Abstract
Motivation: Identifying orthologous genes in multiple genomes is a fundamental task in comparative genomics. Construction of intergenomic symmetrical best matches (SymBets) and joining them into clusters is a popular method of ortholog definition, embodied in several software programs. Despite their wide use, the computational complexity of these programs has not been thoroughly examined.
Results: In this work, we show that in the standard approach of iteration through all triangles of SymBets, the memory scales with at least the number of these triangles, O(g3) (where g = number of genomes), and construction time scales with the iteration through each pair, i.e. O(g6). We propose the EdgeSearch algorithm that iterates over edges in the SymBet graph rather than triangles of SymBets, and as a result has a worst-case complexity of only O(g3log g). Several optimizations reduce the run-time even further in realistically sparse graphs. In two real-world datasets of genomes from bacteriophages (POGs) and Mollicutes (MOGs), an implementation of the EdgeSearch algorithm runs about an order of magnitude faster than the original algorithm and scales much better with increasing number of genomes, with only minor differences in the final results, and up to 60 times faster than the popular OrthoMCL program with a 90% overlap between the identified groups of orthologs.
Availability and implementation: C++ source code freely available for download at ftp.ncbi.nih.gov/pub/wolf/COGs/COGsoft/
Contact: dmk@stowers.org
Supplementary information: Supplementary materials are available at Bioinformatics online.
1 INTRODUCTION
Classification of genes and their products into families of homologs is a key component of any study in comparative genomics. The central problem here is to define orthologous relationships between genes in two or more species. Orthologs, i.e. homologous genes related by speciation (Fitch, 1970, 2000), tend to retain the same function after divergence from their common ancestor, whereas paralogs, i.e. homologous genes related by duplication within a lineage, typically differentiate to perform distinct functions (Kondrashov et al., 2002; Lynch and Conery, 2000; Lynch and Force, 2000; Ohno, 1970). Paralogs are subdivided into in-paralogs, which have diverged after a reference speciation event (often the last speciation event in the minimal clade that includes the two compared lineages) and out-paralogs that diverged before this event (Remm et al., 2001), although in some cases the paralogy status is difficult to resolve in practice, e.g. in the event of differential paralog loss in all examined species (Opazo et al., 2008). Because of the complex interplay of speciation and duplication of genes, a family of in-paralogs in one lineage can be orthologous to a single gene in another lineage, so the problem of identification of orthologs has been redefined to the identification of orthologous groups, i.e. orthologs as well as their lineage-specific duplications (Koonin, 2005). The Clusters of Orthologous Groups (COGs) resource was devised for this purpose soon after the representatives of all three domains of life (Bacteria, Archaea and Eukarya) were sampled by complete genome sequencing (Tatusov et al., 1997).
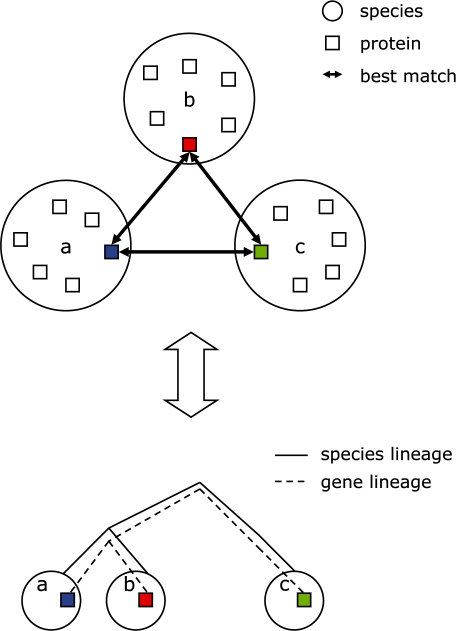
Many methods of ortholog inference have been proposed [reviewed in (Koonin, 2005)], and their comparative performance has been reviewed (Altenhoff and Dessimoz, 2009; Chen et al., 2007). These methods belong to the two major classes of approaches: those that first identify all homologs in a set of species and then attempt to distinguish between orthologs and paralogs by analyzing the distribution of the genes from different species across the tips of the tree (these approaches typically include comparison with another tree representing the consensus view of the evolution of those species), and those that do not reconstruct the trees explicitly, but instead use a heuristic to compile the pairs of genes, each in a different genome, that are each other's best-scoring matches [SymBets, for symmetric best hits (Tatusov et al., 1997), also sometimes called BBH or RBH, for Bidirectional or Reciprocal Best Hits] in their respective genomes (Fig. 1). Examples of automated implementations of the former approach include the publicly available algorithms EnsemblCompara (Vilella et al., 2009), SYNERGY (Wapinski et al., 2007), RIO (Zmasek and Eddy, 2002), Orthostrapper (Storm and Sonnhammer, 2002) and the databases of orthologous protein families HOBACGEN, HOVERGEN and HOGENOME (Dufayard et al., 2005), whereas examples of the latter include OrthoMCL (Li et al., 2003), eggNOG (Jensen et al., 2008), InParanoid and MultiParanoid (Alexeyenko et al., 2006; O'Brien et al., 2005; Remm et al., 2001), MSOAR and MultiMSOAR (Fu and Jiang, 2007; Fu et al., 2007), Homologene (Sayers et al., 2010), RoundUp (Deluca et al., 2006) and OMA (Roth et al., 2008). Still other methods exist that do not fall neatly into either category, such as that described in (Vashist et al., 2007), which uses topological distance in a species tree as a factor in a linkage equation to find dense clusters in a multipartite graph (whose edges are not restricted to SymBets).
Fig. 1.
Schematic of Symmetric Best matches (SymBets) used to build COGs.
The ‘tree-based’ approaches are often considered superior to the ‘pair-linking’ methods because the former utilize explicit algorithms to identify duplication and speciation events on the gene/protein family tree, whereas pair-linking methods use the symmetric-best-match relationship as a surrogate criterion of orthology. An additional objection to using BLAST score for defining SymBets is that it might not be a good estimate of the true evolutionary distance between two homologous sequences, leading to errors in evolutionary inference (Brenner, 1999; Rost, 2002; Todd et al., 2001; Watson et al., 2005). Studies of this problem that we are aware of mostly showcase the examples where the bias leading to errors indeed exists [e.g. (Koski and Golding, 2001)], but there is no evidence to show that the BLAST score is a poor statistical predictor of orthology at the genome scale. On the contrary, the comparisons of tree-based and pair-linking methods of ortholog definition show that the two classes of methods tend to produce similar lists of orthologs, with the differences mostly due to the sensitivity of the homology search and to differential treatment of in-paralogous relationships and the related problem of optimal splitting of large clusters of paralogs (Altenhoff and Dessimoz, 2009; Chen et al., 2007; Koonin, 2005; Li et al., 2003).
Given the generally good correspondence shown by the best methods in the two classes of approaches (Altenhoff and Dessimoz, 2009; Chen et al., 2007), and because pair-linking algorithms are faster and easier to automate than tree-based methods [which, moreover, have the potential to succumb to errors of their own that are intrinsic to construction of phylogenetic trees, especially at large evolutionary distances (Felsenstein, 2004; Forest, 2009)], many practical efforts of genome-scale ortholog identification rely on pair-linking methods. However, despite the prevalence of these approaches in comparative genomics, there has been little examination of their computational complexity and scalability. In this study, we analyze the process of the triangle-merging step of COG construction from a graph-theoretical viewpoint to gain a better understanding of its limitations, and propose a new algorithm that has the complexity of O(g3 log g), where g is the number of genomes, which compares favorably with the high polynomial O(g6) of the more traditional approaches.
2 RESULTS
2.1 Theory
A graph whose vertices are genes and whose edges denote a SymBet relationship between a pair of genes (we call these adjacent if such an edge exists) is called a SymBets graph, G. A subgraph of G is a triangle if it is a set of three vertices such that each pair of them is adjacent to one another. As described elsewhere (Koonin, 2005; Tatusov et al., 1997, 2000), each COG is a subgraph of G that is constructed by using a triangle as a seed and iteratively adding triangles that share a common side, until no new members can be added. This description holds both for the earlier described approach [as implemented, for instance, in the NCBI programs YOG and COGtriangles used for the construction of COGs and publicly available since the year 2007—hereinafter COGtriangles method (ftp.ncbi.nih.gov/pub/wolf/COGs/COGsoft/)] and the new approach proposed here, despite their radically different ways of following these guidelines.
Suppose G has n vertices and m edges. Because G is built of only SymBets between genes in different genomes, it is a g-partite graph, i.e. it has g groups of vertices, each corresponding to genes from the same genome, and edges are only allowed between different groups but not between the vertices in the same group (note that the collection of in-paralogs due to lineage-specific expansions is done separately prior to the main algorithm, and in the graph these are represented by a single vertex). Also, let p be the maximum number of genes in any given genome, which is a constant for a given set of genomes and does not depend on the number of genomes, even though it is possible that the value of p increases as larger genomes are sequenced and added to G.
The number of vertices n grows linearly with the number of genomes g, with the addition of each genome adding at most p new genes to n, so n ≤ gp and the upper bound of the number of vertices O(n) is simply O(g). Since G is a g-partite graph with no edges between genes from the same organism, the number of edges m is:
Thus, the upper bound of m grows with the quadratic term of g that dominates the behavior of this equation, and O(m) = O(g2). Similarly, the number of triangles t grows with the cube of the number of genomes as:
Thus, the upper bound of the number of triangles O(t) = O(g3).
COGs are constructed as subgraphs of G, starting as triangles and growing by iteratively merging triangles if they share a common side, until no more triangles can be added. For instance, the COGtriangles algorithm used to build the most recent available release of NCBI COGs (ftp.ncbi.nih.gov/pub/COG/COG/) proceeds in two stages, by finding all possible triangles in G and then iterating through each pair of them to merge those that share a common side, as follows:
COGtriangles algorithm:
- For each triangle Ta taken from the list of unprocessed triangles
- (2) Initialize a ‘seed’ COG C = Ta
- (3) For each triangle Tb not already part of an existing COG
- (4) If Tb shares a side with a triangle in C, merge Tb into C
- (5) Print C
(6) End
Since there are at most O(g3) triangles and thus O(g3) initial COGs before the merging step, and this algorithm iterates over pairs of triangles (or pairs of a COG and a triangle), it scales with the high polynomial O(g3)*O(g3) = O(g6). Though the space of unused triangles is iteratively reduced as the algorithm progresses, this does not affect the algorithm complexity: if the number of COGs is c, the algorithm complexity is O(c)*O(g3), assuming a O(1) lookup for a common edge between a triangle and a COG, and since O(c) = O(g3) in the worst case, the overall complexity is O(g6).
In practical terms, the COGs were first implemented on seven fully sequenced genomes treated as five lineages (g = 5; see Tatusov et al., 1997), so at most C5,3*p = 10p triangles could exist, and iterating through each pair only cost C10,2*p2 = 45p2 computations. With 10 genomes, these numbers become 120p and 7140p2, respectively, at 20 genomes they rise to 1140p and 6*105 p2, at 50 genomes they are 19 600p and 2*108 p2 and at 100 they became 162 000p and 1010 p2. Building a set of COGs with only bacteria (g ≈ 103 in 2010) produces 108 p triangles, and iterating over each pair would generate 1016 p2 computations.
In this work, we present an algorithm that builds triangles and COGs simultaneously, with the worst-case complexity of only O(g3 log g). The main idea in our approach is to find a specific class of subgraphs, recently called triangularly connected subgraphs (Fan et al., 2008), by iterating over edges instead of triangles, as explained in more detail below.
A triangle-path in G is a sequence of triangles T1, T2,…, Tk in G such that for 1 ≤ i ≤ k − 1, the triangles Ti and Ti+1 share a single edge and for j > i + 1, the triangles Ti and Tj do not share any edges. A connected subgraph C in G is triangularly connected if for any distinct edges e and e′ in C, there is a triangle path T1, T2,…, Tk in C such that e is an edge in T1 and e′ is an edge in Tk. Under these definitions, a subgraph of G is a COG if it is a triangularly connected graph and is not a single edge. Note that such subgraphs of G are edge-disjoint with one another, i.e. no two subgraphs share an edge.
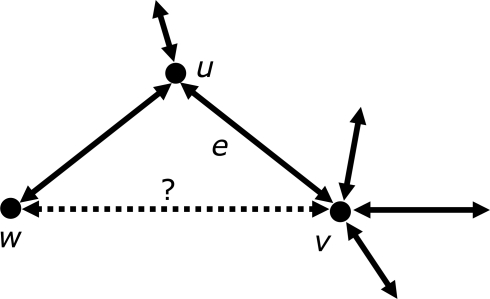
The EdgeSearch algorithm developed in this work is illustrated in Figure 2. Like the COGtriangles algorithm, EdgeSearch starts by initializing a ‘seed’ COG C, but instead of searching the space of all triangles to merge those with common sides, it searches for the pairs of edges that satisfy the following condition:
If the vertices u and v and edge (u, v) are in C, and a third vertex w not in C, and (u, v), (v, w) and (w, u) are edges in G (i.e. vertices u, v and w form a triangle in G), then add the vertex w and edges (v, w) and (w, u) to C.
The above process stops when no more vertices and edges can be added to C. At this point, the algorithm proceeds with another seed edge not contained in any of the previously identified COGs, and this sequence of steps is repeated until all triangularly connected subgraphs (i.e. COGs) with the maximal number of vertices are found. Each such subgraph that is not a single edge is then declared an individual COG and output. In more detail, the EdgeSearch algorithm is:
Fig. 2.
Illustration of EdgeSearch algorithm building triangles and COGs simultaneously while iterating over edges.
EdgeSearch algorithm:
- For each edge taken from the list of unprocessed edges
- (2) Initialize a ‘seed’ COG C with this edge and its vertices
- (3) For each unprocessed edge e(u, v) in C
- (4) For each vertex w such that (u, w) and (v, w) are unprocessed edges in G (at least one of which is not already in C), add this vertex w and edges (u, w) and (w, v) to C (if they are not already part of C)
- (5) Mark e as processed
- (6) Print C if it contains three or more vertices
(7) End
Theorem: —
EdgeSearch finds all maximal triangularly connected subgraphs in G.
Proof. —
This result derives from the following:
The subgraphs output by the algorithm are edge-disjoint—i.e. no two subgraphs share an edge. This is because the iteration through all unprocessed edges in the subgraph C (Step 3) ensures that when triangles that share a common side are merged into C (Step 4), the result is that all triangles that contain a single edge e will become part of that same subgraph C. Then, since the algorithm does not proceed to build another subgraph until all edges of C are processed (Step 3), the subgraphs output in Step 6 are edge-disjoint.
Each triangle in G belongs to a unique output subgraph. Since all edges are processed (introduced in either Step 1 or 3 and triangles formed in Step 4), all triangles in G will eventually be found and assigned to at least one subgraph. Also, since the subgraphs output in Step 6 are edge-disjoint (Statement 1), they are also triangle-disjoint—i.e. no two subgraphs share a triangle, and thus triangles can belong to at most one subgraph. Thus, each triangle in G belongs to a single subgraph.
The output is a set of triangularly connected subgraphs in G. This is because, after forming the first triangle, each subgraph C is expanded by iteratively adding all triangles that share a common edge with an existing triangle in C (Step 4).
These triangularly connected subgraphs are maximal. Since each subgraph is edge-disjoint (Statement 1) and each triangle belongs to a unique subgraph (Statement 2), there cannot be a triangle-path between two different subgraphs. Therefore, each triangularly connected subgraph found by the algorithm (Statement 3) is also maximal.
EdgeSearch finds all maximal triangularly connected subgraphs in G. This follows since all edges are processed (introduced in either Step 1 or 3), and the result is maximal triangularly connected subgraphs (Statements 3 and 4). Q.E.D.
Analysis of the worst-case complexity of the EdgeSearch algorithm gives O(g2)*O(g)*O(log g) = O(g3 log g). This is because the algorithm must: (i) iterate over all edges e(u, v) in C (Step 3), with the worst-case complexity O(m) = O(g2); and for each, (ii) look for a vertex w and edge f (u, w) in G (Step 4), which is at worst O(g) if it must look through all other genomes in the g-partite graph; and finally for each of these, (iii) check whether u and w are adjacent in G, which is an efficient O(log g) lookup from the list of all adjacent vertices of w (or v). The worst-case complexity of EdgeSearch is comparable to the O(V3) (V = number of vertices) of another heuristic method described in Vashist et al. (2007), but uses different topological information, i.e. triangles in a SymBets graph rather than dense clusters (quasi-cliques) in a graph that may include all edges and does not require a species tree.
Our implementation of EdgeSearch, in addition to iterating in the lower-dimensional space of edges rather than in the space of triangles, also takes advantage of optimized data structures. The most important of these are: (i) a list of all edges to iterate through, (ii) a hash of all edges to quickly test the existence of an edge, (iii) a hash of all processed edges to quickly test whether an edge has been processed already and (iv) a list of all adjacent vertices for each vertex in the graph, to avoid searching the entire space of edges (also, as shown in Fig. 2, the vertex with the smaller of the two lists can be chosen to be u in Step 4 of the algorithm). The minimal extra cost of producing these data structures (compared to storing all possible triangles) often gives a large payoff: for example, the knowledge of which edges have already been processed allows EdgeSearch to iterate through edges in O(m)*O(g) = O(g3) time rather than a full pair-wise O(m2) = O(g4). Furthermore, many of these optimizations substantially reduce execution time in realistically sparse graphs, for example by using a list of edges that each gene is adjacent to in order to avoid the search through the entire set of genes to find a third vertex w.
2.2 Additional considerations
The EdgeSearch algorithm deterministically finds triangularly connected subgraphs in G. Iteration over edges reduces its worst-case behavior compared with the COGtriangles algorithm, and several optimizations take advantage of the sparseness of the graph, further reducing its run-time on realistic datasets (see examples below). The question, however, is whether triangularly connected subgraphs are too strict a definition of orthologous groups. For instance, the incompleteness of the SymBets list or an artifact of domain fusion might cause a rare case where a triangle shares a side with a subgraph, but not necessarily with a triangle structure within the subgraph (Supplementary Fig. 1; note that such subgraphs could no longer be called triangularly connected). Expanding the definition of COGs to include such subgraphs can be done by altering COG C to become the subgraph induced by the vertices in C (i.e. C contains all edges connecting its member genes), but this introduces an element of non-determinism to the process of building COGs, where the order of data processing affects the results. In the current implementation of EdgeSearch, we choose to avoid this non-determinism rather than extend the definition to handle these rare events. The alternatives are discussed further in the Supplementary Material.
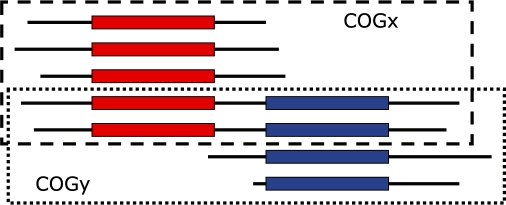
Another concern is whether genes should be allowed to belong to multiple COGs. The first approach is to assign each gene to a single COG and then disallow it from belonging to another COG i.e. COGs are defined as being vertex-disjoint), even though reasons such as differential combination of protein domains or improperly resolved paralogous relationships can make it appear to belong to multiple COGs [the former case can be dealt with by splitting proteins into their component domains (Koonin, 2005; Tatusov et al., 1997)]. Figure 3 illustrates a hypothetical example: in this scenario, the middle two proteins contain both domains, and thus each could be arbitrarily assigned to either the top or bottom COG depending on the order the input is processed in. This phenomenon can affect the overall number of COGs: for instance, in this example, if both proteins are assigned to the top COG, the remaining bottom two proteins are short of the 3-protein requirement to form a full COG. In the current implementation of EdgeSearch, we chose to allow genes to belong to multiple COGs, which is a natural consequence of the SymBets graph structure and preserves this information for future use, such as in domain dissection.
Fig. 3.
Hypothetical scenario illustrating how the presence of multiple domains can complicate the inclusion of proteins into COGs (here, the middle two proteins could be arbitrarily assigned to either the top or bottom COG).
2.3 Performance in construction of phage orthologous groups
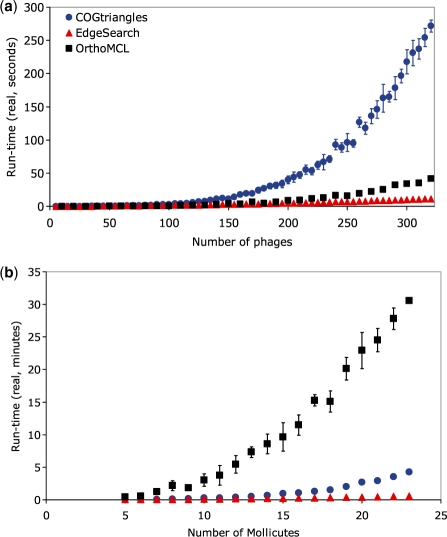
We implemented the algorithm in C++ and tested its ability to make COGs in a real-world dataset of protein-coding genes from 323 double-stranded DNA bacteriophages [these COGs in phages are called Phage Orthologous Groups or POGs (Kristensen et al., 2009; Liu et al., 2006)]. For a more direct comparison of the two algorithms, we integrated the new approach into the old framework by starting with the COGtriangles program, eliminating the makeTriangles() function, and replacing makeCOGs() with an implementation of EdgeSearch that makes triangles and subgraphs simultaneously. Figure 4a demonstrates that, as the number of randomly chosen genomes from this set of phages increases, the time required by the original COGtriangles method (measured on a dual-processor Pentium 3 GHz with 2 GB of RAM) increases from a few seconds to several minutes, whereas EdgeSearch holds steady at <12 s throughout the entire tested range. More important than the actual performance is the shape of the curves, with EdgeSearch handling the increase in number of genomes much more easily than the original approach, indicating that as more genomes become available, the advantage conferred by the new algorithm at handling larger input sizes will become ever stronger.
Fig. 4.
Run-time performance of EdgeSearch (red triangles) compared with the original COGtriangles (blue circles) and OrthoMCL (black squares) in the (a) POGs and (b) MOGs datasets.
Not only is EdgeSearch faster than the original COGtriangles algorithm, but it also outcompetes the newest version (2.0 beta 6) of the popular OrthoMCL approach (Fig. 4a). OrthoMCL is based on a generic clustering MCL algorithm (Enright et al., 2002; Van Dongen, 2000), which uses simulation of stochastic flow on the edges of the graph, with Markov matrices determining the transition probabilities among nodes of the graph. Since the worst-case complexity depends on the input parameters, and further complications arise from the fact that the MCL algorithm is applied iteratively until convergence and that its output is non-deterministic, here we content ourselves with a comparison of its run-time performance rather than an in-depth look into its complexity. Indeed, in the most direct comparison, OrthoMCL (Li et al., 2003) required 44 s to form groups of orthologs from three or more genomes in the 323 dsDNA phage genomes, whereas EdgeSearch required only 12 s and COGtriangles almost 5 min. These numbers (and those in Fig. 4) represent only the clustering step of the respective approaches (performed by the separate MCL program (http://www.micans.org/mcl/) in the case of OrthoMCL), but when the entire pipeline is considered, starting from the BLAST results and continuing all the way to the end point of groups of orthologs, then OrthoMCL (starting with an empty MySQL database to minimize run-time) required 4 min 25 s, EdgeSearch only 41 s and COGtriangles 5 min 9 s (Supplementary Table 1).
In theory, the output of the EdgeSearch algorithm should be identical to that of the COGtriangles algorithm. In practice, differences were observed due to the non-deterministic resolution of several problems by COGtriangles, and to the changes made in EdgeSearch to make its output deterministic (Fig. 3 and Supplementary Fig. 1). For instance, with COGs defined strictly as triangularly connected subgraphs in EdgeSearch but not in the COGtriangles approach, six pairs of POGs (0.6% of the total 2058) are seen to be split in the former but are merged together in the latter. In addition, 1.2% of the proteins were observed to belong to multiple POGs produced by EdgeSearch (regardless of whether proteins were first split into domains prior to POG construction or not, indicating that the major cause is unresolved paralogy), which affected the membership of 93 (5%) of the groups and caused 37 (2%) additional groups to be formed compared to the original approach where proteins are only allowed to belong to a single group.
To further confirm that the EdgeSearch and COGtriangles approaches produce identical results across a wider range of input graphs, 320 additional randomly chosen test sets were constructed from the 323 phage genomes (corresponding to the data points shown in Fig. 4) by randomly sampling an increasing number of genomes, from 5 to 320 in steps of +5, with five independent replicates of each sample. In each case, the two outputs were again the same after accounting for the issues of merging a triangle with a non-triangle within a POG and multiply represented genes.
The results of EdgeSearch and OrthoMCL were less similar. This comes as no surprise given their different underlying rationales [reviewed in (Chen et al., 2007)]. As a result of analysis of the 323 phage genomes, EdgeSearch produces 2058 POGs, whereas OrthoMCL produces 2265 clusters with default parameters. OrthoMCL uses similarity score cutoffs to define SymBets (default e-value of 1e-5), whereas the COG approach requires no cutoffs, but in practice discards matches with e-value greater than the BLAST default of 10 (Tatusov et al., 1997). When these parameters were adjusted, OrthoMCL produces 2250 clusters with an e-value cutoff of 10, and EdgeSearch produces 2062 with an e-value cutoff of 1e-5, indicating that the difference is due to the underlying algorithms rather than the choice of e-value threshold. The majority of the groups produced by the two programs were similar, with 86% of EdgeSearch groups overlapping (sharing three or more genes) with 68% of OrthoMCL's (using default parameters in each program), and 97% of the genes in POGs appearing in an OrthoMCL cluster. However, nearly a third of the clusters and an additional 5800 genes (44% of the shared total) in OrthoMCL's results are not found in POGs, and OrthoMCL's clusters are significantly larger, with a maximum size of 287 genes and average of 8.3 genes per group, compared to a maximum of only 141 genes and an average of 6.5 genes in POGs. OrthoMCL clusters contain a much higher number of paralogs, with the maximum of 13 paralogs in OrthoMCL versus only 4 in POGs, and 796 OrthoMCL clusters (35%) containing at least one paralog versus only 256 (12%) in POGs. The OrthoMCL method has been reported to produce smaller, tighter clusters whereas KOGs (essentially eukaryotic COGs) produces larger, more inclusive groupings. Conceivably, the differences between our results and those of Chen et al. (2007) could stem from different structures of the analyzed datasets, with a considerably greater extent of gene paralogy in the eukaryotic genomes analyzed in their study compared to the bacteriophage genomes that underlie the POGs. For further comparison between the COGs and OrthoMCL approaches (as well as other methods of ortholog identification), see Altenhoff and Dessimoz (2009) and Chen et al. (2007).
2.4 Genomes of cellular organisms: construction of MOGs
To assess the performance of the EdgeSearch approach on larger genomes of cellular organisms, we derived the orthologous groups of 16 726 protein-coding genes from the completely sequenced genomes of 23 bacteria from the Mollicutes class (MOGs). The genome sizes of Mollicutes start at 475 genes in the small parasite Mycoplasma genitalium and reach 1380 genes in the more metabolically complex Acholeplasma laidlawii (Pollack et al., 1996). In addition to encoding larger protein sets than in viruses, these cellular organisms also contain considerably higher levels of paralogy than phages, with a maximum of 39 paralogs in MOGs (a large group of transposases in Mycoplasma mycoides), compared to only 6 in POGs. Other relatively large groups of in-paralogs in MOGs include 13 DNA-binding protein HU in-paralogs of Candidatus Phytoplasma australiense, and 13 ABC transporter ATP-binding protein in-paralogs in Mycoplasma hyopneumoniae. Despite this greater genomic complexity, there were even fewer unresolved paralogy cases in MOGs than in POGs, with only three pairs of MOGs (0.7%) sharing a side with a non-triangle and only 0.5% of the genes belonging to multiple MOGs, which affected the membership of 18 (2%) of the groups and caused 16 (2%) additional groups to be formed.
In this set of cellular genomes, the performance of EdgeSearch is even more striking in comparison to OrthoMCL and COGtriangles (Fig. 4b). While OrthoMCL required >30 min to cluster orthologs, EdgeSearch completed the task in only 28 s (a >60-fold speedup), whereas COGtriangles took nearly 3 min. In the full pipeline starting from the BLAST results, OrthoMCL required nearly an hour (>55 min), whereas EdgeSearch required <3 min and COGtriangles >5 min (Supplementary Table 1). Again, the number of clusters differed, with EdgeSearch producing 833 and OrthoMCL 930 with e-value cutoff of 10.
3 CONCLUSIONS
The graph-theoretical analysis of the process of making COGs frames this problem as partitioning the SymBets graph G into several triangularly connected subgraphs containing genes from three or more genomes. This framework reveals the bottleneck of the earlier approaches at the stage of construction and iteration through all possible triangles, for which memory scales with at least the number of these triangles, O(g3), and construction time scales with the iteration through each pair, or O(g6). Our EdgeSearch algorithm, by iterating over edges rather than triangles, constructs the same COGs with a worst-case complexity of only O(g3 log g), and several optimizations reduce the run-time even further in realistically sparse graphs (at the cost of additional memory, although still less than the cost of storing all possible triangles). Given that EdgeSearch produces the same output as the original COGtriangles (except for cases due to non-determinism, as discussed in the text), and does so much more efficiently, a new version of COGtriangles that uses the EdgeSearch algorithm will replace the original version in the publicly available source code of the COG software at ftp.ncbi.nih.gov/pub/wolf/COGs/COGsoft/.
Funding: Stowers Institute for Medical Research; Intramural Research Program of the National Library of Medicine at the US National Institutes of Health. Funding to pay the Open Access publication charges for this article was provided by the Stowers Institute for Medical Research.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Alexeyenko A, et al. Automatic clustering of orthologs and inparalogs shared by multiple proteomes. Bioinformatics. 2006;22:e9–e15. doi: 10.1093/bioinformatics/btl213. [DOI] [PubMed] [Google Scholar]
- Altenhoff AM, Dessimoz C. Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput. Biol. 2009;5:e1000262. doi: 10.1371/journal.pcbi.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner SE. Errors in genome annotation. Trends Genet. 1999;15:132–133. doi: 10.1016/s0168-9525(99)01706-0. [DOI] [PubMed] [Google Scholar]
- Chen F, et al. Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS One. 2007;2:e383. doi: 10.1371/journal.pone.0000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluca TF, et al. Roundup: a multi-genome repository of orthologs and evolutionary distances. Bioinformatics. 2006;22:2044–2046. doi: 10.1093/bioinformatics/btl286. [DOI] [PubMed] [Google Scholar]
- Dufayard JF, et al. Tree pattern matching in phylogenetic trees: automatic search for orthologs or paralogs in homologous gene sequence databases. Bioinformatics. 2005;21:2596–2603. doi: 10.1093/bioinformatics/bti325. [DOI] [PubMed] [Google Scholar]
- Enright AJ, et al. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, et al. Nowhere-zero 3-flows in triangularly connected graphs. J. Comb. Theory Ser B. 2008;98:1325–1336. [Google Scholar]
- Felsenstein J. Inferring Phylogenies. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- Fitch WM. Distinguishing homologous from analogous proteins. Syst. Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- Fitch WM. Homology a personal view on some of the problems. Trends Genet. 2000;16:227–231. doi: 10.1016/s0168-9525(00)02005-9. [DOI] [PubMed] [Google Scholar]
- Forest F. Calibrating the Tree of Life: fossils, molecules and evolutionary timescales. Ann. Bot. 2009;104:789–794. doi: 10.1093/aob/mcp192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Jiang T. Clustering of main orthologs for multiple genomes. Comput. Syst. Bioinformatics Conf. 2007;6:195–201. [PubMed] [Google Scholar]
- Fu Z, et al. MSOAR: a high-throughput ortholog assignment system based on genome rearrangement. J. Comput. Biol. 2007;14:1160–1175. doi: 10.1089/cmb.2007.0048. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, et al. eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2008;36:D250–D254. doi: 10.1093/nar/gkm796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA, et al. Selection in the evolution of gene duplications. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-2-research0008. RESEARCH0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- Koski LB, Golding GB. The closest BLAST hit is often not the nearest neighbor. J. Mol. Evol. 2001;52:540–542. doi: 10.1007/s002390010184. [DOI] [PubMed] [Google Scholar]
- Kristensen DM, et al. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 2009;18:11–19. doi: 10.1016/j.tim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, et al. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, et al. Protein repertoire of double-stranded DNA bacteriophages. Virus Res. 2006;117:68–80. doi: 10.1016/j.virusres.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien KP, et al. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;33:D476–D480. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by Gene Duplication. New York: Springer; 1970. [Google Scholar]
- Opazo JC, et al. Differential loss of embryonic globin genes during the radiation of placental mammals. Proc. Natl Acad. Sci. USA. 2008;105:12950–12955. doi: 10.1073/pnas.0804392105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack JD, et al. Comparative metabolism of Mesoplasma, Entomoplasma, Mycoplasma, and Acholeplasma. Int. J. Syst. Bacteriol. 1996;46:885–890. doi: 10.1099/00207713-46-4-885. [DOI] [PubMed] [Google Scholar]
- Remm M, et al. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- Rost B. Enzyme function less conserved than anticipated. J. Mol. Biol. 2002;318:595–608. doi: 10.1016/S0022-2836(02)00016-5. [DOI] [PubMed] [Google Scholar]
- Roth AC, et al. Algorithm of OMA for large-scale orthology inference. BMC Bioinformatics. 2008;9:518. doi: 10.1186/1471-2105-9-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38:D5–D16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm CE, Sonnhammer EL. Automated ortholog inference from phylogenetic trees and calculation of orthology reliability. Bioinformatics. 2002;18:92–99. doi: 10.1093/bioinformatics/18.1.92. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, et al. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, et al. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd AE, et al. Evolution of function in protein superfamilies, from a structural perspective. J. Mol. Biol. 2001;307:1113–1143. doi: 10.1006/jmbi.2001.4513. [DOI] [PubMed] [Google Scholar]
- Van Dongen S. PhD Thesis. The Netherlands: University of Utrecht; 2000. Graph clustering by flow simulation. [Google Scholar]
- Vashist A, et al. Ortholog clustering on a multipartite graph. IEEE/ACM Trans. Comput. Biol. Bioinform. 2007;4:17–27. doi: 10.1109/TCBB.2007.1004. [DOI] [PubMed] [Google Scholar]
- Vilella AJ, et al. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski I, et al. Automatic genome-wide reconstruction of phylogenetic gene trees. Bioinformatics. 2007;23:i549–i558. doi: 10.1093/bioinformatics/btm193. [DOI] [PubMed] [Google Scholar]
- Watson JD, et al. Predicting protein function from sequence and structural data. Curr. Opin. Struct. Biol. 2005;15:275–284. doi: 10.1016/j.sbi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Zmasek CM, Eddy SR. RIO: analyzing proteomes by automated phylogenomics using resampled inference of orthologs. BMC Bioinformatics. 2002;3:14. doi: 10.1186/1471-2105-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.