized with the odometry data of the microscope. Correspondence estimation using SIFT descriptors and robust geometric consensus filters performed very well even in presence of significant changes in illumination and sharpness, dirty sections and significant gaps of up to six sections in the stack. Visual inspection of the registered data shows reliable continuity of biologically relevant structures such as axon bundles within and across sections both at low and high scales (Fig. 5a and b).4 Moreover, when the registered dataset is cut perpendicularly, the resulting image, whose lateral resolution is limited by section thickness, resembles electron microscopy data without major discontinuities suggesting that the registration was successful (Fig. 5c).5 For visualization and collaborative annotation, we present the registered dataset on-line through the CATMAID web interface (Saalfeld et al., 2009).6
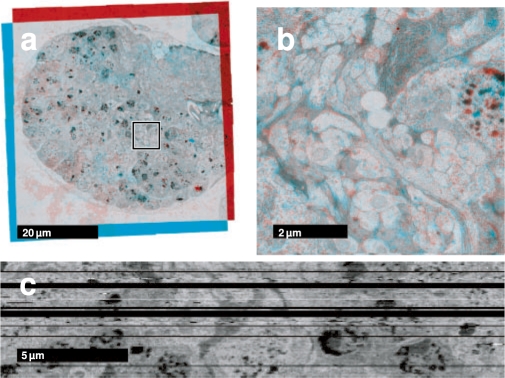
Fig. 5.
Drosophila first instar larval brain ssTEM dataset. Two consecutive registered sections from the dataset as red-cyan color merge. The diameter of the brain is ∼60 μm. (a) The section mosaics as a whole with the areas zoomed in (b) marked. (c) A single perpendicular section through the entire registered volume. Several sections were lost during sectioning and collecting onto the electron microscopy grid and shown here as black rows.
4 DISCUSSION
We presented a fully automated method for registration of large ssTEM image mosaics. The method identifies the optimal configuration of image tiles regardless whether or not an initial guess of the configuration is available. It is capable of correctly placing tiles with only minimal image content provided that this content is connected to the rest of the tile graph. Disconnected graphs of tiles are registered independently. Thus, the approach is ideally suited for alignment of ssTEM data generated either manually or using a robotic setup.
We use SIFT to identify corresponding landmarks and use them as a statistical sample for similar image content in the TEM images. The scale invariance of SIFT is well suited to capture the changing appearance of small structures at low scale levels and the presence of meaningful similar features at multiple scales. Larger, more distinctive local descriptors together with the requirement that correspondence candidates must be geometrically consistent yields reliably large sets of true matches.
The reduction of the registration task to a compact set of corresponding landmarks enables global minimization of square correspondence displacements for very large tile systems. Registering the 6885 tiles Drosophila larval brain dataset took ∼24 h on an Intel® Xeon® computer with two 2.66 GHz dual-core-CPUs and 8 GB of RAM, with the most time-consuming operation in the procedure being the exhaustive nearest neighbor search in the 512 D local descriptor space. Principally, there is no upper limit to the size of the dataset other than the available storage space.
The described methods for robust outlier removal and global optimization are applicable to a wide variety of microscopy image mosaicking tasks. We used it successfully to register light microscopy image mosaics including sets of overlapping 3D confocal image stacks (Preibisch et al., 2009b) and applied it to bead-based registration of Selective Plane Illumination Microscopy (SPIM; Huisken et al., 2004) datasets consisting of 3D stacks of the same specimen taken from different angles (Preibisch et al., 2009a). In Preibisch et al. (2009b), we identify the pairwise 3D translation between overlapping confocal image stacks by normalized cross-correlation and globally minimize the translational offset of large sets of overlapping stacks. In Preibisch et al. (2009a), we proposed the usage of fluorescent beads embedded in the rigid mounting medium as fiduciary markers. We use the local constellations of neighboring beads to automatically identify correspondences invariantly to 3D rotation and scale. The model for the geometric consistency constraint and global minimization of the square transfer errors is a 3D affine transformation with one of the 3D stacks serving as a template.
We generated a unique evaluation dataset designed to resemble the appearance of biological tissue in ssTEM data for quantitative comparison of the registration results. The evaluation framework can be used to assess the performance of any registration scheme that records its transformation model and could become the standard for objectively measuring the performance of various registration approaches. In future work, we will induce artificial deformation and variance in section thickness to the dataset in order to examine the limits of our registration method. As an alternative more realistic ground truth evaluation dataset, we propose to use Serial Block-Face Scanning Electron Microscopy data (Denk and Horstmann, 2004) where consecutive sections are aligned per definition.
The presented registration method is non-rigid but as-rigid-as-possible with a tile being the unit of rigidity for intrasection alignment. This delivers an ssTEM image dataset amenable to quantitative analysis, such as double dissector estimation of synaptic density (Geinisman et al., 1996), which require volumes to be as reliable as possible. On the other hand, rigid-per-tile registration cannot compensate large-scale deformation by tile displacement without introducing noticeable discontinuities at the tile borders. The solution to this problem lies in extending the approach to include arbitrary non-rigid deformation at scales below the tile level preserving the regularization in terms of local rigidity. That means deforming all images minimally. We are currently exploring various ideas of how to express this regularization for a completely non-rigid global registration. The evaluation dataset will be instrumental in assessing the performance of such approaches with respect to reflecting faithfully the ground truth data.
The globally optimal reconstruction of entire brains on TEM level will enable registration of 3D light microscopy data onto electron microscopy volumes. By that it will be possible to establish the connection between brain macro (neuronal lineages) and micro (synaptic connectivity) circuitry (Cardona et al., 2009).
ACKNOWLEDGEMENTS
We want to thank the HHMI Janelia Farm Research Campus for hosting the Janelia Hackathons 2007 and 2008, Mitya Chklovskii for hosting us and sharing ideas, and Johannes Schindelin for maintaining the Fiji platform.
Funding: DIGS-BB PhD stipend (to S.S.).
Conflict of Interest: none declared.
Footnotes
1Movie 1: http://fly.mpi-cbg.de/saalfeld/36.avi
2Movie 2: http://fly.mpi-cbg.de/saalfeld/48.avi
3Evaluation-dataset: http://fly.mpi-cbg.de/saalfeld/tem-evaluation
4Section series as a movie: http://fly.mpi-cbg.de/saalfeld/series.avi
5Resliced series as a movie: http://fly.mpi-cbg.de/saalfeld/resliced.avi
6Registered dataset on-line: http://fly.mpi-cbg.de/first-instar-brain
REFERENCES
- Anderson JR, et al. A computational framework for ultrastructural mapping of neural circuitry. PLoS Biol. 2009;7:e1000074. doi: 10.1371/journal.pbio.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bay H, et al. SURF: Speeded up robust features. Comput. Vis. Image Understand. 2008;110:346–359. [Google Scholar]
- Brown M, Lowe DG. ICCV '03: Proceedings of the Ninth IEEE International Conference on Computer Vision. Washington, DC: IEEE Computer Society; 2003. Recognising panoramas; pp. 1218–1225. [Google Scholar]
- Cardona A. Proceedings of the 1st ImageJ User and Developer Conference. Luxembourg: 2006. TrakEM2: an ImageJ-based program for morphological data mining and 3D modeling; pp. 18–19. May. [Google Scholar]
- Cardona A, et al. Drosophila brain development: Closing the gap between a macroarchitectural and microarchitectural approach. Cold Spring Harb. Symp. Quant. Biol. 2009 doi: 10.1101/sqb.2009.74.037. [Epub ahead of print, doi:10.1101/sqb.2009.74.037] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cason C, et al. POV-ray – the persistance of vision raytracer [release version 3.6.1]. Availabe at http://www.povray.org. (last accessed date August 8, 2007) [Google Scholar]
- Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2:1900–1909. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischler MA, Bolles RC. Random sample consensus: a paradigm for model fitting with applications to image analysis and automated cartography. Commun. ACM. 1981;24:381–395. [Google Scholar]
- Geinisman Y, et al. Unbiased stereological estimation of the total number of synapses in a brain region. J. Neurocytol. 1996;25:805–819. doi: 10.1007/BF02284843. [DOI] [PubMed] [Google Scholar]
- Huisken J, et al. Optical sectioning deep inside live embryos by Selective Plane Illumination Microscopy. Science. 2004;305:1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- Lowe DG. Distinctive image features from scale-invariant keypoints. Int. J. Comput. Vis. 2004;60:91–110. [Google Scholar]
- Matas J, et al. Robust wide baseline stereo from maximally stable extremal regions. In: Rosin PL, Marshall D, editors. Proceedings of the British Machine Vision Conference. Vol. 1. London: BMVA; 2002. pp. 384–393. [Google Scholar]
- Mikolajczyk K, Schmid C. A performance evaluation of local descriptors. International Conference on Computer Vision and Pattern Recognition. 2003;2:257–263. [Google Scholar]
- Mikolajczyk K, Schmid C. Scale and affine invariant interest point detectors. Int. J. Comput. Vis. 2004;60:63–86. [Google Scholar]
- Preibisch S, et al. Pluim J.PW, Dawant BM, editors. Bead-based mosaicing of single plane illumination microscopy images using geometric local descriptor matching. Proceedings of The International Society for Optical Engineering. 2009a;7259 [Google Scholar]
- Preibisch S, et al. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009b;25:1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband W. ImageJ: Image processing and analysis in Java [version 1.43l] Available at http://rsb.info.nih.gov/ij/ (last accessed date November 24, 2009) [Google Scholar]
- Saalfeld S, Tomančák P. Proceedings of the ImageJ User and Developer Conference. Luxembourg: 2008. Automatic landmark correspondence detection for ImageJ; pp. 128–133. [Google Scholar]
- Saalfeld S, et al. CATMAID: Collaborative annotation toolkit for massive amounts of image data. Bioinformatics. 2009;25:1984–1986. doi: 10.1093/bioinformatics/btp266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer S, et al. Image deformation using moving least squares. ACM Trans. on Graph. 2006;25:533–540. [Google Scholar]
- Schindelin JE. Proceedings of the ImageJ User and Developer Conference. Luxembourg: 2008. Fiji is just ImageJ—batteries included; pp. 99–104. [Google Scholar]
- Suloway C, et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Tuytelaars T, Van Gool L. Matching widely separated views based on affine invariant regions. Int. J. Comput. Vis. 2004;59:61–85. [Google Scholar]
- White JG, et al. The structure of the nervous system of the nematode Caenorhaditis elegans. Phil. Trans. R. Soc. Lond. Ser. B Biol. Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]