Abstract
Motivation: Biclustering of transcriptomic data groups genes and samples simultaneously. It is emerging as a standard tool for extracting knowledge from gene expression measurements. We propose a novel generative approach for biclustering called ‘FABIA: Factor Analysis for Bicluster Acquisition’. FABIA is based on a multiplicative model, which accounts for linear dependencies between gene expression and conditions, and also captures heavy-tailed distributions as observed in real-world transcriptomic data. The generative framework allows to utilize well-founded model selection methods and to apply Bayesian techniques.
Results: On 100 simulated datasets with known true, artificially implanted biclusters, FABIA clearly outperformed all 11 competitors. On these datasets, FABIA was able to separate spurious biclusters from true biclusters by ranking biclusters according to their information content. FABIA was tested on three microarray datasets with known subclusters, where it was two times the best and once the second best method among the compared biclustering approaches.
Availability: FABIA is available as an R package on Bioconductor (http://www.bioconductor.org). All datasets, results and software are available at http://www.bioinf.jku.at/software/fabia/fabia.html
Contact: hochreit@bioinf.jku.at
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Recent technologies such as the Affymetrix array plates and next-generation sequencing open up new possibilities for high-throughput expression profiling. These technologies in turn require advanced analysis tools to extract knowledge from the huge amount of data. If the experimental conditions are known, supervised techniques such as support vector machines are suitable to extract the dependencies between conditions and gene expression or to identify condition-indicative genes. However, conditions may not be known or biologists and medical researchers are interested in dependencies within or across conditions. For instance, it could be possible to refine pathways across conditions or to identify new subgroups within one condition. For these tasks, unsupervised methods such as clustering are required, which are usually insufficient, because samples may only be similar on a subset of genes and vice versa. In drug design, for example, researchers want to reveal how compounds affect gene expression; the effects of compounds, however, may be similar only on a subgroup of genes. Under such circumstances, biclustering is the proper unsupervised analysis technique.
A bicluster in a transcriptomic dataset is a pair of a gene set and a sample set for which the genes are similar to each other on the samples and vice versa. If multiple pathways are active in a sample, it belongs to different biclusters. If a gene participates in different pathways for different conditions, it belongs to different biclusters, too. Thus, biclusters can overlap.
A survey of biclustering approaches has been given by Madeira and Oliveira (2004). In principle, there exist four categories of biclustering methods: (1) variance minimization methods, (2) two-way clustering methods, (3) motif and pattern recognition methods and (4) probabilistic and generative approaches. Transcriptomic data are usually supplied as a matrix, where each gene corresponds to one row and each sample to one column; the matrix entries themselves are the expression levels.
Variance minimization methods: define clusters as blocks in the matrix with minimal deviation of their elements. This definition has been already considered by Hartigan (1972) and extended by Tibshirani et al. (1999). The δ-cluster methods search for blocks of elements having a deviation (‘variance’) below δ. One example are δ-ks clusters (Califano et al., 2000), where the maximum and the minimum of each row need to differ less than δ on the selected columns. A second example are δ-pClusters (Wang et al., 2002), which are defined as 2 × 2 submatrices with pairwise edge differences less than δ. A third example are the Cheng and Church (2000) δ-biclusters having a mean squared error below δ after fitting an additive model with a constant, a row and a column effect. FLexible Overlapped biClustering (FLOC; Yang et al., 2005) extend Cheng–Church δ-biclusters by dealing with missing values via an occupancy threshold θ and by using both l1 and l2 norms.
Two-way clustering methods apply conventional clustering to the columns and rows and (iteratively) combine the results. Coupled Two-Way Clustering (CTWC; Getz et al., 2000) iteratively performs standard clustering of the rows (columns) using previously constructed columns (rows) clusters as features. Also Interrelated Two-Way Clustering (ITWC; Tang et al., 2001) using k-means and Double Conjugated Clustering (DCC; Busygin et al., 2002) using self-organizing maps combine column and row clustering.
Motif and pattern recognition methods define a bicluster as samples sharing a common pattern or motif. To simplify this task, some methods discretize the data in a first step, such as xMOTIF (Murali and Kasif, 2003) or Bimax (Prelic et al., 2006), which even binarizes the data and searches for blocks with an enrichment of ones. Order-Preserving SubMatrices (OPSM; Ben-Dor et al., 2003) searches for blocks having the same order of values in their columns. Using partial models, only the column order on subsets must be preserved. Spectral clustering (SPEC; Kluger et al., 2003) performs a singular value decomposition of the data matrix after normalization. SPEC extracts columns (samples) with the same conserved gene expression pattern using the fact that they are linearly dependent and span a subspace associated with a certain singular value. The Iterative Signature Algorithm (ISA; Ihmels et al., 2004) selects samples that have a given gene signature and then uses these samples to define a new sample signature. This sample signature, in turn, is used to select genes and to define a new gene signature. For each bicluster to be extracted, this process is initialized by a randomly selected binary gene signature and repeated iteratively. A related approach uses a Hough transform for identifying groups of linearly dependent genes and samples (Gan et al., 2008). Contiguous column coherent (CCC biclustering; Madeira and Oliveira, 2009; Madeira et al., 2010) is a method for gene expression time series, which finds patterns in contiguous columns.
Probabilistic and generative methods use model-based techniques to define biclusters. Statistical-Algorithmic Method for Bicluster Analysis (SAMBA; Tanay et al., 2002) uses a bi-partitioned graph, where both conditions and genes are nodes. An edge from a gene to a condition means that the gene responds to the condition. With a probabilistic objective, subgraphs are found that have a significantly higher connectivity than the overall graph. In another approach, Sheng et al. (2003) use Gibbs sampling to estimate the parameters of a simple frequency model for the expression pattern of a bicluster. However, the data must first be discretized and then only one bicluster with constant column values at each step can be extracted. Probabilistic Relational Models (PRMs; Getoor et al., 2002) and their extension ProBic (Van den Bulcke, 2009) are fully generative models that combine probabilistic modeling and relational logic. Another generative approach is cMonkey (Reiss et al., 2006), which models biclusters by Markov chain processes. Both PRMs and cMonkey are able to integrate non-transcriptomic data sources.
In the plaid model family (Lazzeroni and Owen, 2002), the i-th bicluster is extracted by row and column indicator variables ρki and κij. The values of each bicluster are explained by a general additive model θkij = μi+αki+βij. Parameters are estimated by a least square fit. Gu and Liu (2008) generalized the plaid models to fully generative models called Bayesian BiClustering model (BBC). To avoid the high percentage of overlap in the plaid models, BBC constrains the overlapping of biclusters to only one dimension. Further it allows different error variances per bicluster. Caldas and Kaski (2008) also extended the plaid model to a fully generative model using a Bayesian framework and found that the plaid model is equivalent to the PRM model for specific parameters.
The latter models (Caldas and Kaski, 2008; Gu and Liu, 2008) are generative models which have the advantage that (i) they select models using well-understood model selection techniques such as maximum likelihood, (ii) hyperparameter selection methods (e.g. to determine the number of biclusters) can rely on the Bayesian framework, (iii) signal-to-noise ratios can be computed, (iv) they can be compared with each other via the likelihood or posterior, (v) tests such as likelihood ratio test are possible and (vi) they produce a global model to explain all data. These models are additive and assume that all effects are Gaussian to utilize Gibbs sampling for parameter estimation. However, after prefiltering, real microarray datasets are not Gaussian distributed and have heavy tails (Hardin and Wilson, 2009), even after log transformation. This can be seen in Supplementary Figures S8, S9 and S19 for gene expression datasets. In this article, we propose a generative multiplicative model tailored to the special characteristics of gene expression data.
This article is organized as follows. Section 2 introduces the multiplicative bicluster model class. Section 3 describes the model selection (training) algorithm for the new model class. Section 4 highlights how biclusters can be ranked according to the information they contained about the data. Section 5 describes how to extract bicluster members from our new models. Finally, Section 6 provides an experimental evaluation of the new method.
2 THE FABIA MODEL
We propose a multiplicative model class for analyzing gene expression datasets for several reasons. First, a multiplicative model allows to model heavy tailed data, as observed in gene expression. Second, it can relate the strength of gene expression patterns to characteristics of the induced condition such as elapsed time or concentration of compounds. After log transformation, exponential dynamics such as decay (mRNA or compound) or saturation can also be modeled. Note that supervised multiplicative models, e.g. support vector machines, were successfully applied to log-transformed gene expression datasets. Further, artificial multiplicative effects are introduced during data preprocessing, for example, if expression values are standardized, then variations stemming from noise scale the signal.
We assume that the gene expression dataset is preprocessed and filtered for genes that contain a signal (e.g. informative call or signal strength). The resulting data is given as a data matrix X ∈ ℝn×l, where every row corresponds to a gene and every column corresponds to a sample; the value xkj corresponds to the expression level of the k-th gene in the j-th sample. The matrix X is the input to biclustering methods.
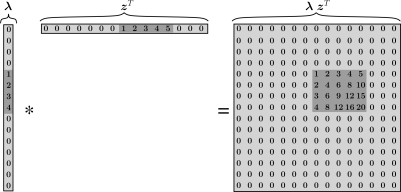
We define a bicluster as a pair of a row (gene) set and a column (sample) set for which the rows are similar to each other on the columns and vice versa. In a multiplicative model, two vectors are similar if one is a multiple of the other, that is, the angle between them is zero or, as realization of random variables, their correlation coefficient is (minus) one. It is clear that such a linear dependency on subsets of rows and columns can be represented as an outer product λ zT of two vectors λ and z. The vector λ corresponds to a prototype column vector that contains zeros for genes not participating in the bicluster, whereas z is a vector of factors with which the prototype column vector is scaled for each sample; clearly z contains zeros for samples not participating in the bicluster. Vectors containing many zeros or values close to zero are called sparse vectors. Figure 1 visualizes this representation by sparse vectors schematically.
Fig. 1.
The outer product λ zT of two sparse vectors results in a matrix with a bicluster. Note that the non-zero entries in the vectors are adjacent to each other for visualization purposes only.
The overall model for p biclusters and additive noise is
| (1) |
where ϒ ∈ ℝn×l is additive noise; λi ∈ ℝn and zi ∈ ℝl are the sparse prototype vector and the sparse vector of factors of the i-th bicluster, respectively. The second formulation above holds if Λ ∈ ℝn×p is the sparse prototype matrix containing the prototype vectors λi as columns and Z ∈ ℝp×l is the sparse factor matrix containing the transposed factors ziT as rows. Note that Equation (1) formulates biclustering as sparse matrix factorization.
According to Equation (1), the j-th sample xj, i.e. the j-th column of X, is
| (2) |
where εj is the j-th column of the noise matrix ϒ and 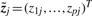 denotes the j-th column of the matrix Z. Recall that ziT = (zi1,…, zil) is the vector of values that constitutes the i-th bicluster (one value per sample), while
denotes the j-th column of the matrix Z. Recall that ziT = (zi1,…, zil) is the vector of values that constitutes the i-th bicluster (one value per sample), while 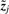 is the vector of values that contribute to the j-th sample (one value per bicluster).
is the vector of values that contribute to the j-th sample (one value per bicluster).
The formulation in Equation (2) facilitates a generative interpretation by a factor analysis model with p factors (Everitt, 1984)
| (3) |
where x is the observation, Λ is the loading matrix, 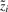 is the value of the i-th factor,
is the value of the i-th factor, 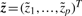 is the vector of factors and ε ∈ ℝn is the additive noise. Standard factor analysis assumes: the noise is independent of
is the vector of factors and ε ∈ ℝn is the additive noise. Standard factor analysis assumes: the noise is independent of 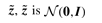 -distributed and ε is 𝒩(0, Ψ)-distributed (the covariance matrix Ψ ∈ ℝn×n is diagonal—expressing independent Gaussian noise). The parameter Λ explains the dependent (common) and Ψ the independent variance in the observations x. Additive noise in gene expression is normally distributed (Hochreiter et al., 2006).
-distributed and ε is 𝒩(0, Ψ)-distributed (the covariance matrix Ψ ∈ ℝn×n is diagonal—expressing independent Gaussian noise). The parameter Λ explains the dependent (common) and Ψ the independent variance in the observations x. Additive noise in gene expression is normally distributed (Hochreiter et al., 2006).
That the covariance matrix for  is the unit matrix means that the biclusters should not be correlated. This assumption ensures that one true bicluster in the data will not be divided into dependent small model biclusters—thereby ensuring maximal model biclusters. Note, however, that this assumption still allows for overlapping biclusters.
is the unit matrix means that the biclusters should not be correlated. This assumption ensures that one true bicluster in the data will not be divided into dependent small model biclusters—thereby ensuring maximal model biclusters. Note, however, that this assumption still allows for overlapping biclusters.
Standard factor analysis does not consider sparse factors and sparse loadings that are essential in our formulation to represent biclusters. Sparseness is obtained by a component-wise independent Laplace distribution (Hyvärinen and Oja, 1999), which is now used as a prior on the factors  instead of the Gaussian:
instead of the Gaussian:
Sparse loadings λi and, therefore sparse Λ, are achieved by two alternative strategies. In the first model, called FABIA, we assume a component-wise independent Laplace prior for the loadings (like for the factors):
| (4) |
The FABIA model contains the product of Laplacian variables which is distributed proportionally to the 0th order modified Bessel function of the second kind (Bithas et al., 2007). For large values, this Bessel function is a negative exponential function of the square root of the random variable. Therefore, the tails of the distribution are heavier than those of the Laplace distribution. The Gaussian noise, however, reduces the heaviness of the tails such that the heaviness is between Gaussian and Bessel function tails—about as heavy as the tails of the Laplacian distribution. These heavy tails are exactly the desired model characteristics.
The second model, called FABIAS, uses a prior distribution for the loadings that is non-zero only in regions where the loadings are sparse. Following (Hoyer, 2004), we define sparseness as
leading to the prior with parameter spL
| (5) |
Relation to Independent Component Analysis (ICA): our models are closely related to ICA (Hyvärinen, 1999). ICA searches for a matrix factorization, where the components of 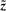 in model Equation (3) without noise ε should be mutually independent. The matrix decomposition for ICA is
in model Equation (3) without noise ε should be mutually independent. The matrix decomposition for ICA is
ICA results in sparse ZICA, whereas ΛICA is not sparse as in our models.
3 MODEL SELECTION
To identify the biclusters, we have to select the model parameters Λ and Ψ that explain the data best. Maximum likelihood is the most common approach for selecting a generative model. Unfortunately, in our case, the likelihood is analytically intractable. The reason is that we aim at generating sparse values, for which we use Laplacian priors (in contrast to the commonly used Gaussian priors). The resulting integral defining the likelihood cannot be computed analytically. In such situations, variational approaches can be applied, where a lower bound of the likelihood is maximized instead of the likelihood itself.
Expectation maximization (EM; Dempster et al., 1977) is the most popular method for maximizing the likelihood. The EM algorithm has been extended to variational EM (Girolami, 2001; Palmer et al., 2006). We follow this approach. However, we also assume a prior on the loadings in order to make the loadings sparse as well. Therefore, we use variational EM for maximizing the posterior—in line with our previous approaches (Hochreiter et al., 2006; Talloen et al., 2007).
3.1 Variational approach for sparse factors
As mentioned above, the likelihood
cannot be computed analytically for a Laplacian prior 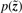 . Girolami (2001) introduces a model family that is parameterized by ξ, where the maximum over models in this family is the true likelihood:
. Girolami (2001) introduces a model family that is parameterized by ξ, where the maximum over models in this family is the true likelihood:
The variational EM algorithm does not only maximize the lower bound on the likelihood with respect to the parameters Λ and Ψ, but also with respect to the variational parameter ξ.
In the following, Λ and Ψ denote the parameter estimates in the current iteration. According to Girolami (2001) and Palmer et al. (2006), we obtain the following variational E-step:
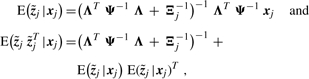 |
where Ξj stands for diag(ξj). The update for ξj is
3.2 New update rules for sparse loadings
The M-step for FABIA (Laplace prior on loadings) is
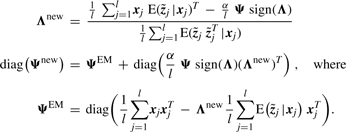 |
(6) |
The M-step for FABIAS updates diag(Ψnew) = ΨEM and Λ according to the standard EM. However, we must take into account that the prior on λi has restricted support. This is ensured by a projection of λi according to Hoyer (2004). The projection is a convex quadratic problem, which minimizes the Euclidean distance to the original vector subject to ‖λi‖ = 1 and sp(λi) = spL, see Equation (5). The final update is
For n > p, the algorithm has a complexity of O(lp2 n) per iteration, i.e. it is linear in n and l.
3.3 Extremely sparse priors
Some microarray data are extremely sparse. For example, we observed a kurtosis larger than 30 for Affymetrix SNP 6 arrays [see copy number variation (CNV) data on FABIA homepage]. We want to generalize our model class to deal with such sparse datasets and define extremely sparse priors both on the factors and the loadings utilizing the following (pseudo) distributions:
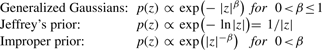 |
The latter may only exist on an interval [ε, a] with sufficiently small ε.
For updating the loadings in the M-step, we need the derivatives of the negative log-priors, which can be expressed proportionally to |z|−spl for a specific exponent spl, where spl = 0 (β = 1) corresponds to the Laplace prior and spl > 0 to sparser priors. The M-step for the loadings is finally as in Equation (6), where sign(Λ) is replaced by |Λ|−spl sign (Λ) with element-wise operations (absolute value, sign, exponentiation and multiplication).
For the factors, we represent the priors by a convex variational form. According to Palmer et al. (2006), this is possible if 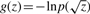 is increasing and concave for z > 0. Our priors fulfill this, because first-order derivatives are positive and second-order derivatives are negative. Then the update for the variational parameter ξj is
is increasing and concave for z > 0. Our priors fulfill this, because first-order derivatives are positive and second-order derivatives are negative. Then the update for the variational parameter ξj is
where spz is the exponent of |z| in the first derivative of g(z); spz = 1/2 (β = 1) represents the Laplace prior and spz> 1/2 leads to sparser priors.
3.4 Data preprocessing and initialization
The data should be centered to zero mean, zero median or zero mode (Supplementary Material). If the correlation of weak signals is of interest too, we recommend to normalize the data.
The iterative model selection procedure requires initialization of the parameters Λ, Ψ and ξj. We initialize the variational parameter vectors ξj by ones, Λ randomly and Ψ = diag(max(δ, covar(x) − Λ ΛT)).
4 INFORMATION CONTENT OF BICLUSTERS
A highly desired property for biclustering algorithms is the ability to rank the extracted biclusters analogously to principal component which are ranked according to the data variance they explain. We rank biclusters according to the information they contain about the data. The information content of 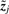 for the j-th observation xj is the mutual information between zj and xj as
for the j-th observation xj is the mutual information between zj and xj as
where H is the entropy. The independence of xj and 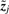 across j gives
across j gives
To assess the information content of one factor, we consider the case that factor  is removed from the final model and, consequently, the explained covariance ξji λi λiT must be considered as noise:
is removed from the final model and, consequently, the explained covariance ξji λi λiT must be considered as noise:
The information of zij given the other factors is
Again independence across j gives
This information content gives that part of the information in x that ziT conveys across all examples. Note that the information content grows with the number of non-zero λi's (size of the bicluster).
5 EXTRACTING MEMBERS OF BICLUSTERS
After model selection and ranking of bicluster, the i-th bicluster has soft gene memberships given by the absolute values of λi and soft sample memberships given by the absolute values of ziT. Soft clustering has the advantage that gradual memberships are able to account for ambiguities that occur in gene expression datasets (where hard memberships can be obscured by noise). However, some applications require hard ‘yes/no’ memberships. We determine the members of the i-th bicluster by selecting absolute values λki and zij above thresholds thresL and thresZ, respectively.
First, the second moment of each factor is normalized to 1 resulting in a factor matrix 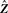 [in accordance with
[in accordance with 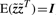 ]. Consequently, Λ is rescaled to
]. Consequently, Λ is rescaled to 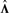 such that
such that 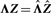 . Now the threshold thresZ can be chosen to determine which percentage of samples will on average belong to a bicluster. For a Laplace prior, this percentage can be computed by
. Now the threshold thresZ can be chosen to determine which percentage of samples will on average belong to a bicluster. For a Laplace prior, this percentage can be computed by 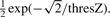 .
.
We extract one bicluster for each factor 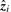 . In gene expression, a gene pattern is either absent or present, but not negatively present. Therefore, the i-th bicluster is either determined by the positive or negative values of
. In gene expression, a gene pattern is either absent or present, but not negatively present. Therefore, the i-th bicluster is either determined by the positive or negative values of 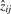 . Which of these two possibilities is chosen is decided by whether the sum over
. Which of these two possibilities is chosen is decided by whether the sum over 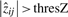 is larger for the positive or negative
is larger for the positive or negative 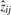 .
.
We may not normalize 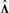 for extracting loadings, since the factors have been normalized already. We suggest to estimate the average contribution of
for extracting loadings, since the factors have been normalized already. We suggest to estimate the average contribution of 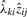 first. Therefore, we compute the standard deviation of
first. Therefore, we compute the standard deviation of 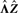 by
by
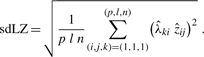 |
Now we choose thresL = sdLZ/thresZ that corresponds to extracting those loadings which have an above-average contribution.
6 EXPERIMENTS
6.1 Evaluating biclustering results
Before comparing biclustering methods, we have to consider how to evaluate the performance of biclustering methods. If the true biclusters are known, the performance of a biclustering method should be evaluated by the consensus between the set of extracted biclusters and the set of true biclusters.
Previous consensus measures such as the one in Gu and Liu (2008) do not take overlapping biclusters into account. Other consensus measures do not consider the numbers of biclusters in both sets (e.g. Prelic et al., 2006, Li et al., 2009). Thus, the set of true biclusters would be in consensus with very large sets of random biclusters. We introduce a novel consensus score for two sets of biclusters which avoids the drawbacks mentioned above as follows:
compute similarities between all pairs of biclusters, where one is from the first set and the other from the second set;
assign the biclusters of one set to biclusters of the other set by maximizing the assignment by the Munkres algorithm (Munkres, 1957); and
divide the sum of similarities of the assigned biclusters by the number of biclusters of the larger set.
Step (3) penalizes different numbers of biclusters as emphasized above.
We use the Jaccard index for computing the similarity of two biclusters. It measures the relative proportion of overlap of two biclusters as the quotient of the number of matrix elements contained in the intersection of the biclusters and the number of matrix elements contained in the union of the biclusters.
The highest consensus is 1 and only obtained for identical sets of biclusters. Further note that the consensus score defined above can be applied analogously to comparing standard clustering results.
6.2 Compared methods
We compare the following 13 biclustering methods:
FABIA: our new method with sparse prior Equation (4).
FABIAS: our new method with sparseness projection Equation (5).
MFSC: matrix factorization with sparseness constraints (Hoyer, 2004).
plaid: plaid model (Lazzeroni and Owen, 2002).
ISA: Ihmels et al. (2004).
OPSM: Ben-Dor et al. (2003).
SAMBA: Tanay et al. (2002).
xMOTIF: conserved motifs (Murali and Kasif, 2003).
Bimax: divide-and-conquer algorithm (Prelic et al., 2006).
CC: Cheng–Church δ-biclusters (Cheng and Church, 2000).
plaid_t: improved plaid model (Turner et al., 2003)
FLOC: a generalization of Cheng–Church δ-biclusters (Yang et al., 2005).
spec: spectral biclustering (Kluger et al., 2003).
We used the following software: for (1)–(3) our R package ‘fabia’, for (4) the software http://www-stat.stanford.edu/∼owen/plaid/, for (5) the R package ‘isa2’, for (6) the software BicAT (Barkow et al., 2006), for (7) the software EXPANDER (Shamir et al., 2005), for (8)–(13) the R package ‘biclust’ (Kaiser and Leisch, 2008).
In all experiments, rows (genes) were standardized to mean 0 and variance 1. For a fair comparison, the parameters of the methods were optimized on auxiliary toy datasets. If more than one setting was close to the optimum, all near optimal parameter settings were tested. In the following, these variants are denoted as method_variant (e.g. plaid_ss). A complete list of all settings and variants is available in the Supplementary Material.
Among the compared methods, not only FABIA and FABIAS but also ISA, OPSM and SPEC are geared to identifying biclusters based on a multiplicative model. Additionally, we included MFSC, although it is not a biclustering method in the strict sense, but it is a standard method for multiplicative factorization and hence provides a baseline for our comparison.
6.3 Simulated datasets with known biclusters
Benchmark datasets published in Prelic et al. (2006) and Li et al. (2009) are small (50 to 100 genes), have low noise, equally sized biclusters, and only simultaneous row and column overlaps. FABIA performed very well on these datasets (see Supplementary, S6.3.1 and S6.3.2). However, we use more realistic simulated datasets that match the characteristics of gene expression data better, especially in terms of the heavy tails. This can be seen in the Supplementary Material by comparing the densities and moments of our simulated datasets (Supplementary Fig. S7) with real gene expression data (Supplementary Figs S8, S9 and S19).
We assumed n = 1000 genes and l = 100 samples and implanted p = 10 multiplicative biclusters with the model given by Equation (1).
The λi's are generated by (i) randomly choosing the number Niλ of genes in bicluster i from {10,…s, 210}, (ii) choosing Niλ genes randomly from {1,…, 1000}, (iii) setting λi components not in bicluster i to 𝒩(0, 0.22) random values and (iv) setting λi components that are in bicluster i to 𝒩(±3, 1) random values, where the sign is chosen randomly for each gene.
The zi's are generated by (i) randomly choosing the number Niz of samples in bicluster i from {5,…, 25}, (ii) choosing Niz samples randomly from {1,…, 100}, (iii) setting zi components not in bicluster i to 𝒩(0, 0.22) random values and (iv) setting zi components that are in bicluster i to 𝒩(2, 1) random values.
Finally, we draw the ϒ entries (additive noise on all entries) according to 𝒩(0, 32) and compute the data X according to Equation (1). Using these settings, noisy biclusters of random sizes between 10 × 5 and 210 × 25 (genes × samples) are generated.
With this procedure, we created 100 independent datasets. Table 1 shows the biclustering results for these datasets. The methods are evaluated by the average consensus score of the extracted biclusters and the true biclusters as defined in Section 6.1. Our new methods FABIA and FABIAS outperform all other methods considerably.
Table 1.
Results on the 100 simulated datasets
| Method | Score | Method | Score |
| FABIA | 0.478 (1e-2) | SAMBA | 0.006 (5e-5) |
| FABIAS | 0.564 (3e-3) | xMOTIF | 0.002 (6e-5) |
| MFSC | 0.057 (2e-3) | Bimax | 0.004 (2e-4) |
| plaid_ss | 0.045 (9e-4) | CC | 0.001 (7e-6) |
| plaid_ms | 0.072 (4e-4) | plaid_t_ab | 0.046 (5e-3) |
| plaid_ms_5 | 0.083 (6e-4) | plaid_t_a | 0.037 (4e-3) |
| ISA_1 | 0.333 (5e-2) | FLOC | 0.006 (3e-5) |
| ISA_2 | 0.299 (6e-2) | spec_1 | 0.032 (5e-4) |
| ISA_3 | 0.188 (4e-2) | spec_2 | 0.011 (5e-4) |
| OPSM | 0.012 (1e-4) |
The numbers denote average consensus scores with the true biclusters as defined in Section 6.1 (standard deviations in parentheses). The best results are highlighted in bold and the second best in italics (‘better’ means significantly better according to both a paired t-test and a McNemar test of correct elements in biclusters).
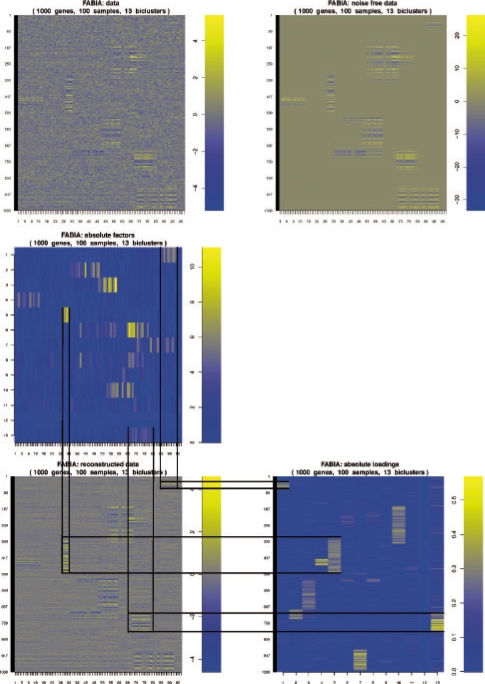
Figure 2 illustrates a FABIA result on a simulated dataset, where, in contrast to our 100 benchmark datasets, the biclusters have been created as contiguous blocks for visualization purposes.
Fig. 2.
An example of FABIA model selection. The data have 10 true biclusters. We have trained the model with 13 biclusters. Only for visualization purposes, the biclusters are generated as contiguous blocks. Top: data (left) and noise-free data (right). Middle: factors Z. Bottom: data reconstructed by the FABIA model as Λ Z (left) and loadings Λ (right). The lines indicate three biclusters and connect each bicluster in the reconstructed data with its corresponding factors (middle) and loadings (bottom right).
We observed the following characteristics of the methods, also confirming earlier findings of Gu and Liu (2008): SAMBA and OPSM excluded many relevant biclusters; SAMBA, Bimax, xMOTIF, CC and FLOC found many small random biclusters (overfitting). spec produces a partition of the samples for each gene set. The plaid models and ISA extract large overlapping clusters.
Ranking by information content: to verify that the information content is useful for ranking the extracted biclusters, we performed a two-sided Spearman rank correlation test comparing (i) the information content and (ii) the Jaccard similarity to the assigned true bicluster. We obtained P-values of 1.7 ×10−5 for FABIA and 6.1 × 10−3 for FABIAS, which shows that true biclusters can indeed be identified by their information content.
Data based on an additive model: we also generated data according to an additive model structure in order to analyze how well FABIA and FABIAS perform on data not satisfying the multiplicative model assumptions. We generated 100 datasets with the above settings, but using the general additive model from Section 1, category (4). Both FABIA and FABIAS outperform all other methods, followed by plaid_ms_5. Specifically, for three different signal levels, FABIAS gave average consensus scores of 0.15–0.27–0.55, FABIA 0.10–0.20–0.48 and plaid_ms_5 0.10–0.14–0.22 (detailed results, also for all other methods, are reported in the Supplementary Material). One would assume plaid methods to perform better than FABIA and FABIAS. We explain the superiority of our methods on datasets that do not even match the data generation model as follows: (i) they construct biclusters simultaneously, thereby, taking overlaps into account; (ii) the decorrelation of factors minimizes redundancy of biclusters; (iii) the low complexity of the model ensures low parameter interdependencies, which facilitates model selection.
6.4 Gene expression datasets
We consider three gene expression datasets that have been provided by the Broad Institute and were previously analyzed by Hoshida et al. (2007). They first clustered the samples using additional datasets and then confirmed the clusters by gene set enrichment analysis. Our goal was to study how well biclustering methods are able to re-identify these clusters without any additional information.
The ‘breast cancer’ dataset (van't Veer et al., 2002) was aimed at a predictive gene signature for the outcome of a breast cancer therapy. We removed the outlier array S54 that leads to a dataset with 97 samples and 1213 genes. After standardization, skewness was 0.45 and excess kurtosis 0.93. In Hoshida et al. (2007), three biologically meaningful subclasses were found that should be re-identified.
The ‘multiple tissue types’ dataset (Su et al., 2002) are gene expression profiles from human cancer samples from diverse tissues and cell lines. The dataset contains 102 samples with 5565 genes. After standardization, skewness was 0.15 and excess kurtosis 1.3. Biclustering should be able to re-identify the tissue types.
The ‘diffuse large-B-cell lymphoma (DLBCL)’ dataset (Rosenwald et al., 2002) was aimed at predicting the survival after chemotherapy. It contains 180 samples and 661 genes, and after standardization the skewness was −0.05 and excess kurtosis 0.35. The three classes found by Hoshida et al. (2007) should be re-identified.
The biclustering results are summarized in Table 2. For the methods assuming a fixed number of biclusters, we chose five biclusters—slightly higher than the number of known clusters to avoid biases toward prior knowledge about the number of actual clusters. The performance was assessed by comparing known classes of samples in the datasets with the sample sets identified by biclustering as defined in Section 6.1, in this case on sample clusters instead of biclusters. For the multiple tissue dataset, plaid performs best and our methods FABIA and FABIAS are second best. For breast cancer and DLBCL datasets, our new methods FABIA and FABIAS detected the clusters most accurately. Further, note that FABIA and FABIAS have considerably fewer genes in their bicluster than the next-best methods.
Table 2.
Results on the breast cancer, multiple tissue samples, DLBCL datasets measured by the consensus score from Section 6.1
| Breast cancer | Multiple tissues | DLBCL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | Score | #bc | #g | #s | Score | #bc | #g | #s | Score | #bc | #g | #s |
| FABIA | 0.52 | 3 | 92 | 31 | 0.53 | 5 | 356 | 29 | 0.37 | 2 | 59 | 62 |
| FABIAS | 0.52 | 3 | 144 | 32 | 0.44 | 5 | 435 | 30 | 0.35 | 2 | 104 | 60 |
| MFSC | 0.17 | 5 | 87 | 24 | 0.31 | 5 | 431 | 24 | 0.18 | 5 | 50 | 42 |
| plaid_ss | 0.39 | 5 | 500 | 38 | 0.56 | 5 | 1903 | 35 | 0.30 | 5 | 339 | 72 |
| plaid_ms | 0.39 | 5 | 175 | 38 | 0.50 | 5 | 571 | 42 | 0.28 | 5 | 143 | 63 |
| plaid_ms_5 | 0.29 | 5 | 56 | 29 | 0.23 | 5 | 71 | 26 | 0.21 | 5 | 68 | 47 |
| plaid_a_ss | 0.37 | 5 | 796 | 35 | 0.65 | 5 | 3711 | 31 | 0.28 | 5 | 389 | 68 |
| plaid_a_ms | 0.34 | 5 | 194 | 35 | 0.58 | 5 | 583 | 34 | 0.27 | 5 | 95 | 61 |
| plaid_a_ms_5 | 0.16 | 5 | 5 | 26 | 0.20 | 5 | 11 | 25 | 0.18 | 5 | 4 | 68 |
| ISA_1 | 0.03 | 25 | 55 | 4 | 0.05 | 29 | 230 | 6 | 0.01 | 56 | 26 | 8 |
| ISA_2 | 0.25 | 2 | 466 | 42 | 0.37 | 3 | 1904 | 28 | 0.22 | 1 | 267 | 74 |
| ISA_3 | 0.22 | 1 | 742 | 33 | 0.35 | 3 | 2856 | 28 | 0.18 | 2 | 385 | 58 |
| OPSM | 0.04 | 12 | 172 | 8 | 0.04 | 19 | 643 | 12 | 0.03 | 6 | 162 | 4 |
| SAMBA | 0.02 | 38 | 37 | 7 | 0.03 | 59 | 53 | 8 | 0.02 | 38 | 19 | 15 |
| SAMBA_01 | 0.01 | 79 | 33 | 8 | 0.01 | 128 | 53 | 9 | 0.01 | 70 | 18 | 14 |
| xMOTIF | 0.07 | 5 | 61 | 6 | 0.11 | 5 | 628 | 6 | 0.05 | 5 | 9 | 9 |
| Bimax | 0.01 | 1 | 1213 | 97 | 0.10 | 4 | 35 | 5 | 0.07 | 5 | 73 | 5 |
| CC | 0.11 | 5 | 12 | 12 | nc | nc | nc | nc | 0.05 | 5 | 10 | 10 |
| plaid_t_ab | 0.24 | 2 | 40 | 23 | 0.38 | 5 | 255 | 22 | 0.17 | 1 | 3 | 44 |
| plaid_t_a | 0.23 | 2 | 24 | 20 | 0.39 | 5 | 274 | 24 | 0.11 | 3 | 6 | 24 |
| spec_1 | 0.12 | 13 | 198 | 28 | 0.37 | 5 | 395 | 20 | 0.05 | 28 | 133 | 32 |
| spec_2 | 0.07 | 14 | 77 | 22 | 0.21 | 1 | 117 | 39 | 0.08 | 8 | 82 | 44 |
| FLOC | 0.04 | 5 | 343 | 5 | nc | nc | nc | nc | 0.03 | 5 | 167 | 5 |
An ‘nc’ entry means that the method did not converge for this dataset. The best results are in bold and the second best in italics (again ‘better’ means significantly better according to a paired t-test). The columns ‘#bc’, ‘#g’ and ‘#s’ provide the numbers of biclusters, their average numbers of genes and their average numbers of samples, respectively.
For the biological interpretation of the FABIA results, we applied gene ontology (GO), Kyoto encyclopedia of genes and genomes (KEGG) pathway and protein interaction network analysis. We provide a summary of these analysis results, details of which can be found in the Supplementary Material.
Breast cancer: GO and KEGG agree that genes in bicluster 1 are related to the cell cycle (KEGG P-value: 9.7 × 10−8; GO P-value: 2.8 × 10−9), especially to M-phase (GO P-value: 2.5 × 10−15). Proteins which drive this bicluster are the cell division control protein CDC2 and the mitosis-related KIF proteins. Genes in bicluster 2 are related to immune response (GO P-value: 1.4 × 10−26) and cytokine–cytokine receptor interaction (KEGG P-value < 10−10), involving cytokine-related proteins such as CCR5, CCL4 and CSF2RB. Note that cytokines are important regulators and mobilizers of the immune response. Bicluster 3 is too small to allow for a reliable biological interpretation.
DLBCL: the most significant GO terms and KEGG pathways found for bicluster 1 are related to the ribosome (GO P-value: 2.2 × 10−6; KEGG P-value: 1.3 × 10−8) and to B-cell receptor signaling (KEGG P-value: 9.6 × 10−8). The latter fits especially well to the kind of cells the data stem from. The most significant GO terms and KEGG pathways for bicluster 2 are immune system-related (GO P-value: 3.2 × 10−6; KEGG P-value: 5.7 × 10−8).
Multiple tissues: this dataset is very heterogeneous and the samples differ in many biological processes; hence, it is difficult to provide a comprehensible biological interpretation.
6.5 Drug design
In a drug design project, Affymetrix GeneChip HT HG-U133+PM array plates with 96 samples (12 × 8) per plate were used to analyze the effect of different compounds on gene expression. The compounds were selected to be active on a cancer cell line and were tested in groups of three replicates.
Raw expression data were summarized with FARMS (Hochreiter et al., 2006) and informative genes are selected by I/NI calls (Talloen et al., 2007). The preprocessed data matrix was 1413 × 95 (one array was missing) with skewness of −0.39 and excess kurtosis larger than 3.0 (i.e. heavier tails than Laplace). We tested FABIA on this dataset. Biclusters were extracted with thresZ = 1.5 to obtain an average of 5–6 samples in a bicluster (note that, for the Laplacian prior, 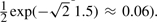 ).
).
FABIA found four biclusters. The first bicluster consisted of two replicate sets (6 arrays), the second consisted of five replicate sets with one replicate missing (14 arrays). The third bicluster consisted of three replicate sets and an additional array (10 arrays). The fourth bicluster consisted of arrays located at the last column of the plate—corresponding to border arrays which dry out. In the meantime, this problem has been fixed by Affymetrix. That replicates are clustered together shows that our biclustering approach works correctly.
The bicluster with highest information content (two sets of replicates) extracted genes related to mitosis (GO analysis gave a P-value <10−13). Regulation of mitosis genes is biologically plausible, as inhibiting cell division would be consistent with an active compound that does not kill the cell. The compounds of this bicluster are now under investigation by Johnson & Johnson Pharmaceutical Research & Development.
7 CONCLUSION
We have introduced a novel biclustering method that is a generative multiplicative model. It assumes realistic non-Gaussian signal distributions with heavy tails. The generative model allows to rank biclusters according to their information content. Model selection is performed by maximum a posteriori via an EM algorithm based on a variational approach.
On 100 simulated datasets with known true biclusters, FABIA clearly outperformed all 11 competing methods. On three gene expression datasets with previously verified subclusters, it was once the second best and twice the best performing method. The biological relevance of the FABIA biclusters has been demonstrated by GO and KEGG analyses. Finally, FABIA has been successfully applied to drug design to find compounds with similar effects on gene expression.
Funding: Janssen Pharmaceutica N.V. and Institute for the Promotion of Innovation by Science and Technology in Flanders (IWT project 80536).
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Barkow S, et al. BicAT: a biclustering analysis toolbox. Bioinformatics. 2006;22:1282–1283. doi: 10.1093/bioinformatics/btl099. [DOI] [PubMed] [Google Scholar]
- Ben-Dor A, et al. Discovering local structure in gene expression data: the order-preserving submatrix problem. J. Comput. Biol. 2003;10:373–384. doi: 10.1089/10665270360688075. [DOI] [PubMed] [Google Scholar]
- Bithas PS, et al. Proceedings of the International Conference on Applied Stochastic Models and Data Analysis. Vol. 12. Chania: 2007. Distributions involving correlated generalized gamma variables. [Google Scholar]
- Busygin S, et al. Proceedings of the 2nd SIAM International Conference on Data Mining/Workshop on Clustering High Dimensional Data. Arlington, VA, USA: 2002. Double conjugated clustering applied to leukemia microarray data. [Google Scholar]
- Caldas J, Kaski S. Proceedings of the IEEE International Workshop on Machine Learning for Signal Processing. XVIII. Cancún, Mexico: 2008. Bayesian biclustering with the plaid model; pp. 291–296. [Google Scholar]
- Califano A, et al. Proceedings of the International Conference on Computational Molecular Biology. Tokyo, Japan: ACM; 2000. Analysis of gene expression microarays for phenotype classification; pp. 75–85. [PubMed] [Google Scholar]
- Cheng Y, Church GM. Proceedings of the International Conference on Intelligent Systems for Molecular Biology. Vol. 8. Tokyo, Japan: ACM; 2000. Biclustering of expression data; pp. 93–103. [PubMed] [Google Scholar]
- Dempster AP, et al. Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. B Met. 1977;39:1–22. [Google Scholar]
- Everitt BS. An Introduction to Latent Variable Models. London: Chapman and Hall; 1984. [Google Scholar]
- Gan X, et al. Discovering biclusters in gene expression data based on high-dimensional linear geometries. BMC Bioinformatics. 2008;9:209. doi: 10.1186/1471-2105-9-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getoor L, et al. Learning probabilistic models of link structure. J. Mach. Learn. Res. 2002;3:679–707. [Google Scholar]
- Getz G, et al. Coupled two-way clustering analysis of gene microarray data. Proc. Natl Acad. Sci. USA. 2000;97:12079–12084. doi: 10.1073/pnas.210134797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girolami M. A variational method for learning sparse and overcomplete representations. Neural Comput. 2001;13:2517–2532. doi: 10.1162/089976601753196003. [DOI] [PubMed] [Google Scholar]
- Gu J, Liu JS. Bayesian biclustering of gene expression data. BMC Genomics. 2008;9(Suppl. 1):S4. doi: 10.1186/1471-2164-9-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardn J, Wilson J. A note on oligonucleotide expression values not being normally distributed. Biostatistics. 2009;10:446–450. doi: 10.1093/biostatistics/kxp003. [DOI] [PubMed] [Google Scholar]
- Hartigan JA. Direct clustering of a data matrix. J. Am. Stat. Assoc. 1972;67:123–129. [Google Scholar]
- Hochreiter S, et al. A new summarization method for Affymetrix probe level data. Bioinformatics. 2006;22:943–949. doi: 10.1093/bioinformatics/btl033. [DOI] [PubMed] [Google Scholar]
- Hoshida Y, et al. Subclass mapping: identifying common subtypes in independent disease data sets. PLoS ONE. 2007;2:e1195. doi: 10.1371/journal.pone.0001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer PO. Non-negative matrix factorization with sparseness constraints. J. Mach. Learn. Res. 2004;5:1457–1469. [Google Scholar]
- Hyvärinen A. Survey on independent component analysis. Neural Comput. Surv. 1999;2:94–128. [Google Scholar]
- Hyvärinen A, Oja E. A fast fixed-point algorithm for independent component analysis. Neural Comput. 1999;9:1483–1492. [Google Scholar]
- Ihmels J, et al. Defining transcription modules using large-scale gene expression data. Bioinformatics. 2004;20:1993–2003. doi: 10.1093/bioinformatics/bth166. [DOI] [PubMed] [Google Scholar]
- Kaiser S, Leisch F. A toolbox for bicluster analysis in R. In: Brito P, editor. Compstat 2008 – Proceedings in Computational Statistics. Heidelberg: Physica Verlag; 2008. pp. 201–208. [Google Scholar]
- Kluger Y, et al. Spectral biclustering of microarray data: coclustering genes and conditions. Genome Res. 2003;13:703–716. doi: 10.1101/gr.648603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzeroni L, Owen A. Plaid models for gene expression data. Stat. Sin. 2002;12:61–86. [Google Scholar]
- Li G, et al. QUBIC: a qualitative biclustering algorithm for analyses of gene expression data. Nucleic Acids Res. 2009;37:e101. doi: 10.1093/nar/gkp491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira SC, Oliveira AL. Biclustering algorithms for biological data analysis: a survey. IEEE ACM Trans. Comput. Biol. 2004;1:24–45. doi: 10.1109/TCBB.2004.2. [DOI] [PubMed] [Google Scholar]
- Madeira SC, Oliveira AL. A polynomial time biclustering algorithm for finding approximate expression patterns in gene expression time series. Algorithm Mol. Biol. 2009;4:8. doi: 10.1186/1748-7188-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira SC, et al. Identification of regulatory modules in time series gene expression data using a linear time biclustering algorithm. IEEE ACM Trans. Comput. Biol. 2010;7:153–165. doi: 10.1109/TCBB.2008.34. [DOI] [PubMed] [Google Scholar]
- Munkres J. Algorithms for the assignment and transportation problems. J. Soc. Ind. Appl. Math. 1957;5:32–38. [Google Scholar]
- Murali TM, Kasif S. Pacific Symposium on Biocomputing. Hawaii, USA: Lihue; 2003. Extracting conserved gene expression motifs from gene expression data; pp. 77–88. [PubMed] [Google Scholar]
- Palmer J, et al. Advances in Neural Information Processing Systems 18. Vancouver, BC, Canada: The MIT Press; 2006. Variational EM algorithms for non-Gaussian latent variable models; pp. 1059–1066. [Google Scholar]
- Prelic A, et al. A systematic comparison and evaluation of biclustering methods for gene expression data. Bioinformatics. 2006;22:1122–1129. doi: 10.1093/bioinformatics/btl060. [DOI] [PubMed] [Google Scholar]
- Reiss DJ, et al. Integrated biclustering of heterogeneous genome-wide datasets for the inference of global regulatory networks. BMC Bioinformatics. 2006;2:280–302. doi: 10.1186/1471-2105-7-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwald A, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. New Engl. J. Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- Shamir R, et al. EXPANDER – an integrative program suite for microarray data analysis. BMC Bioinformatics. 2005;6:232. doi: 10.1186/1471-2105-6-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Q, et al. Biclustering micrarray data by Gibbs sampling. Bioinformatics. 2003;19(Suppl. 2):ii196–ii205. doi: 10.1093/bioinformatics/btg1078. [DOI] [PubMed] [Google Scholar]
- Su AI, et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl Acad. Sci. USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talloen W, et al. I/NI-calls for the exclusion of non-informative genes: a highly effective feature filtering tool for microarray data. Bioinformatics. 2007;23:2897–2902. doi: 10.1093/bioinformatics/btm478. [DOI] [PubMed] [Google Scholar]
- Tanay A, et al. Discovering statistically significant biclusters in gene expression data. Bioinformatics. 2002;18(Suppl. 1):S136–S144. doi: 10.1093/bioinformatics/18.suppl_1.s136. [DOI] [PubMed] [Google Scholar]
- Tang C, et al. Proceedings of the 2nd IEEE International Symposium on Bioinformatics and Bioengineering. Bethesda, MD, USA: IEEE Computer Society; 2001. Interrelated two-way clustering: an unsupervised approach for gene expression data analysis; pp. 41–48. [Google Scholar]
- Tibshirani R, et al. Technical report. Department of Health Research and Policy, Department of Genetics and Department of Biochemestry, Stanford University; 1999. Clustering methods for the analysis of DNA microarray data. [Google Scholar]
- Turner H, et al. Improved biclustering of microarray data demonstrated through systematic performance tests. Comput. Stat. Data Anal. 2003;48:235–254. [Google Scholar]
- Van den Bulcke T. PhD Thesis. Katholieke Universiteit Leuven; 2009. Robust Algorithms for Inferring Regulatory Networks Based on Gene Expression Measurements and Biological Prior Information. Lirias number: 236073. [Google Scholar]
- van't Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Wang H, et al. Proceedings of the 2002 ACM SIGMOD International Conference on Management of Data. 2002. Clustering by pattern similarity in large data sets; pp. 394–405. [Google Scholar]
- Yang J, et al. An improved biclustering method for analyzing gene expression profiles. Int. J. Artif. Intell. T. 2005;14:771–790. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.