Abstract
Nitro-fatty acid products of oxidative inflammatory reactions mediate anti-inflammatory cell signalling responses. LNO2 (nitro-linoleic acid) induces expression of HO-1 (haem oxygenase-1), an enzyme that catabolizes haem into products exhibiting potent anti-inflammatory properties. In the present manuscript, the molecular mechanisms underlying HO-1 induction by LNO2 were examined in HAEC (human aortic endothelial cells), HEK-293 (human embryonic kidney 293) cells, and in transcription factor-deficient MEF (mouse embryonic fibroblasts). LNO2 induced HO-1 expression in Nrf2 [NF-E2 (nuclear factor-erythroid 2)-related factor 2]-deficient MEF and in HEK-293 cells transfected with Nrf2-specific shRNA (small-hairpin RNA), supporting the fact that LNO2-mediated HO-1 induction can be regulated by Nrf2-independent mechanisms. LNO2 activated expression of a − 4.5 kb human HO-1 promoter construct, whereas a − 4.0 kb construct with deletion of 500 bp from the 5′ region was unresponsive. Site-directed mutagenesis of a CRE (cAMP-response element) or of a downstream NF-E2/AP-1 (activating protein-1) element, individually, within this 500 bp region modestly reduced activation of the HO-1 promoter by LNO2. Mutations of both the CRE and the NF-E2/AP-1 site also attenuated LNO2-mediated HO-1 promoter expression, whereas the addition of a third mutation in the proximal E-box sequence completely abolished LNO2-induced HO-1 expression. Chromatin immunoprecipitation assays confirmed CREB (CRE-binding protein)-1 binding to the CRE (located at − 4.0 kb) and E-box regions (located at − 44 bp) of the human HO-1 promoter. A 3C (Chromosome Conformation Capture) assay of intact cells showed LNO2-induced interactions between the CRE- and E-box-containing regions. These observations indicate that regulation of human HO-1 expression by LNO2 requires synergy between CRE, AP-1 and E-box sequences and involves the participation of CREB-1.
Keywords: chromatin conformation, CREB-1, nitrated lipids, transcriptional regulation, upstream stimulatory factor 1 (USF-1), vascular endothelium
INTRODUCTION
Partially reduced oxygen species [superoxide (O2−), H2O2, hydroxyl radical (OH)], NO and its metabolites [nitrite (NO2−), peroxynitrite (ONOO−), nitrogen dioxide (NO2)] and other reactive inflammatory mediators induce the oxidation, nitrosation, and nitration of diverse biomolecules including unsaturated fatty acids [1,2]. Nitroalkene derivatives of fatty acids, including LNO2 (nitrolinoleic acid), are detected as free and esterified species in both cell and animal models of inflammation. Fatty acid nitration products are also present at elevated levels in patients with hyperlipidaemia and are generated via NO-dependent mechanisms in models of inflammation (reviewed in [3]). Current data support that LNO2 and related species function as cytoprotective signalling molecules, eliciting cGMP-independent pleotropic effects predominantly via rapid and reversible electrophilic reactivity. Signalling actions of NO2-FA (nitro-fatty acids) include inhibition of platelet and neutrophil function, inhibition of NF-κB (nuclear factor κB)-dependent signalling and activation of the nuclear lipid receptor PPAR (peroxisome-proliferator-activated receptor)-γ (reviewed in [4]). Fatty acid nitration products also inhibit endotoxin-mediated STAT (signal transducer and activator of transcription) signalling through the induction of MKP-1 [MAPK (mitogen-activated protein kinase) phosphatase 1], a MAPK phosphatase known to contribute to anti-inflammatory signalling [5,6].
The oxygenation of membrane and lipoprotein unsaturated fatty acids occurs during regulated cell signalling responses and, in a more dysregulated fashion, as a consequence of oxidative inflammatory reactions. In concert, cytoprotective responses are induced that include increased superoxide dismutase, glutathione peroxidase, catalase and HO-1 (haem oxygenase-1) expression [7]. HO-1 catalyses the rate-limiting step in the degradation of pro-oxidant haem, liberating CO, iron and biliverdin, and is protective in both acute and chronic disorders, including organ transplant rejection, sepsis, vascular restenosis and atherosclerosis. Mice and humans lacking a functional HO-1 gene display increased atherosclerotic lesion formation and an increased susceptibility to oxidative stress [8,9]. Furthermore, HO-1 induction by chemical manipulation or genetic transfer confers a protective effect against vascular disease [10,11].
The mouse and human HO-1 promoters are subject to complex gene regulation that is controlled bymultiple transcription factors, including Nrf2 (nuclear factor-erythroid 2-related factor 2), heat shock factor proteins, NF-κB and members of the AP-1 (activating protein-1) bZIP (basic region/leucine zipper) family. Much of this regulation occurs at sequences embedded in AREs (antioxidant response elements) [12]. Transcriptional activation of murine HO-1 expression occurs via Nrf2/Bach1-mediated mechanisms [13]. However, activation of human HO-1 is more complex and less well understood, involving the participation of several transcription factors that interact with the HO-1 promoter at sites spanning the − 9.4 kb to − 44 bp region. Transcriptional regulatory roles have been shown for Sp-1 (specificity protein-1), pescadillo, CREB-1 [CRE (cAMP-response element)-binding protein-1] and PPARα and γ [14–18]. The JunB and JunD members of the AP-1 family bind to NF-E2 (nuclear factor-erythroid 2)/AP-1 sequences within classical AREs to antagonistically modulate human HO-1 expression [19]. USF1/2 (upstream stimulatory factors 1 and 2) bind to E-box elements proximal to the transcriptional start site and are required for rat and human HO-1 activation [20–22].
The objective of this study is to define the molecular regulation of human HO-1 gene expression by NO2-FA, since HO-1 transduces many of the adaptive anti-inflammatory protective responses of these and other oxidized fatty acid species and since HO-1 is induced by NO2-FA [23–25]. We reveal that LNO2 can induce HO-1 independently of Nrf2, requires functional CRE, AP-1 and proximal E-box elements, and promotes a unique CREB-1-dependent dynamic interaction between the CRE and proximal E-box elements in the human HO-1 promoter by chromatin looping to activate HO-1 gene expression.
MATERIALS AND METHODS
Reagents
Tissue culture media, serum and supplements were from Lonza and Mediatech. Cadmium chloride was purchased from Sigma (St. Louis, MO, U.S.A.). Anti-HO-1 antibody (SPA-896) was from Stressgen (Vancouver, British Columbia, Canada), and anti-human CREB-1, phospho-CREB, JunD, Nrf2 and USF-1 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA, U.S.A.). Anti-actin antibody was from Sigma. Purification, characterization and quantitation of LNO2 was as previously described in [26].
Cell culture
Primary HAEC (human aortic endothelial cells) (Lonza, Walkersville, MD, U.S.A.) were cultured in endothelial basal medium containing 10%FBS (fetal bovine serum), bovine brain extract (6 µg/ml), epidermal growth factor (10 ng/ml), hydrocortisone (1 µg/ml), gentamycin (30 µg/ml) and amphotericin B (15 ng/ml). HEK-293 (human embryonic kidney 293) cells (A.T.C.C.) and MEF (mouse embryonic fibroblasts, a gift from Dr Jeffrey Chan, University of California, Irvine, CA, U.S.A.) were passaged in DMEM (Dulbecco’s modified Eagle’s medium)/F12 medium (Mediatech) with 10% FBS, 50µM 2-mercaptoethanol, nonessential amino acids (100 µM), gentamycin (30 µg/ml) and amphotericin B (15 ng/ml). All cells were grown at 37°C in 95% air and 5% CO2. Cells were incubated in culture medium containing 1% FBS for ~ 16 h prior to LNO2 treatment. Control cells were treated with vehicle (methanol) at concentrations equivalent to parallel LNO2 treatment groups (<0.1%; v/v).
Real-time and standard PCR
SYBR green-based real-time PCR was performed on cDNA 1st strand synthesis products generated from total RNA (Invitrogen). The reactions were performed in triplicate. Real-time primers are described in Supplementary Table S1 (at http://www.BiochemJ.org/bj/422/bj4220353add.htm). Standard PCR for mouse Nrf2 was performed for 35 cycles at 94°C for 30 s, 58°C for 45 s, and 72°C for 90 s using mouse Nrf2-specific primers as described in Supplementary Table S1.
Western blot analysis
Total cell protein was lysed in RIPA buffer (50 mMTris/HCl, 1% NP-40, 0.25% deoxycholic acid, 150 mM NaCl, 1 mM EGTA, 1 mM sodium orthovanadate and 1 mM sodium fluoride) with protease (Roche Applied Science) and phosphatase (Sigma) inhibitors, and quantified using a Bradford assay (Bio-Rad). Total protein (15 µg) was resolved on a 12%Tris-glycine SDS/PAGE gel. After transfer to a PVDF membrane (Millipore), HO-1 and actin were detected using 1:5000 dilutions of primary rabbit polyclonal antibodies, followed by a horseradish peroxidase-conjugated anti-rabbit IgG. Protein was visualized using the ECL® (enhanced chemiluminescence) detection system (GE Healthcare).
Plasmid constructs and transient transfection analysis
pHOGL3/4.5 and pHOGL3/4.0 luciferase expression plasmids containing the − 4.5 kb (extending from + 80 to − 4.4 kb) and − 4.0 kb regions (extending from + 80 to − 3.85 kb) of the human HO-1 promoter respectively, were constructed as previously described [27]. The plasmid constructs with mutations in the − 4.0 kb region of the human HO-1 promoter (CRE and a single nucleotide in the NF-E2/AP-1 element termed ARE-5) and − 44 bp (mE-box mutation in the E-box element) promoter region (Figure 1A) have been described previously in [19,20]. Dominant-negative CREB-1 (S133A) was constructed as previously described [14]. aUSF (dominant-negative USF-1) was provided by Dr Charles Vinson (National Institutes of Health, Bethesda, MD, U.S.A.) [28]. Plasmids expressing three individual clones of shRNA (small-hairpin RNA) sequences targeted against Nrf2 (1sh, 2sh, 3sh) and their mock control (Msh) were obtained from SABiosciences (Frederick, MD). Transient transfections utilized equimolar amounts of plasmids and diethylaminoethyldextran (for HAEC) and cationic lipid (for HEK-293 cells) methods in a batch transfection protocol. Primers for human Nrf2 were obtained from SABiosciences. Primers for human HO-1 are as described in Supplementary Table S1. Lysates were assayed for luciferase activity [27] using a luciferase assay kit (Promega).
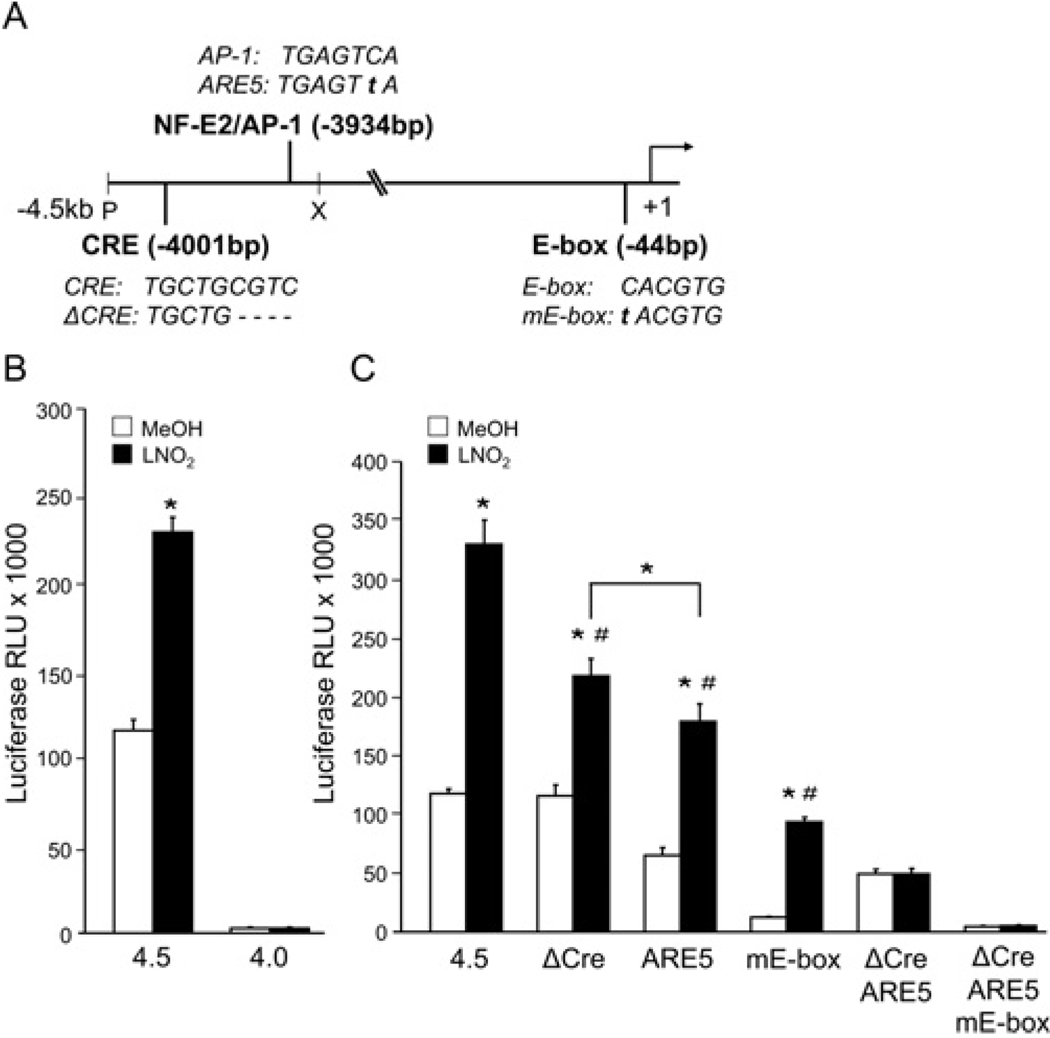
Figure 1. HO-1 promoter activation by LNO2 requires functional CRE, AP-1 and E-box response elements.
(A) Sequences and relative positions of CRE, NF-E2/AP-1 and E-box elements within the human HO-1 promoter. Mutations (ARE5 and mE-box) are denoted by lower case lettering. (B) HAEC were transiently transfected with equimolar amounts of pGL3 luciferase expression plasmids (Promega) under the direction of the − 4.5 kb (4.5; pHOGL3/4.5) or − 3.8 kb (4.0; pHOGL3/4.0) human HO-1 promoter fragments. pHOGL3/4.0 is truncated 500 bp downstream of the − 4.5 kb plasmid. (C) Deletion of the CRE site (ΔCre), and respective single nucleotide mutation of protected guanines within the NF-E2/AP-1 (ARE5) or E-box (mE-box) elements in the pHOGL3/4.5 construct were introduced by site-directed mutagenesis and tested for promoter activation as indicated. After 48 h, cells were exposed to vehicle control (open bars) or 5 µM LNO2 (filled bars) for 16 h, and luciferase assays were performed. Results are derived from 3–6 independent experiments with n = 12/group, *P < 0.01 compared with control; #P < 0.05.
EMSA (electrophoretic mobility-shift assay)
Nuclear protein was harvested [19] and 6 µg of nuclear protein was incubated with 5 pmol of 32P-end-labelled DNA probe (Invitrogen) with or without 2 µg of purified antibody (rabbit IgG sc-2027, CREB-1 sc-271, JunD sc-74x or USF-1 sc-229x). Binding reactions were resolved on 6%acrylamide and subjected to autoradiography. Primer sequences for the human HO-1 CRE, CRE mut, NF-E2/AP-1 and proximal E-box were based on the human HO-1 promoter sequence published in the National Center for Biotechnology Information (Genbank® accession number Z82244) (Supplementary Table S1).
ChIP (chromatin immunoprecipitation) assay
HAEC and HEK-293 cells were grown to approx. 80% confluence on 15 cm plates and induced with LNO2 (5 µM) or vehicle (MeOH) for 2 h and processed for ChIP analysis according to the manufacturer’s instructions (Active Motif). Chromatin–protein interactions were stabilized by formaldehyde fixation for 10 min. After quenching of the fixation reaction, cells were lysed, and nuclei were extracted and homogenized to release chromatin–protein conjugates. Chromatin was sheared by enzymatic digestion for 12 min and then subjected to overnight immunoprecipitation (normal rabbit IgG sc-2027, CREB-1 sc-58, phospho CREB-1 sc-7978, JunD sc-74x and USF-1 sc-229x; Santa Cruz Biotechnology) with Protein G magnetic beads. After multiple washes, chromatin was subjected to proteinase K digestion, purified and analysed for relative binding to protein targets by quantitative real-time PCR. Values were normalized to input PCR levels, and fold induction was compared with IgG and no template values. Primers for the − 4.5 kb region, the proximal E-box and a nonspecific E-box region (+2.2 kb) were as described in [19].
3C (Chromosome Conformation Capture) assay
HEK-293 cells were grown to approx. 80% confluence on 15 cm plates and induced with LNO2 (5 µM) or vehicle (MeOH) for 0.5, 2 or 4 h and processed for 3C analysis as previously described [29]. Chromatin–protein interactions were stabilized by formaldehyde fixation for 10 min. After quenching of the fixation reaction, cells were lysed and nuclei were extracted and homogenized to release chromatin–protein conjugates. Chromatin was digested overnight with SphI restriction enzyme (New England Biolabs) and ligated with 0.3 unit/ml of T4 DNA ligase (New England Biolabs) for 4 h. Chromatin was washed, subjected to Proteinase K digestion, purified and then analysed for relative crosslinking efficiency by real-time PCR, where the internal control, ‘Int’, was set at 100%cross-linking efficiency. A human bacterial artificial chromosome containing the HO-1 gene (Genbank® accession number Z82244) was used as a positive control for the 3C assay. Primer sets for CRE and proximal E-box containing fragments (1R, 3R, 2F and 4F) are as described in Supplementary Table S1.
Statistical analysis
Results are expressed as means ± S.E.M. For luciferase-based assays, analyses were performed using ANOVA and the Student–Newman–Keuls test. All results are considered significant at P < 0.05.
RESULTS
Transcriptional regulation of HO-1 by LNO2 is contained within the − 4.5 kb promoter region and requires functional synergy between CRE, NF-E2/AP-1 and E-box response elements
Previous studies have shown that LNO2 induces HO-1 mRNA and protein at the level of transcription [24]. To determine the minimal responsive region required for LNO2 activation of human HO-1, a − 4.5 kb human HO-1 promoter construct (pHOGL3/4.5) was tested alongside a similar vector containing a 500 bp deletion of the 5′ end of the − 4.5 kb fragment (pHOGL3/4.0). Following transient transfection, HAEC were treated with either vehicle (methanol) or LNO2 (5 µM) for 16h. Figure 1(B) shows that LNO2 specifically transactivated pHOGL3/4.5, but not pHOGL3/4.0, indicating that the minimal responsive element(s) required for HO-1 induction by LNO2 resides within the 500 bp region between the − 4.5 and − 4.0 kb promoter.
Previous studies using DNase I hypersensitivity analysis identified three distal sites (HS-2, -3 and -4) corresponding to approx. − 4.0, − 7.2 and − 9.2 kb respectively of the HO-1 promoter, in addition to one proximal region, HS-1, which is an E-box [20]. Within the − 4.5 kb promoter in the HS-2 site, a putative CRE (‘tgctgcgtc’, located at − 4.0 kb) and an NF-E2/AP-1-binding element (‘tgagtca’, located at − 3934 bp) have been identified to contain protected guanines by in vivo dimethylsulfate footprinting [19]. Site-directed mutagenesis causing a deletion of the CRE element (ΔCRE) or of a downstream single nucleotide mutation of the NF-E2/AP-1 (ARE5) (Figure 1A), individually modestly reduced activation of the HO-1 promoter by LNO2 (Figure 1C). Transfection with a pHOGL3/4.5 expression plasmid containing both the ΔCRE sites and the ARE5 mutation fully inhibited LNO2 activation of the promoter, suggesting that a synergistic role exists for a functional CRE with the NF-E2/AP-1-response element in HO-1 activation by LNO2 (Figure 1C). Triple mutation of CRE, NF-E2/AP-1 and the USF 1/2 responsive proximal E-box element (‘cacgtg’, located at − 44 bp) (mE-box), abolished both basal and LNO2-mediated expression to levels similar to the pHOGL3/4.0 construct. Although transfection of HAEC with the − 4.5 kb fragment, containing only a single nucleotide mutation of the − 44 bp proximal E-box element, reduced overall expression, the fold induction by LNO2 compared with vehicle was 2.7-fold greater than that achieved by the − 4.5 kb wild-type promoter. Together, these data reveal that the cis-acting regulatory sequences containing CRE (− 4.0 kb), NF-E2/AP-1 (− 3934 bp) and proximal E-box (− 44 bp) elements can act in concert to mediate LNO2 activation of the human HO-1 promoter.
LNO2 induction of HO-1 occurs in the absence of Nrf2
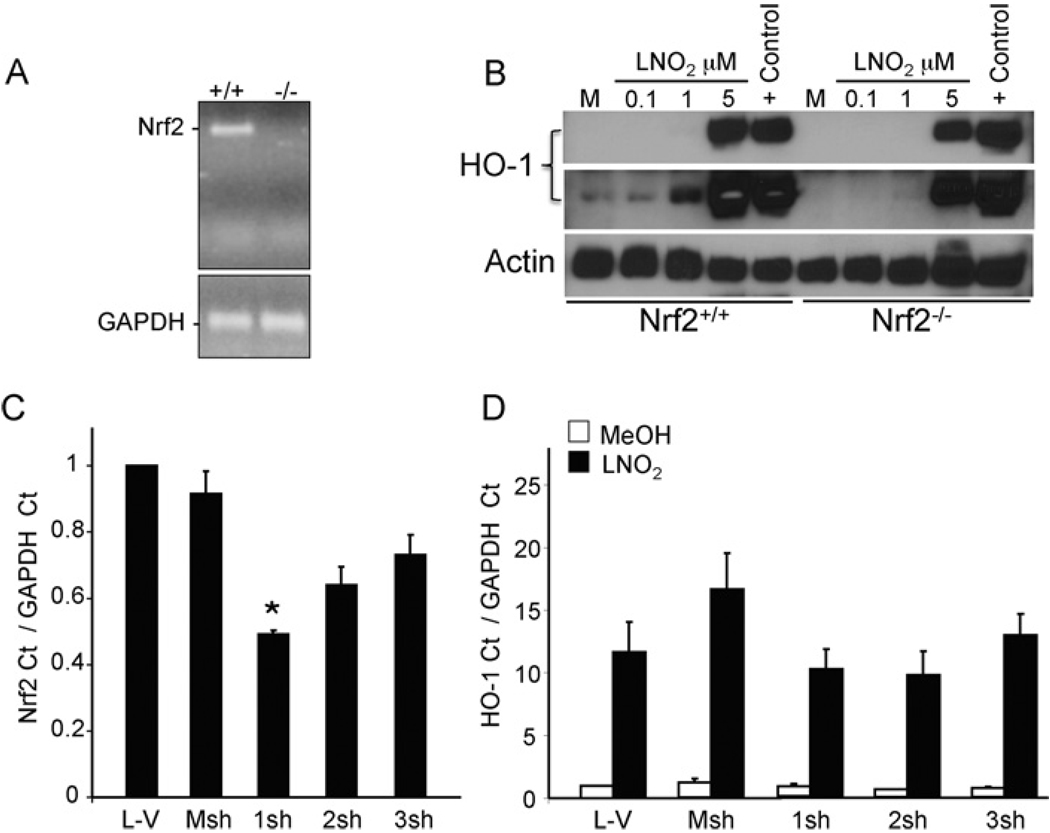
To determine the necessity of Nrf2 for induction of HO-1 by LNO2, wild-type or Nrf2-deficient MEF, cell lines were tested for HO-1 responsiveness to LNO2. The presence or absence of Nrf2 in these cells was verified by PCR (Figure 2A). Nrf2+/+ and Nrf2−/− MEF seeded at similar confluency were treated with 0.1, 1 or 5 µM LNO2 for 16 h and analysed for HO-1 protein levels by Western blot. Although basal levels were lower in Nrf2−/− cells, LNO2 induced HO-1 protein in Nrf2−/− cells, albeit at lower levels, compared with Nrf2+/+ cells at both 1 and 5 µM concentrations (Figure 2B). Cadmium chloride, a known potent inducer of HO-1, was used as a positive control. After determining that LNO2 induces HO-1 protein in HEK-293 cells (Supplementary Figure S1 at http://www.BiochemJ.org/bj/422/bj4220353add.htm), targeted knockdown of Nrf2 was employed to examine the necessity of Nrf2 expression in LNO2-mediated induction of human HO-1. Figure 2(C) shows that a plasmid construct expressing shRNA specific for Nrf2 (RNAi plasmid 1sh) reduced Nrf2 transcript by 50% when compared with vehicle control (L-V) or nonspecific shRNA (Msh). Cells expressing RNAi plasmid 1sh had no significant reduction in HO-1 transcript when treated with 5 µM LNO2 (Figure 2D). These findings show that loss of Nrf2 does not prevent activation of HO-1 by LNO2 and suggests that other pathways may act independently of Nrf2 for LNO2-mediated HO-1 induction.
Figure 2. LNO2 induces HO-1 in the absence of Nrf2.
(A) Wild-type MEF (Nrf2+/+) or Nrf2-deficient (Nrf2−/−) MEF were tested for the presence and absence of Nrf2 by PCR. (B) Nrf2+/+ and Nrf2−/− MEF were incubated with vehicle control (M; methanol) or LNO2 at 0.1, 1 or 5 µM for 16 h. Cells treated with 5 µM cadmium chloride for 16 h were used as a positive control (+Control). HO-1 and actin protein levels were determined by Western blot analysis. The middle panel represents a longer exposure of the same blot showing HO-1 induction at 1 µM concentration of LNO2. (C) HEK-293 cells were transfected with cationic lipid vehicle or plasmids expressing mock shRNA (Msh) or shRNAs (clones 1sh, 2sh, 3sh, and 4sh) targeting human Nrf2. After 48 h, total RNA was harvested and processed for real-time PCR analysis of Nrf2 and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) as described in the Materials and methods section. (D) HEK-293 cells were transfected as in (C). After 48 h, cells were treated with vehicle control (MeOH; methanol) or LNO2 (5 µM) for 4 h. Total RNA was processed for real-time PCR analysis for HO-1 and GAPDH. Nrf2 and HO-1 Cts (cycle thresholds) were normalized to GAPDH Cts. All plasmids were transfected in equimolar concentrations. Results are derived from 2 independent experiments with n = 3/group, *P < 0.05 compared with Msh.
CREB, JunD and USF-1 bind to human HO-1 promoter elements in vitro
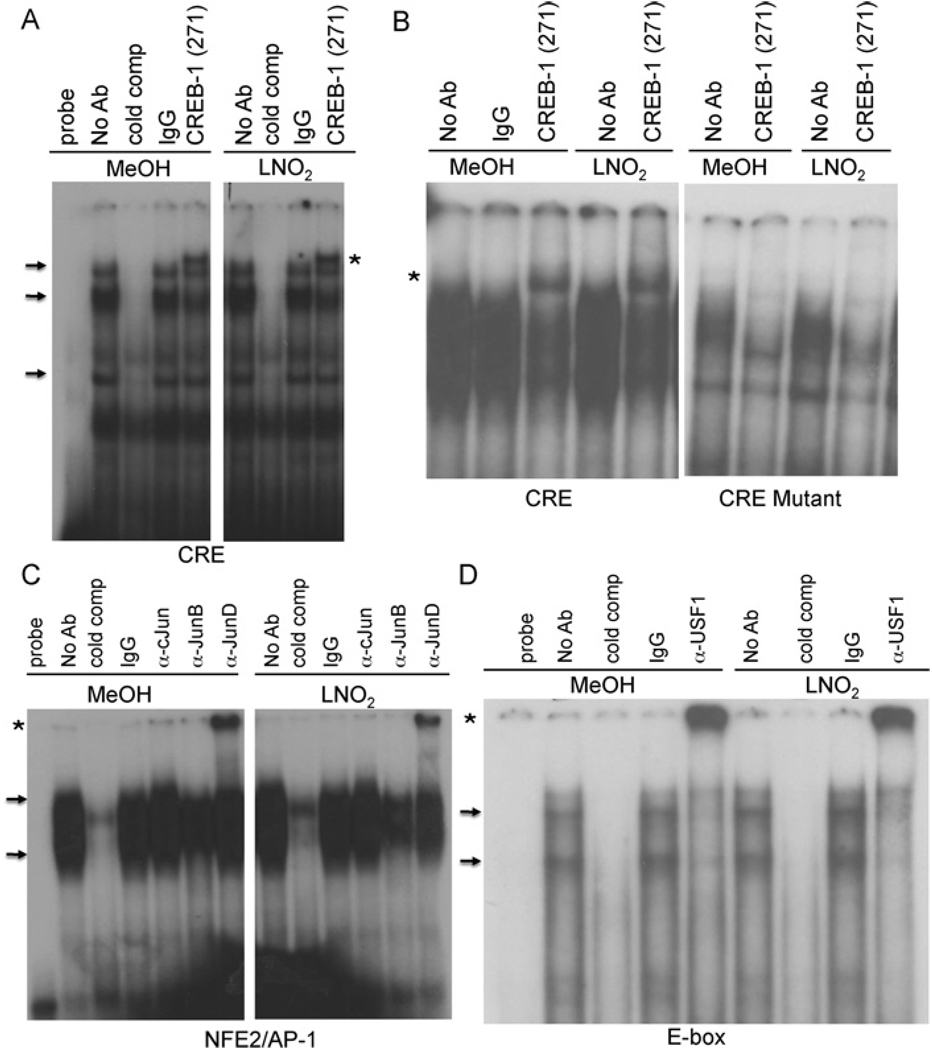
Having determined that CRE, AP-1 and proximal E-box elements are required for LNO2 activation of human HO-1 expression, gel shift analyses were used to examine whether CREB-1, JunD and USF-1 interacted with these elements in vitro. Nuclear extracts from vehicle and LNO2-treated HAEC co-incubated with an anti-CREB-1 antibody retarded mobility of a human HO-1 CRE-containing oligonucleotide (Figure 3A). The loss of the supershift in nuclear extracts incubated with an oligonucleotide containing a mutation of the CRE element supports the specificity of CREB-1 binding to the wild-type CRE (Figure 3B). Antibodies against JunD and USF-1 co-incubated with HAEC nuclear extracts produced supershifts of the human HO-1 NF-E2/AP-1 and proximal E-box elements respectively (Figures 3C and 3D). No discernible supershift was observed with c-Jun or JunB, although a decrease in intensity of DNA–protein complexes was noted with JunB antibody (Figure 3C).
Figure 3. In vitro DNA-protein binding at the CRE, NF-E2/AP-1 and E-box elements in the human HO-1 − 4.5 kb promoter region.
EMSA using nuclear extracts from 5 µM LNO2- or vehicle (MeOH; methanol)-treated HAEC. (A and B) The probe used in these experiments contains either the wild-type human HO-1 CRE or the CRE containing a 5 nucleotide deletion as described in Supplementary Table S1. The CREB-1 antibody was used as indicated. (C and D) The probes used in these experiments contain the human HO-1 AP-1 element (C) and the human HO-1 proximal E-box element (D). USF-1 and JunD antibodies were obtained from Santa Cruz Biotechnology. DNA–protein complexes are indicated by arrows and supershifts are highlighted by asterisks. Non-radiolabelled competitor probe (cold comp) was used in 100-fold excess of the radiolabelled probe. The results are derived from 2–5 independent experiments. No Ab, no antibody; α-protein, anti-protein antibody.
Dominant-negative CREB-1 and dominant-negative USF-1 inhibit HO-1 promoter activation
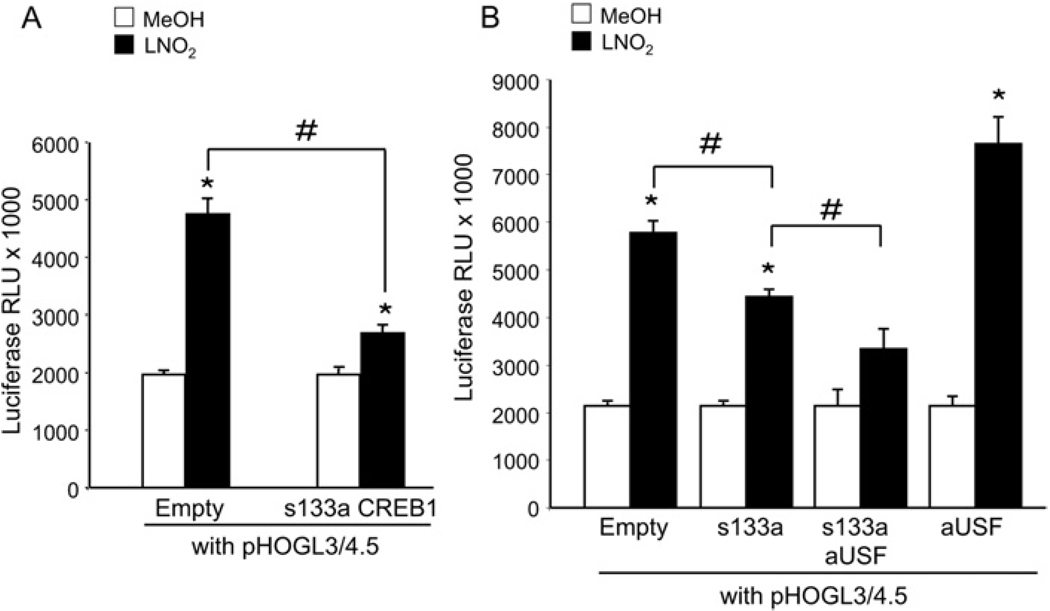
HEK-293 cells were co-transfected with pHOGL3/4.5 and equimolar concentrations of either the dominant-negative S133A CREB-1 expression vector or its backbone control (empty). The empty vector did not affect LNO2 activation of pHOGL3/4.5, whereas co-expression of S133A CREB-1 reduced LNO2 activation of the human HO-1 promoter (Figure 4A). Cells transfected with pHOGL3/4.5, S133A CREB-1 and the dominant-negative USF-1 vector, aUSF, showed a sequential decrease in HO-1 promoter activation by LNO2 compared with cells transfected with equimolar amounts of pHOGL3/4.5 and S133A CREB-1 alone (Figure 4B). Co-expression of aUSF alone induced a significantly greater extent of promoter activation (Figure 4B), supporting that CREB-1 in part mediates the induction of HO-1 by LNO2, and suggests that CREB-1 may work in concert with USF-1 to modulate HO-1 promoter expression.
Figure 4. Dominant-negative CREB-1 mediated inhibition of HO-1 promoter activation by LNO2 in HEK-293 cells.
(A) HEK-293 cells were transiently transfected with pHOGL3/4.5 (2 µg) and equimolar amounts of either pCMV empty vector or pCMV S133A CREB-1. After 48 h, cells were exposed to vehicle control (MeOH; methanol, open bars) or 5 µMLNO2 (filled bars) for 16 h, and luciferase assay was performed. (B) HEK-293 cells were transfected as described in (A), and included equimolar co-transfection of dominant-negative USF expression vector (aUSF) with either pCMV empty vector or pCMV S133A CREB-1. Results are derived from 2–3 independent experiments with n = 12/group, *P < 0.01 compared with control methanol-treated groups, #P < 0.05 compared with indicated groups. RLU, relative light units.
LNO2 induces CREB-1 binding to the HO-1 CRE and proximal E-box element
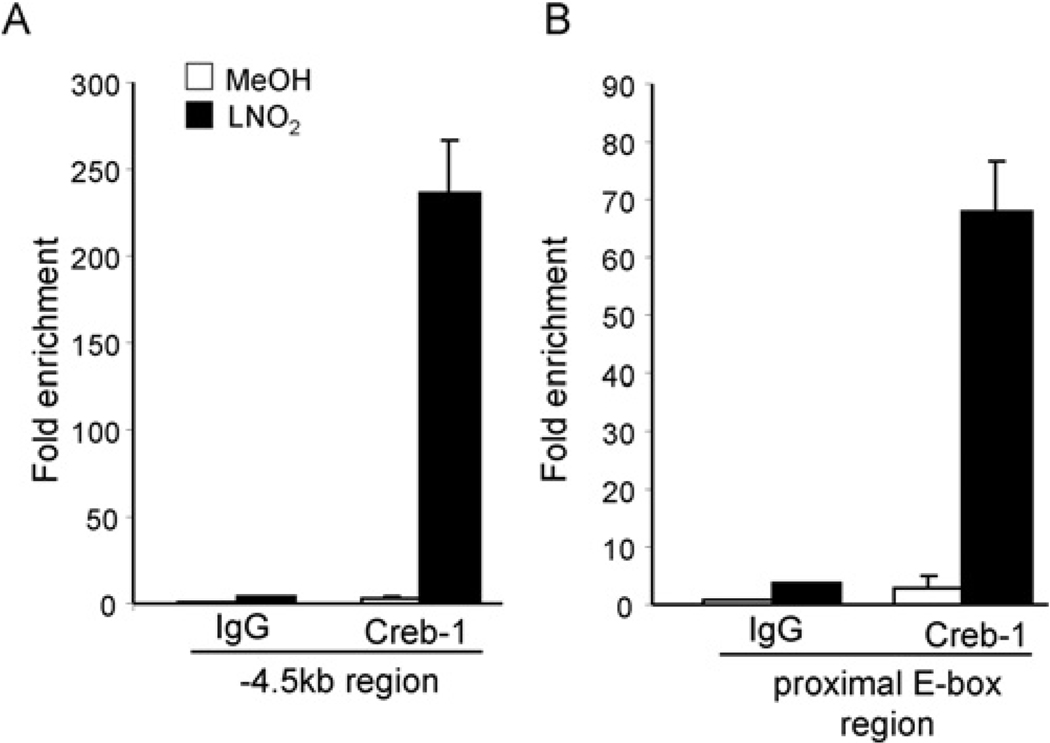
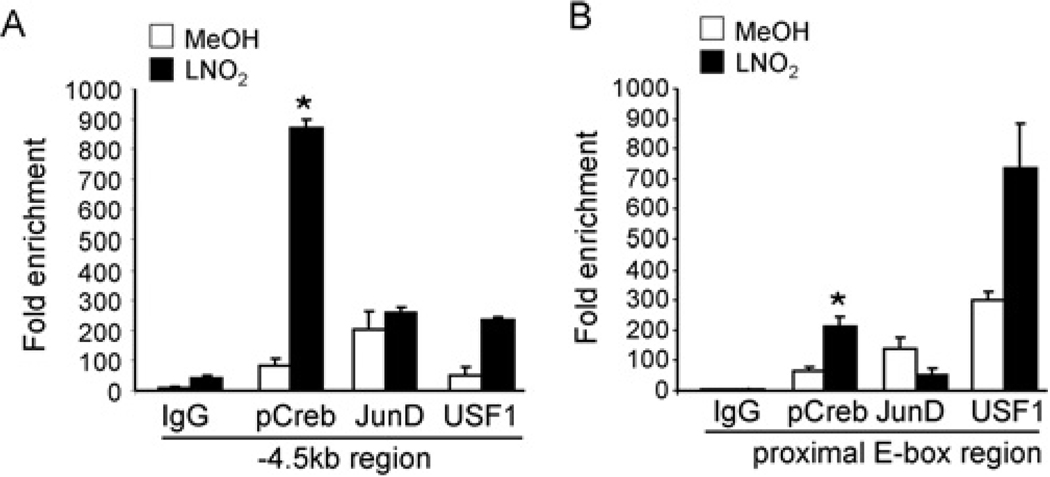
To explore binding of CREB-1 to regions of the human HO-1 promoter, ChIP analysis was performed in HAEC and HEK-293 cells treated with vehicle (MeOH) or LNO2 (5 µM) for 2 h. Quantitative real-time PCR analysis of the ChIP data shows increased CREB-1 binding at both the − 4.5 kb CRE and − 44 bp proximal E-box regions inHAEC(Figures 5A and 5B) in response to LNO2. ChIP analysis showed that phospho-CREB-1 binding to the− 4.5 kb CRE-containing and− 44 bp proximal E-box regions within the human HO-1 promoter was significantly increased in LNO2-treated cells (Figures 6A and 6B). USF-1 binding was also significantly increased in response to LNO2 at both the − 4.5 kb region and the proximal E-box, supporting that USF-1 interacts with the HO-1 CRE. The decrease in JunD binding in response to LNO2 at the proximal E-box (Figure 6B) is consistent with our previous findings, with reduced binding at this region observed with haemin stimulation of human renal epithelial cells, and supports the concept that JunD is a negative regulator of human HO-1 [19]. The observation that LNO2 promotes CREB-1 association with the − 44 bp proximal E-box element and the USF-1 associated with the CRE (located at − 4.0 kb) indicates that the CRE and proximal E-box regions are in close proximity upon stimulation with LNO2.
Figure 5. LNO2 mediates interaction of CRE and E-box elements with CREB-1 in HAEC.
(A) HAEC were treated with vehicle (MeOH; methanol, open bars) or 5 µM LNO2 (filled bars) for 2 h, cross-linked, and analysed by ChIP. The − 4.5 kb CRE-containing, and − 44 bp proximal E-box-containing regions of the human HO-1 promoter were amplified by real-time PCR following ChIP with antibodies (2 µg) against control IgG and CREB-1. Results are interpreted as fold enrichment in specific antibody treated Ct (cycle threshold) IgG treated Ct values.
Figure 6. LNO2 mediates interaction of CRE and E-box elements with CREB-1, USF-1 and Jun-D in HEK-293 cells.
(A) HEK-293 cells were treated with vehicle (open bars) or 5 µM LNO2 (filled bars) for 2 h, cross-linked, and analysed by ChIP. The − 4.5 kb and − 44 bp proximal E-box regions of the human HO-1 promoter were amplified by real-time PCR following ChIP with antibodies (2 µg) against control IgG, phospho-CREB-1, JunD and USF-1. Data are interpreted as fold enrichment in antibody treated Ct (cycle threshold) IgG template Ct. Results are derived from 2 independent experiments with n = 3/group, *P < 0.05 compared with control
LNO2 promotes dynamic interactions between the CRE and proximal E-box element in vivo
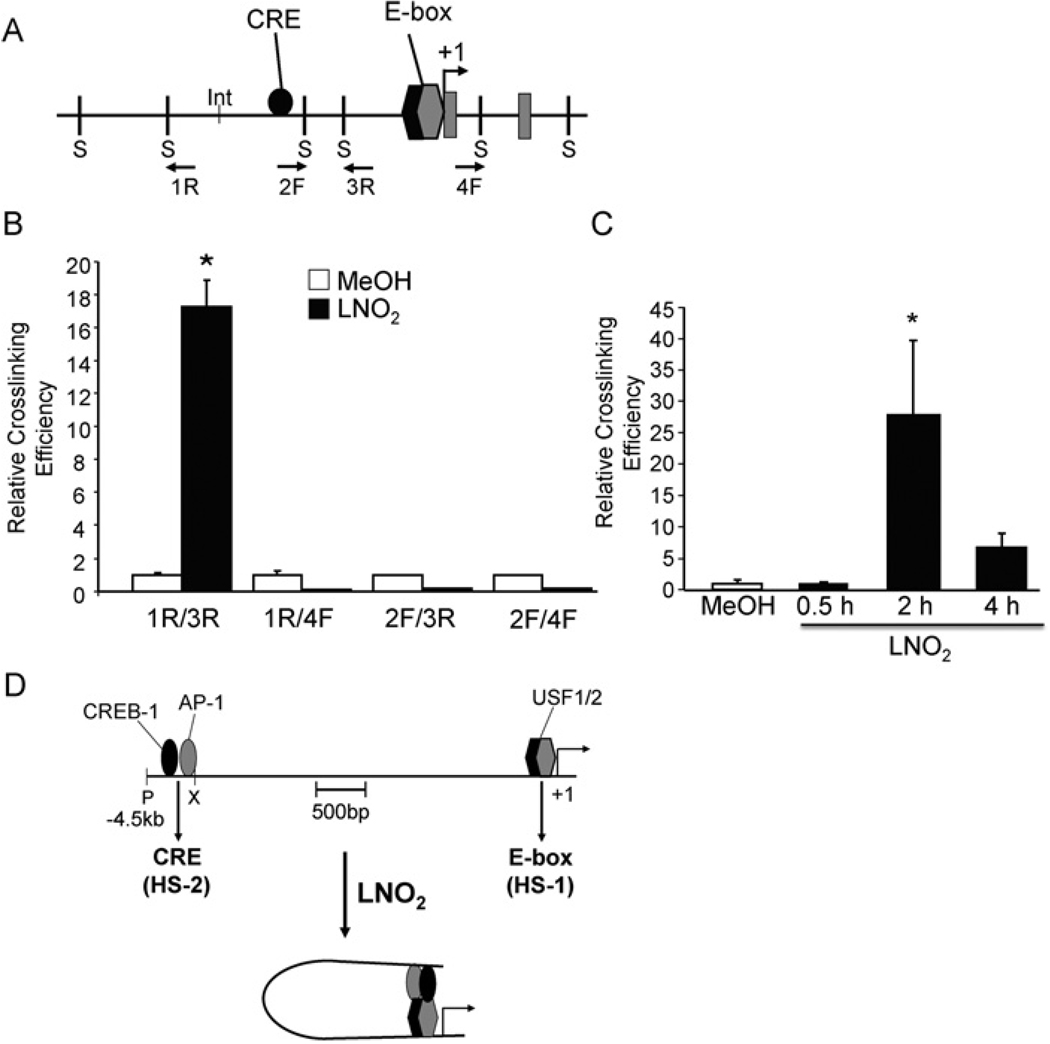
The relative physical proximity of the − 4.0 kb CRE and − 44 bp E-box during LNO2-mediated activation of human HO-1 was determined via 3C analysis in HEK-293 cells. Chromatin was cross-linked after treatment with vehicle (methanol) or LNO2 for 2 h, digested with SphI restriction enzyme and subjected to low stringency ligation. The crosslinking efficiency relative to an undigested internal control set at 100% revealed relative proximity of SphI-digested fragments. There was significantly increased cross-linking efficiency between the CRE and proximal E-box-containing fragments in LNO2-treated cells as indicated by the 1R/3R primer pairs, whereas no significant interactions were observed with the other primer sets (Figures 7A and 7B). The interaction between CRE and E-box elements was further confirmed by the 3C assay performed on cells treated with LNO2 for 30 min, 2 and 4 h, which showed a time-dependent increase in cross-linking efficiency between the 1R/3R primer sets (Figure 7C). Positive controls for the 3C assay were performed using a human HO-1-containing BAC (Bacterial Artificial Chromosome) clone (results not shown). These data indicate that the CRE and E-box regions of the HO-1 promoter may dynamically associate by chromatin looping and are in close proximity in response to LNO2 activation.
Figure 7. LNO2 treatment promotes physical association between CRE and proximal E-box-containing regions in HEK-293 cells.
(A) Map of the human HO-1 promoter, where S denotes Sph1 restriction sites flanking the CRE and E-box elements and 1R, 2F, 3R and 4F denote primer direction. (B and C) 3C assay was performed on cross-linked genomic DNA isolated from cells incubated with vehicle control or LNO2 as described in the Materials and methods section. SphI restriction enzyme was used and the genomic DNA was ligated under dilute conditions (0.3 units/ml). Ligation products were amplified by real-time PCR. Results are expressed as cross-linking efficiency compared with internal control. *P < 0.05 compared with vehicle control (MeOH; methanol). (D) Working model showing the molecular mechanism of HO-1 gene activation by LNO2. CREB-1, AP-1 and USF1/2 proteins bound to the HS-1 and HS-2 sites in the human HO-1 promoter. The lower panel shows chromatin loop formation bringing the HS-1 region (containing the E-box) into close proximity with the HS-2 region (containing the CRE and AP-1) following LNO2 stimulation, resulting in HO-1 gene activation.
DISCUSSION
Oxidatively-modified lipids, typically viewed as pro-inflammatory and pathogenic mediators, can also regulate the expression of genes that mitigate inflammatory responses [30–32]. Low, clinically relevant concentrations of unsaturated fatty acid oxidation and nitration products mediate a diversity of adaptive and anti-inflammatory signalling actions via the modulation of inflammatory and metabolic-related transcription factor and receptor function [33,34]. We report a novel and unprecedented mode of regulation for human HO-1 expression by LNO2 and demonstrate that such induction requires co-ordinated interactions between cis-acting regulatory CRE, NF-E2/AP-1, and proximal E-box response elements. This occurs via a dynamic interaction between CREB-1 and the proximal E-box that is a consequence of chromatin looping between the − 4.0 kb CRE-containing region and the − 44 bp E-box region upon activation by a nitroalkene derivative of linoleic acid.
The data presented here suggests that LNO2-mediated activation of the human HO-1 promoter relies on a functional CRE element acting in concert with theNF-E2/AP-1 and proximal E-box elements. Loss of either the CRE or NF-E2/AP-1 element significantly attenuated but did not abrogate activation of the − 4.5 kb promoter fragment by LNO2. However, mutation of both CRE and NF-E2/AP-1 fully reversed LNO2 activation of the promoter, and the triple mutation of CRE, NF-E2/AP-1 and the proximal E-box abolished promoter activity (Figure 1C). These data are in agreement with human HO-1 promoter responsiveness to Ox-PAPC (oxidized 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphorylcholine), in which double mutations of the CRE and NF-E2/AP-1 elements resulted in loss of Ox-PAPC-mediated HO-1 induction [14].
Notably, activation of the human HO-1 promoter–reporter construct containing only a proximal E-box single nucleotide mutation resulted in both overall lower promoter activity and significantly greater fold induction by LNO2 when compared with the wild-type − 4.5 kb construct (Figure 1C). The significantly greater fold induction than that observed in the wild-type promoter was also observed in cells transfected with the dominant-negative aUSF vector (Figure 4B), suggesting that USFs exert a dual effect on HO-1 expression depending on which interacting partners it can dimerize with. In this regard, USFs can act as transcriptional activators and repressors in a variety of human promoters via recruitment of complex transcriptional machinery. The specific roles of USF-1 and USF-2 in modulating HO-1 warrant further exploration, since Figures 1(C) and 4(B) suggest that either USF-1 or USF-2 may play a repressive role in human HO-1 promoter activation. It is unknown if USF-1 or USF-2 dimerizes with JunD in vivo. JunD associates with the USF-binding site of the human HO-1 promoter and represses HO-1 gene expression in renal epithelial cells [19] and may in part provide an explanation for the effects of aUSF in this study.
Previous studies demonstrated that transcription factor binding to the human HO-1 promoter by dimethylsulfate footprinting and gel-shift assays is constitutive with respect to JunB- and JunD-mediated regulation of HO-1 expression. However, ChIP and promoter activation of HO-1 transcription was dependent on specific induction by haemin [19]. The present manuscript shows by gel-shift analysis a similar mechanism of induction, in which CREB-1, JunD and USF-1 binding to the CRE, NF-E2/AP-1 and proximal E-box regions respectively, is constitutive (Figure 3). Of interest, ChIP analysis of the HO-1 promoter revealed LNO2-specific augmentation of transcription factor binding to the CRE- and NF-E2/AP-1-containing and proximal E-box-containing regions (Figures 5 and 6) in both HAEC and HEK-293 cells.
Epigenetic factors influencing gene regulation include changes in phosphorylation, methylation, chromatin assembly machinery and dynamic chromatin–chromatin interaction, and may account for the observed discrepancy in constitutive transcription factor binding, but not LNO2-dependent transcriptional activation. Use of the 3C assay has been an effective tool for determining the role that changes in long- and short-range chromatin conformation may have on gene activation, including endotoxin-mediated molecular regulation of the inflammatory response gene, inducible NO synthase, through chromatin looping of AP-1 and NF-κB sites [35]. In the present manuscript, 3C analysis reveals a specific short-range interaction between the CRE and E-box regions in cells treated with LNO2 over distances spanning 4.0 kb. Results in Figure 7 show that activation of HO-1 by LNO2-mediated changes in chromatin brings the CRE-containing region, located at − 4.0 kb in the HS-2 region, and the E-box-containing region, located at − 44 bp in the proximal promoter, into relatively close proximity by chromatin loop formation. It is conceivable that changes in HO-1 promoter conformation are responsible for the LNO2-specific activation of human HO-1 transcription by constitutively bound CREB-1, JunD and USF-1.
One mechanism underlying LNO2-mediated induction of HO-1 expression can include electrophilic adduction to signalling proteins involved in HO-1 regulation. For example, LNO2 binding of Keap-1 promotes Nrf2 nuclear localization and transactivation of Nrf2-responsive genes and inhibition of vascular smooth muscle cell proliferation in an Nrf2-dependent manner [36]. Although Nrf2 is a potent activator of HO-1 and is activated in response to other electrophilic lipids including 15-deoxyprostaglandin J2 [37], LNO2 can induce HO-1 in an Nrf2-independent manner (Figure 2). Related to this, Nrf2-deficient MEF are responsive to arsenite-dependent oxidative stress, with HO-1 induction occurring via AP-1-dependent mechanisms. Moreover, the induction occurred in a manner distinct from that of MEF expressing Nrf2; Nrf2+/+ MEF induced HO-1 in response to arsenite in an acute and rapid biphasic manner, whereas Nrf2−/− MEF elicited a lower and more sustained up-regulation of HO-1 expression [38]. A previous study has shown that Nrf2 does not interact with the NF-E2/AP-1 regulatory element within the human HO-1 promoter in renal proximal tubule cells [19]. Figure 2 provides support for a limited role of Nrf2 in human cells in that an observed 50% transient knockdown of Nrf2 did not mitigate LNO2-mediated induction of HO-1 RNA when compared with mock shRNA, sh2 or sh3 controls (Figure 2C).
Electrophilic fatty acids such as LNO2 may contribute to the up-regulation of human HO-1 activity by modulating protein kinase and phosphatase activity [6]. PKA (protein kinase A), PKC (protein kinase C), PI3K (phosphoinositide 3-kinase), p38 MAPK, p44 ERK (extracellular-signal-regulated kinase), JNK (c-Jun N-terminal kinase)/AP-1 and STAT all contribute to induction of human and mouse HO-1 [14,17,38–41]. The cAMP pathway regulates the phospho-activation of CREB-1 primarily through PKA. However, stress-response pathways, such as PI3K/Akt, p38 MAPK, and MSK-1 (mitogen and stress activated protein kinase-1) also target CREB-1 for phosphorylation [42–45]. In this regard, LNO2-mediated activation of HO-1 in HEK-293 cells is regulated, in part, by p38 MAPK and PKCα/β pathways (not shown).
The activation of HO-1 expression is a central element of the response of tissues to metabolic and inflammatory stress, and thus is regulated by multiple transcription factors sensitive to functional changes induced by oxidative and electrophilic species. These transcription factors may act either synergistically or antagonistically to modulate HO-1 expression. Based on site-directed mutagenesis, ChIP and 3C analyses, a model is supported where LNO2 induces human HO-1 gene transcription via chromatin looping between the proximal E-box region, the − 4.0 kb distal HO-1 promoter and the proteins (CREB-1, Jun and USF-1) associated with this interaction (Figure 7D). These results reveal (i) the complex regulatory factors and mediator interactions that modulate human HO-1 gene expression, and (ii) new molecular targets within the CREB-1 pathway for modulating HO-1 expression in vascular inflammatory disorders where redox-modified lipid mediators are generated.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Jeffrey Chan for the Nrf2 wild-type and knockout mouse embryonic fibroblasts and to Dr Charles Vinson for the dominant-negative aUSF expression vector [28].
FUNDING
This work was supported by the National Institutes for Heath [grant numbers HL068157, DK59600, DK75532 and 1P30 DK79337 (to A.A.)].
Abbreviations used
- AP-1
activating protein-1
- ARE
antioxidant response element
- ChIP
chromatin immunoprecipitation
- CRE
cAMP-response element
- CREB
CRE-binding protein
- 3C assay
Chromatin Conformation Capture assay
- EMSA
electrophoretic mobility-shift assay
- FBS
fetal bovine serum
- HAEC
human aortic endothelial cells
- HEK-293
human embryonic kidney 293
- HO-1
haem oxygenase-1
- LNO2
nitrolinoleic acid
- MAPK
mitogen-activated protein kinase
- MEF
mouse embryonic fibroblasts
- NO2-FA
nitro-fatty acids
- NF-E2
nuclear factor-erythroid 2
- NF-κB
nuclear factor κB
- Nrf2
NF-E2-related factor 2
- Ox-PAPC
oxidized 1-palmitoyl-2-arachidonoyl-sn-glycero-3 phosphorylcholine
- PI3K
phosphoinositide 3-kinase
- PKA
protein kinase A
- PKC
protein kinase C
- PPAR
peroxisome-proliferator-activated receptor
- shRNA
small-hairpin RNA
- STAT
signal transducer and activator of transcription
- USF
upstream stimulatory factor
- aUSF
dominant-negative USF-1
Footnotes
AUTHOR CONTRIBUTION
Marcienne Wright and Junghyun Kim performed the experiments, Bruce Freeman, Thomas Hock and Norbert Leitinger contributed reagents/analytic tools, and Bruce Freeman, Marcienne Wright and Anupam Agarwal contributed to the experimental design, execution, data interpretation and writing of the manuscript.
REFERENCES
- 1.Rubbo H, Radi R. Protein and lipid nitration: role in redox signaling and injury. Biochim. Biophys. Acta. 2008;1780:1318–1324. doi: 10.1016/j.bbagen.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Trostchansky A, Rubbo H. Nitrated fatty acids: mechanisms of formation, chemical characterization, and biological properties. Free Radical Biol. Med. 2008;44:1887–1896. doi: 10.1016/j.freeradbiomed.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Baker PR, Schopfer FJ, O’Donnell VB, Freeman BA. Convergence of nitric oxide and lipid signaling: anti-inflammatory nitro-fatty acids. Free Radical Biol. Med. 2009;46:989–1003. doi: 10.1016/j.freeradbiomed.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freeman BA, Baker PR, Schopfer FJ, Woodcock SR, Napolitano A, d’Ischia M. Nitro-fatty acid formation and signaling. J. Biol. Chem. 2008;283:15515–15519. doi: 10.1074/jbc.R800004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abraham SM, Lawrence T, Kleiman A, Warden P, Medghalchi M, Tuckermann J, Saklatvala J, Clark AR. Antiinflammatory effects of dexamethasone are partly dependent on induction of dual specificity phosphatase 1. J. Exp. Med. 2006;203:1883–1889. doi: 10.1084/jem.20060336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ichikawa T, Zhang J, Chen K, Liu Y, Schopfer FJ, Baker PR, Freeman BA, Chen YE, Cui T. Nitroalkenes suppress lipopolysaccharide-induced signal transducer and activator of transcription signaling in macrophages: a critical role of mitogen-activated protein kinase phosphatase 1. Endocrinology. 2008;149:4086–4094. doi: 10.1210/en.2007-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leopold JA, Loscalzo J. Oxidative enzymopathies and vascular disease. Arterioscler. Thromb. Vasc. Biol. 2005;25:1332–1340. doi: 10.1161/01.ATV.0000163846.51473.09. [DOI] [PubMed] [Google Scholar]
- 8.Kawashima A, Oda Y, Yachie A, Koizumi S, Nakanishi I. Heme oxygenase-1 deficiency: the first autopsy case. Hum. Pathol. 2002;33:125–130. doi: 10.1053/hupa.2002.30217. [DOI] [PubMed] [Google Scholar]
- 9.Yet SF, Layne MD, Liu X, Chen YH, Ith B, Sibinga NE, Perrella MA. Absence of heme oxygenase-1 exacerbates atherosclerotic lesion formation and vascular remodeling. FASEB J. 2003;17:1759–1761. doi: 10.1096/fj.03-0187fje. [DOI] [PubMed] [Google Scholar]
- 10.Lin CC, Liu XM, Peyton K, Wang H, Yang WC, Lin SJ, Durante W. Far infrared therapy inhibits vascular endothelial inflammation via the induction of heme oxygenase-1. Arterioscler. Thromb. Vasc. Biol. 2008;28:739–745. doi: 10.1161/ATVBAHA.107.160085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abraham NG. Therapeutic applications of human heme oxygenase gene transfer and gene therapy. Curr. Pharm. Des. 2003;9:2513–2524. doi: 10.2174/1381612033453758. [DOI] [PubMed] [Google Scholar]
- 12.Alam J, Cook JL. How many transcription factors does it take to turn on the heme oxygenase-1 gene? Am. J. Respir. Cell Mol. Biol. 2007;36:166–174. doi: 10.1165/rcmb.2006-0340TR. [DOI] [PubMed] [Google Scholar]
- 13.Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, Cook JL. Nrf2, a Cap’n’Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J. Biol. Chem. 1999;274:26071–26078. doi: 10.1074/jbc.274.37.26071. [DOI] [PubMed] [Google Scholar]
- 14.Kronke G, Bochkov VN, Huber J, Gruber F, Bluml S, Furnkranz A, Kadl A, Binder BR, Leitinger N. Oxidized phospholipids induce expression of human heme oxygenase-1 involving activation of cAMP-responsive element-binding protein. J. Biol. Chem. 2003;278:51006–51014. doi: 10.1074/jbc.M304103200. [DOI] [PubMed] [Google Scholar]
- 15.Kronke G, Kadl A, Ikonomu E, Bluml S, Furnkranz A, Sarembock IJ, Bochkov VN, Exner M, Binder BR, Leitinger N. Expression of heme oxygenase-1 in human vascular cells is regulated by peroxisome proliferator-activated receptors. Arterioscler. Thromb. Vasc. Biol. 2007;27:1276–1282. doi: 10.1161/ATVBAHA.107.142638. [DOI] [PubMed] [Google Scholar]
- 16.Sikorski EM, Uo T, Morrison RS, Agarwal A. Pescadillo interacts with the cadmium response element of the human heme oxygenase-1 promoter in renal epithelial cells. J. Biol. Chem. 2006;281:24423–24430. doi: 10.1074/jbc.M602287200. [DOI] [PubMed] [Google Scholar]
- 17.Traylor A, Hock T, Hill-Kapturczak N. Specificity protein 1 and Smad-dependent regulation of human heme oxygenase-1 gene by transforming growth factor-beta1 in renal epithelial cells. Am. J. Physiol. Renal Physiol. 2007;293:F885–F894. doi: 10.1152/ajprenal.00519.2006. [DOI] [PubMed] [Google Scholar]
- 18.Sikorski EM, Hock T, Hill-Kapturczak N, Agarwal A. The story so far: molecular regulation of the heme oxygenase-1 gene in renal injury. Am. J. Physiol. Renal Physiol. 2004;286:F425–F441. doi: 10.1152/ajprenal.00297.2003. [DOI] [PubMed] [Google Scholar]
- 19.Hock TD, Liby K, Wright MM, McConnell S, Schorpp-Kistner M, Ryan TM, Agarwal A. JunB and JunD regulate human heme oxygenase-1 gene expression in renal epithelial cells. J. Biol. Chem. 2007;282:6875–6886. doi: 10.1074/jbc.M608456200. [DOI] [PubMed] [Google Scholar]
- 20.Hock TD, Nick HS, Agarwal A. Upstream stimulatory factors, USF1 and USF2, bind to the human haem oxygenase-1 proximal promoter in vivo and regulate its transcription. Biochem. J. 2004;383:209–218. doi: 10.1042/BJ20040794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muraosa Y, Shibahara S. Identification of a cis-regulatory element and putative trans-acting factors responsible for 12-O-tetradecanoylphorbol-13-acetate (TPA)-mediated induction of heme oxygenase expression in myelomonocytic cell lines. Mol. Cell. Biol. 1993;13:7881–7891. doi: 10.1128/mcb.13.12.7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato M, Fukushi Y, Ishizawa S, Okinaga S, Muller RM, Shibahara S. Transcriptional control of the rat heme oxygenase gene by a nuclear protein that interacts with adenovirus 2 major late promoter. J. Biol. Chem. 1989;264:10251–10260. [PubMed] [Google Scholar]
- 23.Iles KE, Wright MM, Cole MP, Welty NE, Ware LB, Matthay MA, Schopfer FJ, Baker PR, Agarwal A, Freeman BA. Fatty acid transduction of nitric oxide signaling: nitrolinoleic acid mediates protective effects through regulation of the ERK pathway. Free Radical Biol. Med. 2009;46:866–875. doi: 10.1016/j.freeradbiomed.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright MM, Schopfer FJ, Baker PR, Vidyasagar V, Powell P, Chumley P, Iles KE, Freeman BA, Agarwal A. Fatty acid transduction of nitric oxide signaling: nitrolinoleic acid potently activates endothelial heme oxygenase 1 expression. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4299–4304. doi: 10.1073/pnas.0506541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui T, Schopfer FJ, Zhang J, Chen K, Ichikawa T, Baker PR, Batthyany C, Chacko BK, Feng X, Patel RP, et al. Nitrated fatty acids: Endogenous anti-inflammatory signaling mediators. J. Biol. Chem. 2006;281:35686–35698. doi: 10.1074/jbc.M603357200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baker PR, Schopfer FJ, Sweeney S, Freeman BA. Red cell membrane and plasma linoleic acid nitration products: synthesis, clinical identification, and quantitation. Proc. Natl. Acad. Sci. U.S.A. 2004;101:11577–11582. doi: 10.1073/pnas.0402587101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hill-Kapturczak N, Voakes C, Garcia J, Visner G, Nick HS, Agarwal A. A cis-acting region regulates oxidized lipid-mediated induction of the human heme oxygenase-1 gene in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2003;23:1416–1422. doi: 10.1161/01.ATV.0000081656.76378.A7. [DOI] [PubMed] [Google Scholar]
- 28.Qyang Y, Luo X, Lu T, Ismail PM, Krylov D, Vinson C, Sawadogo M. Cell-type-dependent activity of the ubiquitous transcription factor USF in cellular proliferation and transcriptional activation. Mol. Cell. Biol. 1999;19:1508–1517. doi: 10.1128/mcb.19.2.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 30.Hong HY, Jeon WK, Kim BC. Up-regulation of heme oxygenase-1 expression through the Rac1/NADPH oxidase/ROS/p38 signaling cascade mediates the anti-inflammatory effect of 15-deoxy-delta 12,14-prostaglandin J2 in murine macrophages. FEBS Lett. 2008;582:861–868. doi: 10.1016/j.febslet.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 31.Jozkowicz A, Huk I, Nigisch A, Weigel G, Weidinger F, Dulak J. Effect of prostaglandin-J(2) on VEGF synthesis depends on the induction of heme oxygenase-1. Antioxid. Redox Signal. 2002;4:577–585. doi: 10.1089/15230860260220076. [DOI] [PubMed] [Google Scholar]
- 32.Anwar AA, Li FY, Leake DS, Ishii T, Mann GE, Siow RC. Induction of heme oxygenase 1 by moderately oxidized low-density lipoproteins in human vascular smooth muscle cells: role of mitogen-activated protein kinases and Nrf2. Free Radical Biol. Med. 2005;39:227–236. doi: 10.1016/j.freeradbiomed.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 33.Serhan CN, Chiang N, Van Dyke TE. Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat. Rev. Immunol. 2008;8:349–361. doi: 10.1038/nri2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ceaser EK, Moellering DR, Shiva S, Ramachandran A, Landar A, Venkartraman A, Crawford J, Patel R, Dickinson DA, Ulasova E, et al. Mechanisms of signal transduction mediated by oxidized lipids: the role of the electrophile-responsive proteome. Biochem. Soc. Trans. 2004;32:151–155. doi: 10.1042/bst0320151. [DOI] [PubMed] [Google Scholar]
- 35.Guo H, Mi Z, Kuo PC. Characterization of short range DNA looping in endotoxin-mediated transcription of the murine inducible nitric-oxide synthase (iNOS) gene. J. Biol. Chem. 2008;283:25209–25217. doi: 10.1074/jbc.M804062200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Villacorta L, Zhang J, Garcia-Barrio MT, Chen XL, Freeman BA, Chen YE, Cui T. Nitro-linoleic acid inhibits vascular smooth muscle cell proliferation via the Keap1/Nrf2 signaling pathway. Am. J. Physiol. Heart Circ. Physiol. 2007;293:H770–H776. doi: 10.1152/ajpheart.00261.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong P, Stewart D, Hu B, Li N, Cook J, Nel A, Alam J. Activation of the mouse heme oxygenase-1 gene by 15-deoxy-Delta(12,14)-prostaglandin J(2) is mediated by the stress response elements and transcription factor Nrf2. Antioxid. Redox Signal. 2002;4:249–257. doi: 10.1089/152308602753666307. [DOI] [PubMed] [Google Scholar]
- 38.Harada H, Sugimoto R, Watanabe A, Taketani S, Okada K, Warabi E, Siow R, Itoh K, Yamamoto M, Ishii T. Differential roles for Nrf2 and AP-1 in upregulation of HO-1 expression by arsenite in murine embryonic fibroblasts. Free Radical Res. 2008;42:297–304. doi: 10.1080/10715760801975735. [DOI] [PubMed] [Google Scholar]
- 39.Xu C, Yuan X, Pan Z, Shen G, Kim JH, Yu S, Khor TO, Li W, Ma J, Kong AN. Mechanism of action of isothiocyanates: the induction of ARE-regulated genes is associated with activation of ERK and NK and the phosphorylation and nuclear translocation of Nrf2. Mol. Cancer Ther. 2006;5:1918–1926. doi: 10.1158/1535-7163.MCT-05-0497. [DOI] [PubMed] [Google Scholar]
- 40.Ning W, Song R, Li C, Park E, Mohsenin A, Choi AM, Choi ME. TGF-beta1 stimulates HO-1 via the p38 mitogen-activated protein kinase in A549 pulmonary epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2002;283:L1094–L1102. doi: 10.1152/ajplung.00151.2002. [DOI] [PubMed] [Google Scholar]
- 41.Pischke SE, Zhou Z, Song R, Ning W, Alam J, Ryter SW, Choi AM. Phosphatidylinositol 3-kinase/Akt pathway mediates heme oxygenase-1 regulation by lipopolysaccharide. Cell. Mol. Biol. 2005;51:461–470. [PubMed] [Google Scholar]
- 42.Arthur JS. MSK activation and physiological roles. Front. Biosci. 2008;13:5866–5879. doi: 10.2741/3122. [DOI] [PubMed] [Google Scholar]
- 43.Deak M, Clifton AD, Lucocq LM, Alessi DR. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 1998;17:4426–4441. doi: 10.1093/emboj/17.15.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du K, Montminy M. CREB is a regulatory target for the protein kinase Akt/PKB. J. Biol. Chem. 1998;273:32377–32379. doi: 10.1074/jbc.273.49.32377. [DOI] [PubMed] [Google Scholar]
- 45.Montminy M. Transcriptional regulation by cyclic AMP. Annu. Rev. Biochem. 1997;66:807–822. doi: 10.1146/annurev.biochem.66.1.807. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.