Abstract
An analysis of the distribution of the Na+-translocating ATPases/ATP synthases among microbial genomes identified an atypical form of the F1Fo-type ATPase that is present in the archaea Methanosarcina barkeri and M.acetivorans, in a number of phylogenetically diverse marine and halotolerant bacteria and in pathogens Burkholderia spp. In complete genomes, representatives of this form (referred to here as N-ATPase) are always present as second copies, in addition to the typical proton-translocating ATP synthases. The N-ATPase is encoded by a highly conserved atpDCQRBEFAG operon and its subunits cluster separately from the equivalent subunits of the typical F-type ATPases. N-ATPase c subunits carry a full set of sodium-binding residues, indicating that most of these enzymes are Na+-translocating ATPases that likely confer on their hosts the ability to extrude Na+ ions. Other distinctive properties of the N-ATPase operons include the absence of the delta subunit from its cytoplasmic sector and the presence of two additional membrane subunits, AtpQ (formerly gene 1) and AtpR (formerly gene X). We argue that N-ATPases are an early-diverging branch of membrane ATPases that, similarly to the eukaryotic V-type ATPases, do not synthesize ATP.
Contact: galperin@ncbi.nlm.nih.gov; amulkid@uos.de
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
F1Fo-type (F-type) and V/A-type ATPases are membrane-anchored rotary enzymes that couple translocation of H+ or Na+ ions across the membrane to the synthesis or hydrolysis of ATP. Although most of their subunits are evolutionarily related, the two classes of ATPases have clear differences in structure and phylogenetic distribution: F-type ATPases are found in bacteria, mitochondria and chloroplasts, whereas V/A-type ATPases are found in eukaryotic cell membranes (in particular, vacuoles), as well as in all archaea and in some bacteria (Hilario and Gogarten, 1998; Forgac, 2007; von Ballmoos et al., 2008; Mulkidjanian et al., 2009). V/A-type ATPases are often subdivided into two classes based (i) on the ability of the prokaryotic (A-type) enzyme to function in the direction of ATP synthesis and (ii) presence of additional subunits in the eukaryotic (V-type) enzyme that normally functions only as an ion-translocating ATPase, using the energy of ATP hydrolysis to acidify cellular compartments (Hilario and Gogarten, 1998; Forgac, 2007).
Structural characterization of the Na+-binding sites in the c-subunits of F- and V-type ATPases (Meier et al., 2005; Murata et al., 2005) allowed unequivocal assignment of the cation specificity for the membrane ATPases encoded in numerous sequenced genomes: only those c-subunits that contain full sets of Na+ ligands appeared to transport Na+, the loss of at least one of those ligands correlated with the loss of Na+ specificity (Mulkidjanian et al., 2008a; Meier et al., 2009).
In the view of several reports on the sodium dependence of energy-coupled reactions in cyanobacteria (Willey et al., 1987; Skulachev, 1988; Pogoryelov et al., 2003), we have searched cyanobacterial genomes for the Na+-translocating ATPases. We found several apparently Na+-dependent cyanobacterial ATPases, but always as second copies, in addition to the H+-translocating ATP synthases. Here, we report the common properties of these ATPases, which are encoded in apparently highly mobile operons and have a set of specific traits that qualify them as a separate subfamily of the F-type ATPases. Since these ATPases, besides forming a novel subfamily, are always encoded next to the typical rotary ATPases and are predominantly Na+-dependent, we refer to them hereafter as N-ATPases.
2 METHODS
Phylogenetic distribution of the N-ATPase operons was deduced from protein BLAST (Altschul et al., 1997) searches against the NCBI's RefSeq database (Pruitt et al., 2009) (last searched February 1, 2010) and verified by examining gene neighborhoods of the retrieved ORFs and by checking for the presence of the N-ATPase-specific subunit AtpR. Phylogenetic trees were constructed using the neighbor-joining algorithm with MEGA (Kumar et al., 2008). Multiple alignments were constructed from BLAST outputs with manual editing. Transmembrane segments were predicted using TMHMM (Krogh et al., 2001). Sequence logos were drawn with WebLogo (Crooks et al., 2004).
3 RESULTS
Search of the NCBI's RefSeq database (Pruitt et al., 2009) for cyanobacterial c (proteolipid) subunits that would have a full set of Na+-binding ligands (Mulkidjanian et al., 2008a; Meier et al., 2009) returned five hits, all coming from marine cyanobacteria (Supplementary Fig. S1a). In each case, the operon encoding the Na+-binding c subunit was present in the genome along with another ATPase operon, encoding an H+-translocating F-type ATPase. As depicted below for Synechococcus sp. PCC 7002, an alignment of the Na+- and H+-binding c-subunits (top and bottom, respectively) from the same cyanobacteria revealed a Glu substitution of the Gln residue (shown in blue) that serves as a Na+ ligand (uncharged residues of the transmembrane segments are highlighted in yellow, Na+ and H+ ligands are labeled with asterisks) Among the 4084

c-subunit sequences currently listed in the Pfam family ATP-synt_C (PF00137; Finn et al., 2010), only 227 (5.5%) contain a Glu residue in that position and only ∼1% combine it with a typical ESTxxY Na+-binding motif (Supplementary Fig. S1). The Na+-dependent ATPase operons in all cyanobacteria had a similar gene order, with a single gene insertion in Acaryochloris marina and a two-gene insertion in the two strains of Cyanothece sp. (Supplementary Fig. S2).
3.1 Always the second: conservation and distinctive properties of the N-ATPase
A search of the complete genome database identified homologous N-ATPase operons in some representatives of the bacterial phyla Aquificae, Chlorobi and Planctomycetes, in certain members of α-, β-, γ- and δ-subdivisions of Proteobacteria, and in two archaea, Methanosarcina barkeri and M.acetivorans (see Supplementary Fig. S2). All these operons had the same atpDCI-urf2-atpBEFAG gene order, encoding, respectively, β-, ε-, 1-, Urf2-, a-, c-, b−, α- and γ-subunits of this particular F-type ATPase, first described in M.barkeri by Sumi et al. (1997). The only exception outside of cyanobacteria was Persephonella marina, a member of the Aquificae, which has an N-ATPase operon with the urf2-atpBEFAGDCI gene order and also harbors operons for F- and A-type ATPases (Supplementary Fig. S2).
Identification of the N-ATPase operons in diverse microorganisms was simplified by the fact that these operons are always present in the genomes as second copies alongside operons that encode typical H+-transporting F-type ATPases (in M.barkeri and M.acetivorans, A-type ATPases). In contrast, most N-ATPase c-subunits contain full sets of Na+ ligands, indicating that these enzymes are specific for Na+ ions. Just like the cyanobacterial Na+-binding c-subunits, c-subunits of most N-ATPases had Glu residues in the middle of both transmembrane helices (Supplementary Fig. S1).
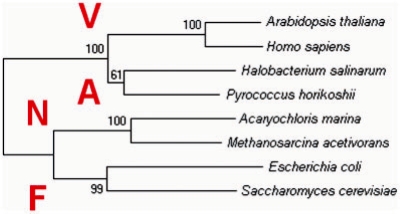
The same subunits of the N-ATPases from diverse bacteria are closely related and form distinct branches on the phylogenetic trees, well separated from the equivalent subunits of the F- and V-type ATPases (Fig. 1 and Supplementary Fig. S3, see also Supplementary Fig. 5 in Swingley et al., 2008). Phylogenetic trees built for individual N-ATPase subunits showed similar topologies, so that the whole N-ATPase operons appeared to co-evolve (Supplementary Fig. S4).
Fig. 1.
A neighbor-joining tree comparing α-subunits of N-ATPases with α-subunits of F-type ATPases and B-subunits of A- and V-type ATPases. See Supplementary Fig. S3 for the full tree.
Another distinct trait of the N-ATPase operons was the presence of an extra gene, urf2 (Sumi et al., 1997). Its product is currently annotated as ‘F1/F0 ATPase, Methanosarcina type, subunit 2’ (F1F0_chp_2, TIGR03165, DH Haft, December 4, 2006) in the JCVI's Comprehensive Microbial Resource (Davidsen et al., 2010) and as ‘ATPase_F1/F0-cplx_su2_Meth-typ’ (IPR017581) in the InterPro database (Hunter et al., 2009), whereas UniProt uniformly annotates these proteins as ‘Putative uncharacterized protein’. The urf2 (hereafter atpR) gene was found in every N-ATPase operon and could be used as a tell-tale sign of these operons. Its product is a hydrophobic protein with three predicted transmembrane segments, two of which carry conserved Arg residues (Supplementary Fig. S5). Presence of charged residues in the hydrophobic core of the membrane is rare and usually indicative of a functional role.
The N-ATPase operons also include so-called gene 1 (hereafter atpQ) that encodes another distinct membrane protein. Although this gene was originally marked as atpI (Sumi et al., 1997) and is occasionally still labeled this way (Saum et al., 2009), its product shows no statistically significant sequence similarity to the genuine AtpI (UncI) proteins, previously described in the genomes of Escherichia coli and other model organisms. In accordance with their distinct sequences, AtpI and AtpQ proteins are assigned to two different Pfam families, PF03899 and PF09527, respectively, and to two different COGs, COG3312 and COG5536 in the CDD database (Marchler-Bauer et al., 2009). In contrast to atpR, the atpQ gene is not limited to the N-ATPase operons.
While the N-ATPase operon lacks the atpH gene that encodes the δ-subunit of the F-type ATPase, its atpF gene (atpFN) is unusually long (Sumi et al., 1997). We were able to align C-terminal regions of the AtpFN and AtpH gene products (Supplementary Fig. S6). This showed that N-ATPase contains at least part of the δ-subunit, in agreement with the earlier analyses (Saum et al., 2009).
3.2 Lateral transfer of N-ATPase genes
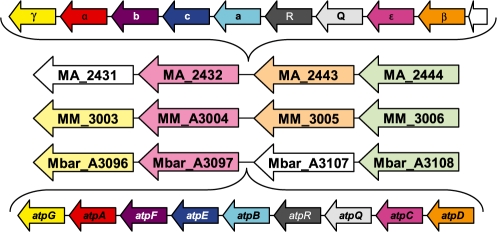
Presence of the N-ATPase operon in the genomes of M.barkeri and M.acetivorans, but not in the closely related M.mazei, suggested that this operon had been acquired via lateral gene transfer. This suggestion is consistent with the gene neighborhoods of the atpDCQRBEFG operons in M.barkeri and M.acetivorans (Fig. 2) and the absence of these operons in any other archaeal genomes sequenced so far. Gene neighborhoods of the N-ATPase operons in various bacteria are also consistent with the insertion of this operon (Supplementary Fig. S7). The widespread presence of the N-ATPase genes among diverse bacteria deprecates the historical designation of these enzymes as ‘archaebacterial F1Fo-ATPases’. The strict conservation of the gene order and co-linearity of the phylogenetic trees for distinct N-ATPase subunits suggests that the whole operon is being transferred as a single unit. However, the GC content of the N-ATPase operons shows a good correlation with the average GC content of the host genomes (Supplementary Fig. S8), indicating either a relatively ancient gene transfer or a rapid adaptation of the N-ATPase genes to their host environment. An additional indication of the lateral mobility of the N-ATPase operon is its presence on plasmids, pREB4 in A.marina and pAQ7 in Synechococcus sp. PCC 7002. These data might be related to the earlier functional evidence of the presence of two ion-translocating ATPases in M.mazei Gö1. While the A-type ATP synthase was apparently H+-dependent, the second, F-type ATPase appeared to be Na+-translocating (Becher and Müller, 1994; Pisa et al., 2007). Although this second ATPase has not been found in the sequenced genome of M.mazei (Fig. 2), the respective genes could be plasmid-borne, as is the case of N-ATPases in at least two cyanobacteria.
Fig. 2.
Conserved gene neighborhoods in M.acetivorans, M.mazei and M.barkeri, indicating the points of insertion of the N-ATPase operons in the former (top line, showing the subunit names) and the latter (bottom line, showing the gene names). Orthologous genes are indicated with the same colors. The arrows do not reflect the lengths of the genes.
4 DISCUSSION
Following the description of an ‘archaebacterial F1Fo-ATPase’ in M.barkeri (Sumi et al., 1997), the presence of a ‘Methanosarcina-like’ F-ATPase operon was repeatedly noted in bacterial genomes (Glöckner et al., 2003; McInerney et al., 2007; Swingley et al., 2008), although the exact function(s) of these enzymes and their cation specificity remained obscure. McInerney et al. (2007) noted two F-type ATPases encoded in the genome of Syntrophus aciditrophicus and suggested that both of them were Na+-translocating (incidentally, the F-type ATPase of this organism is definitely H+-specific, whereas the cation specificity of its N-ATPase is unknown; it might be specific for Na+, see below). In their analysis of the A.marina genome, Swingley et al. (2008) noted the presence of a plasmid-encoded ‘set of ATP synthase genes that were arranged into a unique operon … conserved with full synteny in a remarkable array of organisms, including cyanobacteria, archaea, planctomycetes, chlorobi and proteobacteria’. The authors noted that the α-subunits encoded in these operons clustered together on a phylogenetic tree and suggested that these enzymes formed a separate new family of ATP synthases. However, they slightly overstated their case by claiming that its ‘individual proteins do not clearly fit into any of the described families’ (Swingley et al., 2008). They also disputed the idea that these enzymes were Na+-translocating. In addition, the key observation by Daniel Haft that these enzymes always ‘represent a second F1/Fo ATPase system’ has only been published online in the JCVI's Comprehensive Microbial Resource (Davidsen et al., 2010). As a result, there still exists a significant confusion as to the phylogenetic distribution, organization and the functional role(s) of these enzymes.
The alignment in Supplementary Fig. S1 shows that, despite the doubts of Swingley et al. (2008), the c subunit of the A.marina N-ATPase has a full set of Na+-binding ligands, including the recently recognized additional Thr residue of the ESTxxY motif (Mulkidjanian et al., 2008a; Meier et al., 2009). While this residue is missing in c-subunits of several N-ATPases, including the one from S.aciditrophicus, a Glu residue is present instead of the Na+-coordinating Gln residue in the first transmembrane helix of the c subunit of nearly all N-ATPases (Supplementary Fig. S1). As has been noted previously (Meier et al., 2009; Saum et al., 2009), this Glu residue could potentially provide two ligands for the Na+ ion and thereby complete the Na+ coordination shell. If so, all these N-ATPases would end up being capable of binding Na+ ions.
A hallmark of the N-ATPase operons is the presence of the atpR gene. Because of the low dielectric permittivity of the membrane, the strategic positioning of two Arg residues of AtpR in the hydrophobic core of the membrane (http://bioinformatics.oxfordjournals.org/cgi/content/full/btq234/DC1) implies the presence of negatively charged residues in their vicinity. Given the absence in the N-ATPase operons of the atpI gene, whose product was recently shown to interact with the c-ring (Suzuki et al., 2007), we suggest that the product of the AtpR gene serves essentially the same function, regulating N-ATPase assembly and/or its activity. Just like AtpI assists c-ring assembly by directly interacting with the c-subunits (Suzuki et al., 2007), AtpR could do that through the interaction of its two Arg residues with N-ATPase-specific c-subunits, most of which carry two Glu residues in the middle of their transmembrane helices (Supplementary Fig. S1).
The observation that N-ATPases are always found alongside typical F- or A-type ATPases suggests that the N-ATPases cannot functionally replace those enzymes in their role as ATP synthases. Indeed, in M.acetivorans, deletion of the N-ATPase operon had no visible effect on cell growth or ATP synthesis, whereas a mutant lacking the A-ATP synthase genes could not be obtained (Saum et al., 2009). We conclude that, similarly to the eukaryotic V-ATPases, the N-ATPases do not catalyze ATP synthesis, which leaves ATP-driven ion pumping as the most plausible function for these enzymes. By analogy with V-ATPases, the N-ATPase c-oligomer ring can be expected to consist of a smaller number of c-subunits than the c-oligomer ring of F-type ATP synthases.
Acquisition of an operon capable of extrusion of Na+ ions would be beneficial to the marine bacteria and other organisms living in high-salt environments. Accordingly, many N-ATPase-encoding bacteria are either marine organisms or grow in the presence of salt. Since Na+-translocating ATPases can also translocate protons (von Ballmoos et al., 2008), the N-ATPases could in principle function as outward proton pumps in low-sodium and/or acidic environments. A typical N-ATPase operon (with untranslated AtpR) has been found in an industrial strain of Pseudomonas veronii growing on 2-butanone (Onaca et al., 2007). The presence of N-ATPase genes in such pathogens as Burkholderia mallei appears to be inherited from their free-living relatives and might be related to their survival in blood.
Using a larger set of sequences than the one used by Swingley et al. (2008), we have confirmed that N-ATPases branch separately from other F-type ATPases (Fig. 1, Supplementary Fig. S3). This separate branching suggests a possible early divergence of the N-ATPases, which is compatible with the following, supposedly ancestral traits of these enzymes:
Both AtpFN subunit of the N-ATPase and the E subunit of the peripheral stalk of the V/A-type ATPases correspond to a fusion of b- and δ-subunits of the typical F-ATPases (Pallen et al., 2006).
The presence of the second membrane-embedded Glu residue is consistent with the evolution of the c-subunit from a duplication of an amphiphilic helix that contained a Glu residue in the middle (Davis, 2002). Similar two-Glu c-subunits are found in the A-type ATPases of methanogens and F-type ATPases of Thermotogae.
Outward pumping of Na+ ions, the predicted function of N-ATPases, appears to be an ancient trait and has been previously suggested as a function of the common ancestor of the F- and V-type ATPases (Mulkidjanian et al., 2007; 2008a, b; 2009).
All these features, which N-ATPases share either with V/A-ATPases—to the exclusion of most F-ATPases—or with the putative common ancestor of all rotary ATPases, suggest that N-ATPases represent a distinct early-diverging family of rotary ATPases. Thus, the Na-translocating common ancestor of all F-type ATPases apparently gave rise to two different families of ATPases: (i) the reversible ATPases/ATP synthases (‘genuine’ F-ATPases) and (ii) ATP-driven ion pumps (N-ATPases).
In conclusion, the N-ATPases (until now usually referred to as ‘archaebacterial F1Fo-ATPases’) are encoded in an apparently highly mobile operon that, most likely, confers on its hosts the ability of ATP-driven outward pumping of Na+ ions, which complements the H+ specificity of the native chromosome-encoded F-ATPase (or A-ATPase). We predict that, similarly to the eukaryotic V-ATPases, the N-ATPases do not catalyze ATP synthesis, which is why an N-ATPase is never found alone in a genome, only as the second enzyme in cells that already encode an F-type or a A-type ATP synthase. Accordingly, caution should be exercised when referring to their components as subunits of the ATP synthase. We believe that experimental verification of the predicted functions of the N-ATPases would be useful for the proper annotation of these interesting enzymes and could also help in understanding the evolution of energy conservation mechanisms.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Eugene Koonin and Thomas Meier for discussion.
Funding: Deutsche Forschungsgemeinschaft (to D.V.D., A.Y.M.); German Academic Exchange Service (to D.V.D.); Intramural Research Program of the NLM at the National Institutes of Health (to M.Y.G.).
Conflict of Interest: none declared.
REFERENCES
- Altschul SF, et al. Gapped BLAST and PSI-BLAST—a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher B, Müller V. ΔμNa+drives the synthesis of ATP via an ΔμNa+-translocating F1Fo-ATP synthase in membrane vesicles of the archaeon Methanosarcina mazei Gö1. J. Bacteriol. 1994;176:2543–2550. doi: 10.1128/jb.176.9.2543-2550.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, et al. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidsen T, et al. The comprehensive microbial resource. Nucleic Acids Res. 2010;38:D340–D345. doi: 10.1093/nar/gkp912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BK. Molecular evolution before the origin of species. Prog. Biophys. Mol. Biol. 2002;79:77–133. doi: 10.1016/s0079-6107(02)00012-3. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- Glöckner FO, et al. Complete genome sequence of the marine planctomycete Pirellulasp. strain 1. Proc. Natl Acad. Sci. USA. 2003;100:8298–8303. doi: 10.1073/pnas.1431443100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilario E, Gogarten JP. The prokaryote-to-eukaryote transition reflected in the evolution of the V/F/A-ATPase catalytic and proteolipid subunits. J. Mol. Evol. 1998;46:703–715. doi: 10.1007/pl00006351. [DOI] [PubMed] [Google Scholar]
- Hunter S, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, et al. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kumar S, et al. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney MJ, et al. The genome of Syntrophus aciditrophicus: life at the thermodynamic limit of microbial growth. Proc. Natl Acad. Sci. USA. 2007;104:7600–7605. doi: 10.1073/pnas.0610456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier T, et al. Structure of the rotor ring of F-type Na+-ATPase from Ilyobacter tartaricus. Science. 2005;308:659–662. doi: 10.1126/science.1111199. [DOI] [PubMed] [Google Scholar]
- Meier T, et al. Complete ion-coordination structure in the rotor ring of Na+-dependent F-ATP synthases. J. Mol. Biol. 2009;391:498–507. doi: 10.1016/j.jmb.2009.05.082. [DOI] [PubMed] [Google Scholar]
- Mulkidjanian AY, et al. Inventing the dynamo machine: the evolution of the F-type and V-type ATPases. Nat. Rev. Microbiol. 2007;5:892–899. doi: 10.1038/nrmicro1767. [DOI] [PubMed] [Google Scholar]
- Mulkidjanian AY, et al. Evolutionary primacy of sodium bioenergetics. Biol. Direct. 2008a;3:13. doi: 10.1186/1745-6150-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkidjanian AY, et al. The past and present of the sodium energetics: may the sodium-motive force be with you. Biochim. Biophys. Acta. 2008b;1777:985–992. doi: 10.1016/j.bbabio.2008.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkidjanian AY, et al. Co-evolution of primordial membranes and membrane proteins. Trends Biochem. Sci. 2009;34:206–215. doi: 10.1016/j.tibs.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T, et al. Structure of the rotor of the V-type Na+-ATPase from Enterococcus hirae. Science. 2005;308:654–659. doi: 10.1126/science.1110064. [DOI] [PubMed] [Google Scholar]
- Onaca C, et al. Degradation of alkyl methyl ketones by Pseudomonas veroniiMEK700. J. Bacteriol. 2007;189:3759–3767. doi: 10.1128/JB.01279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallen MJ, et al. Evolutionary links between FliH/YscL-like proteins from bacterial type III secretion systems and second-stalk components of the FoF1 and vacuolar ATPases. Protein Sci. 2006;15:935–941. doi: 10.1110/ps.051958806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisa KY, et al. The coupling ion in the methanoarchaeal ATP synthases: H+vs. Na+in the A1AoATP synthase from the archaeon Methanosarcina mazeiGö1. FEMS Microbiol. Lett. 2007;277:56–63. doi: 10.1111/j.1574-6968.2007.00939.x. [DOI] [PubMed] [Google Scholar]
- Pogoryelov D, et al. Sodium dependency of the photosynthetic electron transport in the alkaliphilic cyanobacterium Arthrospira platensis. J. Bioenerg. Biomembr. 2003;35:427–437. doi: 10.1023/a:1027339814544. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, et al. NCBI reference sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saum R, et al. The F1FoATP synthase genes in Methanosarcina acetivoransare dispensable for growth and ATP synthesis. FEMS Microbiol. Lett. 2009;300:230–236. doi: 10.1111/j.1574-6968.2009.01785.x. [DOI] [PubMed] [Google Scholar]
- Skulachev VP. Membrane Bioenergetics. Berlin: Springer; 1988. [Google Scholar]
- Sumi M, et al. FoF1-ATPase genes from an archaebacterium Methanosarcina barkeri. Biochem. Biophys. Res. Commun. 1997;241:427–433. doi: 10.1006/bbrc.1997.7809. [DOI] [PubMed] [Google Scholar]
- Suzuki T, et al. The product of uncIgene in F1Fo-ATP synthase operon plays a chaperone-like role to assist c-ring assembly. Proc. Natl Acad. Sci. USA. 2007;104:20776–20781. doi: 10.1073/pnas.0708075105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swingley WD, et al. Niche adaptation and genome expansion in the chlorophyll d-producing cyanobacterium Acaryochloris marina. Proc. Natl Acad. Sci. USA. 2008;105:2005–2010. doi: 10.1073/pnas.0709772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Ballmoos C, et al. Unique rotary ATP synthase and its biological diversity. Annu. Rev. Biophys. 2008;37:43–64. doi: 10.1146/annurev.biophys.37.032807.130018. [DOI] [PubMed] [Google Scholar]
- Willey JM, et al. Sodium-coupled motility in a swimming cyanobacterium. J. Bacteriol. 1987;169:3429–3434. doi: 10.1128/jb.169.8.3429-3434.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.