Abstract
Marek’s disease virus (MDV) is an alphaherpesvirus that causes lethal T-cell lymphomas in chickens. MDV is unique in that it harbors two copies of a viral telomerase RNA subunit (vTR) in its genome exhibiting 88% sequence identity to the chicken orthologue, chTR. The minimal telomerase ribonucleoprotein complex consists of a protein subunit with reverse transcriptase activity (TERT) and TR. Physiologically, the complex compensates for the progressive telomere shortening that occurs during mitosis and is involved in the process of cellular immortalization. Previous studies showed that MDV vTR performes an auxiliary function during oncogenesis. Comparative in vitro analysis of the viral and chicken TR promoters revealed that the vTR promoter (PvTR) was up to 3-fold more efficient than the chTR promoter (PchTR) in avian cells and that the stronger transcriptional activity of PvTR resulted largely from an E-box located two nucleotides downstream of the transcriptional start site of the vTR gene. To test the hypothesis that PvTR is required for vTR expression and, hence, efficient tumor formation, we generated a recombinant virus, vPchTR+/+, in which the vTR promoter was replaced by that of chTR. In vivo, growth of vPchTR+/+ was indistinguishable from that of parental virus; however, tumor induction was reduced by > 50% and lymphomas were smaller and less disseminated when compared to those induced by parental virus. We concluded that PvTR is not required for lytic replication in vivo but is essential for efficient transcription of vTR and thereby critical for efficient MDV lymphoma formation.
Keywords: MDV, lymphoma, viral telomerase RNA, promoter control
1. INTRODUCTION
Marek’s disease virus (MDV) is an avian herpesvirus that causes Marek’s disease (MD), a highly contagious and malignant T-cell lymphoma in chicken. MDV is a member of the genus Mardivirus within the Alphaherpesvirinae subfamily [4, 5]. The MDV genome is approximately 180 kb in length and consists of a unique long (UL) and a unique-short (US) segment, each of which are flanked by inverted repeats, called terminal and internal repeats long (TRL and IRL), and terminal and internal repeats short (TRS and IRS), respectively [17, 37]. MD is characterized by immunosuppression, neurological symptoms and the induction of T-cell lymphomas in the viscera as early as 2 weeks post-infection (p.i.). According to the current model of pathogenesis, infection is initiated in the respiratory tract after inhalation of infectious virus where phagocytic cells, macrophages and/or dendritic cells (DC), become infected. Within 24 h after initial uptake, virus is detectable in the bursa of Fabricius. Lytic replication occurs in many cell types, including B and T cells, but only T cells, predominantly of the CD4+ phenotype, harbor latent MDV that can ultimately result in transformation [2, 29].
Apart from its significance to animal health, MDV provides a good model to study virus-induced oncogenesis. It is well established that MDV encodes the oncoprotein Meq, which has been shown to directly cause transformation of cells. Recently, another viral product encoding a telomerase RNA component (TR), termed vTR, has been reported to play a role in the development, maintenance, and dissemination of MDV-induced tumors [36]. MDV contains in its genome two identical copies of vTR with one copy contained in both the TRL and IRL [7].
TERT, a reverse transcriptase, and its partner TR represent the catalytic core of the telomerase, a eukaryotic ribonucleoprotein complex that compensates by the addition of telomeres for the inability of DNA polymerase to copy the 3′ end of linear chromosomes [1]. Telomeres are essential nucleoprotein structures of all eukaryotic chromosomes, protecting them from exonuclease degradation and end-to-end chromosomal fusions. Telomerase replicates telomeres by utilizing TERT and its intrinsic RNA template TTAGGG within TR to add multiple repeats onto the ends of chromosomes. This DNA elongation compensates for the erosion of telomeres during each round of DNA replication [1, 40]. Telomerase activity is not detectable in normal somatic cells, but is strongly expressed during fetal development and constitutively expressed in highly proliferative cells such as germ line cells, stem cells, and lymphocytes [11, 38]. Hence, telomere erosion limits the replicative capacity of somatic cells [9, 20] and when telomeres shorten to a critical length, cells enter a stage termed replicative senescence where cell division is prevented [10].
Telomerase levels are elevated in more than 85% of human cancers. High telomerase activity and stabilization of telomere lengths have also been detected in spontaneously-induced tumors or virus-transformed cell lines [16, 31, 32]. In cells infected by oncogenic viruses, an increase in telomerase activity has been associated with an up regulation of the TERT component through a complex network of transcription factors and cis-acting sequences [22].
MDV is unique among viruses because it is the only known virus encoding its own telomerase RNA. The MDV RB-1B strain vTR is a 443 nt RNA that displays 88% sequence homology to that of the chicken TR gene (chTR), suggesting that it was pirated from the chicken genome [7]. Functionality of vTR has been demonstrated in TR deficient murine cells in which vTR is capable of supporting telomerase activity by interacting with TERT more efficiently than chTR [8]. The role of vTR in MDV lymphomagenesis was also demonstrated as deletions of the CR1-2 region in both copies of vTR in the oncogenic strain MDV RB-1B resulted in a 60% reduction of tumors in infected chickens [36]. Lymphomas in chickens challenged with vTR deletion viruses were smaller in size and less widely disseminated. Thus, vTR exhibits auxiliary functions during oncogenesis, enhancing the incidence and severity of lymphoma caused by MDV [36]. Also, up regulation of telomerase activity during MDV infection was associated with an increase in vTR gene expression [33]. In vitro analyses showed that up regulation of vTR expression is in fact related to a stronger transcriptional activity of the vTR promoter (PvTR), that was associated to an E-box located two nucleotides downstream from the transcriptional start site of the vTR gene. The c-Myc oncoprotein has been implicated in the transcriptional regulation of vTR, which in turn can be controlled through Meq activity [19, 24, 33].
Here, we tested the hypothesis that the authentic PvTR is required for efficient and timely coordinated expression of vTR and, hence, efficient tumor formation. By generating a recombinant virus, vPchTR+/+, in which PvTR was replaced by its chicken counterpart, we observed that lytic replication of vPchTR+/+ in vivo was indistinguishable from that of parental virus. However, tumor induction was reduced by more than 50% and those lymphomas were smaller in size and less widely disseminated when compared to those induced by parental virus. These data indicated that the PvTR is also essential for efficient MDV lymphoma formation, most likely through high level vTR expression.
2. MATERIALS AND METHODS
2.1. Cells and viruses
Chicken embryo cells (CEC) were prepared from 11-day-old embryos as previously described [30] and maintained in minimal essential medium (Biochrom, Berlin, Germany) supplemented with 10% fetal bovine serum (Biochrom) and grown at 37 °C in a 5% CO2 atmosphere.
The mutant in which the PvTR was exchanged with the chTR promoter (PchTR) was termed vPchTR+/+ and the revertant virus in which the vTR promoter was reinserted was termed vPvTR+/+. The mutants were generated from pRB-1B, an infectious bacterial artificial chromosome (BAC) containing the genome of the highly virulent RB-1B strain [26] that had been restored for horizontal transmission [13]. Parental vRB-1B and recombinant viruses vPchTR+/+ and vPvTR+/+ were reconstituted by CaPO4 co-transfection of BAC DNA with pCAGGS-NLS/Cre [12, 15], a plasmid expressing the Cre enzyme for excision of mini-F sequences using loxP sites, into CEC. After 2 to 4 passages, virus stocks were prepared and frozen in liquid nitrogen until further use.
2.2. Bacteria, plasmids and BAC
All plasmids were constructed and maintained in Escherichia coli DH10B cells grown at 37 °C in Luria-Bertani (LB) medium unless otherwise stated. BAC maintenance and Red recombinations were performed in E. coli strain GS1783 (kindly provided by Dr Gregory Smith, Northwestern University, Chicago, IL, USA) harboring pRB-1B and containing a λ prophage encoding the recombination enzymes Exo, Beta, and Gam [35]. The PvTR sequences were replaced by the PchTR sequences in pRB-1B genome by two-step Red mutagenesis, following the strategy described for introduction of long sequence stretches [35]. DNA extracted from CEC was used as a template to PCR amplify the PchTR with primers PchTR-F and PchTR-R (Tab. I). The PCR product was digested with HindIII and EcoRI and cloned into the corresponding sites of plasmid pUC18 resulting in recombinant plasmid pUC18-PchTR. The second step was the construction of the universal transfer plasmid pEPkan-PchTR, which harbors a transfer cassette, termed PchTR-I-SceI-aphAI, and contains the sequences of interest (PchTR) with an aphAI sequence that confers kanamycin resistance (kan r), an adjoining I-SceI site, and a duplication of a short sequence of PchTR. To this end, the I-SceI-aphAI fragment was PCR amplified from plasmid pEPkan-S [35] using primers pEPchTRpromIN-F and pEPchTRpromIN-R (Tab. I). After gel purification, the PCR product was cloned into plasmid pUC18-PchTR using a singular PstI site located in the PchTR fragment. The cassette PchTR-I-SceI-aphAI was amplified from the plasmid pEPkan-PchTR using primers PvTRexch-Forw and PvTRexch-Rev and then introduced into pRB-1B by a first Red recombination after electroporation of 100 ng of the purified fragment into GS1783 harboring the BAC. The insertion of the transfer cassette resulted in duplication of a short sequence stretch within PchTR, which was used to remove the aphAI resistance gene by a second Red recombination after induction of the restriction endonuclease I-SceI [35]. The exact same procedure was repeated in the second repeat-long region, resulting in introduction of two copies of PchTR into pRB-1B; the final construct was termed pPchTR+/+.
Table I.
Primers used in this study.
| Primers | Sequence 5′→3′ | |
|---|---|---|
| PchTR | AAAAAAGCTTCAAGAGATCGGCGTTGCTTTC | Underlined HindIII site |
| PchTR-R | AAAAGAATTCGCCTCCACCCGCCACGCGTG | Underlined EcoRI site |
| pEPchTRpromIN-F | AAAACTGCAGCTAGCTTGCTGGCGGTGTCCGGAGCAAA TCCGCATCTCGAACTGTAGGGATAACAGGGTAATCGATTT | Underlined PstI site |
| pEPchTRpromIN-R | AAATCTGCAGGCCAGTGTTACAACCAATTAACC | Underlined PstI site |
| PvTRexch-Forw | CCCCAAAAACCTCTCGCGGCCGACGCGCCCCCAGGGATGTAATTACGTCCCTCCCCCCTAGGGGGCAGCAGGCCAAGAGATCGGCGTTGCTTTC | |
| PvTRexch-Rev | G ACACAGCGGAGCCTTCCACCCGCCACGTGTGCCTTCCACCCGCCACGCGTG | |
| Resc-PvTR-F | AAGCTTGACAGGCAGTTGTACACCTG | Underlined HindIII |
| Resc-PvTR-R | GAATTCCTGAAAGTCAGCGAGTAAAC | Underlined EcoRI site |
| pEP-PvTR-F | GGATCCGATCCCGCAGACCCCGGCCCACAGGAAGGGGCGGGGCACGTGCAGGATGACGACGATAAGTAGGGATAAC | |
| pEP-PvTR-R | GGAT CCGGGTAATGCCAGTGTTACAACCA | |
| EPrescPvTR-F | CCAAAAACCTCTCGCGGCCGACAGGCAGTTGTACAC CTG | |
| Kan-F | TAGGGATAACAGGGTAATCGATTTATTC | |
| Kan-R | GCCAGTGTTACA ACCAATTAACC | |
| SB-PchTR-forw | CAAGAGATCGGCGTTGCTTT | |
| SB-PchTR-rev | CAAGCTAG CTGCAGTCTCAG | |
| qRT-PCRvTR-Forw | CCCTAGCCCGCTGAAAGTC | |
| qRT-PCRvTR-Rev | CCTAATCGGAGGTATT GATGGTACTG | |
| vTR-probe | Fam-CCCTCCGCCCGCTGTTTACTCG-BHQ-1 |
To reintroduce PvTR sequences into the pPchTR+/+ genome, PvTR was amplified with primers Resc-PvTR-F and Resc-PvTR-R (Tab. I). The PvTR fragment was digested with EcoRI and HindIII and cloned into pcDNA3 (Invitrogen, Darmstadt, Germany) generating recombinant vector pcDNA3-PvTR. Primers pEP-PvTR-F and pEP-PvTR-R (Tab. I) were used for PCR amplification of the I-SceI-aphAI cassette from pEPkan-S and transferred into the unique BamHI site of pcDNA3-PvTR generating the transfer plasmid pEPkanPvTR. From this vector, a fragment containing the PvTR sequence and the I-SceI-aphAI cassette was amplified with primers EPrescPvTR-F and Resc-PvTR-R (Tab. I) and used for the repair of pPchTR+/+ by two-step Red recombination exactly as described above.
2.3. Southern blot analysis
Probes used in Southern blot analysis were prepared using the PCR DIG Probe Synthesis kit (Roche Applied Science, Mannheim, Germany) according to the manufacturer’s protocols. The DIG-labeled kan r probe was amplified from plasmid pEPkan-S using specific primers Kan-F and Kan-R (Tab. I). The DIG-labeled PchTR probe was amplified with primers SB-PchTR-forw and SB-PchTR-rev (Tab. I). For Southern blot analyses, DNA digested with PstI was separated by 0.8% agarose gel electrophoresis, gels were stained with ethidium bromide, and DNA was transferred to nylon membranes (Amersham, Freiburg, Germany) according to the manufacturer’s instructions using 20 × SSC.
2.4. Measurement of plaque areas
One-million CEC seeded in six-well plates were infected with 100 plaque-forming units (PFU) of the parental virus vRB-1B, the mutant virus vPchTR+/+ or the rescuant virus vPvTR+/+. At day 5 p.i., cells were fixed with 3% paraformaldehyde for 15 min then permeabilized with 0.5% Triton X-100 for 5 min. Cells were incubated in a blocking solution (0.1% Triton X-100, 3% bovine serum albumin in phosphate buffered saline) at room temperature for 20 min. Indirect immunofluorescence (IIF) was performed with an anti-pp38 mouse antibody 011 [28, 30] followed by a secondary goat anti-mouse antibody labeled with Alexa Fluor 568 (Invitrogen). Digital images of 75 individual plaques were obtained using an epifluorescence Axiovert S100 inverted microscope and an AxioCam HRc digital camera (Zeiss, Jena, Germany). Plaque areas were measured using the National Institutes of Health ImageJ software1, and average plaque sizes and standard deviations were determined for each virus.
2.5. In vitro viral replication
Virus growth curves were determined as previously described [25]. Briefly, 1 × 106 CEC were infected with approximately 100 PFU of parental or mutant viruses. At 0, 24, 48, 72, 96, 120 and 144 h p.i., monolayers were harvested and inoculated onto fresh CEC in triplicate dishes. At day 5 p.i., plaques were stained by IIF as described above. The mean number of plaques was determined for each virus at each time point. Two independent experiments were performed.
2.6. Animal experiments
Commercial white leghorn specific pathogen-free (SPF) chickens (Lohmann Tierzucht, Cuxhaven, Germany) were housed in isolation units. Three groups of 1-day old chickens (n = 13) were inoculated subcutaneously with 1 000 PFU of vRB-1B (group 1), the mutant vPchTR+/+ (group 2) or the rescue virus vPvTR+/+ (group 3). Chickens were evaluated daily for symptoms of MD, euthanized when clinical signs were evident, and examined for gross MD lesions. The experiment was terminated 8 weeks after infection.
The number of viral DNA copies in chicken blood was determined using quantitative PCR (qPCR) assays. Whole blood obtained from the wing vein of infected chickens on days 4, 7, 11, 18, 24, 32 and 42 p.i. was used for DNA extraction using the E-Z 96 blood DNA kit (OMEGA bio-tek). Quantification of MDV genomic copies using qPCR was performed as previously described [12, 39].
2.7. Relative quantification of vTR expression by qRT-PCR
Total RNA was extracted from mock-infected CEC or cells infected with either vRB-1B, vPchTR+/+, vPvTR+/+ or a virus in which both vTR copies were deleted (vCR1-4−/−, [36] using the RNeasy Plus Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer instructions. RNA integrity was electrophoretically verified by ethidium bromide staining and by OD260/OD280 nm absorption ratio > 1.95. RNA was then used for qRT-PCR analysis of vTR expression using the TaqMan RNA-to-Ct 1-step kit (Applied Biosystems, Darmstadt, Germany). The sequences of primers (qRT-PCRvTR-Forw, qRT-PCRvTR-Rev) and sequence of the vTR-probe are listed in Table I. The qRT-PCR reaction consisted of 4 μL RNA (1 μg), 1× TaqMan RT-PCR mix, 1× TaqMan RT enzyme mix, 900 nM of each primer and 250 nM of probe in a final volume of 20 μL. The following experimental run protocol was used: RT step (48 °C for 15 min), enzyme in activation (95 °C for 10 min), denaturation, amplification/extension program repeated 40 times (95 °C for 15 s, 60 °C for 1 min). All qRT-PCR assays were performed in an ABI Prism 7500 Fast Real-Time PCR System (Applied Biosystems), and the results were analyzed with the Sequence Detection Systems version V2.0.3 software using the comparative Ct method (2−∆∆Ct) of relative quantification.
3. RESULTS
3.1. Construction of recombinant MDV
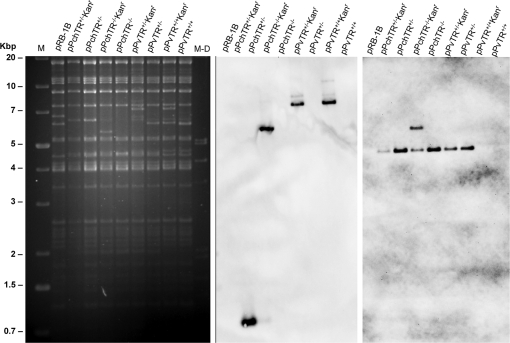
MDV contains in its genome two copies of the vTR gene located in the BamHI L fragment, which is found in both the TRL and IRL region [7]. An MDV mutant, in which both copies of the PvTR were replaced by the PchTR, was generated from pRB-1B [26] using a two-step Red-mediated mutagenesis strategy that allows for markerless manipulation of BAC DNA as previously described [35]. Genomic DNA of each clone generated during each recombination step was then analyzed by PCR, restriction fragment length polymorphism (RFLP) using BamHI (data not shown) or PstI digestion and by Southern blotting using probes specific for the kanamycin cassette or the PchTR sequence. Theoretical genomic modifications of BAC DNA were confirmed for each BAC clone (Fig. 1). We could show that no spurious rearrangements occurred during the genetic manipulation as shown by RFLP, although minor sequence modifications or rearrangements cannot formally be excluded. Similarly, revertant BAC pPvTR+/+ with two copies of PvTR reinserted into the mutant pPchTR+/+ was generated following the same procedure and analyzed with the same methods (Fig. 1).
Figure 1.
Restriction fragment length polymorphism and Southern blot analysis of recombinant MDV BAC. DNA of pRB-1B and mutant BAC derived thereof was prepared, digested with PstI, and separated on a 0.8% agarose gel together with 1-Kb marker (M, Promega) and a Dig-labeled marker (M-D). Southern blot analysis was performed using PCR-generated digoxigenin-labeled probes using oligonucleotide primers specific for the kanamycin cassette (middle panel) or the PchTR sequence (right panel).
3.2. PchTR induces low levels of vTR in vitro
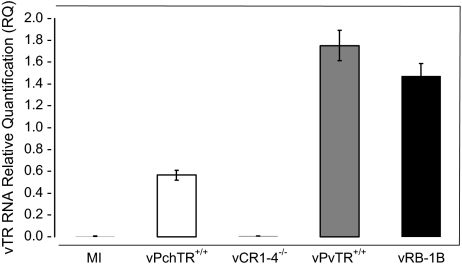
In order to determine the effect on vTR expression when PvTR is replaced with its chicken counterpart PchTR, CEC were infected with either vRB-1B, the mutant virus vPchTR+/+ or the rescuant vPvTR+/+ virus. At 48 h p.i., total RNA was extracted from infected cells and used as a template for a qRT-PCR analysis of vTR expression. At 48 h p.i., vTR expression was 3-fold lower in cells infected with the mutant virus when compared to that in cells infected with parental vRB-1B or the rescuant virus (Fig. 2). In mock-infected cells and in cells infected with a virus in which both vTR copies were deleted, vCR1-4−/− [36], vTR expression was not detectable (Fig. 2). We concluded that the PchTR has lower transcriptional activity in avian cells than PvTR and that the promoter was therefore not under the control of a virus-derived factor.
Figure 2.
Relative quantification of vTR RNA in infected CEC. CEC were mock infected (MI) or infected with either the parental virus vRB-1B, the mutant virus vPchTR+/+, the rescuant virus vPvTR+/+ or a virus deleted for both vTR copies (vCR1-4−/−). At 48 h p.i., total RNA was extracted and used for qRT-PCR analysis of vTR expression. Relative expression of vTR gene was normalized to cellular GAPDH expression levels. Error bars represent standard error of the mean of two independent experiments at the 48 h time point.
3.3. vTR levels do not affect MDV replication in vitro
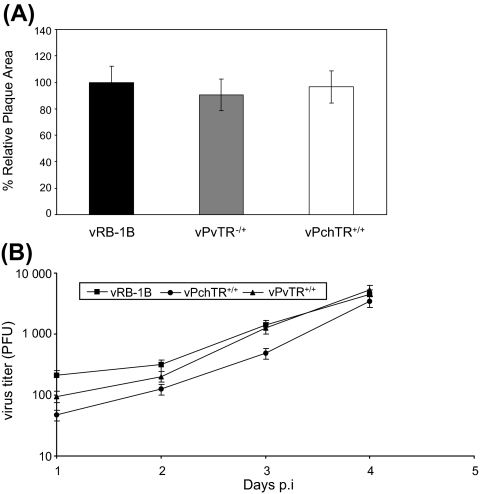
Parental vRB-1B, mutant vPchTR+/+ and the vPvTR+/+ viruses were assessed for in vitro growth properties by analyzing plaque sizes (Fig. 3A) and growth kinetics (Fig. 3B). Determination of the mean plaque areas demonstrated that there was no discernable difference when vPchTR and vPvTR+/+ were compared to vRB-1B (p = 0.25 and p = 0.7, respectively) (Fig. 3A). Similarly, growth kinetics of the mutant vPchTR+/+ virus were virtually indistinguishable from those of the parental vRB-1B or the rescuant vPvTR+/+ virus (Fig. 3B). From the results of the in vitro growth properties of the generated mutant viruses we concluded that the mutant vPchTR+/+ virus was similar to parental vRB-1B with respect to the growth characteristics in cultured cells and that vTR expression levels do not influence MDV replication in vitro.
Figure 3.
(A) Relative plaque sizes induced by vRB-1B, vPchTR+/+ or rescuant virus vPvTR+/+. CEC were infected with the viruses and at day 5 p.i. cells were analyzed by IIF with anti-pp38 MAb 011 and Alexa Fluor 568 goat anti-mouse IgG (Molecular Probes). For each virus, 75 random plaques were photographed with a digital camera (Axiovert 25 with Axiocam, Zeiss) and plaque sizes were determined using ImageJ software. The mean plaque area of the wild type vRB-1B virus was set to 100%, and average relative plaque areas of vPchTR+/+ and the vPvTR+/+ were calculated. Means and standard deviations (error bars) are given. (B) Multi-step growth kinetics of vRB-1B, vPchTR+/+ and vPvTR+/+. One-million CEC were infected with approximately 100 PFU of the indicated virus. At the given times p.i., cells were trypsinized, titrated and coseeded with fresh CEC. At day 5 p.i., cells were fixed; permeabilized and viral plaques were counted after IIF. Mean virus titers and standard deviations (error bars) of the results of two independent experiments are shown.
3.4. vTR levels do not affect MDV cytolytic replication in vivo
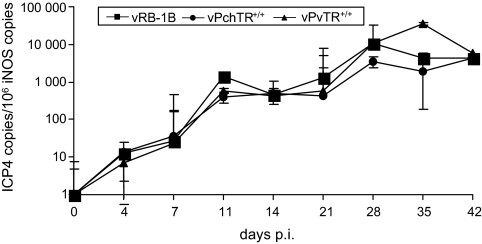
The next series of experiments addressed in vivo replication of vPchTR+/+ in the natural host. Three groups of 1-day-old chickens were infected subcutaneously with either vRB-1B, vPchTR+/+ or vPvTR+/+. On days 4, 7, 11, 14, 21, 28, 35 and 42 after infection, whole blood samples were collected from infected animals. Copy numbers of viral genomes were determined and the results are summarized in Figure 4. Both vPchTR+/+ and vPvTR+/+ viruses maintained virus replication rates virtually similar to those of vRB-1B, except that on day 35 p.i. viremia levels were slightly higher in birds infected with vPvTR+/+ when compared with those infected with the mutant or the parental virus. However, the difference was not statistically significant (p = 0.3) and vPvTR+/+ viral loads reached values virtually identical to those of wild-type and mutant viruses at day 42 p.i. Taken together, the results showed that replacement of PchTR in place of PvTR in the genome of MDV does not impair the ability of the mutant virus to replicate in vivo.
Figure 4.
High levels of vTR are dispensable for MDV cytolytic replication in vivo. Total DNA isolated from whole blood samples taken at days 4, 7, 11, 14, 21, 28, 35 and 42 p.i. from chickens infected with vPchTR+/+, vPvTR+/+ or the parental vRB-1B were used for qPCR analysis of the viral ICP4 gene and a host gene (iNOS). Mean viral loads are given as copies of ICP4 per 1 × 106 copies of chicken iNOS. No statistically significant differences in viremia levels were observed between groups infected with vPchTR+/+, vPvTR+/+ or vRB-1B. Graphs represented the means of 13 chickens per group and standard deviations are given (error bars).
3.5. Mutant MDV harboring the chTR promoter is impaired in lymphoma formation and dissemination
To investigate the role of vTR in MDV induced lymphomagenesis, chickens infected with either the vPchTR+/+ mutant virus, the parental virus vRB-1B or the rescuant virus vPvTR+/+ were monitored for clinical signs and gross pathological lesions. MD incidence was evaluated up to 56 days p.i. at which time all surviving chickens were sacrificed and examined for gross MD lesions. Infection with vPchTR+/+ induced a significant reduction in tumor incidence (46%, p < 0.05) in comparison to vRB-1B (92%) or vPvTR+/+ (85%). The difference in the percentage of birds developing tumors after infection with vRB-1B (92%) and vPvTR+/+ (85%) was not significant (p = 0.56).
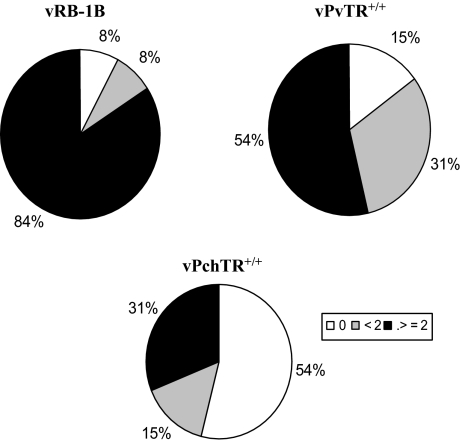
As regards to the pattern of lymphoma distribution, we observed that tumors in the vPchTR+/+ group were less disseminated since only 31% of chickens developed tumors at more than two distinct organ sites relative to 84% in the vRB-1B group and 54% in the revertant group (Fig. 5). These results showed that vPchTR+/+ differs from vRB-1B and vPvTR+/+ in terms of tumorigenic potential in the natural host. Moreover, we could demonstrate that high levels of vTR controlled by the virus promoter are essential for MDV pathogenesis and can influence disease progression.
Figure 5.
Mutant virus harboring two copies of the chTR promoter is attenuated. Animals were inoculated subcutaneously with 1 000 PFU of either vRB-1B, vPchTR+/+ or vPvTR+/+. During the course of the experiment, moribund birds were sacrificed and necropsied. At the end of the experiment (8 weeks p.i.), surviving birds were also necropsied and evaluated for MD. Morbidity and lymphoma incidences were recorded. Lymphomas induced by mutant viruses harboring two copies of the chTR promoter showed an altered dissemination pattern within individual chickens. The values correspond to the percentage of birds infected with the indicated viruses, which failed to develop lymphoma (0), in which less than (< 2) or equal to or more than two sites of lymphoma manifestation (greater or equal to 2) were observed.
4. DISCUSSION
MDV is among the most oncogenic herpesviruses known [3] and the only virus known to date that encodes its own telomerase RNA (vTR). Evidence regarding the role of vTR in pathogenesis [36] highlighted the importance of telomerase in cancer development but raised new questions regarding the mechanism by which expression of vTR contributes to oncogenesis.
Functional analyses have shown that vTR can reconstitute telomerase activity by interacting with TERT more efficiently than chTR does [7, 8]. Interestingly, telomerase activity during MDV infection is increased in lymphocytes, the target cells for transformation, and was associated with high levels of vTR expression [33]. These data suggested that chTR is the limiting factor and that high copy numbers of vTR are a precondition for increasing telomerase activity and promoting cell immortalization.
Regulation of telomerase activity varies among vertebrates and is achieved through TR and TERT expression at various levels such as transcriptional control and subcellular localization of each component. Modulation of telomerase, which involves a complex network of transcription factors and cis-acting sequences, was mainly studied in model organisms and man [22]. With respect to telomerase regulation, the chicken has many features in common with man such as age being associated with telomere shortening [6] or telomerase downregulation in most somatic cells and tissues during embryonic and postnatal development [34]. As described for the hTR promoter, sequence analysis of the chTR and vTR promoter revealed common transcription factor binding sites that are involved in maintaining basal TR expression levels. Importantly, this analysis identified cis elements specifically involved in vTR transcriptional regulation including three E-boxes and an EBS site [7, 33]. Existence of additional transcription factor binding sites on the vTR promoter implied other levels of control for vTR expression.
Our study aimed at determining whether vTR promoter sequences are required for lymphomagenesis. To test this hypothesis, we used the highly oncogenic RB-1B strain of MDV to generate a mutant virus termed vPchTR+/+ harboring the chicken TR promoter in lieu of the vTR promoter. We showed by qRT-PCR that, when compared with parental and revertant viruses, vPchTR+/+ had reduced levels of vTR expression in culture cells confirming that vTR promoter has a stronger transcriptional activity than its chicken counterpart (Fig. 2). Additionally, we showed that vPchTR+/+ replication was not impaired in vitro since vPchTR+/+ replicates with levels similar to those of the parental and revertant viruses (Fig. 3). This result further suggested that no spurious rearrangement occurred during genetic manipulation of the BAC DNA. Importantly, in vivo replication of vPchTR+/+ was also similar to the wild-type and the rescuant viruses (Fig. 4). Thus, our results correlate well with those of Trapp et al. [36] who showed that vTR expression is dispensable for MDV lytic replication and the establishment of latency, indicating that vTR serves a role supporting tumorigenesis rather than affecting tumorigenesis through a failure to efficiently replicate lytically. Consistent with this interpretation, our in vivo experiment revealed that high levels of vTR are essential for MDV pathogenesis and for disease progression. Lymphomagenesis in chickens infected with the vPchTR+/+ mutant was reduced by 54% in comparison to the wild-type and rescuant viruses (Fig. 5). Moreover, lesions in animals infected with the mutant virus were smaller in size and less disseminated, results virtually identical to those obtained earlier with vTR deletion mutants [36]. Our data thus confirm that vTR expression is not only important for MDV lymphomagenesis but expression levels controlled by the virus are necessary for its tumorigenic function.
vTR can result in increased telomerase activity in concert with chTERT in vitro [8]. In addition to the role of vTR in the telomerase complex, some of its pro-oncogenic functions seem to be independent of telomerase action. Correlation between expression of vTR and its oncogenic properties have been demonstrated in DF-1 cells, in which telomerase activity is undetectable. vTR expression led to morphological alterations in DF-1 cells similar to those observed after expression of the bona fide MDV oncoprotein, Meq [36]. Meq is a nuclear oncoprotein essential for MDV lymphomagenesis that acts as a regulator of transcription [21]. Depending on its dimerization partner, it can induce either transcriptional activation or suppression [18]. Like vTR, Meq is expressed during both lytic and latent infection and higher levels of Meq have been detected in tumor cells and in lymphoid cell line transformed by MDV [14, 27]. Interestingly, higher transcriptional levels of Meq have also been associated with higher levels of telomerase activity in lymphocytes and tumor formation in chickens. A previous mutational study of AP-1 site in the vTR promoter did not have any effect on the transcriptional activity of vTR promoter in MSB-1 cells expressing Meq [33]. Consistent with this data, attempts to identify Meq binding sites on MDV genome through ChIP experiments failed to identify Meq binding in the vTR promoter [18]. Based on these results, it is conceivable that Meq might indirectly transactivate vTR expression through dimerization with one of its cellular partners. It is notable that Meq/c-Jun heterodimers transactivate c-Myc expression [18, 19, 23] and that c-Myc binding sites are present on the vTR promoter. The presence of E-Boxes in the vTR promoter is compatible with a direct effect of proteins of the bHLH-Zip superfamily like the Myc/Mad/Max transcription factors that regulate many cellular functions, including proliferation, differentiation, and apoptosis. The ability of c-Myc to bind to the vTR promoter and transactivate vTR gene expression has been revealed in vitro by overexpression assays in LMH cells [33]. ChIP experiments also showed that endogenously expressed c-Myc can interact with the vTR promoter region containing E-Boxes 2 and 3 in MSB-1 cells [33].
As mentioned earlier, MDV vTR is both a lytic and a latent gene since it is expressed in MDV-infected CEC and in MDV-transformed lymphoblastoid cell lines [33, 36]. Semi-quantitative RT-PCR analysis of vTR expression showed that the level of vTR in lymphoid cells is 17-fold higher than in CEC undergoing lytic replication. The fact that c-Myc is also expressed in both lytically and latently infected cells strongly suggests that additional factors are involved in vTR transcriptional activation. Shkreli et al. suggested that Ets-1 might be a factor involved in vTR transcriptional regulation and analysis of the EBS site located on vTR promoter sequence revealed the presence of specific binding domain for three Ets proteins, PU.1, Ets-1 and PEA3 [33]. However, only Ets-1 appeared to be a potential candidate because of high levels of Ets-1 expression in lymphoid tissues in mature birds.
To conclude, we have shown here that expression of vTR through its endogenous promoter (PvTR) is essential for MDV lymphomagenesis. Further studies identifying the specific vTR promoter elements causing increased levels of vTR expression and their importance in vivo will help delineate the pathogenesis of MDV-induced deadly lymphoma.
Acknowledgments
This study was supported by a grant from the National Institutes of Health (5R01CA127238) to N. Osterrieder and K.W. Jarosinski and by unrestricted funds made available to N. Osterrieder from the Freie Universität Berlin.
Footnotes
References
- 1.Blackburn E.H., Switching and signaling at the telomere, Cell (2001) 106:661–673 [DOI] [PubMed] [Google Scholar]
- 2.Calnek B.W., Marek’s disease – a model for herpesvirus oncology, Crit. Rev. Microbiol. (1986) 12:293–320 [DOI] [PubMed] [Google Scholar]
- 3.Calnek B.W., Pathogenesis of Marek’s disease virus infection, Curr. Top. Microbiol. Immunol. (2001) 255:25–55 [DOI] [PubMed] [Google Scholar]
- 4.Davison A., Comments on the phylogenetics and evolution of herpesviruses and other large DNA viruses, Virus Res. (2002) 82:127–132 [DOI] [PubMed] [Google Scholar]
- 5.Davison A.J., Evolution of the herpesviruses, Vet. Microbiol. (2002) 86:69–88 [DOI] [PubMed] [Google Scholar]
- 6.Delany M.E., Krupkin A.B., Miller M.M., Organization of telomere sequences in birds: evidence for arrays of extreme length and for in vivo shortening, Cytogenet. Cell Genet. (2000) 90:139–145 [DOI] [PubMed] [Google Scholar]
- 7.Fragnet L., Blasco M.A., Klapper W., Rasschaert D., The RNA subunit of telomerase is encoded by Marek’s disease virus, J. Virol. (2003) 77:5985–5996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fragnet L., Kut E., Rasschaert D., Comparative functional study of the viral telomerase RNA based on natural mutations, J. Biol. Chem. (2005) 280:23502–23515 [DOI] [PubMed] [Google Scholar]
- 9.Harley C.B., Futcher A.B., Greider C.W., Telomeres shorten during ageing of human fibroblasts, Nature (1990) 345:458–460 [DOI] [PubMed] [Google Scholar]
- 10.Harley C.B., Vaziri H., Counter C.M., Allsopp R.C., The telomere hypothesis of cellular aging, Exp. Gerontol. (1992) 27:375–382 [DOI] [PubMed] [Google Scholar]
- 11.Hiyama K., Hirai Y., Kyoizumi S., Akiyama M., Hiyama E., Piatyszek M.A., et al., Activation of telomerase in human lymphocytes and hematopoietic progenitor cells, J. Immunol. (1995) 155:3711–3715 [PubMed] [Google Scholar]
- 12.Jarosinski K.W., Osterrieder N., Nair V.K., Schat K.A., Attenuation of Marek’s disease virus by deletion of open reading frame RLORF4 but not RLORF5a, J. Virol. (2005) 79:11647–11659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarosinski K.W., Margulis N.G., Kamil J.P., Spatz S.J., Nair V.K., Osterrieder N., Horizontal transmission of Marek’s disease virus requires US2, the UL13 protein kinase, and gC, J. Virol. (2007) 81:10575–10587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones D., Lee L., Liu J.L., Kung H.J., Tillotson J.K., Marek disease virus encodes a basic-leucine zipper gene resembling the fos/jun oncogenes that is highly expressed in lymphoblastoid tumors, Proc. Natl. Acad. Sci. USA (1992) 89:4042–4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamil J.P., Tischer B.K., Trapp S., Nair V.K., Osterrieder N., Kung H.J., vLIP, a viral lipase homologue, is a virulence factor of Marek’s disease virus, J. Virol. (2005) 79:6984–6996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim N.W., Piatyszek M.A., Prowse K.R., Harley C.B., West M.D., Ho P.L., et al., Specific association of human telomerase activity with immortal cells and cancer, Science (1994) 266:2011–2015 [DOI] [PubMed] [Google Scholar]
- 17.Kingham B.F., Zelnik V., Kopacek J., Majerciak V., Ney E., Schmidt C.J., The genome of herpesvirus of turkeys: comparative analysis with Marek’s disease viruses, J. Gen. Virol. (2001) 82:1123–1135 [DOI] [PubMed] [Google Scholar]
- 18.Levy A.M., Izumiya Y., Brunovskis P., Xia L., Parcells M.S., Reddy S.M., et al al., Characterization of the chromosomal binding sites and dimerization partners of the viral oncoprotein Meq in Marek’s disease virus-transformed T cells, J. Virol. (2003) 77:12841–12851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy A.M., Gilad O., Xia L., Izumiya Y., Choi J., Tsalenko A., et al., Marek’s disease virus Meq transforms chicken cells via the v-Jun transcriptional cascade: a converging transforming pathway for avian oncoviruses, Proc. Natl. Acad. Sci. USA (2005) 102:14831–14836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindsey J., McGill N.I., Lindsey L.A., Green D.K., Cooke H.J., In vivo loss of telomeric repeats with age in humans, Mutat. Res. (1991) 256:45–48 [DOI] [PubMed] [Google Scholar]
- 21.Lupiani B., Lee L.F., Cui X., Gimeno I., Anderson A., Morgan R.W., et al., Marek’s disease virus-encoded Meq gene is involved in transformation of lymphocytes but is dispensable for replication, Proc. Natl. Acad. Sci. USA (2004) 101:11815–11820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mergny J.L., Riou J.F., Mailliet P., Teulade-Fichou M.P., Gilson E., Natural and pharmacological regulation of telomerase, Nucleic Acids Res. (2002) 30:839–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nair V.K., Kung H.J., Marek’s disease virus oncogenicity: molecular mechanisms, in: Davison F., Nair V. (Eds.), Marek’s disease, an evolving problem, Elsevier Academic Press, London, 2004 [Google Scholar]
- 24.Osterrieder N., Kamil J.P., Schumacher D., Tischer B.K., Trapp S., Marek’s disease virus: from miasma to model, Nat. Rev. Microbiol. (2006) 4:283–294 [DOI] [PubMed] [Google Scholar]
- 25.Parcells M.S., Anderson A.S., Cantello J.L., Morgan R.W., Characterization of Marek’s disease virus insertion and deletion mutants that lack US1 (ICP22 homolog), US10, and/or US2 and neighboring short-component open reading frames, J. Virol. (1994) 68:8239–8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petherbridge L., Brown A.C., Baigent S.J., Howes K., Sacco M.A., Osterrieder N., Nair V.K., Oncogenicity of virulent Marek’s disease virus cloned as bacterial artificial chromosomes, J. Virol. (2004) 78:13376–13380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ross N., O’Sullivan G., Rothwell C., Smith G., Burgess S.C., Rennie M., et al., Marek’s disease virus EcoRI-Q gene (meq) and a small RNA antisense to ICP4 are abundantly expressed in CD4+ cells and cells carrying a novel lymphoid marker, AV37, in Marek’s disease lymphomas, J. Gen. Virol. (1997) 78:2191–2198 [DOI] [PubMed] [Google Scholar]
- 28.Rudolph J., Seyboldt C., Granzow H., Osterrieder N., The gene 10 (UL49.5) product of equine herpesvirus 1 is necessary and sufficient for functional processing of glycoprotein M, J. Virol. (2002) 76:2952–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schat K.A., Chen C.L., Calnek B.W., Char D., Transformation of T-lymphocyte subsets by Marek’s disease herpesvirus, J. Virol. (1991) 65:1408–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schumacher D., Tischer B.K., Fuchs W., Osterrieder N., Reconstitution of Marek’s disease virus serotype 1 (MDV-1) from DNA cloned as a bacterial artificial chromosome and characterization of a glycoprotein B-negative MDV-1 mutant, J. Virol. (2000) 74:11088–11098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shay J.W., Bacchetti S., A survey of telomerase activity in human cancer, Eur. J. Cancer (1997) 33:787–791 [DOI] [PubMed] [Google Scholar]
- 32.Shay J.W., Wright W.E., Telomerase: a target for cancer therapeutics, Cancer Cell. (2002) 2:257–265 [DOI] [PubMed] [Google Scholar]
- 33.Shkreli M., Dambrine G., Soubieux D., Kut E., Rasschaert D., Involvement of the oncoprotein c-Myc in viral telomerase RNA gene regulation during Marek’s disease virus-induced lymphomagenesis, J. Virol. (2007) 81:4848–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor H.A., Delany M.E., Ontogeny of telomerase in chicken: impact of downregulation on pre- and postnatal telomere length in vivo, Dev. Growth Differ. (2000) 42:613–621 [DOI] [PubMed] [Google Scholar]
- 35.Tischer B.K., von Einem J., Kaufer B., Osterrieder N., Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli, Biotechniques (2006) 40:191–197 [DOI] [PubMed] [Google Scholar]
- 36.Trapp S., Parcells M.S., Kamil J.P., Schumacher D., Tischer B.K., Kumar P.M., et al., A virus-encoded telomerase RNA promotes malignant T cell lymphomagenesis, J. Exp. Med. (2006) 203:1307–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tulman E.R., Afonso C.L., Lu Z., Zsak L., Rock D.L., Kutish G.F., The genome of a very virulent Marek’s disease virus, J. Virol. (2000) 74:7980–7988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wright W.E., Piatyszek M.A., Rainey W.E., Byrd W., Shay J.W., Telomerase activity in human germline and embryonic tissues and cells, Dev. Genet. (1996) 18:173–179 [DOI] [PubMed] [Google Scholar]
- 39.Yunis R., Jarosinski K.W., Schat K.A., Association between rate of viral genome replication and virulence of Marek’s disease herpesvirus strains, Virology (2004) 328:142–150 [DOI] [PubMed] [Google Scholar]
- 40.Zakian V.A., Telomeres: beginning to understand the end, Science (1995) 270:1601–1607 [DOI] [PubMed] [Google Scholar]