Abstract
The rapid replication of HIV-1 and the errors made during viral replication, cause the virus to evolve rapidly in patients, making the problems of vaccine development and drug therapy particularly challenging. In the absence of an effective vaccine, drugs are the only useful treatment. Anti-HIV drugs work; so far drug therapy has saved more than three million years of life. Unfortunately, HIV-1 develops resistance to all of the available drugs. Although a number of useful anti-HIV drugs have been approved for use in patients, the problems associated with drug toxicity and the development of resistance means that the search for new drugs is an ongoing process. The three viral enzymes, reverse transcriptase (RT), integrase (IN), and protease (PR) are all good drug targets. Two distinct types of RT inhibitors, both of which block the polymerase activity of RT, have been approved to treat HIV-1 infections, nucleoside analogs (NRTIs) and nonnucleosides (NNRTIs), and there are promising leads for compounds that either block the RNase H activity or block the polymerase in other ways. A better understanding of the structure and function(s) of RT and of the mechanism(s) of inhibition can be used to generate better drugs; in particular drugs that are effective against the current drug-resistant strains of HIV-1.
In the absence of an effective vaccine, drugs are the only therapeutic tools that can be used to treat HIV-1 infections. Unfortunately, HIV-1 infections cannot be cured, so that drug therapy, once initiated, must be continued for the life of the patient. This places a special burden on the design of anti-HIV drugs: They need to be relatively nontoxic so that they can be used in long-term therapy. HIV-1 replication is error prone (1) (and references within) and the errors that arise during the viral life cycle, together with the rapid replication of the virus in patients, allows the virus to escape the host’s immune system and develop resistance to all of the available drugs (2). The virus evolves sufficiently rapidly that, unless the therapy is well-designed, resistance will develop in all treated patients. The only way to stop the development of resistance is to completely block viral replication; this, in turn, stops the evolution of resistance. It takes a combination of drugs (usually three) to completely block viral replication; this is the reason that three-drug regimens are used in standard HIV-1 therapies. Of the approved drugs, most target two of the three virally encoded enzymes that carry out viral replication, reverse transcriptase (RT) and protease (PR). A new drug, raltegravir, that targets the third enzyme integrase (IN) has recently been approved (http://www.fda.gov/oashi/aids/virals.html). In addition to the drugs that target the viral enzymes, there are two approved drugs, enfurvitide and maraviroc, that target different aspects of viral entry.
Because standard anti-HIV therapies include anti-RT drugs, it is important to understand the drug target (RT) itself, the role(s) RT plays in the viral life cycle, the ways the drugs act to inhibit the normal functions of RT, and the mechanisms that the virus, and more importantly RT, uses to evade the available drugs. Armed with a better understanding of RT, and the mechanisms of inhibition and drug resistance that directly involve RT, it should be possible to develop drugs that will be more effective; in particular to develop drugs that can block the replication of viruses that are resistant to the currently available drugs.
1. Role of HIV-1 reverse rranscriptase in viral replication
Viral infections are initiated by the fusion of the viral and cellular membranes; this fusion reaction is caused by the interactions of the viral envelope glycoprotein with its receptor (CD4) and a co-receptor, usually either CCR5 or CXCR4 (for a review of the retroviral life cycle, and an overview of reverse transcription, see Coffin, Hughes and Varmus, Retroviruses, 1997 (1)). Binding the receptor and co-receptor causes changes in the structure of the envelope glycoprotein, which leads to membrane fusion. Membrane fusion places the viral core, which contains RT, into the cytoplasm of the cell. A poorly understood process called “uncoating” modifies the core in ways that promote reverse transcription.
The HIV-1 virion contains, in addition to the viral proteins, two copies of a single-stranded RNA genome. RT has two enzymatic activities, a DNA polymerase that can copy either a DNA or an RNA template, and an RNase H that cleaves RNA only if the RNA is part of an RNA/DNA duplex. The two enzymatic functions of RT, polymerase and RNase H, cooperate to convert the RNA into a double-stranded linear DNA. This conversion takes place in the cytoplasm of the infected cell; after DNA synthesis has been completed, the resulting linear double-stranded viral DNA is translocated to the nucleus where the viral DNA is inserted into the host genome by IN. This inserted DNA copy, called a provirus, is the source of both viral genomic and viral messenger RNAs, which are generated by the host DNA-dependent RNA polymerase. Although other viral proteins (notably the nucleic acid chaperone nucleocapsid, and perhaps IN) and probably some cellular factors, help RT carry out the reactions that converts the viral RNA into DNA, RT contains all the necessary enzymatic activities for the conversion.
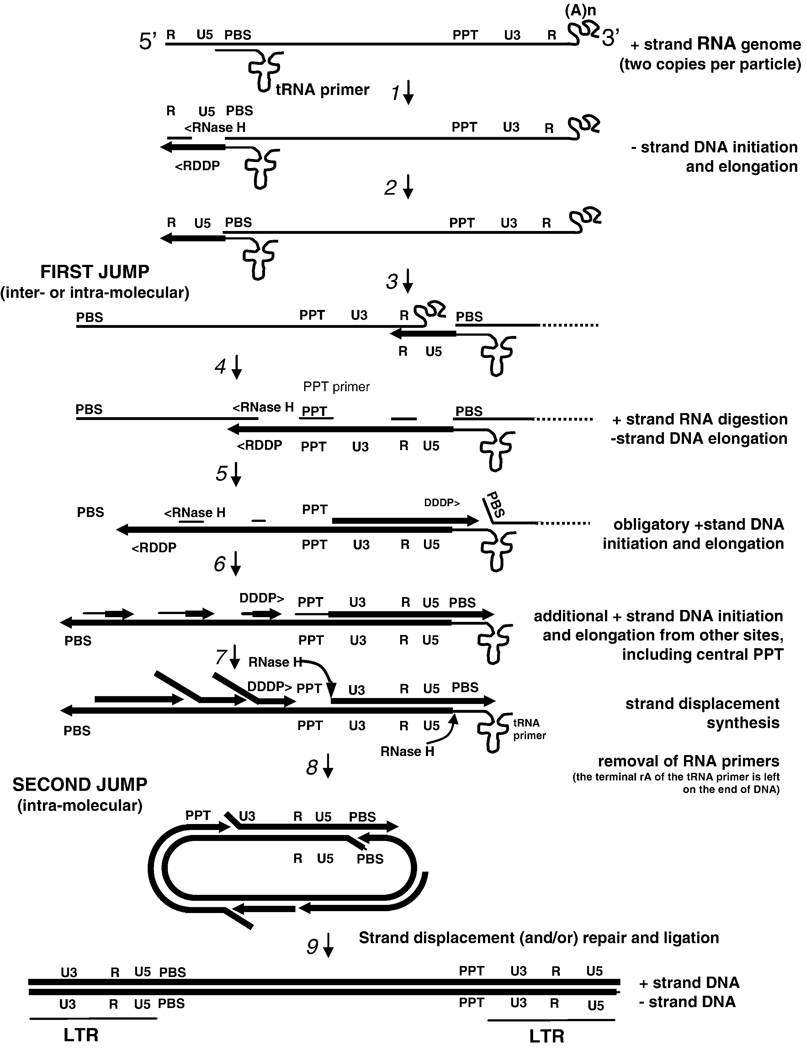
Like many other DNA polymerases, RT requires both a primer and a template. DNA synthesis is initiated from a host tRNA primer (in HIV-1 the primer is tRNAlys3). There is, near the 5’ end of the viral genome, a segment 18 nucleotides long, called the primer binding site (PBS) that is complementary to the 18 nucleotides at the 3’ end of tRNAlys3. The tRNA primer is hybridized to the PBS (Figure 1). The viral RNA genome, which serves as the template, is plus-strand. First (minus) strand DNA synthesis is initiated from the tRNA primer, allowing RT to copy the 5’ end of the viral RNA genome. Synthesis of the minus-strand DNA generates an RNA/DNA hybrid that is a substrate for RNase H. RNase H degrades the RNA strand, leaving the nascent minus-strand DNA single stranded. The sequences at the 5’ and 3’ ends of the viral RNA genome are identical (R, or repeat, see Figure 1). This allows the minus-strand DNA to hybridize with the R sequence at the 3’ end of one of the two viral RNAs in the virion, a step that is called the first jump, or the first, or minus-strand transfer. After the nascent DNA hybridizes to R, minus-strand synthesis can continue along the viral RNA. As DNA synthesis proceeds, RNase H degrades the RNA strand. Although most of the RNase H cleavages do not appear to be sequence specific, there is a purine-rich sequence (called the polypurine tract or PPT) near the 3’ end of the viral RNA (Figure 1). The polypurine tract is relatively resistant to RNase H cleavage, and it serves as the primer for second (plus) strand DNA synthesis. Plus-strand DNA synthesis proceeds until RT starts to copy the tRNA primer. The first 18 nucleotides can be copied; however the next nucleotide in the tRNA is a modified A that cannot be copied by RT. Once the 3’ end of the tRNA has been reverse transcribed, an RNA/DNA hybrid that is a substrate for RNase H is created. In the reverse transcription of most retroviral genomes, the RNase H of RT removes the entire tRNA primer. HIV-1 is the exception; its RNase H cleaves precisely one nucleotide from the tRNA/DNA junction, leaving a ribo-A on the 3’ end of the viral minus-strand DNA. This sets the stage for the second jump, also called the second or plus-strand transfer. The removal of the tRNA primer exposes a single-stranded portion of the plus-strand DNA that has the same sequence as the PBS. Exposure of the 3’ end of the plus-strand DNA allows the 5’ end of the minus-strand (once the PBS has been copied) to be transferred to the plus-strand (Figure 1). Once this second transfer happens, both the minus- and plus- strands are extended until the entire DNA is double stranded, creating a DNA that has the same sequences at both ends (these repeats are called the long terminal repeats or LTRs). As Figure 1 shows, the DNA is longer than the RNA genome(s) from which it derives, allowing the viral DNA, once integrated, to serve as the template from which new copies of the viral genome (and the viral messenger RNAs) can be copied by the host enzyme DNA-dependent RNA polymerase.
Figure 1.
Reverse transcription of the HIV-1 genome (with permission from Annual Review of Biochemistry). Each retroviral particle contains two copies of the RNA genome. Minus-strand DNA synthesis starts near the 5’ end of the plus-stand RNA genome using as a primer a host tRNAlys3 that anneals at the primer-binding site (PBS).
Step 1: Synthesis proceeds to the 5’ end of the RNA genome through the U5 region, ending at the R region at the 5’end, forming the “minus-strand strong stop DNA.” Step 2: DNA synthesis is accompanied by RNase H digestion of the RNA portion of the RNA-DNA hybrid product, thus exposing the single-strand DNA product. Step 3: This exposure facilitates hybridization with the R region at the 3’ end of the same, or the second RNA genome, a strand-transfer reaction known as the “first jump”. Step 4: When minus-strand elongation passes a polypurine rich region called the polypurine tract (PPT) region, a unique plus-strand RNA primer is formed by RNase H cleavage at its borders. Plus-strand synthesis then continues back to the U5 region using the minus-strand DNA as a template. Step 5: Meanwhile, minus-strand synthesis continues through the genome using the plus-strand RNA as a template, and removing the RNA template in its wake via RNase H activity. Step 6: The RNase H digestion products formed are presumed to provide additional primers for plus-strand synthesis at a number of internal locations along the minus-strand DNA. Step 7: PPT-initiated plus-strand DNA synthesis stops after copying the annealed portion of the tRNA to generate the plus-strand DNA form o f the PBS, forming the “plus-strand strong stop” product. The tRNA is then removed by the RNase H activity of RT. Step 8: This may facilitate annealing to the PBS complement on the minus-strand DNA, providing the complementarity for the “second jump.” DNA synthesis then continues. Step 9: Strand displacement synthesis by RT to the PBS and PPT ends, and/or repair and ligation of a circular intermediate produces a linear duplex with long terminal repeats (LTRs) at both ends.
Although there are complexities to the reverse transcription process that are beyond the scope of this review, there are a few additional points that should be made here. As has already been mentioned, the end product of reverse transcription process is the substrate for IN. As such, the ends of the linear viral DNA need to be relatively precise. As is shown in Figure 1, it is the RNase H cleavages that generate and remove the PPT primer that define the U3 end of the linear viral DNA, and it is the removal of the tRNA primer that defines the U5 end. Although RNase H has no mechanism that allows it to recognize specific sequences, it carries out these particular cleavage reactions with absolute specificity; and the ends of the linear viral DNA genome are defined to the exact nucleotide.
There is also the question why virions contain two copies of the viral RNA genome instead of one. In theory, the reactions outlined in Figure 1 could be carried out with only one copy of the viral RNA genome. The obvious explanation is that, if there was only one copy of the viral RNA in a virion, a single break in the RNA would be fatal, preventing the synthesis of a complete DNA copy. However, if there is a second copy of the RNA, and minus-strand DNA synthesis is blocked by a break in the RNA template, synthesis can be continued if minus-strand DNA synthesis is transferred to the second RNA genome. In fact, if two copies of the RNA genome are present, a complete DNA copy can be made even if both RNA genomes are extensively nicked, so long as there are no sites at which both RNA copies are nicked. If a virion is produced by a cell that contains only one integrated DNA genome, the two RNA copies will be identical (unless RNA polymerase makes an error), and the fact that, during minus-strand DNA synthesis, RT often shifts back and forth between the two RNA templates will have no significant consequences. However, if a host cell contains two different integrated viral DNA genomes, then virions can be produced that contain two related RNA genomes that have different sequences. This sets the stage for the generation of viral recombinants. If RT makes a double-stranded viral DNA by copying from two different RNA genomes, the resulting DNA will contain sequences that derive from both of the parental genomes. In HIV-1, recombination during reverse transcription is the rule, not the exception. This can have important consequences. For example recombination can lead to the generation of viruses that are resistant to multiple drugs from parental viruses that are each resistant to only a single drug.
2. Structure of HIV-1 reverse transcriptase
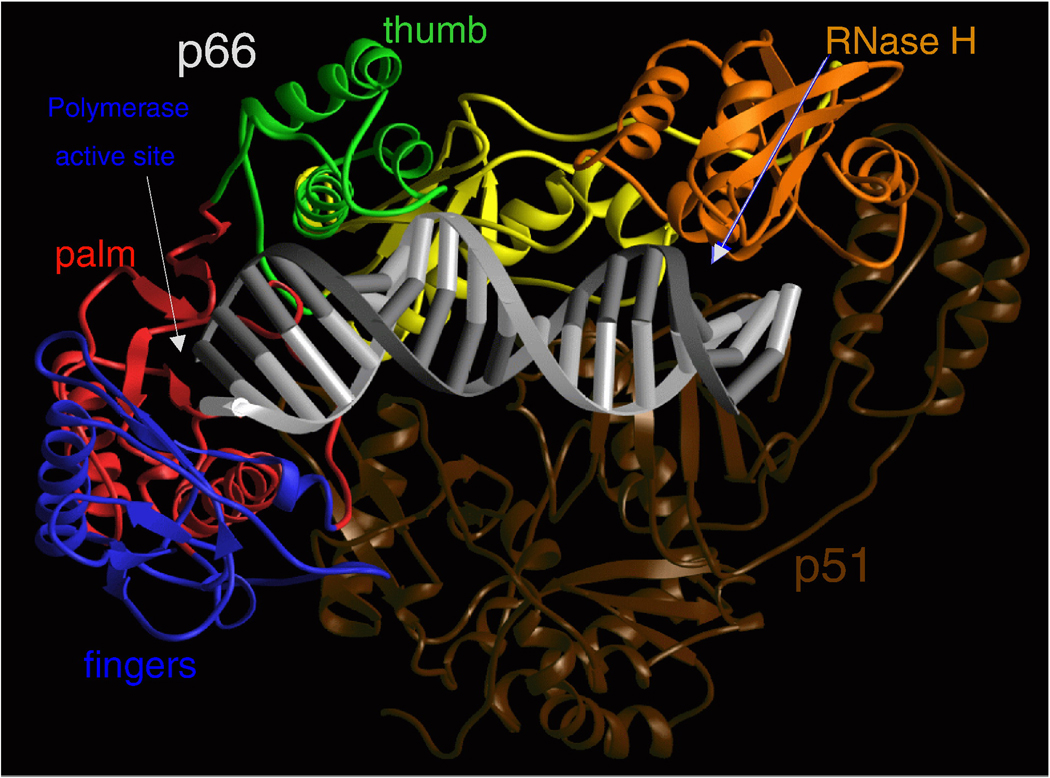
RT of HIV-1 is an asymmetric heterodimer composed of two related subunits, p66 and p51. Both subunits derive, by cleavage by the viral protease (PR), from a Gag-Pol polyprotein that is synthesized from unspliced viral RNA (3, 4). p66 and p51 share a common amino terminus; p66 is 560 amino acids in length, p51 is 440 amino acids long. As has been discussed briefly, both of the enzymatic functions of RT, the DNA polymerase and RNase H, are essential for the copying of the single stranded RNA genome found in virions into the double-stranded DNA that is inserted into the host genome by IN (for a review of the retroviral life cycle, and an overview of reverse transcription, see Coffin, Hughes and Varmus, Retroviruses, 1997 (1)) . The larger subunit of the RT heterodimer, p66, contains the active sites for both of the enzymatic activities of RT (polymerase and RNase H); the smaller subunit plays a structural role (Figure 2). Important structural features of RT were elucidated by the first crystallographic studies (5, 6). p66 is composed of two spatially distinct domains, polymerase and RNase H. The polymerase domain is composed of four subdomains: fingers (residues 1–85 and 118–155), palm (residues 86–117 and 156–236), thumb (237–318), and connection (319–426) (5,6). p51 folds into the same four subdomains as the polymerase domain of p66 (fingers, palm, thumb, and connection); however the positions of the subdomains relative to each other are different in p66 and p51 (see Figure 2).
Figure 2.
Ribbon representation of HIV-1 RT in a complex with nucleic acid. The fingers, palm, thumb, connection, and RNase H subdomains of the p66 subunit are shown in blue, red, green, yellow, and orange, respectively. The p51 subunit is shown in dark brown. The template and primer DNA strands are shown in light and dark gray, respectively.
The nucleic-acid binding cleft is formed primarily by the p66 fingers, palm, thumb, connection, and RNase H subdomains of p66. The connection and thumb subdomains of p51 form the floor of the binding cleft (Figure 2). The binding cleft is configured so that the nucleic acid contacts both the polymerase and the RNase H active sites; these are located about 17–18 base pairs apart on the nucleic acid substrate. The αH and αI helices of the p66 thumb help to properly position the nucleic acid through interactions that involve both the primer and template strands. The DNA “primer grip”, a highly conserved structural motif (7) that consists of the p66 β12-β13 hairpin in HIV-1 RT (6) helps position the 3’-OH end of the primer strand at the polymerase active site. Mutational studies have shown that changes in the DNA primer grip alter nucleic acid binding and may affect both polymerase and RNase H activities of RT (8–16).
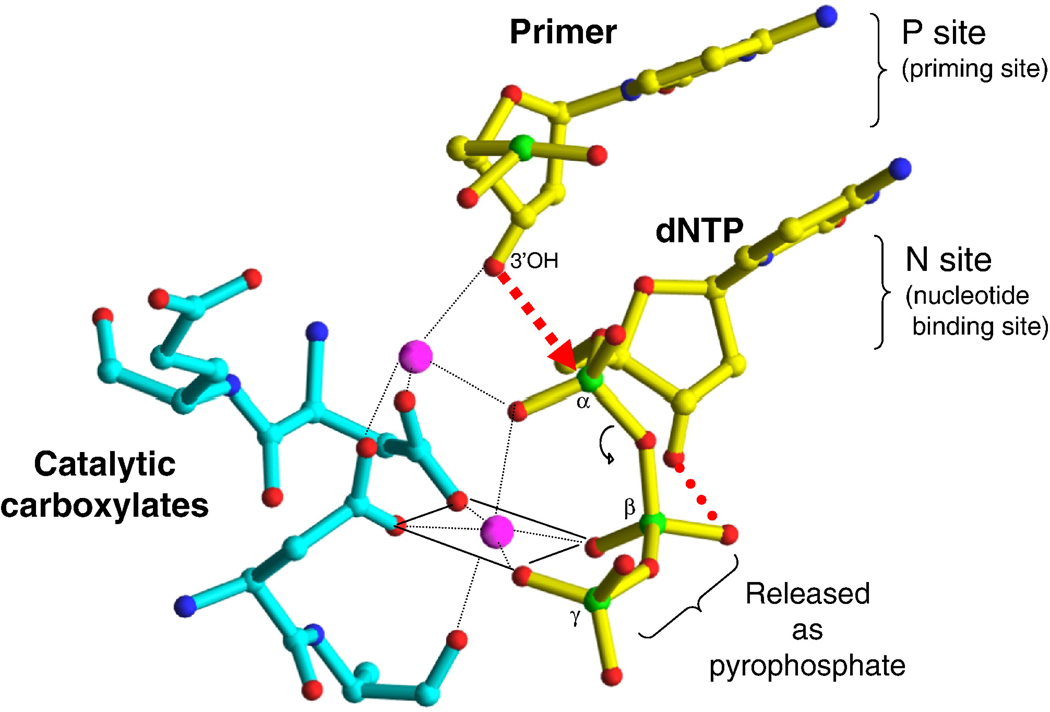
The polymerase active site is composed of three catalytic carboxylates in the palm subdomain of p66 (D110, D185, and D186) that bind two divalent ions (Figure 3) (Mg2+ appears to be the divalent cation used in vivo; Mn2+ can support polymerization in vitro) that are required for catalysis (17). D185 and D186 are part of the YXDD motif which is highly conserved in retroviral RTs (X is Met in HIV RT and Val, Leu, or Ala in other retroviruses) (18–20) .Other conserved residues that help form the dNTP binding site of RT include: a) R72 and K65 that are involved in the binding the β- and γ- phosphates respectively in the incoming dNTP (21); b) residue Y115 that contributes to the binding of the deoxyribose ring of the incoming dNTP and has been termed to be a steric gate that discriminates between deoxy and ribonucleoside triphosphates (22–24); and Q151, a residue that interacts directly with the 3’-OH of the incoming dNTP (21).
Figure 3.
Metal chelation in the polymerase active site of an enzyme/DNA/dNTP ternary complex. The nucleotide binding site (N site or pre-translocation site) and the priming site (P site or post-translocation site) are shown.
3. Molecular mechanisms
3.a. Molecular mechanism of polymerization
The mechanism of DNA polymerization by HIV RT is reasonably well understood; extensive biochemical and crystallographic data have helped define the individual steps of the process (Figure 4). The reaction begins with the binding of RT to the nucleic acid substrate, which results in a conformational change in the position of the p66 thumb, from a “closed” to an “open” conformation. Like many other DNA polymerases, RT requires both a primer and a template. In most sequence contexts, RT preferentially binds a double-stranded nucleic acid so that the 3’ end of the primer strand is bound at the priming site (P site), adjacent to the polymerase active site (6, 25–27). The initial step in nucleotide incorporation is the binding of the incoming dNTP at the nucleotide binding site (N site) to form a ternary complex (21). The rate-limiting step in the polymerization reaction is a conformational change in which a portion of the p66 fingers subdomain closes down on the incoming dNTP, which helps to precisely align the 3’-OH of the primer, the α-phosphate of the dNTP, and the polymerase active site (21, 28, 29). The chemical step that follows leads to the formation of a phosphodiester bond between the newly incorporated nucleoside and the primer with the concomitant generation of pyrophosphate. The fingers open to allow the pyrophosphate to leave the active site. In processive DNA synthesis, the nucleic acid substrate must translocate relative to RT to free the nucleotide-binding site so that RT can bind the next incoming dNTP.
Figure 4.
Reaction steps during the mechanism of DNA polymerization by HIV-1 RT.
Both steady state and pre-steady state kinetic experiments have been used to characterize dNTP binding and nucleotide incorporation (28, 29). Generally, dNTP binding has Kd in the low µM range. It was shown that dNTP binding is slightly more efficient on a DNA/DNA template-primer (Kd = 4µM) than to an RNA/DNA substrate (Kd = 14µM). The rate of incorporation of the bound nucleotide (kpol) is slightly increased when RNA/DNA is the substrate (∼2 fold increase compared to DNA/DNA). Transient kinetics studies have shown that the rate-limiting step for single nucleotide incorporation is a conformational change (fingers “closing down” on the active site). However, steady state experiments have shown that the overall reaction is limited by the dissociation rate of RT from the nucleic acid substrate (28, 30).
The approved NRTIs all lack a 3’-OH and act as chain terminators when they are incorporated into viral DNA by RT. If sufficient dNTPs are present, incorporation of a chain-terminator into DNA may result in the formation of a stable “dead-end complex” (DEC), where RT has incorporated the NRTI, the end of the primer (with the NRTI at the 3’ end) has been translocated, and the incoming dNTP is bound in a stable closed complex. This tighter binding of the nucleic acid substrate in the closed conformation can be demonstrated by electrophoresis in a native polyacrylamide gel (31).
The chemical step requires two divalent metal ions, there is good reason to believe that the normal in vivo metals are both Mg2+. The metals coordinate the oxygens of all three phosphates of the incoming dNTP and the side chains of the three catalytic Asp residues (110, 185, and 186). Metal coordination at the polymerase site facilitates the attack of the 3’-OH on the a-phosphate of the incoming nucleotide by activating the hydroxyl group (Figure 3). The two metal ions also stabilize the charge of the reaction intermediates (32).
Insights into the molecular details of the translocation step that follows incorporation of the nucleotide during processive DNA synthesis have been provided by crystal structures of RT covalently cross-linked to nucleic acid in pre- and post-translocation states (N and P complexes, respectively) (33). A comparison of these structures indicates unfavorable interactions between the conserved YMDD loop of the polymerase active site and the terminal phosphate of the extended primer terminus that may contribute to translocation after dNMP incorporation. It has not been possible to determine whether translocation precedes the release of pyrophosphate product. Recently, using a site-specific foot-printing assay, it was demonstrated that phosphonoformic acid, an analog of pyrophosphate, stabilizes the pre-translocation complex (N complex), suggesting that the pyrophosphate product is released before translocation of RT (26).
3.b. Molecular mechanisms of HIV-1 reverse transcriptase inhibition
HIV-1 RT inhibitors
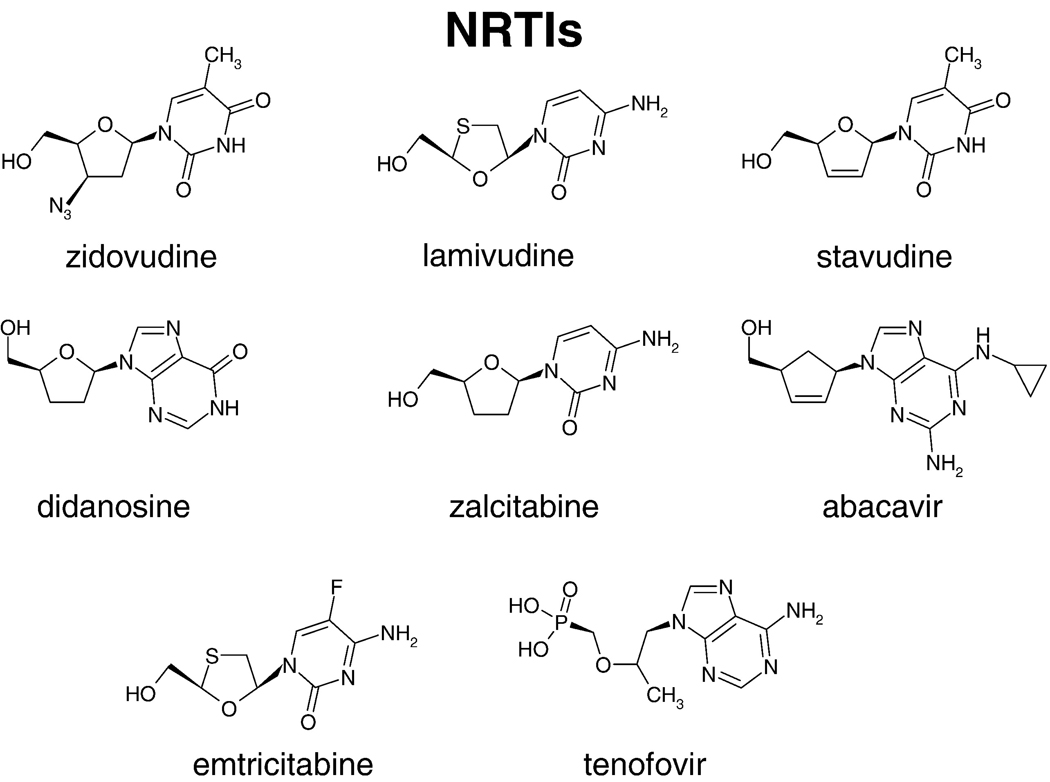
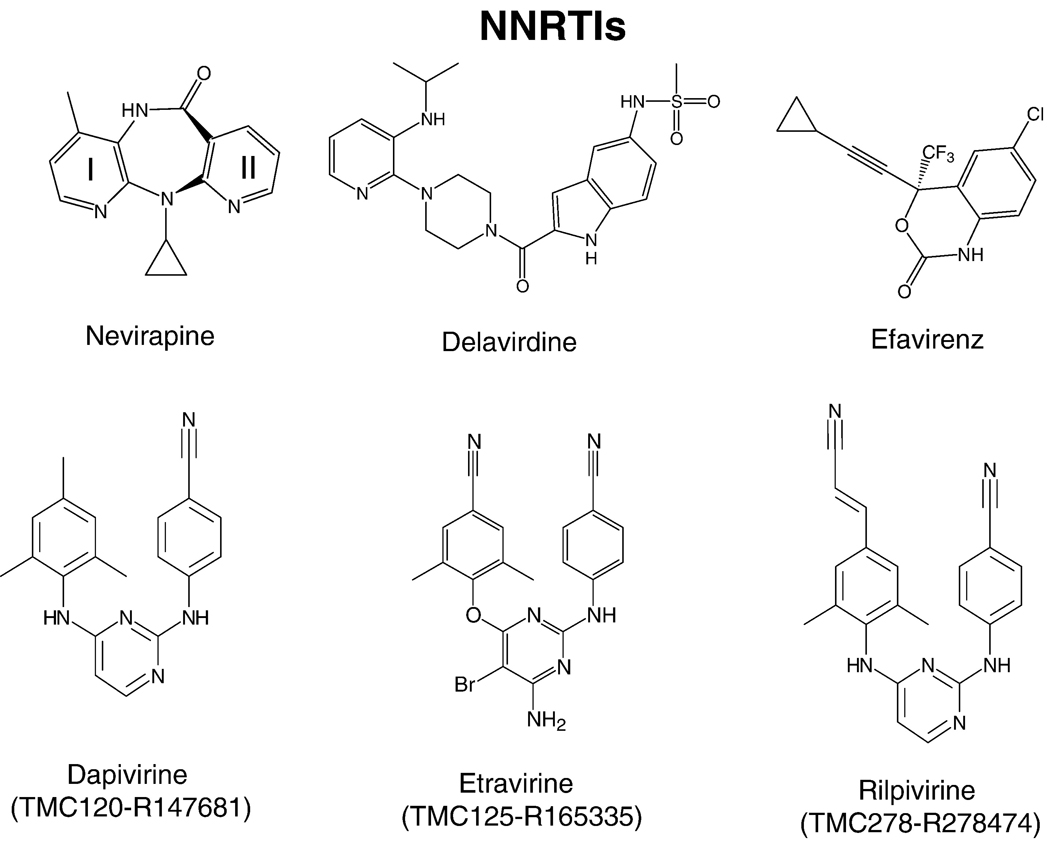
Because of its central role in the life cycle of HIV, RT has been a key target for the development of antiviral therapies. Nearly half of the anti-AIDS drugs target the polymerase activity of RT (http://www.fda.gov/oashi/aids/virals.html). The approved anti-RT drugs belong to one of two broad classes: nucleoside RT inhibitors (NRTIs) (Figure 5) and nonnucleoside RT inhibitors (NNRTIs) (Figure 6). Most standard three-drug regimens involve two NRTIs combined with either a PR inhibitor or an NNRTI; other combination therapies, some of which involve the entry/fusion inhibitors, are used to treat some of the patients who have failed the standard therapies. The following NRTIs have been approved by the US Food and Drug Administration: AZT-zidovudine, 3TC-lamivudine, FTC-emtricitabine, ddI-didanosine, and ABC-abacavir. d4T-stavudine and ddC-zalcitabine are not used in therapies any more. TFV-tenofovir is a nucleotide RT inhibitor and, for simplicity, it will be discussed with the nucleoside inhibitors (NRTIs). Some of these inhibitors are available as combinations, for example TFV+FTC (truvada), AZT+3TC (combivir), and ABC+3TC+AZT (trizivir) (http://www.hivandhepatitis.com/hiv_and_aids/hiv_treat.html). Four NNRTIs have been approved for the treatment of AIDS [the first generation NNRTIs are nevirapine and delavirdine, the second generation NNRTI is efavirenz, and the third generation is etravirine (TMC125)]. Additional NNRTIs are currently in late stage clinical trials. These include TMC278 (rilpivirine), UK-453,061, and RDEA-806; these third generation NNRTIs have increased effectiveness against drug-resistant HIV strains. Both tenofovir and TMC120 (dapivirine) are in large-scale trials as a chemopreventives; specific antiretroviral chemoprevention strategies have great potential to reduce the sexual transmission of HIV. While some specific inhibitors of the RNase H activity of RT have been described, none has yet been approved for the treatment of HIV infections.
Figure 5.
Chemical structures of NRTIs.
Figure 6.
Chemical structures of NNRTIs.
Inhibition by nucleoside reverse transcriptase inhibitors (NRTIs)
NRTIs are structurally diverse analogs of the natural substrates of DNA synthesis (Figure 5). As has already been discussed, the approved NRTIs all lack the 3’-OH and act as chain terminators when incorporated in viral DNA by RT. For NRTIs to be effective against HIV, they must be taken up by the host cell and then phosphorylated by cellular enzymes to convert them to their active form, the NRTI triphosphates. The efficiency of this conversion to the active metabolite and the stability of NRTIs (and their triphosphates) in the presence of catabolic enzymes are important considerations in antiviral therapies, because these factors help determine the concentration of the inhibitor in the bloodstream that is required for the NRTI to be effective. Although all NRTIs are analogs of normal nucleosides, not all NRTIs are good substrates for the cellular kinases (34). For example, AZTTP levels are low because the addition of the second and third phosphates is limiting (35). However, for other NRTIs, the addition of the first phosphate is the limiting step. For this reason it can be helpful to develop prodrug versions of NRTIs that contain the first phosphate. Prodrugs are modified by the addition of hydrophobic moieties that allow them to enter cells. The hydrophobic groups are designed to be removed once the compound enters the cell. For example, TFV is given to patients as a prodrug phosphonate nucleotide. As such, TFV needs the addition of two, instead of three, phosphates (Figure 5). The NRTIs ABC, and ddI (Figure 5) are modified by cellular enzymes to carbovir-triphosphate and dideoxyadenosine-triphosphate (ddATP), the respective active metabolites. RT incorporates NRTI triphosphates with variable efficiencies. For example, RT incorporates AZTTP, a thymidine analog, almost as efficiently as dTTP (36). In contrast, ddCTP is incorporated less efficiently than dCTP (37). 3TC-TP, another cytidine analog, is given to patients as the L rather than the D enantiomer and is incorporated less efficiently than either dCTP or ddCTP (12).
Inhibition by non-nucleoside reverse transcriptase inhibitors (NNRTIs)
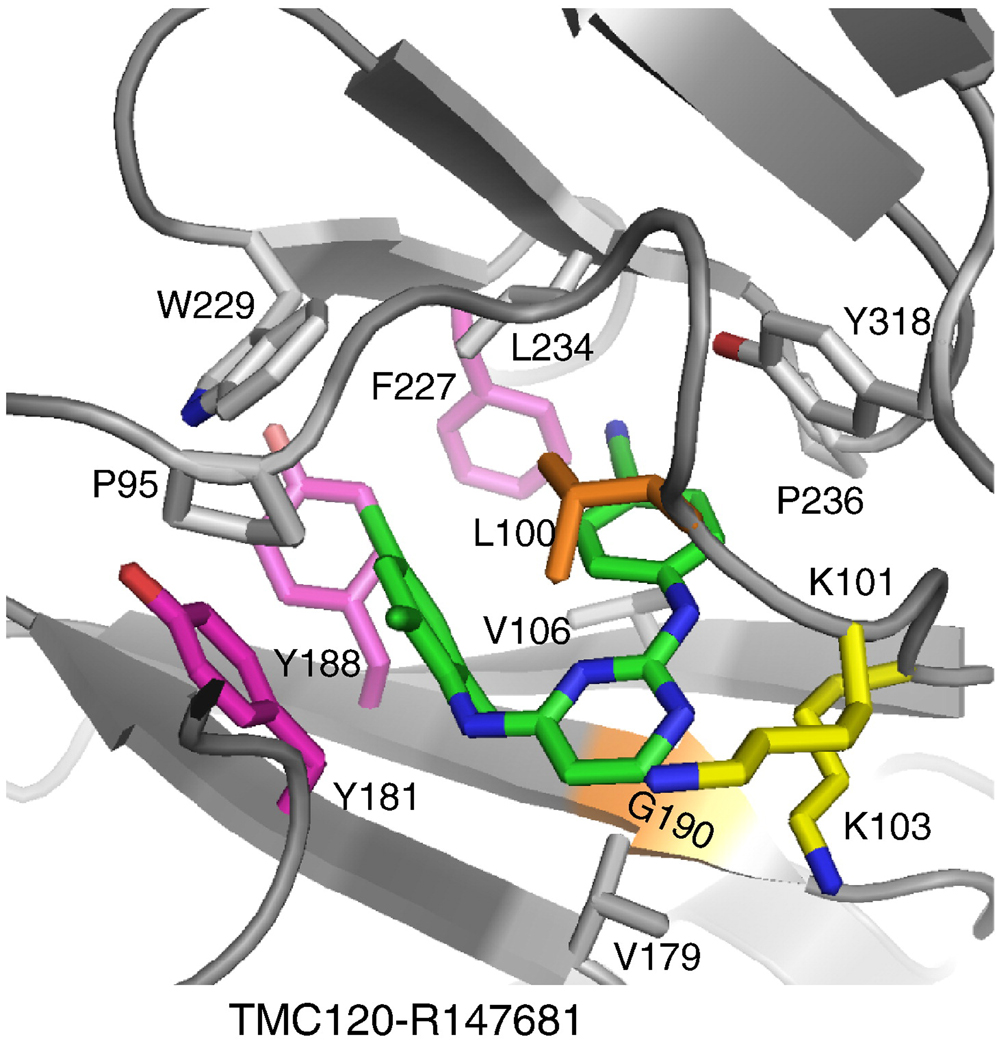
NNRTIs (Figure 6) bind at the NNRTI-binding pocket (NNIBP), a hydrophobic pocket adjacent to the polymerase active site (∼10 Å) (Figure 7). The NNIBP consists of residues L100, K101, K103, V106, T107, V108, V179, Y181, Y188, V189, G190, F227, W229, L234, and Y318 of p66 and E138 of p51 (5, 38).
Figure 7.
Ribbon representation of the NNRTI-binding pocket, showing the residues where NNRTI-resistance mutations occur.
Biochemical data have shown that NNRTIs are non-competitive inhibitors and do not directly interfere with the binding of either the dNTP or the nucleic acid substrates of RT. Pre-steady state kinetic analysis of single nucleotide addition in the presence of NNRTIs has shown that binding of NNRTI interferes with the chemical step of DNA synthesis (28, 29). However, the molecular details of NNRTI inhibition are not clearly understood.
Structural studies of HIV-1 RT have shown that: 1) the NNIBP does not exist in structures of HIV-1 RT that have no bound NNRTI; 2) the NNIBP is created by large structural rearrangements, particularly an extended conformation of the primer grip and rearrangements of the aromatic-ring containing residues Y181 and Y188 (Figure 7)-this structural rearrangement locks the p66 thumb and fingers in their hyper-extended conformations (5, 38–41); 3) the NNIBP is created primarily from the p66 subunit (near the polymerase active site); 4) p51, which contains amino acids identical to the polymerase domain of p66, does not have a second NNIBP; 5) the NNRTI-resistance mutations are located in and around the NNIBP (5, 38, 42); and 6) there are no metal co-factors bound at the active site with persuasive coordination geometry when an NNRTI is bound to RT (43).
The structural data suggest that NNRTI-binding distorts the position of the primer grip, thus affecting the alignment of the primer terminus with the polymerase active site, which could affect the chemical step of viral DNA synthesis. There is also structural evidence to suggest that NNRTI-binding affects the conformation of the catalytic carboxylates that bind the metal cofactors (43). Specifically, the β9–β10 loop that contains the conserved YMDD motif (Figure 3) assumes different conformations during the course of DNA polymerization (33). In the RT/DNA/dNTP complex the YMDD loop flexes and moves “down” by 2 Å to bind dNTP and the metal ions. However, in the RT/NNRTI complexes, the YMDD loop always assumes the opposite conformation (“up”). This suggests that NNRTI binding restricts a conformational change of the YMDD loop that is needed to attain the metal-binding conformation and that his restriction could affect the translocation of the nucleic acid that normally occurs after the incorporation of each nucleotide (43).
3.c. Molecular mechanisms of resistance
NRTI resistance
Drug resistance remains a central challenge in HIV therapies. Because resistant viruses can be transmitted, the prevalence of resistant viruses is increasing in untreated HIV-1 patients. For the virus to replicate and be transmitted, HIV-1 RT must be able to complete viral DNA synthesis, and NRTI-resistant RTs must retain the ability to incorporate normal dNTPs with reasonable efficiency. This means that NRTI resistance involves enhanced discrimination between normal nucleosides and NRTIs. Two basic types of NRTI-resistance mechanisms are known for HIV-1 RT: 1) one resistance mechanism (exclusion) involves enhanced discrimination at the time the NRTI-TP is incorporated. The M184V/I mutations provide a clear example of the exclusion mechanism, M184V/I selectively reduce the incorporation of 3TC and FTC by steric hindrance (21, 44, 45). 2) The second mechanism involves the selective removal of the NRTI from the end of the viral DNA after it has been incorporated by RT (46–49). This is the excision mechanism; a well-studied example involves AZT resistance caused by a set of mutations including M41L, D67N, K70R, L210W, T215F/Y, K219E/Q (these mutations will be collectively referred to as AZT resistant or AZTr; they are also referred to as thymidine-analog mutations, TAMs,or excision-enhancing mutations, EEMs). Although the HIV-1 RTs isolated from AZT-resistant viruses in patients do not ordinarily have all of the AZTr mutations, combinations of these mutations give rise to high levels of resistance to AZT and to much lower levels of resistance to some other NRTIs (50–53). HIV-1 RTs carrying the AZTr mutations can acquire additional mutations that enhance their ability to excise AZT, and allow these RTs to excise most other NRTIs more efficiently.
NRTI exclusion
In addition to the M184V/I mutations that cause resistance to 3TC and FTC by the exclusion mechanism (which has already been discussed), the mutations L74V (ddI resistance) (54), K65R (TFV and ddI resistance) (55–57) and a complex set of several mutations that involves Q151M (resistance to most NRTIs) cause resistance by the exclusion mechanism (54, 58). All of the amino acids involved in resistance by the exclusion mechanism are in the fingers or the palm of RT; all are in positions that could affect the binding of an incoming dNTP. We do not yet have as detailed a picture for the exact mechanisms of resistance for these mutations as we do for the M184V/I mutations. For K65R, biochemical studies have shown that it has a decreased rate of nucleotide incorporation and markedly decreased rate of TFV incorporation (59, 60). We also know that the K65R mutation causes hypersusceptibility to AZT by reducing the efficiency of ATP-mediated excision, which counteracts a decreased rate of AZT incorporation (61, 62). Finally, it has also been shown that in the background of K65R, M184V causes partial resensitization to TFV (63). Q151 interacts with the 3’-OH of a normal triphosphate (21). It appears that the Q151M mutation, and the additional mutations that usually accompany the Q151M mutation, cause differences in the hydrogen-bonding network between the deoxyribose of an incoming dNTP and the enzyme, and that these changes enhance the importance of the interactions of the enzyme with the 3’OH (64). This allows the mutant enzyme to better discriminate between normal dNTPs, which have the 3’-OH, and NRTIs, which do not have a 3’-OH. Presumably L74V (65) (and the less common mutation V75T (66)) both disrupt the hydrogen bonding network that involves the 3’-OH, although the details by which this occurs have not been well defined (67), and the discrimination is selective for ddI (which is converted to ddA in cells) and, to a lesser degree, for ddC. What is striking is that, despite the fact that the mechanism apparently involves enhanced recognition of the 3’-OH, the L74V enzyme is not resistant to other NRTIs (68).
NRTI excision
The first example of NRTI resistance to be described was resistance to AZT. However, it took more than 10 years for the underlying molecular resistance mechanism to be understood. This was, at least in part, because the mechanism was unexpected. An AZT-resistant RT incorporates AZT-TP as efficiently as wild-type HIV-1 RT. However, AZT-resistant RTs have an enhanced ability to remove the incorporated AZT-MP from the 5’ end of the template strand. The underlying mechanism involves ATP-dependent excision; the excision mechanism is related to the normal mechanism of polymerization run in reverse (46–48). Although there was some initial controversy about whether ATP or PPi is the pyrophosphate donor in the excision reaction that gives rise to AZT resistance, it is now clear that the excision reaction that gives rise to AZT resistance in vivo uses ATP as the pyrophosphate donor, rather than PPi, which is the normal product of the polymerization reaction. These proposals answered some important questions but left other questions unanswered. 1) If the mechanism is ATP-dependent excision, why is AZT excised so much more efficiently than other NRTIs? 2) How do the AZT-resistance mutations cause enhanced excision? In 2001, Boyer et al. (49) proposed a specific model in which ATP is the pyrophosphate donor, and at least some of the mutations that cause AZT resistance enhance the appropriate binding of ATP. In particular, a key mutation, T215F/Y, is selected because an F or Y at 215 can stack with the adenine ring of ATP. Although previous studies have shown little or no difference in ATP binding in the presence of AZT resistance mutations (69, 70), Dharmasena et al (71) show, in experiments using the excision product AZTMP-PPPA, that the primary mutations that give rise to AZT resistance enhance the ability of RT to bind ATP. For the excision reaction to occur, the end of the primer strand must be at the polymerase active site (also called the N, or nucleotide binding site) (Figure 3). Normally, translocation moves an incorporated dNTP or NRTI-TP from the N site to the P or priming site so that the next incoming dNTP can bind. In most cases the concentration of dNTPs in cells is high enough that the incoming dNTP can be bound. NRTIs lack the 3’-OH; so that, if a NRTI-TP has been incorporated and the end of the primer is translocated to the P site, the next incoming dNTP can be bound, but cannot be incorporated. At reasonable concentrations of dNTPs this is the inhibited state of an RT that has just incorporated an NRTI: The recently incorporated NRTI-MP is in the P site and the enzyme forms a closed complex with the incoming dNTP bound at the N site. The closed complex is relatively stable and, as was just discussed, excision requires that the NRTI be at the N, and not the P site. This is why an AZTr RT cannot efficiently excise most NRTI-MPs from the end of the primer strand: The NRTI-MP at the end of the primer cannot be translocated from the P to the N site because the bound incoming dNTP blocks the N site. In the Boyer et al. model, AZT-MP is excised more efficiently than are other NRTI-MPs because, when an AZT-MP is the nucleotide at the 3’ end of the primer, the long azido group of the AZT-MP interferes with the appropriate binding of the incoming dNTP, which destabilizes the closed complex. Because the closed complex is less stable, an AZT-MP terminated primer has better access to the N site (active site), where it can be excised. There are structural data which support this part of the model (33). In addition, the model is supported both by the observations that the aromaticity of the nucleobase on the pyrophosphate donor determines how well it functions in an excision reaction and by the fact that diphosphate nucleotides are poor excision substrates (presumably because when the nucleobase binds to T215F/Y the phosphates are not in position to react with the end of the primer)(72).
There are complexities to the excision reaction that are still not completely understood. For example, many other nucleoside analogs are excised less efficiently than AZTMP even in the absence of an incoming dNTP (73). One possibility is that an AZT terminated primer is a good substrate for excision not only because the incoming dNTP binds poorly (31, 48, 49), but because, in the absence of an incoming dNTP, an AZT terminated primer preferentially binds to the N site (25, 27). However, this may not be the only explanation, and it is entirely possible that there are other factors that affect the efficiency of excision. For example, it was shown that not all 3'-azido analogs are good substrates for excision which suggests that other components of the primer terminus, such as the base, also contribute to the efficiency of excision (74).
As has already been mentioned, AZTr RTs can acquire additional mutations that allow the RT to excise a much broader set of NRTIs than does AZTr RT. One set of mutations that enhance the breadth and efficiency of NRTI excision by AZTr RTs involves the insertion of amino acids near position 69 of the fingers subdomain of HIV-1 RT, these insertions are accompanied by mutations of the adjacent amino acids (75–78). There are a number of related “fingers insertion” mutations (79–85). The insertions are in the β3-β4 loop of the fingers, which is the loop that closes down on the incoming dNTP to form the closed complex. The insertion mutations apparently broadly destabilize the formation of dNTP-induced stable closed complexes for primers terminated with a variety of NRTI-NMPs so that, in the presence of physiological dNTP concentrations, excision of these NRTIs would be inhibited to a lesser extent than with WT RT (75, 76). However, the finger mutations significantly enhance excision in the absence of dNTPs; there are data to show that insertion of SG or AG can increase the rate of excision directly (76). RTs only acquire the fingers insertion mutations if they already have AZTr mutations, and are able to bind ATP efficiently and appropriately for the excision. The addition of the fingers insertion mutations allows a much broader array of incorporated NRTI-MPs access to the N site where they can be excised (25). Conversely, there are NRTI and NNRTI resistance mutations that reduce the ability of HIV-1 RT to excise AZT (and other NRTIs). For example, the M184V/I mutations that give rise to 3TC/FTC resistance also reduce the excision efficiency of WT and drug-resistant HIV-1 RTs (86, 87), although another study that used a different template-primer sequence did not show a significant reduction in excision (88). One of the connection subdomain mutations, N348I, was shown to confer moderate cross-resistance to both NRTIs and NNRTIs (89, 90). This mutation is present in a significant number of clinical isolates, usually in the presence of other NRTI-resistance mutations.
NNRTIs and NNRTI resistance
There are four NNRTI drugs [nevirapine, delavirdine (first generation), efavirenz (second generation), and etravirine (third generation)] that are currently approved for treating HIV-1 infections and several other potent NNRTIs that inhibit HIV-1 at nanomolar concentrations (EC50) are in clinical trials. However, there are mutations in RT that can cause resistance to all of the approved NNRTIs. Most of the NNRTI resistance mutations are found in and around the NNIBP. K103N and Y181C are the most frequently observed resistance mutations in patients treated with the approved NNRTIs. Other NNRTI-resistance mutations observed in patients include L100I, K101E, V106A, V179D, Y188L, G190A, and P236L; the NNRTI-resistance mutations can occur singly, or in combinations. Unfortunately, resistance to first and second generation NNRTIs can evolve relatively quickly. The most promising of the third generation NNRTIs are effective against HIV strains that carry most common single and double mutations; however, viral strains carrying multiple NNRTI-resistance mutations can exhibit significant levels of drug resistance.
Extensive crystallographic, molecular modeling, and biochemical studies have contributed towards understanding NNRTI drug resistance and the development of better NNRTIs, which is an ongoing effort. Our current understanding suggests that there are at least three broad classes of NNRTI-resistance mechanisms.
Loss/change of key hydrophobic interactions
Amino acid residues Y181, Y188, and F227 are located in the hydrophobic core of the NNIBP (Figure 7) (5, 39, 91, 92). Specific residues in this core have extensive interactions with NNRTIs. Mutations in some of the key residues (Y181C, Y188L, F227L) cause significant resistance through the loss of the aromatic ring interactions with NNRTIs, which are generally hydrophobic (5, 93–96). This causes high levels of resistance to the first generation NNRTIs, which are relatively rigid. More advanced NNRTIs, however, are designed with so-called "strategic flexibility". This intrinsic flexibility makes it possible for the newer drugs to have compensatory interactions with RTs that have mutations that cause resistance to the first-generation NNRTIs (97). This flexibility in the binding has been called “wiggling” and “jiggling”, and its structural basis has been described in recent structural studies of wild-type, K103N/Y181C, and L100I/K103N HIV-1 RT complexes with TMC278/rilpivirine (97). Wiggling and jiggling allow NNRTIs to adapt to changes in the NNIBP caused by resistance mutations; the side chains of the pocket residues adjust to accommodate inhibitor binding in a “shrink-wrap” mode.
Steric Hindrance
Amino acid residues L100 and G190 are in the central region of the NNIBP. Mutations in either of these residues cause high levels of resistance to many NNRTIs. The L100I mutation confers resistance by changing the shape of the pocket (the amino acid is β-branched instead of γ-branched) (96), whereas G190A introduces a bulge (67). For example, HBY 097, which effectively inhibits viruses carrying the Y181C mutation, cannot inhibit viruses that carry the G190A mutation (39). In the crystal structures of the wild-type RT/HBY 097 complex (92), the bulky and rigid quinoxaline ring of HBY 097 is near the β6–β10–β9 sheet (that contains G190). The G190A mutation would cause the Cβ-atom of A190 to have a steric clash with the quinoxaline moiety and reduce the binding of HBY 097.
Pocket Entrance Mutations
The K103N and K101E mutations are two NNRTI-resistance mutations that frequently cause resistance to first generation NNRTIs. Amino acid residues K101 and K103 are located at the rim of the entrance to the NNIBP with their side chains pointing out. These mutations apparently cause resistance by interfering with the entry of NNRTIs into the pocket (98, 99) . Second generation NNRTIs were designed to overcome this problem. For example, DAPY NNRTIs are able to inhibit K103N mutant because they interact with the side chain of the mutated N103 residue (97, 100). However, new drug-resistance mutations, or combinations of mutations, will be selected when the more advanced NNRTIs are used to treat HIV-1 infected patients.
Recently, it has been reported that a number of mutations in the connection, or RNase H subdomains of RT, can enhance resistance to both NRTI and NNRTI inhibitors of RT (89, 90, 101, 102). The biochemical mechanism of this effect is still the subject of investigation; however, it does appear that the reduction in RNase H activity reduces the degradation of the RNA template, which allows more time for the excision mechanism to occur, increasing resistance to AZT (103). In addition, it was shown that connection domain mutations cause accumulation of short AZT-terminated RNase H cleavage products that are better substrates for the excision reaction which results in AZT resistance (104).
4. Molecular mechanism of RNase H activity
As mentioned earlier, the RNase H is responsible for the degradation of the RNA portion of the RNA/DNA substrate that is formed during minus strand synthesis. It is also responsible for the removal of the priming tRNA (105, 106) and the polypurine tract (PPT) (107). Viruses deficient in RNase H activity are non-infectious (108, 109), making the RNase H an interesting target for anti-HIV inhibitors. The HIV-1 RNase H was the first segment of RT to be crystallized (110). It was shown to be structurally similar to other RNase H such as RNase HI from Escherichia coli and Thermus thermophilus. Crystal structures of full-length HIV-1 RT with a DNA/DNA (6, 38) or RNA/DNA (10) substrate show a distance of ∼60Å between the polymerase active site and the RNase H active site. In terms of nucleotides, the distance is 17 base pairs for a DNA/DNA substrate and 18 base pairs for a RNA/DNA substrate, which is in agreement with biochemical analysis (111–118).
HIV-1 RNase H was shown to function in a two-metal-ion mechanism similar to other polymerase associated nucleases. The crystal structure of the RNase H fragment shows 2 Mn2+ ions bound the active site at a distance of ∼4Å of each other and interacting with conserved residues D442, E478, D498 and D549 (110). Calorimetry and solution NMR experiments also demonstrated the presence of 2 Mn2+ or 2 Mg2+ ions bound to the RNase H (119, 120). Biochemical assays suggested that the optimal RNase H activity is obtained in the presence of 1 Mg2+ ion and 1 Mn2+ ion (121); however, it is likely that, in vivo, both metals are Mg2+. Recently, the crystal structure of an RNase H complexed with an RNA/DNA hybrid confirmed a mechanism for substrate recognition and two-metal-ion-dependent catalysis that has been proposed to be common between the functionally similar transposases, retroviral integrases, Holliday junction resolvases, and RISC nuclease Argonaute (122)
It has been suggested that the RNase H specificity for RNA/DNA substrate relies on the width of the minor groove (123–130). This proposal comes from the observation that RNA/DNA has an intermediate conformation between A- and B-form double-stranded nucleic acids named H-form, which has a minor groove width of ∼9–10Å. The HIV-1 RNase H cleaves substrates with different minor groove widths much less efficiently. These include the junction of an RNA-DNA hybrid annealed to a DNA template where the minor groove can be as narrow as 4.5–5.5Å (130). The crystal structure of RT bound to an RNA/DNA substrate supports the idea that it is the width of the minor groove, in combination with the RNase H primer grip, which binds and positions the primer strand on the enzyme, that is responsible for cleavage specificity (10). Mutagenesis suggests that the residues in the RNase H that are in contact with the primer strand are more important than the residues contacting the template strand (9, 131).
As has already been described, the polypurine tract (PPT) is used as primer for the initiation of the plus-strand synthesis (1) (and references within). A crystal structure of HIV-1 RT in complex with the PPT as RNA template annealed to a DNA primer provides insights regarding the resistance of the PPT to RNase H degradation (10). The structure shows a narrower than expected minor groove in the RNA/DNA substrate near to the RNase H active site (∼7Å), which positions the RNA template approximately 3Å away from the RNase H active site. This distance of 3Å is approximately the same as the difference between the minor groove width in the PPT RNA/DNA structure and the predicted minor groove width in H-form nucleic acid. The difference in width of the minor groove is thought to be due to the presence of A-tracts in the PPT; A-tracts are known to have a narrower minor groove and to provide more stiffness to the nucleic acid.
As has already been discussed, RNase H is also responsible for the removal of the PPT from the 5’ end of the genome, and the tRNA used as primer from the 3’ end of the genome, these cleavages define the ends of the linear DNA that is the substrate for integration. The RNase H cleaves at the junction between the PPT RNA and the DNA transcript made by RT (1) (and references within). Using a single molecule fluorescence resonance energy transfer assay, Abbondanzieri et al. recently showed that the preferred orientation of RT on a nucleic acid substrate depends on the both the composition of substrate (RNA/DNA vs. DNA/DNA) and on the sequence of the nucleic acid. They also showed that RT can rapidly switch between orientations when it binds to polypurine RNA primers for plus-strand DNA synthesis (132).
In most retroviruses, the RNase H removes the priming tRNA by cleaving at the junction of the RNA and the DNA genome. In most cases, IN removes two nucleotides from each of the 3’ end of viral DNA, exposing a conserved CA dinucleotide that is then joined to the host DNA. In the case of HIV-1, there is only a single nucleotide between the CA and the primer binding site (PBS), where viral DNA synthesis is initiated. Sequence analysis of the 2-LTR circle junctions which are derived from the ligation of the ends of the viral linear DNA, suggested that RNase H cleaves one nucleotide from the 3’ end of the tRNA, giving rise to viral DNA substrate that would allow IN to remove two nucleotides (133, 134); and in vitro analysis of the RNase H cleavage on model substrates supports this interpretation (105, 106).
4b. Inhibitors of RNase H
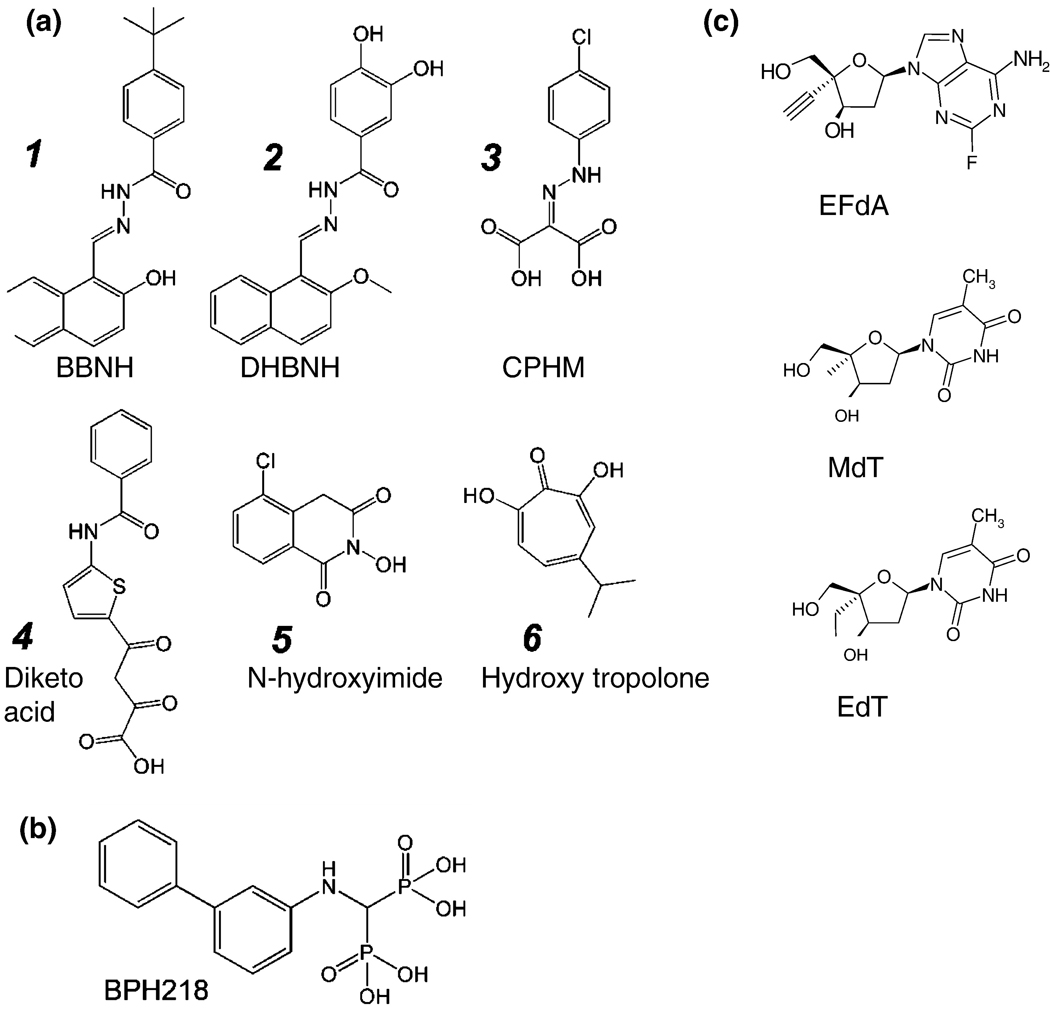
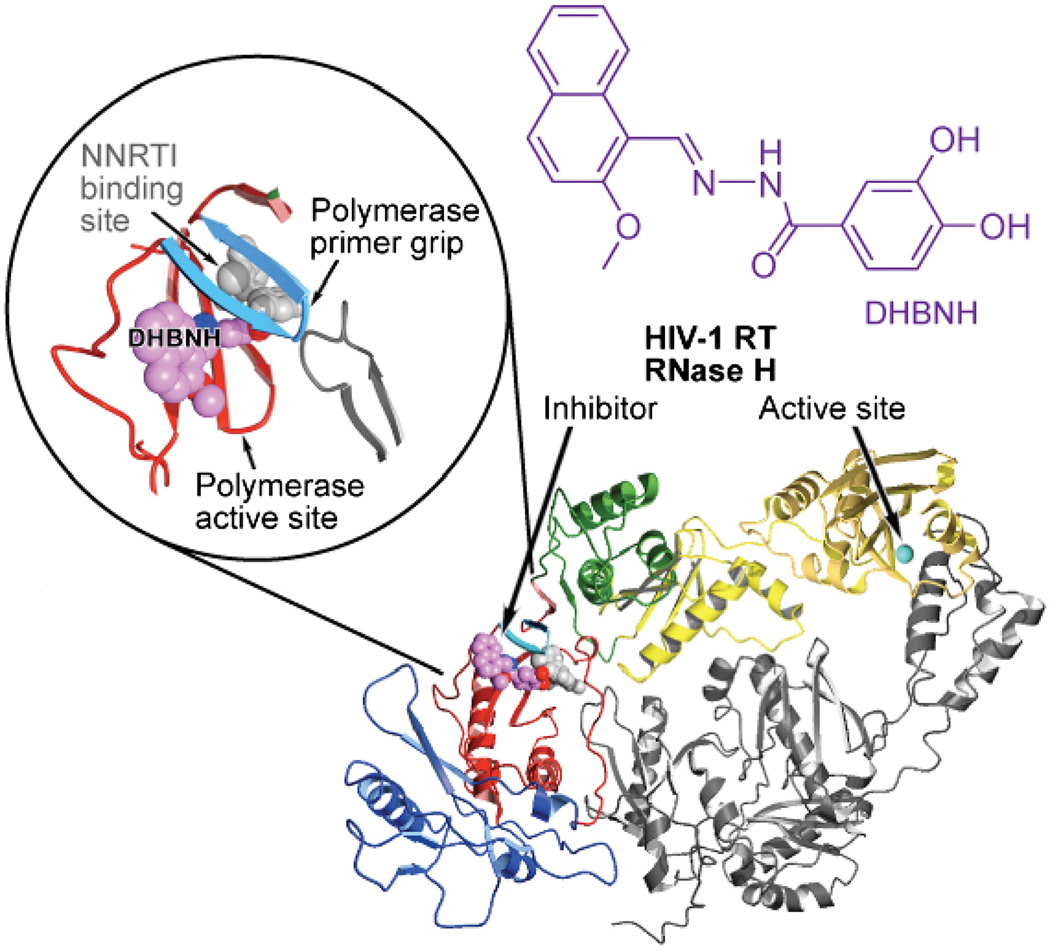
As has been mentioned earlier, RT RNase H activity is essential for HIV-1 replication. For that reason, RNase H is a promising target for the development of antiretroviral drugs (135, 136). RNase H inhibitors (RNHIs) are likely to be active against all of the current drug-resistant HIV-1 variants. Only six RNHIs with reasonable in vitro potencies have been described in any detail (Figure 8A, structures 1 – 6), and most of these have minimal therapeutic value because of limited cell uptake or cytotoxicity. Although all of the available inhibitors of the polymerase activity of RT are bound either at or near the polymerase active site, not all the RNHIs are bound at or near the RNase H active site. The acylhydrazone N-(4-tert-butylbenzoyl)-2-hydroxy-1-naphthaldehyde hydrazone (BBNH; Figure 8A, structure 1) is an RNHI with reasonable potency (137). BBNH is a bifunctional inhibitor of HIV-1 RT; it inhibits both the RT DNA polymerase and RNase H activities of the enzyme with similar potency (IC50 ≈ 3 µM). A crystal structure of HIV-1 RT complexed with an analog of BBNH, dihydroxy benzoyl naphthyl hydrazone (DHBNH) has been published (138). Unlike BBNH, DHBNH (Figure 8A, structure 2) is a specific inhibitor of RT RNase H activity (IC50 ≈ 0.5 µM) and has relatively limited activity against the DNA polymerase activity of RT. DHBNH is non-cytotoxic and inhibits the replication of a variety of drug-resistant HIV-1 RT mutants (138). Surprisingly, in the crystal structure, DHBNH binds more than 50 Å away from the RNase H active site, at a site that is in some sense between the polymerase active site and the NNIBP (Figure 9). The mechanism by which DHBNH binding inhibits RT RNase H is not clear, but may involve repositioning of the polymerase primer grip and the thumb of RT in a manner that alters the trajectory of the nucleic acid near the RNase H active site, thereby preventing RNase H-catalyzed cleavage (138). Alternatively, DHBNH (and other acylhydrazone analogs) may also bind to a second site in or near the RNase H domain that was not seen in the crystal (139). Recent NMR studies of the interaction of DHBNH with an active RNase H fragment that does not contain the polymerase domain are consistent with this possibility (Parniak & Ishima, unpublished observations).
Figure 8.
Chemical structure of RNase H inhibitors (8A): 1, N-(4-t-butylbenzoyl)-2-hydroxy-1-naphthaldehyde hydrazone (BBNH); 2, dihydroxy benzoyl naphthyl hydrazone (DHBNH); 3, 4-chlorophenylhydrazone of mesoxalic acid (CPHM); 4, diketo acid; 5, N-hydroxyimide; 6, hydroxyl tropolone (2,7-dihydroxy-4-1(methylethyl)-2,4,6-cycloheptatrien-1-one, or β-thujaplicinol); excision inhibitors (8B), and 4’-substituted nucleoside analogs (8C): 1, 4’ethynyl-2Fluoro deoxyadenosine (EFdA); 2, 4’methyl-deoxythymidine (MdT), 4’ethynyl-deoxythymidine (EdT).
Figure 9.
Ribbon representation of HIV-1 RT with DHBNH bound (with permission from ACS Chem Biol). In the crystal structure of DHBNH, an RNase H inhibitor binds >50 Å away from the RNase H subdomain, at a site that partially overlaps the NNIBP. The subdomains of the p66 subunit are color-coded (fingers in blue, palm in red, thumb in green, connection in yellow, and RNase H in gold). Upper left inset: a close-up of the DHBNH binding site. The inhibitor is shown in magenta. The pocket that is occupied by NNRTIs is shown in gray (the NNRTI pocket is not occupied in this structure).
The aryl hydrazone 4-chlorophenyl hydrazone mesoxalate (CPHM; Figure 8A, structure 3) inhibits the RNase H of HIV-1 RT with potency similar to that of BBNH (140). Unlike BBNH, CPHM is specific for RNase H and does not inhibit the DNA polymerase activity of RT. CPHM has no antiviral activity, and it is possible that this charged molecule cannot enter cells. The Merck group has described a diketo acid inhibitor of RNase H, 4-[5-benzoylamino) thien-2-yl]-2,4-dioxobutanoic acid (Figure 8A, structure 4) that was discovered in their integrase inhibitor discovery program (141). Like CPHM, this molecule has no antiviral activity. Structural information for the interaction of CPHM and RT is not yet available. Based on the proposed mechanism for the diketo acid inhibitors of integrase, it seems likely that CPHM and diketo acids inhibit RNase H by binding to the active site and chelating the essential Mg2+ ions. Other RNHIs include the N-hydroxyimides (121, 142) (Figure 8A, structure 5) and hydroxy tropolones (143) (Figure 8A, structure 6). These RNHIs also possess functional groups spaced to allow the coordination of the two metal cations in the RNase H active site. Neither of these compounds are therapeutic candidates because they are cytotoxic.
Future perspectives
Delayed chain terminators
Because excision is a major pathway for HIV-1 NRTI resistance, it is important to develop NRTIs that are relatively resistant to excision. Conventional NRTIs are susceptible to excision because they remain at the 3’ end of the primer strand, where they could be excised, if resistance mutations in HIV-1 RT allow the 3’ end of the primer good access to the N site. Nucleoside analogs that cause DNA chain termination only after additional normal nucleotides have been incorporated (delayed chain terminators) should be less susceptible to excision. The conformationally locked nucleosides North methanocarbathymidine (N-MCT) and North methanocarba-2’-deoxyadenosine (N-MCdA) act as delayed chain terminators in an in vitro HIV-1 RT polymerization reaction. The N-MC compounds are poorly excised by excision-proficient RTs. Unfortunately N-MCT is poorly phosphorylated in cultured cells (144, 145); however, the compound can be tested against HIV-1 (or HIV-1 based vectors) in cell lines that express herpes simplex virus thymidine kinase (HSV-TK), which allows N-MCT to be phosphorylated reasonably well. N-MCT potently inhibits HIV-1 replication in cells that express HSV-TK and efficiently blocks the replication of HIV-1 variants that replicate using excision-proficient RTs (146). Although the N-MC compounds are not drug candidates, these results suggested that this is a promising approach to the problem posed by excision proficient NRTI-resistant mutants.
The results obtained with the N-MC compounds illustrate a key point: To be active in cells, nucleoside analogs must be phosphorylated by host cell kinases. The deoxyribose rings of nucleosides are not flat, and can exist in various conformations. Two preferred conformations have been called North (found in A-form nucleic acids) and South (in B-form nucleic acids). Most DNA polymerases (including HIV-1 RT) prefer to incorporate dNTPs that are in the North conformation (21, 146–149); however, cellular kinases prefer the South conformation (150–153). The N-MC compounds are poorly phosphorylated because they are locked in the North conformation.
4’ substituted NRTIs
Several groups have concentrated on developing compounds that have 4’ modifications on the pseudosugar ring and interfere with viral DNA synthesis. Mitsuya and colleagues have done extensive work on 4’-modified nucleosides and identified promising compounds (154). The most potent of the compounds described thus far have a 4’-ethynyl (4’E) substitution and have antiviral activity against wild-type and drug-resistant HIV strains (155). Second generation 4’-ethynyl substituted compounds have an additional 2-fluoro (2F) substitution on the adenine ring, and inhibit HIV at picomolar concentrations (156, 157) . 4’E-2FdA (Figure 8C) retains significant activity against multi-drug resistant viruses isolated from patients. The compound appears to have low cytotoxicity and it does not appear to inhibit human DNA polymerases a and β, or mitochondrial DNA polymerase ? (158).
Marquez and colleagues have shown that the triphosphates of 4’ methyl T and 4’-ethyl thymidine (Figure 8C) are potent inhibitors of HIV RT in vitro (159). While these nucleosides were not efficiently activated by host cell kinases, they were highly effective against both wild-type HIV-1 and a number of NRTI-resistant mutants in the presence of HSV-TK.
Interestingly, the 4’-substituted compounds shown in Figure 8C have a 3’-OH. How then do they inhibit viral replication? None of these compounds appear to be a delayed chain terminator. 4’-methyl TTP was incorporated into DNA reasonably efficiently by HIV-1 RT; however, in many cases, the next (normal) nucleotide, which would be joined to the 4’-methyl T at the 3’ end of the primer, was incorporated slowly. The degree of pausing depended on both the sequence and the nature of the template (DNA or RNA). The underlying mechanisms of RT inhibition by N-MC or 4’E compounds appear to be distinct. The N-MC compounds presumably cause delayed chain termination because of a steric clash between the cyclopropane ring on the pseudosugar ring of the N-MC compounds and HIV-1 RT at a position near the base of the thumb of the p66 subunit (146). The molecular details of RT inhibition by 4’-substituted dNTPs that retain the 3’OH are currently under investigation. Notably, addition of a 4’-ethynyl group to the classical chain-terminator d4T improves its potency and reduces its cytotoxicity (160, 161).
Inhibitors of NRTI excision
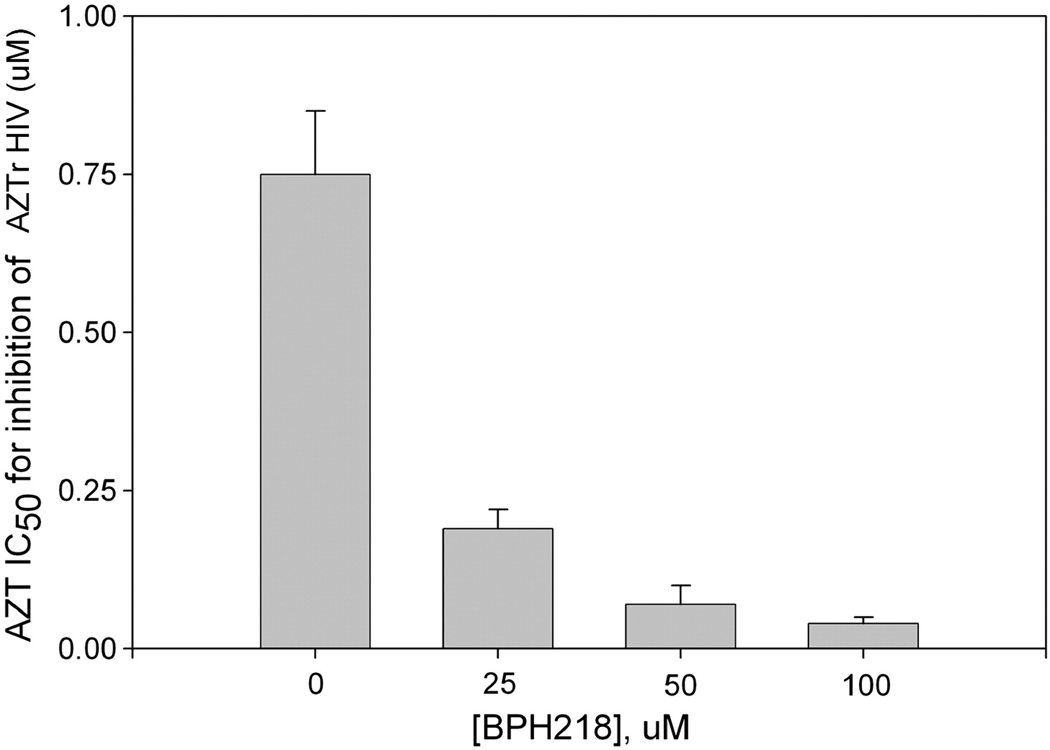
As discussed earlier (46–48), excision is an important mechanism of NRTI resistance. Specific inhibitors of NRTI excision could be therapeutically useful if they could be used to prevent the excision of an incorporated NRTI, which should restore the potency of an NRTI that would otherwise be excised. Optimally, such inhibitors should not reduce the incorporation of the NRTI because this would antagonize the antiretroviral activity of the NRTI. Parniak and colleagues have found that certain bisphosphonate compounds such as BPH218 (Figure 8B) are potent inhibitors of ATP-mediated phosphorolytic excision of 3’-terminal AZT-MP (in vitro IC50 ∼ 2 µM). Bisphosphonates are highly charged at neutral pH and are unable to traverse the plasma membrane of cells, thus BPH218 has no antiviral efficacy either in the presence or the absence of an NRTI. However, copolymer carriers greatly facilitate cell uptake of bisphosphonates (Parniak et al, unpublished data). BPH218 administered to cells in this manner has no effect on HIV-1 replication; however, the compound provides up to 30-fold potentiation of the antiviral activity of AZT against AZTr HIV-1 (Figure 10). This has potential clinical significance in that substantial numbers of treatment-experienced patients are infected with AZTr-HIV. Although bisphosphonates such as BPH218 are not therapeutics, these findings show that RT-catalyzed NRTI excision is a target for the development of adjuvant drugs that could restore the efficacy of NRTIs in patients infected with excision-proficient NRTI-resistant HIV.
Figure 10.
Potentiation of AZT antiviral activity against AZT-resistant HIV-1 by BPH218.
In conclusion, structural studies of wild-type and drug-resistant mutant HIV RTs in complex with inhibitors or substrates, together with biochemical and virological experiments have provided valuable insights into the mechanisms of DNA polymerization, inhibition and drug resistance. Importantly, these studies continue to guide the design of better inhibitors that offer promise for improved treatments of HIV infections.
Acknowledgement
S.G.S. acknowledges support by NIH (grants AI076119, AI079801, and AI074389). Support for B.M. comes from an amfAR Mathilde Kim Fellowship grant. S.H.H. was supported by the Intramural Research Program of NIH, NCI, Center for Cancer Research, and NIGMS. M.A.P. was supported by NIH grants AI060452, AI73975, AI076119, AI07980. E.A. is grateful to NIH (grants AI27690 MERIT Award and P01 GM 066671).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 2.Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 3.di Marzo Veronese F, Copeland TD, DeVico AL, Rahman R, Oroszlan S, Gallo RC, Sarngadharan MG. Characterization of highly immunogenic p66/p51 as the reverse transcriptase of HTLV-III/LAV. Science. 1986;231:1289–1291. doi: 10.1126/science.2418504. [DOI] [PubMed] [Google Scholar]
- 4.Lowe DM, Aitken A, Bradley C, Darby GK, Larder BA, Powell KL, Purifoy DJ, Tisdale M, Stammers DK. HIV-1 reverse transcriptase: crystallization and analysis of domain structure by limited proteolysis. Biochemistry. 1988;27:8884–8889. doi: 10.1021/bi00425a002. [DOI] [PubMed] [Google Scholar]
- 5.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 6.Jacobo-Molina A, Ding J, Nanni RG, Clark AD, Jr, Lu X, Tantillo C, Williams RL, Kamer G, Ferris AL, Clark P, et al. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci U S A. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh M, Jacques PS, Rodgers DW, Ottman M, Darlix JL, Le Grice SF. Alterations to the primer grip of p66 HIV-1 reverse transcriptase and their consequences for template-primer utilization. Biochemistry. 1996;35:8553–8562. doi: 10.1021/bi952773j. [DOI] [PubMed] [Google Scholar]
- 9.Julias JG, McWilliams MJ, Sarafianos SG, Arnold E, Hughes SH. Mutations in the RNase H domain of HIV-1 reverse transcriptase affect the initiation of DNA synthesis and the specificity of RNase H cleavage in vivo. Proc Natl Acad Sci U S A. 2002;99:9515–9520. doi: 10.1073/pnas.142123199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarafianos SG, Das K, Tantillo C, Clark AD, Jr, Ding J, Whitcomb JM, Boyer PL, Hughes SH, Arnold E. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. Embo J. 2001;20:1449–1461. doi: 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wisniewski M, Palaniappan C, Fu Z, Le Grice SF, Fay P, Bambara RA. Mutations in the primer grip region of HIV reverse transcriptase can increase replication fidelity. J Biol Chem. 1999;274:28175–28184. doi: 10.1074/jbc.274.40.28175. [DOI] [PubMed] [Google Scholar]
- 12.Krebs R, Immendorfer U, Thrall SH, Wohrl BM, Goody RS. Single-step kinetics of HIV-1 reverse transcriptase mutants responsible for virus resistance to nucleoside inhibitors zidovudine and 3-TC. Biochemistry. 1997;36:10292–10300. doi: 10.1021/bi970512z. [DOI] [PubMed] [Google Scholar]
- 13.Powell MD, Ghosh M, Jacques PS, Howard KJ, Le Grice SF, Levin JG. Alanine-scanning mutations in the "primer grip" of p66 HIV-1 reverse transcriptase result in selective loss of RNA priming activity. J Biol Chem. 1997;272:13262–13269. doi: 10.1074/jbc.272.20.13262. [DOI] [PubMed] [Google Scholar]
- 14.Palaniappan C, Wisniewski M, Jacques PS, Le Grice SF, Fay PJ, Bambara RA. Mutations within the primer grip region of HIV-1 reverse transcriptase result in loss of RNase H function. J Biol Chem. 1997;272:11157–11164. doi: 10.1074/jbc.272.17.11157. [DOI] [PubMed] [Google Scholar]
- 15.Ghosh M, Williams J, Powell MD, Levin JG, Le Grice SF. Mutating a conserved motif of the HIV-1 reverse transcriptase palm subdomain alters primer utilization. Biochemistry. 1997;36:5758–5768. doi: 10.1021/bi963045e. [DOI] [PubMed] [Google Scholar]
- 16.Jacques PS, Wohrl BM, Ottmann M, Darlix JL, Le Grice SF. Mutating the "primer grip" of p66 HIV-1 reverse transcriptase implicates tryptophan-229 in template-primer utilization. J Biol Chem. 1994;269:26472–26478. [PubMed] [Google Scholar]
- 17.Larder BA, Purifoy DJ, Powell KL, Darby G. Site-specific mutagenesis of AIDS virus reverse transcriptase. Nature. 1987;327:716–717. doi: 10.1038/327716a0. [DOI] [PubMed] [Google Scholar]
- 18.Delarue M, Poch O, Tordo N, Moras D, Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990;3:461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- 19.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. Embo J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson MS, McClure MA, Feng DF, Gray J, Doolittle RF. Computer analysis of retroviral pol genes: assignment of enzymatic functions to specific sequences and homologies with nonviral enzymes. Proc Natl Acad Sci U S A. 1986;83:7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 22.Martin-Hernandez AM, Domingo E, Menendez-Arias L. Human immunodeficiency virus type 1 reverse transcriptase: role of Tyr115 in deoxynucleotide binding and misinsertion fidelity of DNA synthesis. Embo J. 1996;15:4434–4442. [PMC free article] [PubMed] [Google Scholar]
- 23.Gao G, Orlova M, Georgiadis MM, Hendrickson WA, Goff SP. Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. Proc Natl Acad Sci U S A. 1997;94:407–411. doi: 10.1073/pnas.94.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyer PL, Sarafianos SG, Arnold E, Hughes SH. Analysis of mutations at positions 115 and 116 in the dNTP binding site of HIV-1 reverse transcriptase. Proc Natl Acad Sci U S A. 2000;97:3056–3061. doi: 10.1073/pnas.97.7.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchand B, Gotte M. Site-specific footprinting reveals differences in the translocation status of HIV-1 reverse transcriptase. Implications for polymerase translocation and drug resistance. J Biol Chem. 2003;278:35362–35372. doi: 10.1074/jbc.M304262200. [DOI] [PubMed] [Google Scholar]
- 26.Marchand B, Tchesnokov EP, Gotte M. The pyrophosphate analogue foscarnet traps the pre-translocational state of HIV-1 reverse transcriptase in a Brownian ratchet model of polymerase translocation. J Biol Chem. 2007;282:3337–3346. doi: 10.1074/jbc.M607710200. [DOI] [PubMed] [Google Scholar]
- 27.Meyer PR, Rutvisuttinunt W, Matsuura SE, So AG, Scott WA. Stable complexes formed by HIV-1 reverse transcriptase at distinct positions on the primer-template controlled by binding deoxynucleoside triphosphates or foscarnet. J Mol Biol. 2007;369:41–54. doi: 10.1016/j.jmb.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kati WM, Johnson KA, Jerva LF, Anderson KS. Mechanism and fidelity of HIV reverse transcriptase. J Biol Chem. 1992;267:25988–25997. [PubMed] [Google Scholar]
- 29.Reardon JE. Human immunodeficiency virus reverse transcriptase: steady-state and pre-steady-state kinetics of nucleotide incorporation. Biochemistry. 1992;31:4473–4479. doi: 10.1021/bi00133a013. [DOI] [PubMed] [Google Scholar]
- 30.Rittinger K, Divita G, Goody RS. Human immunodeficiency virus reverse transcriptase substrate-induced conformational changes and the mechanism of inhibition by nonnucleoside inhibitors. Proc Natl Acad Sci U S A. 1995;92:8046–8049. doi: 10.1073/pnas.92.17.8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong W, Lu CD, Sharma SK, Matsuura S, So AG, Scott WA. Nucleotide-induced stable complex formation by HIV-1 reverse transcriptase. Biochemistry. 1997;36:5749–5757. doi: 10.1021/bi962410z. [DOI] [PubMed] [Google Scholar]
- 32.Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 33.Sarafianos SG, Clark AD, Jr, Das K, Tuske S, Birktoft JJ, Ilankumaran P, Ramesha AR, Sayer JM, Jerina DM, Boyer PL, Hughes SH, Arnold E. Structures of HIV-1 reverse transcriptase with pre- and post-translocation AZTMP-terminated DNA. Embo J. 2002;21:6614–6624. doi: 10.1093/emboj/cdf637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perno CF, Yarchoan R, Cooney DA, Hartman NR, Gartner S, Popovic M, Hao Z, Gerrard TL, Wilson YA, Johns DG, et al. Inhibition of human immunodeficiency virus (HIV-1/HTLV-IIIBa-L) replication in fresh and cultured human peripheral blood monocytes/macrophages by azidothymidine and related 2',3'-dideoxynucleosides. J Exp Med. 1988;168:1111–1125. doi: 10.1084/jem.168.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Furman PA, Fyfe JA, St Clair MH, Weinhold K, Rideout JL, Freeman GA, Lehrman SN, Bolognesi DP, Broder S, Mitsuya H, et al. Phosphorylation of 3'-azido-3'-deoxythymidine and selective interaction of the 5’-triphosphate with human immunodeficiency virus reverse transcriptase. Proc Natl Acad Sci U S A. 1986;83:8333–8337. doi: 10.1073/pnas.83.21.8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kerr SG, Anderson KS. Pre-steady-state kinetic characterization of wild type and 3'-azido-3'-deoxythymidine (AZT) resistant human immunodeficiency virus type 1 reverse transcriptase: implication of RNA directed DNA polymerization in the mechanism of AZT resistance. Biochemistry. 1997;36:14064–14070. doi: 10.1021/bi9713862. [DOI] [PubMed] [Google Scholar]
- 37.Feng JY, Anderson KS. Mechanistic studies comparing the incorporation of (+) and (-) isomers of 3TCTP by HIV-1 reverse transcriptase. Biochemistry. 1999;38:55–63. doi: 10.1021/bi982340r. [DOI] [PubMed] [Google Scholar]
- 38.Ding J, Das K, Hsiou Y, Sarafianos SG, Clark AD, Jr, Jacobo-Molina A, Tantillo C, Hughes SH, Arnold E. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 A resolution. J Mol Biol. 1998;284:1095–1111. doi: 10.1006/jmbi.1998.2208. [DOI] [PubMed] [Google Scholar]
- 39.Ren J, Esnouf R, Garman E, Somers D, Ross C, Kirby I, Keeling J, Darby G, Jones Y, Stuart D, et al. High resolution structures of HIV-1 RT from four RT-inhibitor complexes. Nat Struct Biol. 1995;2:293–302. doi: 10.1038/nsb0495-293. [DOI] [PubMed] [Google Scholar]
- 40.Rodgers DW, Gamblin SJ, Harris BA, Ray S, Culp JS, Hellmig B, Woolf DJ, Debouck C, Harrison SC. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1995;92:1222–1226. doi: 10.1073/pnas.92.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsiou Y, Ding J, Das K, Clark AD, Jr, Hughes SH, Arnold E. Structure of unliganded HIV-1 reverse transcriptase at 2.7 A resolution: implications of conformational changes for polymerization and inhibition mechanisms. Structure. 1996;4:853–860. doi: 10.1016/s0969-2126(96)00091-3. [DOI] [PubMed] [Google Scholar]
- 42.Tantillo C, Ding J, Jacobo-Molina A, Nanni RG, Boyer PL, Hughes SH, Pauwels R, Andries K, Janssen PA, Arnold E. Locations of anti-AIDS drug binding sites and resistance mutations in the three-dimensional structure of HIV-1 reverse transcriptase Implications for mechanisms of drug inhibition and resistance. J Mol Biol. 1994;243:369–387. doi: 10.1006/jmbi.1994.1665. [DOI] [PubMed] [Google Scholar]
- 43.Das K, Sarafianos SG, Clark AD, Jr, Boyer PL, Hughes SH, Arnold E. Crystal structures of clinically relevant Lys103Asn/Tyr181Cys double mutant HIV-1 reverse transcriptase in complexes with ATP and non-nucleoside inhibitor HBY 097. J Mol Biol. 2007;365:77–89. doi: 10.1016/j.jmb.2006.08.097. [DOI] [PubMed] [Google Scholar]
- 44.Sarafianos SG, Das K, Clark AD, Jr, Ding J, Boyer PL, Hughes SH, Arnold E. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids. Proc Natl Acad Sci U S A. 1999;96:10027–10032. doi: 10.1073/pnas.96.18.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao HQ, Boyer PL, Sarafianos SG, Arnold E, Hughes SH. The role of steric hindrance in 3TC resistance of human immunodeficiency virus type-1 reverse transcriptase. J Mol Biol. 2000;300:403–418. doi: 10.1006/jmbi.2000.3823. [DOI] [PubMed] [Google Scholar]
- 46.Arion D, Kaushik N, McCormick S, Borkow G, Parniak MA. Phenotypic mechanism of HIV-1 resistance to 3'-azido-3'-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry. 1998;37:15908–15917. doi: 10.1021/bi981200e. [DOI] [PubMed] [Google Scholar]
- 47.Meyer PR, Matsuura SE, So AG, Scott WA. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc Natl Acad Sci U S A. 1998;95:13471–13476. doi: 10.1073/pnas.95.23.13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyer PR, Matsuura SE, Mian AM, So AG, Scott WA. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol Cell. 1999;4:35–43. doi: 10.1016/s1097-2765(00)80185-9. [DOI] [PubMed] [Google Scholar]
- 49.Boyer PL, Sarafianos SG, Arnold E, Hughes SH. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J Virol. 2001;75:4832–4842. doi: 10.1128/JVI.75.10.4832-4842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Larder BA, Kemp SD. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT) Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 51.Rooke R, Parniak MA, Tremblay M, Soudeyns H, Li XG, Gao Q, Yao XJ, Wainberg MA. Biological comparison of wild-type and zidovudine-resistant isolates of human immunodeficiency virus type 1 from the same subjects: susceptibility and resistance to other drugs. Antimicrob Agents Chemother. 1991;35:988–991. doi: 10.1128/aac.35.5.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kellam P, Boucher CA, Larder BA. Fifth mutation in human immunodeficiency virus type 1 reverse transcriptase contributes to the development of high-level resistance to zidovudine. Proc Natl Acad Sci U S A. 1992;89:1934–1938. doi: 10.1073/pnas.89.5.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harrigan PR, Kinghorn I, Bloor S, Kemp SD, Najera I, Kohli A, Larder BA. Significance of amino acid variation at human immunodeficiency virus type 1 reverse transcriptase residue 210 for zidovudine susceptibility. J Virol. 1996;70:5930–5934. doi: 10.1128/jvi.70.9.5930-5934.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin JL, Wilson JE, Haynes RL, Furman PA. Mechanism of resistance of human immunodeficiency virus type 1 to 2',3'-dideoxyinosine. Proc Natl Acad Sci U S A. 1993;90:6135–6139. doi: 10.1073/pnas.90.13.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winters MA, Shafer RW, Jellinger RA, Mamtora G, Gingeras T, Merigan TC. Human immunodeficiency virus type 1 reverse transcriptase genotype and drug susceptibility changes in infected individuals receiving dideoxyinosine monotherapy for 1 to 2 years. Antimicrob Agents Chemother. 1997;41:757–762. doi: 10.1128/aac.41.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harrigan PR, Stone C, Griffin P, Najera I, Bloor S, Kemp S, Tisdale M, Larder B. Resistance profile of the human immunodeficiency virus type 1 reverse transcriptase inhibitor abacavir (1592U89) after monotherapy and combination therapy. CNA2001 Investigative Group. J Infect Dis. 2000;181:912–920. doi: 10.1086/315317. [DOI] [PubMed] [Google Scholar]
- 57.Margot NA, Lu B, Cheng A, Miller MD. Resistance development over 144 weeks in treatment-naive patients receiving tenofovir disoproxil fumarate or stavudine with lamivudine and efavirenz in Study 903. HIV Med. 2006;7:442–450. doi: 10.1111/j.1468-1293.2006.00404.x. [DOI] [PubMed] [Google Scholar]
- 58.Ueno T, Shirasaka T, Mitsuya H. Enzymatic characterization of human immunodeficiency virus type 1 reverse transcriptase resistant to multiple 2',3'-dideoxynucleoside 5'-triphosphates. J Biol Chem. 1995;270:23605–23611. doi: 10.1074/jbc.270.40.23605. [DOI] [PubMed] [Google Scholar]
- 59.White KL, Chen JM, Feng JY, Margot NA, Ly JK, Ray AS, Macarthur HL, McDermott MJ, Swaminathan S, Miller MD. The K65R reverse transcriptase mutation in HIV-1 reverses the excision phenotype of zidovudine resistance mutations. Antivir Ther. 2006;11:155–163. doi: 10.1177/135965350601100209. [DOI] [PubMed] [Google Scholar]
- 60.Gu Z, Arts EJ, Parniak MA, Wainberg MA. Mutated K65R recombinant reverse transcriptase of human immunodeficiency virus type 1 shows diminished chain termination in the presence of 2',3'-dideoxycytidine 5'-triphosphate and other drugs. Proc Natl Acad Sci U S A. 1995;92:2760–2764. doi: 10.1073/pnas.92.7.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parikh UM, Zelina S, Sluis-Cremer N, Mellors JW. Molecular mechanisms of bidirectional antagonism between K65R and thymidine analog mutations in HIV-1 reverse transcriptase. AIDS. 2007;21:1405–1414. doi: 10.1097/QAD.0b013e3281ac229b. [DOI] [PubMed] [Google Scholar]
- 62.White KL, Margot NA, Ly JK, Chen JM, Ray AS, Pavelko M, Wang R, McDermott M, Swaminathan S, Miller MD. A combination of decreased NRTI incorporation and decreased excision determines the resistance profile of HIV-1 K65R RT. Aids. 2005;19:1751–1760. doi: 10.1097/01.aids.0000189851.21441.f1. [DOI] [PubMed] [Google Scholar]
- 63.Deval J, White KL, Miller MD, Parkin NT, Courcambeck J, Halfon P, Selmi B, Boretto J, Canard B. Mechanistic basis for reduced viral and enzymatic fitness of HIV-1 reverse transcriptase containing both K65R and M184V mutations. J Biol Chem. 2004;279:509–516. doi: 10.1074/jbc.M308806200. [DOI] [PubMed] [Google Scholar]
- 64.Feng JY, Myrick FT, Margot NA, Mulamba GB, Rimsky L, Borroto-Esoda K, Selmi B, Canard B. Virologic and enzymatic studies revealing the mechanism of K65R- and Q151M-associated HIV-1 drug resistance towards emtricitabine and lamivudine. Nucleosides Nucleotides Nucleic Acids. 2006;25:89–107. doi: 10.1080/15257770500379157. [DOI] [PubMed] [Google Scholar]
- 65.Deval J, Navarro JM, Selmi B, Courcambeck J, Boretto J, Halfon P, Garrido-Urbani S, Sire J, Canard B. A loss of viral replicative capacity correlates with altered DNA polymerization kinetics by the human immunodeficiency virus reverse transcriptase bearing the K65R and L74V dideoxynucleoside resistance substitutions. J Biol Chem. 2004;279:25489–25496. doi: 10.1074/jbc.M313534200. [DOI] [PubMed] [Google Scholar]
- 66.Selmi B, Boretto J, Navarro JM, Sire J, Longhi S, Guerreiro C, Mulard L, Sarfati S, Canard B. The valine-to-threonine 75 substitution in human immunodeficiency virus type 1 reverse transcriptase and its relation with stavudine resistance. J Biol Chem. 2001;276:13965–13974. doi: 10.1074/jbc.M009837200. [DOI] [PubMed] [Google Scholar]
- 67.Sarafianos SG, Das K, Hughes SH, Arnold E. Taking aim at a moving target: designing drugs to inhibit drug-resistant HIV-1 reverse transcriptases. Curr Opin Struct Biol. 2004;14:716–730. doi: 10.1016/j.sbi.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 68.Miranda LR, Gotte M, Liang F, Kuritzkes DR. The L74V mutation in human immunodeficiency virus type 1 reverse transcriptase counteracts enhanced excision of zidovudine monophosphate associated with thymidine analog resistance mutations. Antimicrob Agents Chemother. 2005;49:2648–2656. doi: 10.1128/AAC.49.7.2648-2656.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ray AS, Murakami E, Basavapathruni A, Vaccaro JA, Ulrich D, Chu CK, Schinazi RF, Anderson KS. Probing the molecular mechanisms of AZT drug resistance mediated by HIV-1 reverse transcriptase using a transient kinetic analysis. Biochemistry. 2003;42:8831–8841. doi: 10.1021/bi034435l. [DOI] [PubMed] [Google Scholar]
- 70.Marchand B, White KL, Ly JK, Margot NA, Wang R, McDermott M, Miller MD, Gotte M. Effects of the translocation status of human immunodeficiency virus type 1 reverse transcriptase on the efficiency of excision of tenofovir. Antimicrob Agents Chemother. 2007;51:2911–2919. doi: 10.1128/AAC.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dharmasena S, Pongracz Z, Arnold E, Sarafianos SG, Parniak MA. 3'-Azido-3'-deoxythymidine-(5')-tetraphospho-(5')-adenosine, the product of ATP-mediated excision of chain-terminating AZTMP, is a potent chain-terminating substrate for HIV-1 reverse transcriptase. Biochemistry. 2007;46:828–836. doi: 10.1021/bi061364s. [DOI] [PubMed] [Google Scholar]
- 72.Meyer PR, Matsuura SE, Tolun AA, Pfeifer I, So AG, Mellors JW, Scott WA. Effects of specific zidovudine resistance mutations and substrate structure on nucleotide-dependent primer unblocking by human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 2002;46:1540–1545. doi: 10.1128/AAC.46.5.1540-1545.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meyer PR, Smith AJ, Matsuura SE, Scott WA. Effects of primer-template sequence on ATP-dependent removal of chain-terminating nucleotide analogues by HIV-1 reverse transcriptase. J Biol Chem. 2004;279:45389–45398. doi: 10.1074/jbc.M405072200. [DOI] [PubMed] [Google Scholar]
- 74.Sluis-Cremer N, Arion D, Parikh U, Koontz D, Schinazi RF, Mellors JW, Parniak MA. The 3’-azido group is not the primary determinant of 3'-azido-3'-deoxythymidine (AZT) responsible for the excision phenotype of AZT-resistant HIV-1. J Biol Chem. 2005;280:29047–29052. doi: 10.1074/jbc.M503166200. [DOI] [PubMed] [Google Scholar]
- 75.Boyer PL, Sarafianos SG, Arnold E, Hughes SH. Nucleoside analog resistance caused by insertions in the fingers of human immunodeficiency virus type 1 reverse transcriptase involves ATP-mediated excision. J Virol. 2002;76:9143–9151. doi: 10.1128/JVI.76.18.9143-9151.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meyer PR, Lennerstrand J, Matsuura SE, Larder BA, Scott WA. Effects of dipeptide insertions between codons 69 and 70 of human immunodeficiency virus type 1 reverse transcriptase on primer unblocking, deoxynucleoside triphosphate inhibition, and DNA chain elongation. J Virol. 2003;77:3871–3877. doi: 10.1128/JVI.77.6.3871-3877.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.White KL, Chen JM, Margot NA, Wrin T, Petropoulos CJ, Naeger LK, Swaminathan S, Miller MD. Molecular mechanisms of tenofovir resistance conferred by human immunodeficiency virus type 1 reverse transcriptase containing a diserine insertion after residue 69 and multiple thymidine analog-associated mutations. Antimicrob Agents Chemother. 2004;48:992–1003. doi: 10.1128/AAC.48.3.992-1003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mas A, Parera M, Briones C, Soriano V, Martinez MA, Domingo E, Menendez-Arias L. Role of a dipeptide insertion between codons 69 and 70 of HIV-1 reverse transcriptase in the mechanism of AZT resistance. Embo J. 2000;19:5752–5761. doi: 10.1093/emboj/19.21.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Andreoletti L, Weiss L, Si-Mohamed A, Piketty C, Prazuck T, Calamy G, Malkin JE, Matta M, Mbopi-Keou FX, Clavel F, Kazatchkine MD, Belec L. Multidrug-resistant HIV-1 RNA and proviral DNA variants harboring new dipeptide insertions in the reverse transcriptase pol gene. J Acquir Immune Defic Syndr. 2002;29:102–104. doi: 10.1097/00126334-200201010-00015. [DOI] [PubMed] [Google Scholar]
- 80.Bonfanti P, Faggion I, La Seta Catamancio S, Violin M, Balotta C, Rusconi S. Response to antiretroviral therapy in a patient with an uncommon codon 69 insertion in the human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 2000;44:1767. doi: 10.1128/aac.44.6.1767-1768.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.De Antoni A, Foli A, Lisziewicz J, Lori F. Mutations in the pol gene of human immunodeficiency virus type 1 in infected patients receiving didanosine and hydroxyurea combination therapy. J Infect Dis. 1997;176:899–903. doi: 10.1086/516511. [DOI] [PubMed] [Google Scholar]
- 82.de Jong JJ, Goudsmit J, Lukashov VV, Hillebrand ME, Baan E, Huismans R, Danner SA, ten Veen JH, de Wolf F, Jurriaans S. Insertion of two amino acids combined with changes in reverse transcriptase containing tyrosine-215 of HIV-1 resistant to multiple nucleoside analogs. Aids. 1999;13:75–80. doi: 10.1097/00002030-199901140-00010. [DOI] [PubMed] [Google Scholar]
- 83.Larder BA, Bloor S, Kemp SD, Hertogs K, Desmet RL, Miller V, Sturmer M, Staszewski S, Ren J, Stammers DK, Stuart DI, Pauwels R. A family of insertion mutations between codons 67 and 70 of human immunodeficiency virus type 1 reverse transcriptase confer multinucleoside analog resistance. Antimicrob Agents Chemother. 1999;43:1961–1967. doi: 10.1128/aac.43.8.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rakik A, Ait-Khaled M, Griffin P, Thomas TA, Tisdale M, Kleim JP. A novel genotype encoding a single amino acid insertion and five other substitutions between residues 64 and 74 of the HIV-1 reverse transcriptase confers high-level cross-resistance to nucleoside reverse transcriptase inhibitorsAbacavir CNA2007 International Study Group. J Acquir Immune Defic Syndr. 1999;22:139–145. doi: 10.1097/00126334-199910010-00005. [DOI] [PubMed] [Google Scholar]
- 85.Winters MA, Coolley KL, Girard YA, Levee DJ, Hamdan H, Shafer RW, Katzenstein DA, Merigan TC. A 6-basepair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers resistance to multiple nucleoside inhibitors. J Clin Invest. 1998;102:1769–1775. doi: 10.1172/JCI4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gotte M, Arion D, Parniak MA, Wainberg MA. The M184V mutation in the reverse transcriptase of human immunodeficiency virus type 1 impairs rescue of chain-terminated DNA synthesis. J Virol. 2000;74:3579–3585. doi: 10.1128/jvi.74.8.3579-3585.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boyer PL, Sarafianos SG, Arnold E, Hughes SH. The M184V mutation reduces the selective excision of zidovudine 5'-monophosphate (AZTMP) by the reverse transcriptase of human immunodeficiency virus type 1. J Virol. 2002;76:3248–3256. doi: 10.1128/JVI.76.7.3248-3256.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Naeger LK, Margot NA, Miller MD. Increased drug susceptibility of HIV-1 reverse transcriptase mutants containing M184V and zidovudine-associated mutations: analysis of enzyme processivity, chain-terminator removal and viral replication. Antivir Ther. 2001;6:115–126. [PubMed] [Google Scholar]
- 89.Yap SH, Sheen CW, Fahey J, Zanin M, Tyssen D, Lima VD, Wynhoven B, Kuiper M, Sluis-Cremer N, Harrigan PR, Tachedjian G. N348I in the connection domain of HIV-1 reverse transcriptase confers zidovudine and nevirapine resistance. PLoS Med. 2007;4:e335. doi: 10.1371/journal.pmed.0040335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hachiya A, Kodama EN, Sarafianos SG, Schuckmann MM, Sakagami Y, Matsuoka M, Takiguchi M, Gatanaga H, Oka S. Amino acid mutation N348I in the connection subdomain of human immunodeficiency virus type 1 reverse transcriptase confers multiclass resistance to nucleoside and nonnucleoside reverse transcriptase inhibitors. J Virol. 2008;82:3261–3270. doi: 10.1128/JVI.01154-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ding J, Das K, Moereels H, Koymans L, Andries K, Janssen PA, Hughes SH, Arnold E. Structure of HIV-1 RT/TIBO R 86183 complex reveals similarity in the binding of diverse nonnucleoside inhibitors. Nat Struct Biol. 1995;2:407–415. doi: 10.1038/nsb0595-407. [DOI] [PubMed] [Google Scholar]
- 92.Hsiou Y, Das K, Ding J, Clark AD, Jr, Kleim JP, Rosner M, Winkler I, Riess G, Hughes SH, Arnold E. Structures of Tyr188Leu mutant and wild-type HIV-1 reverse transcriptase complexed with the non-nucleoside inhibitor HBY 097: inhibitor flexibility is a useful design feature for reducing drug resistance. J Mol Biol. 1998;284:313–323. doi: 10.1006/jmbi.1998.2171. [DOI] [PubMed] [Google Scholar]
- 93.Das K, Ding J, Hsiou Y, Clark AD, Jr, Moereels H, Koymans L, Andries K, Pauwels R, Janssen PA, Boyer PL, Clark P, Smith RH, Jr, Kroeger Smith MB, Michejda CJ, Hughes SH, Arnold E. Crystal structures of 8-Cl and 9-Cl TIBO complexed with wild-type HIV-1 RT and 8-Cl TIBO complexed with the Tyr181Cys HIV-1 RT drug-resistant mutant. J Mol Biol. 1996;264:1085–1100. doi: 10.1006/jmbi.1996.0698. [DOI] [PubMed] [Google Scholar]
- 94.Ren J, Esnouf R, Hopkins A, Ross C, Jones Y, Stammers D, Stuart D. The structure of HIV-1 reverse transcriptase complexed with 9-chloro-TIBO: lessons for inhibitor design. Structure. 1995;3:915–926. doi: 10.1016/S0969-2126(01)00226-X. [DOI] [PubMed] [Google Scholar]
- 95.Ren J, Nichols C, Bird L, Chamberlain P, Weaver K, Short S, Stuart DI, Stammers DK. Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors. J Mol Biol. 2001;312:795–805. doi: 10.1006/jmbi.2001.4988. [DOI] [PubMed] [Google Scholar]
- 96.Ren J, Nichols CE, Chamberlain PP, Weaver KL, Short SA, Stammers DK. Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors. J Mol Biol. 2004;336:569–578. doi: 10.1016/j.jmb.2003.12.055. [DOI] [PubMed] [Google Scholar]
- 97.Das K, Clark AD, Jr, Lewi PJ, Heeres J, De Jonge MR, Koymans LM, Vinkers HM, Daeyaert F, Ludovici DW, Kukla MJ, De Corte B, Kavash RW, Ho CY, Ye H, Lichtenstein MA, Andries K, Pauwels R, De Bethune MP, Boyer PL, Clark P, Hughes SH, Janssen PA, Arnold E. Roles of conformational and positional adaptability in structure-based design of TMC125-R165335 (etravirine) and related non-nucleoside reverse transcriptase inhibitors that are highly potent and effective against wild-type and drug-resistant HIV-1 variants. J Med Chem. 2004;47:2550–2560. doi: 10.1021/jm030558s. [DOI] [PubMed] [Google Scholar]
- 98.Hsiou Y, Ding J, Das K, Clark AD, Jr, Boyer PL, Lewi P, Janssen PA, Kleim JP, Rosner M, Hughes SH, Arnold E. The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance. J Mol Biol. 2001;309:437–445. doi: 10.1006/jmbi.2001.4648. [DOI] [PubMed] [Google Scholar]
- 99.Ren J, Nichols CE, Chamberlain PP, Weaver KL, Short SA, Chan JH, Kleim JP, Stammers DK. Relationship of potency and resilience to drug resistant mutations for GW420867X revealed by crystal structures of inhibitor complexes for wild-type, Leu100Ile, Lys101Glu, and Tyr188Cys mutant HIV-1 reverse transcriptases. J Med Chem. 2007;50:2301–2309. doi: 10.1021/jm061117m. [DOI] [PubMed] [Google Scholar]
- 100.Janssen PA, Lewi PJ, Arnold E, Daeyaert F, de Jonge M, Heeres J, Koymans L, Vinkers M, Guillemont J, Pasquier E, Kukla M, Ludovici D, Andries K, de Bethune MP, Pauwels R, Das K, Clark AD, Jr, Frenkel YV, Hughes SH, Medaer B, De Knaep F, Bohets H, De Clerck F, Lampo A, Williams P, Stoffels P. In search of a novel anti-HIV drug: multidisciplinary coordination in the discovery of 4-[[4-[[4-[(1E)-2-cyanoethenyl]-2,6-dimethylphenyl]amino]-2- pyrimidinyl]amino]benzonitrile (R278474, rilpivirine) J Med Chem. 2005;48:1901–1909. doi: 10.1021/jm040840e. [DOI] [PubMed] [Google Scholar]
- 101.Nikolenko GN, Delviks-Frankenberry KA, Palmer S, Maldarelli F, Fivash MJ, Jr, Coffin JM, Pathak VK. Mutations in the connection domain of HIV-1 reverse transcriptase increase 3'-azido-3'-deoxythymidine resistance. Proc Natl Acad Sci U S A. 2007;104:317–322. doi: 10.1073/pnas.0609642104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Delviks-Frankenberry KA, Nikolenko GN, Barr R, Pathak VK. Mutations in human immunodeficiency virus type 1 RNase H primer grip enhance 3'-azido-3'-deoxythymidine resistance. J Virol. 2007;81:6837–6845. doi: 10.1128/JVI.02820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Delviks-Frankenberry KA, Nikolenko GN, Boyer PL, Hughes SH, Coffin JM, Jere A, Pathak VK. HIV-1 reverse transcriptase connection subdomain mutations reduce template RNA degradation and enhance AZT excision. Proc Natl Acad Sci U S A. 2008;105:10943–10948. doi: 10.1073/pnas.0804660105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ehteshami M, Beilhartz GL, Scarth BJ, Tchesnokov EP, McCormick S, Wynhoven B, Harrigan PR, Gotte M. Connection domain mutations N348I and A360V in HIV-1 reverse transcriptase enhance resistance to 3'-azido-3'-deoxythymidine through both RNase H-dependent and -independent mechanisms. J Biol Chem. 2008;283:22222–22232. doi: 10.1074/jbc.M803521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Furfine ES, Reardon JE. Human immunodeficiency virus reverse transcriptase ribonuclease H: specificity of tRNA(Lys3)-primer excision. Biochemistry. 1991;30:7041–7046. doi: 10.1021/bi00243a001. [DOI] [PubMed] [Google Scholar]
- 106.Pullen KA, Ishimoto LK, Champoux JJ. Incomplete removal of the RNA primer for minus-strand DNA synthesis by human immunodeficiency virus type 1 reverse transcriptase. J Virol. 1992;66:367–373. doi: 10.1128/jvi.66.1.367-373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Huber HE, Richardson CC. Processing of the primer for plus strand DNA synthesis by human immunodeficiency virus 1 reverse transcriptase. J Biol Chem. 1990;265:10565–10573. [PubMed] [Google Scholar]
- 108.Tanese N, Goff SP. Domain structure of the Moloney murine leukemia virus reverse transcriptase: mutational analysis and separate expression of the DNA polymerase and RNase H activities. Proc Natl Acad Sci U S A. 1988;85:1777–1781. doi: 10.1073/pnas.85.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schatz O, Cromme FV, Gruninger-Leitch F, Le Grice, F S. Point mutations in conserved amino acid residues within the C-terminal domain of HIV-1 reverse transcriptase specifically repress RNase H function. FEBS Lett. 1989;257:311–314. doi: 10.1016/0014-5793(89)81559-5. [DOI] [PubMed] [Google Scholar]
- 110.Davies JF, 2nd, Hostomska Z, Hostomsky Z, Jordan SR, Matthews DA. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science. 1991;252:88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- 111.Schatz O, Mous J, Le Grice SF. HIV-1 RT-associated ribonuclease H displays both endonuclease and 3'----5' exonuclease activity. EMBO J. 1990;9:1171–1176. doi: 10.1002/j.1460-2075.1990.tb08224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wohrl BM, Moelling K. Interaction of HIV-1 ribonuclease H with polypurine tract containing RNA-DNA hybrids. Biochemistry. 1990;29:10141–10147. doi: 10.1021/bi00496a001. [DOI] [PubMed] [Google Scholar]
- 113.Gopalakrishnan V, Peliska JA, Benkovic SJ. Human immunodeficiency virus type 1 reverse transcriptase: spatial and temporal relationship between the polymerase and RNase H activities. Proc Natl Acad Sci U S A. 1992;89:10763–10767. doi: 10.1073/pnas.89.22.10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ghosh M, Howard KJ, Cameron CE, Benkovic SJ, Hughes SH, Le Grice F. Truncating alpha-helix E' of p66 human immunodeficiency virus reverse transcriptase modulates RNase H function and impairs DNA strand transfer. J Biol Chem. 1995;270:7068–7076. doi: 10.1074/jbc.270.13.7068. [DOI] [PubMed] [Google Scholar]
- 115.Gotte M, Fackler S, Hermann T, Perola E, Cellai L, Gross HJ, Le Grice SF, Heumann H. HIV-1 reverse transcriptase-associated RNase H cleaves RNA/RNA in arrested complexes: implications for the mechanism by which RNase H discriminates between RNA/RNA and RNA/DNA. Embo J. 1995;14:833–841. doi: 10.1002/j.1460-2075.1995.tb07061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gao HQ, Boyer PL, Arnold E, Hughes SH. Effects of mutations in the polymerase domain on the polymerase, RNase H and strand transfer activities of human immunodeficiency virus type 1 reverse transcriptase. J Mol Biol. 1998;277:559–572. doi: 10.1006/jmbi.1998.1624. [DOI] [PubMed] [Google Scholar]
- 117.Gao HQ, Sarafianos SG, Arnold E, Hughes SH. Similarities and differences in the RNase H activities of human immunodeficiency virus type 1 reverse transcriptase and Moloney murine leukemia virus reverse transcriptase. J Mol Biol. 1999;294:1097–1113. doi: 10.1006/jmbi.1999.3325. [DOI] [PubMed] [Google Scholar]
- 118.Gotte M, Maier G, Gross HJ, Heumann H. Localization of the active site of HIV-1 reverse transcriptase-associated RNase H domain on a DNA template using site-specific generated hydroxyl radicals. J Biol Chem. 1998;273:10139–10146. doi: 10.1074/jbc.273.17.10139. [DOI] [PubMed] [Google Scholar]
- 119.Cowan JA, Ohyama T, Howard K, Rausch JW, Cowan SM, Le Grice FS. Metal-ion stoichiometry of the HIV-1 RT ribonuclease H domain: evidence for two mutually exclusive sites leads to new mechanistic insights on metal-mediated hydrolysis in nucleic acid biochemistry. J Biol Inorg Chem. 2000;5:67–74. doi: 10.1007/s007750050009. [DOI] [PubMed] [Google Scholar]
- 120.Pari K, Mueller GA, DeRose EF, Kirby TW, London RE. Solution structure of the RNase H domain of the HIV-1 reverse transcriptase in the presence of magnesium. Biochemistry. 2003;42:639–650. doi: 10.1021/bi0204894. [DOI] [PubMed] [Google Scholar]
- 121.Klumpp K, Hang JQ, Rajendran S, Yang Y, Derosier A, Wong Kai In P, Overton H, Parkes KE, Cammack N, Martin JA. Two-metal ion mechanism of RNA cleavage by HIV RNase H and mechanism-based design of selective HIV RNase H inhibitors. Nucleic Acids Res. 2003;31:6852–6859. doi: 10.1093/nar/gkg881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 123.Fedoroff O, Salazar M, Reid BR. Structure of a DNA:RNA hybrid duplex. Why RNase H does not cleave pure RNA. J Mol Biol. 1993;233:509–523. doi: 10.1006/jmbi.1993.1528. [DOI] [PubMed] [Google Scholar]
- 124.Salazar M, Fedoroff O, Zhu L, Reid BR. The solution structure of the r(gcg)d(TATACCC):d(GGGTATACGC) Okazaki fragment contains two distinct duplex morphologies connected by a junction. J Mol Biol. 1994;241:440–455. doi: 10.1006/jmbi.1994.1519. [DOI] [PubMed] [Google Scholar]
- 125.Zhu L, Salazar M, Reid BR. DNA duplexes flanked by hybrid duplexes: the solution structure of chimeric junctions in [r(cgcg)d(TATACGCG)]2. Biochemistry. 1995;34:2372–2380. doi: 10.1021/bi00007a033. [DOI] [PubMed] [Google Scholar]
- 126.Horton NC, Finzel BC. The structure of an RNA/DNA hybrid: a substrate of the ribonuclease activity of HIV-1 reverse transcriptase. J Mol Biol. 1996;264:521–533. doi: 10.1006/jmbi.1996.0658. [DOI] [PubMed] [Google Scholar]
- 127.Fedoroff OY, Ge Y, Reid BR. Solution structure of r(gaggacug):d(CAGTCCTC) hybrid: implications for the initiation of HIV-1 (+)-strand synthesis. J Mol Biol. 1997;269:225–239. doi: 10.1006/jmbi.1997.1024. [DOI] [PubMed] [Google Scholar]
- 128.Han GW, Kopka ML, Cascio D, Grzeskowiak K, Dickerson RE. Structure of a DNA analog of the primer for HIV-1 RT second strand synthesis. J Mol Biol. 1997;269:811–826. doi: 10.1006/jmbi.1997.1085. [DOI] [PubMed] [Google Scholar]
- 129.Bachelin M, Hessler G, Kurz G, Hacia JG, Dervan PB, Kessler H. Structure of a stereoregular phosphorothioate DNA/RNA duplex. Nat Struct Biol. 1998;5:271–276. doi: 10.1038/nsb0498-271. [DOI] [PubMed] [Google Scholar]
- 130.Szyperski T, Gotte M, Billeter M, Perola E, Cellai L, Heumann H, Wuthrich K. NMR structure of the chimeric hybrid duplex r(gcaguggc).r(gcca)d(CTGC) comprising the tRNA-DNA junction formed during initiation of HIV-1 reverse transcription. J Biomol NMR. 1999;13:343–355. doi: 10.1023/a:1008350604637. [DOI] [PubMed] [Google Scholar]
- 131.Rausch JW, Lener D, Miller JT, Julias JG, Hughes SH, Le Grice FS. Altering the RNase H primer grip of human immunodeficiency virus reverse transcriptase modifies cleavage specificity. Biochemistry. 2002;41:4856–4865. doi: 10.1021/bi015970t. [DOI] [PubMed] [Google Scholar]
- 132.Abbondanzieri EA, Bokinsky G, Rausch JW, Zhang JX, Le Grice SF, Zhuang X. Dynamic binding orientations direct activity of HIV reverse transcriptase. Nature. 2008;453:184–189. doi: 10.1038/nature06941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Whitcomb JM, Kumar R, Hughes SH. Sequence of the circle junction of human immunodeficiency virus type 1: implications for reverse transcription and integration. J Virol. 1990;64:4903–4906. doi: 10.1128/jvi.64.10.4903-4906.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Katz RA, Merkel G, Kulkosky J, Leis J, Skalka AM. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 135.Klumpp K, Mirzadegan T. Recent progress in the design of small molecule inhibitors of HIV RNase H. Curr Pharm Des. 2006;12:1909–1922. doi: 10.2174/138161206776873653. [DOI] [PubMed] [Google Scholar]
- 136.Tramontano E. HIV-1 RNase H: recent progress in an exciting, yet little explored, drug target. Mini Rev Med Chem. 2006;6:727–737. doi: 10.2174/138955706777435733. [DOI] [PubMed] [Google Scholar]
- 137.Borkow G, Fletcher RS, Barnard J, Arion D, Motakis D, Dmitrienko GI, Parniak MA. Inhibition of the ribonuclease H and DNA polymerase activities of HIV-1 reverse transcriptase by N-(4-tert-butylbenzoyl)-2-hydroxy-1-naphthaldehyde hydrazone. Biochemistry. 1997;36:3179–3185. doi: 10.1021/bi9624696. [DOI] [PubMed] [Google Scholar]
- 138.Himmel DM, Sarafianos SG, Dharmasena S, Hossain MM, McCoy-Simandle K, Ilina T, Clark AD, Jr, Knight JL, Julias JG, Clark PK, Krogh-Jespersen K, Levy RM, Hughes SH, Parniak MA, Arnold E. HIV-1 reverse transcriptase structure with RNase H inhibitor dihydroxy benzoyl naphthyl hydrazone bound at a novel site. ACS Chem Biol. 2006;1:702–712. doi: 10.1021/cb600303y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Arion D, Sluis-Cremer N, Min KL, Abram ME, Fletcher RS, Parniak MA. Mutational analysis of Tyr-501 of HIV-1 reverse transcriptase. Effects on ribonuclease H activity and inhibition of this activity by N-acylhydrazones. J Biol Chem. 2002;277:1370–1374. doi: 10.1074/jbc.M110254200. [DOI] [PubMed] [Google Scholar]
- 140.Davis WR, Tomsho J, Nikam S, Cook EM, Somand D, Peliska JA. Inhibition of HIV-1 reverse transcriptase-catalyzed DNA strand transfer reactions by 4-chlorophenylhydrazone of mesoxalic acid. Biochemistry. 2000;39:14279–14291. doi: 10.1021/bi0015764. [DOI] [PubMed] [Google Scholar]
- 141.Shaw-Reid CA, Munshi V, Graham P, Wolfe A, Witmer M, Danzeisen R, Olsen DB, Carroll SS, Embrey M, Wai JS, Miller MD, Cole JL, Hazuda DJ. Inhibition of HIV-1 ribonuclease H by a novel diketo acid, 4-[5-(benzoylamino)thien-2-yl]-2,4-dioxobutanoic acid. J Biol Chem. 2003;278:2777–2780. doi: 10.1074/jbc.C200621200. [DOI] [PubMed] [Google Scholar]
- 142.Hang JQ, Rajendran S, Yang Y, Li Y, In PW, Overton H, Parkes KE, Cammack N, Martin JA, Klumpp K. Activity of the isolated HIV RNase H domain and specific inhibition by N-hydroxyimides. Biochem Biophys Res Commun. 2004;317:321–329. doi: 10.1016/j.bbrc.2004.03.061. [DOI] [PubMed] [Google Scholar]
- 143.Budihas SR, Gorshkova I, Gaidamakov S, Wamiru A, Bona MK, Parniak MA, Crouch RJ, McMahon JB, Beutler JA, Le Grice SF. Selective inhibition of HIV-1 reverse transcriptase-associated ribonuclease H activity by hydroxylated tropolones. Nucleic Acids Res. 2005;33:1249–1256. doi: 10.1093/nar/gki268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Marquez VE, Ben-Kasus T, Barchi JJ, Jr, Green KM, Nicklaus MC, Agbaria R. Experimental and structural evidence that herpes 1 kinase and cellular DNA polymerase(s) discriminate on the basis of sugar pucker. J Am Chem Soc. 2004;126:543–549. doi: 10.1021/ja037929e. [DOI] [PubMed] [Google Scholar]
- 145.Marquez VE, Hughes SH, Sei S, Agbaria R. The history of N-methanocarbathymidine: the investigation of a conformational concept leads to the discovery of a potent and selective nucleoside antiviral agent. Antiviral Res. 2006;71:268–275. doi: 10.1016/j.antiviral.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 146.Boyer PL, Julias JG, Marquez VE, Hughes SH. Fixed conformation nucleoside analogs effectively inhibit excision-proficient HIV-1 reverse transcriptases. J Mol Biol. 2005;345:441–450. doi: 10.1016/j.jmb.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 147.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 148.Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 149.Eom SH, Wang J, Steitz TA. Structure of Taq polymerase with DNA at the polymerase active site. Nature. 1996;382:278–281. doi: 10.1038/382278a0. [DOI] [PubMed] [Google Scholar]
- 150.Maltseva T, Usova E, Eriksson S, Milecki J, Foldesi A, Chattopadhayaya J. The NMR conformation study of the complexes of deoxycytidine kinase (dCK) and 2'-deoxycytidine/2'-deoxyadenosine. Nucleosides Nucleotides Nucleic Acids. 2001;20:1225–1228. doi: 10.1081/NCN-100002523. [DOI] [PubMed] [Google Scholar]
- 151.Lee K, Chong Y, Chu CK. Understanding the mode of action of L-nucleosides as antiviral agents: a molecular modeling approach. Nucleosides Nucleotides Nucleic Acids. 2001;20:385–388. doi: 10.1081/NCN-100002311. [DOI] [PubMed] [Google Scholar]
- 152.Xu Y, Sellam O, Morera S, Sarfati S, Biondi R, Veron M, Janin J. X-ray analysis of azido-thymidine diphosphate binding to nucleoside diphosphate kinase. Proc Natl Acad Sci U S A. 1997;94:7162–7165. doi: 10.1073/pnas.94.14.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Eriksson S, Munch-Petersen B, Johansson K, Eklund H. Structure and function of cellular deoxyribonucleoside kinases. Cell Mol Life Sci. 2002;59:1327–1346. doi: 10.1007/s00018-002-8511-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ohrui H, Mitsuya H. 4'-C-substituted-2'-deoxynucleosides: a family of antiretroviral agents which are potent against drug-resistant HIV variants. Curr Drug Targets Infect Disord. 2001;1:1–10. doi: 10.2174/1568005013343218. [DOI] [PubMed] [Google Scholar]
- 155.Kodama EI, Kohgo S, Kitano K, Machida H, Gatanaga H, Shigeta S, Matsuoka M, Ohrui H, Mitsuya H. 4'-Ethynyl nucleoside analogs: potent inhibitors of multidrug-resistant human immunodeficiency virus variants in vitro. Antimicrob Agents Chemother. 2001;45:1539–1546. doi: 10.1128/AAC.45.5.1539-1546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kawamoto A, Kodama E, Sarafianos SG, Sakagami Y, Kohgo S, Kitano K, Ashida N, Iwai Y, Hayakawa H, Nakata H, Mitsuya H, Arnold E, Matsuoka M. 2'-Deoxy-4'-C-ethynyl-2-halo-adenosines active against drug-resistant human immunodeficiency virus type 1 variants. Int J Biochem Cell Biol. 2008 doi: 10.1016/j.biocel.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 157.Ohrui H, Kohgo S, Hayakawa H, Kodama E, Matsuoka M, Nakata T, Mitsuya H. 2’-Deoxy-4’-C-ethynyl-2-fluoroadenosine: a nucleoside reverse transcriptase inhibitor with highly potent activity against all HIV-1 strains, favorable toxic profiles and stability in plasma. Nucleic Acids Symp Ser (Oxf) 2006:1–2. doi: 10.1093/nass/nrl001. [DOI] [PubMed] [Google Scholar]
- 158.Nakata H, Amano M, Koh Y, Kodama E, Yang G, Bailey CM, Kohgo S, Hayakawa H, Matsuoka M, Anderson KS, Cheng YC, Mitsuya H. Activity against Human Immunodeficiency Virus Type 1, Intracellular Metabolism, and Effects on Human DNA Polymerases of 4'-Ethynyl-2-Fluoro-2'-Deoxyadenosine. Antimicrob Agents Chemother. 2007;51:2701–2708. doi: 10.1128/AAC.00277-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Boyer PL, Julias JG, Ambrose Z, Siddiqui MA, Marquez VE, Hughes SH. The nucleoside analogs 4'C-methyl thymidine and 4'C-ethyl thymidine block DNA synthesis by wild-type HIV-1 RT and excision proficient NRTI resistant RT variants. J Mol Biol. 2007;371:873–882. doi: 10.1016/j.jmb.2007.05.043. [DOI] [PubMed] [Google Scholar]
- 160.Tanaka H, Haraguchi K, Kumamoto H, Baba M, Cheng YC. 4’-Ethynylstavudine (4’-Ed4T) has potent anti-HIV-1 activity with reduced toxicity and shows a unique activity profile against drug-resistant mutants. Antivir Chem Chemother. 2005;16:217–221. doi: 10.1177/095632020501600402. [DOI] [PubMed] [Google Scholar]
- 161.Yang G, Dutschman GE, Wang CJ, Tanaka H, Baba M, Anderson KS, Cheng YC. Highly selective action of triphosphate metabolite of 4'-ethynyl D4T: a novel anti-HIV compound against HIV-1 RT. Antiviral Res. 2007;73:185–191. doi: 10.1016/j.antiviral.2006.10.002. [DOI] [PubMed] [Google Scholar]