Abstract
Motivation: The identification of putative ligand-binding sites on proteins is important for the prediction of protein function. Knowledge-based approaches using structure databases have become interesting, because of the recent increase in structural information. Approaches using binding motif information are particularly effective. However, they can only be applied to well-known ligands that frequently appear in the structure databases.
Results: We have developed a new method for predicting the binding sites of chemically diverse ligands, by using information about the interactions between fragments. The selection of the fragment size is important. If the fragments are too small, then the patterns derived from the binding motifs cannot be used, since they are many-body interactions, while using larger fragments limits the application to well-known ligands. In our method, we used the main and side chains for proteins, and three successive atoms for ligands, as fragments. After superposition of the fragments, our method builds the conformations of ligands and predicts the binding sites. As a result, our method could accurately predict the binding sites of chemically diverse ligands, even though the Protein Data Bank currently contains a large number of nucleotides. Moreover, a further evaluation for the unbound forms of proteins revealed that our building up procedure was robust to conformational changes induced by ligand binding.
Availability: Our method, named ‘BUMBLE’, is available at http://bumble.hgc.jp/
Contact: kasahara@cb.k.u-tokyo.ac.jp
Supplementary information: Supplementary Material is available at Bioinformatics online.
1 INTRODUCTION
Structural information of proteins has been explosively increasing, mainly due to structural genomics projects. On the other hand, the molecular functions of many proteins still remain uncharacterized. Therefore, computational methods that can predict the molecular functions are required (Kinoshita and Nakamura, 2003; Thornton et al., 2000). Since many proteins conduct their molecular functions through the specific recognition of small molecules (ligands), information about the ligand-binding sites can provide insights into their molecular functions (Campbell et al., 2003; Sotriffer and Klebe, 2002).
Currently, the most successful prediction methods adopt the similarity-based approach. This approach searches databases for proteins that are similar to a query protein in one or more properties, such as sequence, fold and physicochemical properties, and predicts the functions of the query protein by transferring the annotations from similar proteins (Juncker et al., 2009; Kinoshita and Nakamura, 2005; Lee et al., 2007; Loewenstein et al., 2009). However, this approach fails if there is no protein with any detectable similarities.
To overcome these intrinsic limitations, several methods for binding-site prediction that are based only on the 3D structure of the query protein have been proposed. Most of these methods fall into two groups: the geometry-based and force field-based approaches. The former does not consider the physicochemical properties of the protein surface, because it focuses only on the shapes of the surface (i.e. sizes and depths of clefts; Brady and Stouten, 2000; Huang and Schroeder, 2006). On the other hand, the latter method, which considers the stability of probes positioned around the protein surface, does not adequately consider complicated effects, such as solvent effects (Laurie and Jackson, 2005; Morita et al., 2008).
The knowledge-based approach is an appealing alternative. This approach can include a wide range of complicated effects by taking advantage of the statistics of molecular interactions obtained from structure databases, such as the Protein Data Bank (Berman et al., 2003). The potential of this approach is growing fast, because of the recent rapid accumulation of structure data. Existing knowledge-based methods are based on interactions on one of two levels: interatomic and fragment-level interactions.
The approach based on the interatomic interactions uses pair-wise potential functions constructed from the statistics of interatomic contacts observed in the databases. This approach has been mainly used for scoring functions in molecular docking studies (Gohlke et al., 2000; Muegge and Martin, 1999; Zhou and Zhou, 2002). However, the approach does not consider the patterns of interactions derived from binding motifs that are known to appear among unrelated proteins (Denessiouk and Johnson, 2000; Denessiouk et al., 2001; Kinoshita et al., 1999; Kobayashi and Go, 1997), because many-body interactions, such as those in binding motifs, cannot be described by pair-wise interatomic interactions.
On the other hand, the approach based on fragment-level interactions can incorporate the binding-motif information by using the spatial distributions of atoms around a fragment, but very large fragments can only be used for some specific ligands. In this approach, a structural definition of the fragments is very important. A method proposed by Shionyu-Mitsuyama et al. (2003) and its extension by Saito et al. (2006) manually defined the fragments for carbohydrates and nucleotide bases, respectively, but these fragments, such as glucose, galactose, guanine, adenine and others, only correspond to a few specific ligands. Since a knowledge-based approach requires repeated appearances of the fragments to obtain statistics, large fragments can only be used for ligands that are frequently observed in the database. Therefore, these methods cannot be utilized with chemically diverse ligands.
As described above, there is a trade-off in defining the unit of interactions. If the unit is too small (atomic level), then structural motifs cannot be considered. On the other hand, when the fragment is too large (residue level), the fragment will specify a ligand and result in the limitation of the applicable ligands to those frequently appearing in the database.
We now propose a new knowledge-based method to address this problem. In our method, the unit of interactions is defined as a pair of fragments; that is, a main or side chain of an amino acid and three covalently linked atoms in a ligand. Since one ligand atom can belong to more than one fragment in this definition, the patterns of the interactions in larger parts of molecules, i.e. those derived from binding motifs, can be considered by focusing on the consensus of the fragment interactions through atoms that are shared by more than one fragment. Furthermore, our method can be applied to chemically diverse ligands, because the fragments are not manually defined as large units that may specify ligands. In our method, the favorable positions, or ‘interaction hotspots’, are first predicted for all atoms of the ligand. The binding sites are then predicted by building the energetically favorable ligand conformations from the predicted interaction hotspots. Evaluations of the bound structures revealed that our method could predict 90% of binding sites as partially correct binding sites, correct binding sites or correct conformations, among which 53% were for correct conformations. Moreover, an evaluation of the unbound structures revealed that the prediction performance was unaffected by the degree of conformational change occurring upon ligand binding, which is a very important feature in the function prediction of uncharacterized proteins.
2 METHODS
2.1 Dataset construction
Five datasets were constructed in this study: (i) the background knowledge dataset, which was used for the pre-processing step described below; (ii) the parameter tuning dataset, which was used to determine some adjustable parameters; (iii) the nucleotide dataset; (iv) the chemically diverse dataset; and (v) the unbound dataset. The latter three datasets were used for evaluation studies.
These datasets were obtained by the following procedure. The background knowledge dataset was composed of all complexes in the sc-PDB database (5524 complexes in 2007; Kellenberger et al., 2006). Next, in order to construct datasets (ii) and (iii), we focused on 11 kinds of nucleotides that frequently appear in the database: AMP (adenosine monophosphate), ADP (adenosine diphosphate), ATP (adenosine triphosphate), ANP (phosphoaminophosphonic acid-adenylate ester), GDP (guanosine diphosphate), GTP (guanosine triphosphate), GNP (phosphoaminophosphonic acid-guanylate ester), FMN (flavin mononucleotide), FAD (flavine-adenine dinucleotide), NAD (nicotine-adenine dinucleotide) and NAP (nicotinamide-adenine dinucleotide phosphate), because of their biological importance and the abundance of known complexes of the nucleotides. The database contained 1006 complexes with these nucleotides, which represented ∼18% of the total. After eliminating the redundancy with a threshold of 30% sequence identity, 754 complexes were obtained. The parameter tuning dataset (ii) was constructed by choosing 10 complexes for each nucleotide (110 complexes), and the remaining complexes were used as the nucleotide dataset (644 complexes). For the chemically diverse dataset (iv), 147 complexes with ligands that were >500 daltons, other than nucleotides, peptides and sugar were selected from the sc-PDB. The unbound dataset (v) consisting of 35 pairs of protein structures in the bound and unbound forms, was developed by Laurie and Jackson (2005).
In the calculations for the parameter tuning and evaluations, entries of proteins similar to the query (≥30% sequence identity) were removed from the background knowledge dataset.
2.2 Method overview
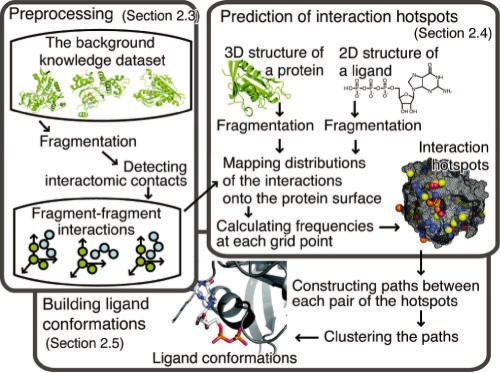
An overview of our method is shown in Figure 1. Our method is composed of three steps: pre-processing (Section 2.3), prediction of interaction hotspots (Section 2.4), and building ligand conformations (Section 2.5). First, information about the fragment–fragment interactions is extracted from the background knowledge dataset. Second, interaction hotspots that are favorable positions for each ligand atom are predicted based on the interaction information. Third, binding sites are predicted by building the conformations of the ligands, based on the interaction hotspots.
Fig. 1.
Overview of our method named ‘BUMBLE’. This method is composed of three steps: pre-processing (Section 2.3), prediction of interaction hotspots (Section 2.4), and building ligand conformations (Section 2.5). In this method, the proteins and ligands are divided into fragments by the ‘fragmentation’ process. The predictions are based on information about the fragment–fragment interactions.
2.3 Pre-processing
In the first step, the information about interactions between protein and ligand fragments is extracted from the 3D structures of protein–ligand complexes in the background knowledge dataset.
In each entry, at first, a protein and a ligand are divided into fragments. The fragments of the protein are defined as the main and side chain moieties of the 20 regular amino acids, while the fragments of the ligand consist of three successive or covalently linked atoms. Next, protein–ligand interatomic contacts are detected by using a threshold of the sum of the van der Waals radii and an offset value (1 Å) as the maximum interatomic distance. When protein and ligand fragment pair contains at least one contacting atom pair, it is recognized as interacting. For each interacting pair of fragments, the types of fragments and the coordinates of the atoms of the ligand fragment, in a coordination system defined by three predefined representative atoms of the protein fragment (Supplementary Table 1), are recorded. The types of protein fragments are defined by the amino acid type and either the main or side chain moiety. For ligand fragments, the types are defined by the force field atom types in the Tripos 5.2 force field (Clark et al., 1989) of the three atoms.
The application of the procedure to all entries in the background knowledge dataset generates the spatial distributions of the ligand fragments around the protein fragments for each combination of fragment types. Then, for each distribution, the coordinates of the ligand fragments are clustered by the complete linkage method, using the RMSD value among them as the clustering radius. The average coordinates in each cluster are used in the following steps.
2.4 Prediction of interaction hotspots
In this step, the interaction hotspots are predicted by using the spatial distributions obtained in the previous step. First, the query protein and the ligand are divided into fragments, as in the pre-processing step. For all pairs of protein fragments that are accessible to solvent and ligand fragments, the spatial distributions are mapped on the query protein surface, by superimposing the protein fragments for the three representative atoms (Supplementary Table S1). Next, the space around the query protein is divided into a 3D grid, and the propensities for interactions at each grid point j are estimated by the following calculation, which is similar to SuperStar (Boer et al., 2001; Verdonk et al., 1999). Each atom ki in the mapped distributions is assigned to eight surrounding grid points j, and the weight w(i, j, ki) is calculated by
where i denotes the unique number assigned to each atom of the query ligand, ki = {1, 2,…, Ni} is the unique number for each mapped atom that is labeled with the atom ID i, Ni is the total number of mapped atoms that are labeled with i, r(ki, j) is the distance between the mapped atom ki and the grid point j, and the summation over j' means the sum of the eight surrounding grid points. In each grid point j- and i-th atom, the frequency f(i, j) is calculated by obtaining the sum over the contributions of all mapped ligand atoms, as follows:
The f(i, j) value is normalized by the Z-score by using the mean and standard deviation of f(i, j), and thus j denotes all of the grid points, and we refer to this as the ‘interaction propensity score’ hereafter. Subsequently, a certain number of grid points are chosen according to the interaction propensity scores for each ligand atom i. The chosen grid points are clustered with their neighbors by a single-linkage method. The grid points with the highest interaction propensity score in each cluster, for each ligand atom i, are regarded as ‘interaction hotspots’.
2.5 Building ligand conformations
In the next step, the ligand conformations are built from the predicted interaction hotspots. For all pairs of interaction hotspots, the shortest paths on a molecular graph of the ligand, between two interaction hotspots, are identified. The paths that do not meet the following three conditions are removed. (i) The path length should be equal to or less than a predefined threshold, and not zero. (ii) The Euclid distance between the two interaction hotspots should be in a predefined range (0.5 Å–1.5 Å per edge). (iii) The path should not be contained in any other paths.
For each generated path, the coordinates of the intervening atoms are simply interpolated and optimized based on the downhill simplex method, one by one. When the total energy of the path is less stable than the predefined threshold, the path is removed. Then, the paths are clustered by the complete linkage method, using a distance that is the RMSD value of the common atoms in each path. In each cluster, the average coordinates of each atom ID i are calculated. If there are deficit atoms in the clusters, then the favorable positions of each deficit atom are screened from the grid points, in the order of their interaction propensity score. When a path between the grid point and the nearest atom in the cluster satisfies the conditions mentioned above, the deficit atom is placed on this grid point. Finally, the conformations are optimized in the Tripos 5.2 force field (Clark et al., 1989) by the simulated annealing method.
The generated ligand conformations are ranked in the order of the sum of the interaction propensity scores of the atoms.
2.6 Parameter tuning
We optimized some adjustable parameters by using the tuning dataset to maximize the prediction accuracy. The prediction accuracy was evaluated by the following three measures: (i) the minimum interatomic distance; (ii) the center distance; and (iii) the RMSD value, between the native and predicted ligands. Threshold maximum values of 3 Å, 5 Å and 5 Å in measures (i), (ii) and (iii), respectively, were used for the success criteria. When only the first criterion was satisfied, it means that the binding site was partially well predicted. In the same way, the third criterion indicates the correct conformations, while the second one means that the binding site was correctly found.
We performed a series of calculations to optimize the parameters by maximizing the prediction accuracy. The determined parameters were the RMSD as the clustering criterion of the fragment interactions in the background knowledge dataset (the following values were tested: 0.0 Å, 1.0 Å, 2.0 Å and 3.0 Å; the optimal value was 2.0 Å), the number of the interaction hotspots for each atom (6, 8, 10, 12, 14; 12), the clustering criterion of the interaction hotspots (1.0 Å, 1.5 Å, 2.0 Å, 2.5 Å, 3.0 Å; 2.0 Å), the maximum number of covalent bonds for the valid path (8, 10, 12; 10), the maximum potential energy (100, 1000, 10 000, 100 000 kJ/mol; 1000 kJ/mol), and the RMSD value as a clustering criterion of the paths (1.0 Å, 2.0 Å, 3.0 Å; 2.0 Å). In this process, all of the parameters, except for the number and the clustering criterion of the interaction hotspots, are considered as independent. Among the various parameter combinations, the most successful calculation resulted in success rates of 89%, 69% and 50% for the first ranked predictions, and 99%, 92% and 75% for the best 10 predictions, in each threshold.
3 RESULTS
3.1 Overview of the test for bound structures
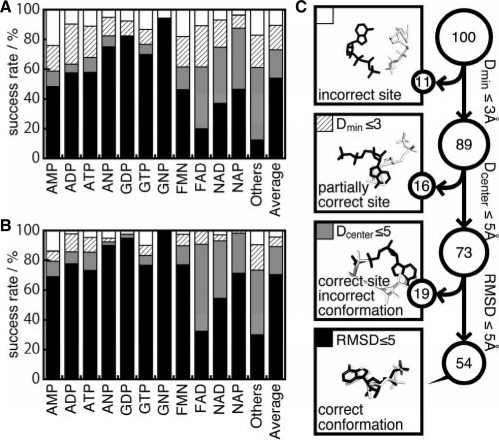
As the first evaluation of our method, we applied it to the nucleotides and the chemically diverse dataset. On the basis of the criteria mentioned in Section 2.6, the prediction results are summarized in Figure 2. The averages success rates were 90%, 71% and 53% for the first-ranked-predicted conformations, and 97%, 90% and 70% in the top 10 conformations. In spite of the fact that we did not use complexes of proteins similar to the query (sequence identity ≥30%) in the background knowledge dataset, our method predicted the binding sites and the conformations reasonably well. In addition, the prediction performances were almost independent of the threshold value used to eliminate the similar proteins in the background knowledge dataset (Supplementary Figure S2). This result may indicate that our method is not strongly influenced by the similarity of the global folds of proteins, since it focuses on the local structural elements represented by the fragment–fragment interactions.
Fig. 2.
Summary of the prediction accuracies for the nucleotide and the chemically diverse datasets. (A and B) Success rates of the first-ranked conformation, and the best in the top 10 conformations, respectively. The first 11 bars indicate the results of each ligand in the nucleotide dataset. ‘Others’ is the result of the chemically diverse dataset. ‘Average’ is the mean value among the other 12 bar plots. (C) Schematic representation of the success rates for ‘Average’ in panel A). All of the predicted conformations can be classified in four categories. Among the predictions, 11% were categorized as predicted conformations located far from the native binding site, 16% were predicted to partially use the same binding sites, 19% were predicted as using the same binding sites but with different binding conformations, and the remaining 54% were correctly predicted, with the same binding sites and conformations as those in the top-ranked prediction. For the top 10 ranked predictions, these ratios were 3%, 7%, 19% and 71%, respectively.
3.2 Evaluation for nucleotide prediction
As summarized in Figure 2A and B, the performances of the predictions were clearly different among the ligand types. In particular, the binding sites and conformations for ligands that contain a guanine moiety were quite accurately predicted. The success rates of the binding conformations for GNP, GDP and GTP were 94%, 83% and 70%, respectively, as a first-ranked candidate. Among the 89 entries involving a guanine moiety, there were only three entries that failed to predict one of the top 10 conformations. These complexes were uracil phosphoribosyl transferase (PDB: 1jlr), ornithine decarboxylase (PDB: 1c4k) and RNA polymerase (PDB: 1s49). In the case of 1jlr, the predicted conformations were located at the binding sites for another ligand (uracil), as shown in Supplementary Figure S1A, B and C. Although the entry 1c4k contains only one subunit, the GTP-binding site was identified at the interface of two subunits, in the original report describing this structure (Vitali et al., 1999). In the entry 1s49, the query protein was RNA-polymerase with a large binding cavity, and the native conformation of GTP in 1s49 was highly exposed to the solvent. The predicted conformations were located at the binding pockets for a template chain of RNA and a nucleotide triphosphate. As described later in detail, our method tends to be weak with exposed ligands.
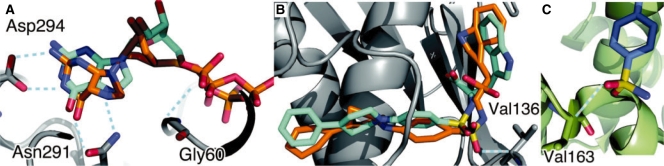
On the other hand, Figure 3A shows the results of the prediction for the Gα13-GDP complex (PDB: 1zcb) as an example of successful prediction with the strictest criterion, where the RMSD value between the native and predicted ligands was 2.05 Å (Fig. 3A). In order to investigate the diversity of the known fragment interactions that contributed to the prediction, we counted the number of interactions assigned to the nearest grid points from the positions of the predicted atoms. The predicted conformation of 1zcb was supported by 54 237 known fragment interactions in 2479 complexes from the background knowledge dataset. In particular, 19 966 known fragment interactions supported the prediction of the interactions of Gly-60, which is the first residue of the P-loop motifs (Kinoshita et al., 1999). In addition, the interactions of Asn-291 and of Asp-294 were supported by 3233 and 2971 known interactions, respectively. These interactions mainly corresponded to hydrogen bonds between the side chains and the guanine base, and were described as the G1 and G3 motifs by Saito et al. (2006). As shown here, this prediction was primarily supported by the interactions derived from well-known binding motifs.
Fig. 3.
(A and B) Screen shots of the prediction results of 1zcb and 1ciz. The molecules shown in cyan and orange depict the native and predicted conformations, respectively. The cyan dashed lines indicate hydrogen bonds described in the manuscript. (C) 2byi, as an example of the fact that background knowledge from different protein folds can contribute to the correct prediction of 1ciz.
In the case of FAD, the success rate of binding conformations (black part of the bar in Fig. 2A and B) was relatively low. Due to the size and the complexity of its chemical structure, it can be difficult to build an appropriate ligand structure. However, the success rates for the binding site (hatched and gray parts of the bars in Fig. 2A and B) were comparable with the average value among all ligands. Therefore, these results may indicate that the interaction hotspots and the binding sites were correctly characterized, in spite of the size and the complexity of the ligand.
3.3 Evaluation for the chemically diverse ligands
It is more difficult to predict the binding sites of chemically diverse ligands than those of nucleotides, due to the bias of the background knowledge dataset [dataset (i) in Section 2.1]. In the dataset, 18% of the entries were complexes with one of the 11 kinds of nucleotides in the nucleotide dataset. In spite of this drawback, the predictions for the non-nucleotide ligands (shown in Fig. 2 as ‘others’) were still accurate. The success rates for the partially correct binding site predictions were 83% and 90% in the first-ranked predictions for the non-nucleotides and for the average of the nucleotides, respectively.
Here, we discuss the prediction for stromelysin-1 with an inhibitor (PDB: 1ciz), where the RMSD value between the native and predicted ligands was 1.86 Å (Fig. 3B). The predicted conformation of 1ciz was supported by 5238 known fragment interactions in 1790 complexes from the background knowledge dataset. Among them, 129 interactions were derived from 14 complexes with the same fold as the query protein, according to the CATH classification (3.40.390.10). This means that the successful prediction of 1ciz was mainly supported by many unrelated proteins, rather than a few similar proteins. For instance, in spite of the dissimilarity between the complex 2byi (HSP90α with an inhibitor) and 1ciz, in terms of its sequence, fold and ligand chemistry, the information of the complex 2byi contributed to the prediction. The hydrogen bond between a Cα atom of Val-163 and an oxygen atom of a sulfoxide in the predicted conformation was similar to the interaction involving Val-136 in 2byi (Fig. 3B and C).
On the other hand, the predictions of 14 entries (10%) in the chemically diverse dataset were not successful by any criteria. An analysis of these complexes revealed that most of the native binding conformations were highly exposed to the solvent. For example, in two cases, interleukin-2 (PDB: 1m49) and acetyl-CoA carboxylase (PDB: 1w96), the ligands bound to the interfaces of protein–protein interactions. In another case, glycogen phosphorylase (PDB: 1z6q), the structure data were for the monomeric protein, but the binding site is formed at the interface of two subunits (Kristiansen et al., 2004). In some other cases, the predictions failed even though the ligand was not highly solvent-exposed. For example, NAD(P)H nitroreductase (PDB: 1oon) bound two molecules in one binding site simultaneously (Supplementary Fig. S1D, E and F). Such complicated binding situations are difficult to predict, because this method does not model the interactions between ligands. In the case of calcium ATPase 1 (PDB: 1wpg), the ligand and the binding site are highly hydrophobic (Toyoshima et al., 2004). In this case, the shape complementarity of the molecular surfaces may be more important than the chemical complementarity.
3.4 Relation between the solvent accessibility of the ligand and the prediction accuracy
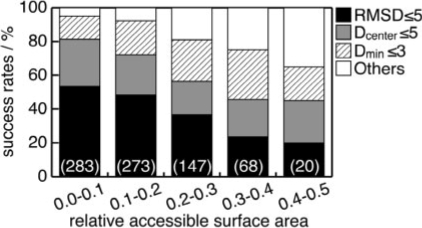
As described above, our method does not adequately predict the binding modes that are highly exposed to the solvent. We investigated this problem quantitatively, by dividing the nucleotide and the chemically diverse datasets into five subsets. These subsets were discriminated by the ratio of the accessible surface areas (ASA) of the ligands in the complex to those in the isolated form, by intervals of 0.1. Their success rates are shown in Figure 4. As a result, the accuracy was found to be strongly affected by the relative ASA value. The success rate of the most exposed ligands was ∼65%, under the partially correct binding site criterion (hatched + gray + black part of the bar, Fig. 4), while that for the most buried ligands was 95% for the top prediction. In short, our method was relatively weak for exposed ligand binding modes. However, it is noteworthy that 97.5% of the entries in the dataset have relative ASA values of <0.4, and thus this characteristic was a minor concern.
Fig. 4.
Dependence of the prediction performance on the relative ASA. The relative ASA is defined as the ratio of the ligand ASA in the isolated form to that in the complex state. The numbers in parentheses indicate the number of entries in each subset.
3.5 Application to unbound structures
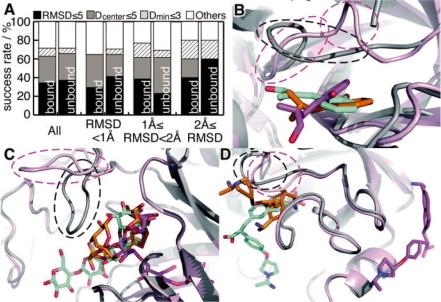
We have applied our method to the dataset consisting of 35 pairs of protein structures in bound and unbound forms. Figure 5A shows a comparison of the success rates between the predictions for bound and unbound forms as query proteins. Surprisingly, the prediction accuracy for the unbound forms was almost the same as that for the bound forms, regardless of differences in the protein conformations. In order to clarify the sensitivity of our prediction to the conformational changes of proteins, we divided the dataset into three subsets, according to the all-atom RMSD values of the binding site residues between the bound and unbound structures; that is, (i) RMSD < 1.0 Å (17 pairs), (ii) 1.0 Å ≤ RMSD < 2.0 Å (13 pairs) and (iii) 2.0 Å ≤ RMSD (five pairs). However, we could not find any significant differences between the success rates for the bound and unbound forms. This was unexpected, but further analysis revealed that the difficulty caused by the conformational changes depended on the manner of change, rather than the amount. For example, in the case of chymotrypsin (PDB: 3gch/1chg, RMSD = 2.97 Å), the binding conformations were correctly predicted in the bound and unbound cases (Fig. 5B). Similarly, in the case of amylase (PDB: 1byb/1bya, RMSD = 3.10 Å), the binding sites of two of the four sugar residues were correctly predicted with the criterion of ‘partially correct binding site’ (Fig. 5C). In contrast to the above two examples, the prediction for trypsinogen (PDB: 1mtw/2tga, RMSD = 1.16 Å) failed due to conformational changes, in which the binding pocket was filled by a loop located near the pocket in the unbound form, although the RSMD value was rather small, as compared with the former two cases (Fig. 5D). In the two successful cases, the binding pockets were open in the unbound forms, but in the last failed case, the binding pocket was closed by the conformational change.
Fig. 5.
The prediction results for the unbound dataset. (A) Success rates of the prediction as a first-ranked prediction, in same manner as Figure 2. ‘All’ means the results for all of the entries in the dataset. The others show the results for each subset, which were divided according to the RMSD value of all atoms in their binding site residues between the bound and unbound forms. Panels B, C and D are screen shots of the prediction results of 3gch/1chg, 1byb/1bya and 1mtw/2tga, respectively. The protein structures in the bound and unbound states are shown by gray and pink ribbons, respectively. The predicted ligand conformations for the proteins in the bound and unbound states are shown by orange and purple sticks, respectively. The native conformations of the ligands are shown by cyan sticks. Remarkable conformational changes induced by ligand binding are highlighted by dashed circles.
3.6 Comparison with existing methods
A comparison of the performance of our method with those of other methods is not straightforward, because of the different presumptions. For example, the existing methods for binding site prediction usually do not require a ligand structure as a query, and many methods search for binding site-like cavities without considering the binding conformations and complementarities. In contrast, our method predicts the binding sites by considering the binding conformations of the query ligand. In addition, the aim of the existing fragment-based methods, which try to predict the binding conformations of ligands by placing a numerous fragments and linking them, is different from ours, since they assume that the binding site is known (Caflisch et al., 1993; Schubert and Stultz, 2009) and they try to predict the precise conformations in the similar way than the docking methods used in AutoDock. Here, we will only discuss the differences between the cases that can and cannot be predicted by our method and others.
Morita et al. (2008) developed a binding-site-prediction method, and evaluated it by comparison with Q-site Finder and Pocket Finder (Laurie and Jackson, 2005). As a result, there were five proteins for which all three methods could not find the binding sites correctly; that is, 6ins, 2tga 1bya, 3app and 1chg. The former two cases also failed in our method possibly because their ligands were highly exposed (relative ASA = 0.34 and 0.35 for 3mth/6ins and 1mtw/2tga, respectively). Moreover, there were significant conformational changes in 2tga from the bound state (Fig. 5D), as described above. In the cases of 1chg (Fig. 5B), 1bya (Fig. 5C) and 3app (RMSD = 1.17 Å to the bound state), the binding sites were successfully predicted by our method, although there were large conformational changes. Our method was more robust to the conformational changes, but more sensitive to the exposure of the binding ligands.
We also compared our method with the AutoDock program (Morris et al., 2009). As a result, when the binding sites were successfully predicted by both methods, the binding conformations predicted by our method tended to be less precise than those predicted by AutoDock. On the other hand, our method predicted the binding sites more accurately than AutoDock did (Supplementary Fig. S3).
3.7 Computation time and limitations
The computation times required for the pre-processing step, the prediction of interaction hotspots, and the building ligand conformations are shown in Table 1. The computation time for the pre-processing linearly increases with the size of the dataset. In the prediction of the hotspots step, the computation time linearly increases with the product of the size of the background knowledge dataset, the number of fragments of the query protein, and that of the ligand. In the building conformation step, the time depends on the number of pairs of interaction hotspots within the defined distance range.
Table 1.
Computation times for pre-processing and predictions for the nucleotide dataset
| Pre-processing | Prediction of interaction hotspots | Building ligand conformations |
|---|---|---|
| 1304 s/5524 complexes | 78 s/complex | 1843 s/complex |
The values for the prediction of interaction hotspots and building ligand conformations are the mean values among 644 entries in the nucleotide dataset. The calculations were performed with an Intel Quad Core Xeon E5450 (3.0 GHz).
In principle, this method does not have upper limits for the sizes of the query protein and ligand. However, the sizes of proteins and ligands are technically restricted by the computation time. In addition, as a lower limitation, the query protein and ligand must contain at least one fragment: an amino acid and three successive atoms, respectively.
4 CONCLUSION
We have proposed a new knowledge-based method for predicting binding sites, by building the ligand conformations from the predicted interaction hotspots. Evaluations revealed that our method could reasonably predict the binding sites not only for nucleotides but also for chemically diverse ligands, although the background knowledge dataset contained a large number of nucleotides. In addition, the robustness to the conformational changes of proteins was shown by a further evaluation with protein structures in the unbound form. An important point is that the predictions were accomplished by using the information about the patterns of fragment interactions that are common among various proteins, as well as the binding motifs.
Our method is available on the web server named ‘BUMBLE’, which means ‘building up molecules for binding location estimation’, at the following address: http://bumble.hgc.jp/.
Funding: Global COE program ‘Deciphering Biosphere from Genome Big Bang’ and KAKENHI (Grant-in-Aid for Scientific Research) on Priority Areas ‘Systems Genomics’ from the Ministry of Education, Culture, Sports, Science and Technology of Japan; Research Fellowships from the Japan Society for the Promotion of Science for Young Scientists (to K.K.); Computation time was provided by the Super Computer System, Human Genome Center, Institute of Medical Science, The University of Tokyo.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Berman H, et al. Announcing the worldwide Protein Data Bank. Nature Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- Boer DR, et al. SuperStar: Comparison of CSD and PDB-based interaction fields as a basis for the prediction of protein-ligand interactions. J. Mol. Biol. 2001;312:275–287. doi: 10.1006/jmbi.2001.4901. [DOI] [PubMed] [Google Scholar]
- Brady GP, Stouten P.FW. Fast prediction and visualization of protein binding pockets with PASS. J. Comp. Aid. Mol. Des. 2000;14:383–401. doi: 10.1023/a:1008124202956. [DOI] [PubMed] [Google Scholar]
- Caflisch A, et al. Multiple copy simultaneous search and construction of ligands in binding-sites–application to inhibitors of hiv-1 aspartic proteinase. J. Med. Chem. 1993;36:2142–2167. doi: 10.1021/jm00067a013. [DOI] [PubMed] [Google Scholar]
- Campbell SJ, et al. Ligand binding: functional site location, similarity and docking. Curr. Opin. Struct. Biol. 2003;13:389–395. doi: 10.1016/s0959-440x(03)00075-7. [DOI] [PubMed] [Google Scholar]
- Clark M, et al. Validation of the general-purpose tripos 5.2 force-field. J. Comput. Chem. 1989;10:982–1012. [Google Scholar]
- Denessiouk KA, Johnson MS. When fold is not important: A common structural framework for adenine and AMP binding in 12 unrelated protein families. Protein Struct. Funct. Genet. 2000;38:310–326. [PubMed] [Google Scholar]
- Denessiouk KA, et al. Adenine recognition: A motif present in ATP-, CoA-, NAD-, NADP-, and FAD-dependent proteins. Protein Struct. Funct. Genet. 2001;44:282–291. doi: 10.1002/prot.1093. [DOI] [PubMed] [Google Scholar]
- Gohlke H, et al. Knowledge-based scoring function to predict protein-ligand interactions. J Mol. Biol. 2000;295:337–356. doi: 10.1006/jmbi.1999.3371. [DOI] [PubMed] [Google Scholar]
- Huang BD, Schroeder M. LIGSITE(csc): predicting ligand binding sites using the Connolly surface and degree of conservation. BMC Struct. Biol. 2006;6 doi: 10.1186/1472-6807-6-19. Article no. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juncker AS, et al. Sequence-based feature prediction and annotation of proteins. Genome Biol. 2009;10:6. doi: 10.1186/gb-2009-10-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger E, et al. sc-PDB: an annotated database of druggable binding sites from the protein data bank. J. Chem. Inform. Model. 2006;46:717–727. doi: 10.1021/ci050372x. [DOI] [PubMed] [Google Scholar]
- Kinoshita K, Nakamura H. Protein informatics towards function identification. Curr. Opin. Struct. Biol. 2003;13:396–400. doi: 10.1016/s0959-440x(03)00074-5. [DOI] [PubMed] [Google Scholar]
- Kinoshita K, Nakamura H. Identification of the ligand binding sites on the molecular surface of proteins. Protein Sci. 2005;14:711–718. doi: 10.1110/ps.041080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K, et al. Structural motif of phosphate-binding site common to various protein superfamilies: all-against-all structural comparison of protein-mononucleotide complexes. Protein Engineer. 1999;12:11–14. doi: 10.1093/protein/12.1.11. [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Go N. ATP binding proteins with different folds share a common ATP-binding structural motif. Nat. Struct. Biol. 1997;4:6–7. doi: 10.1038/nsb0197-6. [DOI] [PubMed] [Google Scholar]
- Kristiansen M, et al. Identification, synthesis, and characterization of new glycogen phosphorylase inhibitors binding to the allosteric AMP site. J. Med. Chem. 2004;47:3537–3545. doi: 10.1021/jm031121n. [DOI] [PubMed] [Google Scholar]
- Laurie A.TR, Jackson RM. Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites. Bioinformatics. 2005;21:1908–1916. doi: 10.1093/bioinformatics/bti315. [DOI] [PubMed] [Google Scholar]
- Lee D, et al. Predicting protein function from sequence and structure. Nat. Rev. Mol. Cell Biol. 2007;8:995–1005. doi: 10.1038/nrm2281. [DOI] [PubMed] [Google Scholar]
- Loewenstein Y, et al. Protein function annotation by homology-based inference. Genome Biol. 2009;10:207. doi: 10.1186/gb-2009-10-2-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, et al. Highly accurate method for ligand-binding site prediction in unbound state (apo) protein structures. Protein Struct. Funct. Bioinform. 2008;73:468–479. doi: 10.1002/prot.22067. [DOI] [PubMed] [Google Scholar]
- Morris GM, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muegge I, Martin YC. A general and fast scoring function for protein-ligand interactions: a simplified potential approach. J. Med. Chem. 1999;42:791–804. doi: 10.1021/jm980536j. [DOI] [PubMed] [Google Scholar]
- Saito M, et al. An empirical approach for detecting nucleotide-binding sites on proteins. Protein Engineer. Des. Select. 2006;19:67–75. doi: 10.1093/protein/gzj002. [DOI] [PubMed] [Google Scholar]
- Schubert CR, Stultz CM. The multi-copy simultaneous search methodology: a fundamental tool for structure-based drug design. J. Comput. Aid. Mol. Des. 2009;23:475–489. doi: 10.1007/s10822-009-9287-y. [DOI] [PubMed] [Google Scholar]
- Shionyu-Mitsuyama C, et al. An empirical approach for structure-based prediction of carbohydrate-binding sites on proteins. Protein Engineer. 2003;16:467–478. doi: 10.1093/protein/gzg065. [DOI] [PubMed] [Google Scholar]
- Sotriffer C, Klebe G. Identification and mapping of small-molecule binding sites in proteins: computational tools for structure-based drug design. Farmaco. 2002;57:243–251. doi: 10.1016/s0014-827x(02)01211-9. [DOI] [PubMed] [Google Scholar]
- Thornton JM, et al. From structure to function: approaches and limitations. Nat. Struct. Biol. 2000;7:991–994. doi: 10.1038/80784. [DOI] [PubMed] [Google Scholar]
- Toyoshima C, et al. Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues. Nature. 2004;432:361–368. doi: 10.1038/nature02981. [DOI] [PubMed] [Google Scholar]
- Verdonk ML, et al. SuperStar: A knowledge-based approach for identifying interaction sites in proteins. J. Mol. Biol. 1999;289:1093–1108. doi: 10.1006/jmbi.1999.2809. [DOI] [PubMed] [Google Scholar]
- Vitali J, et al. Three-dimensional structure of the Gly121Tyr dimeric form of ornithine decarboxylase from Lactobacillus 30a. Acta Crystallogr. D. Biol. Crystallogr. 1999;55:1978–1985. doi: 10.1107/s0907444999010756. [DOI] [PubMed] [Google Scholar]
- Zhou HY, Zhou YQ. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11:2714–2726. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.