Abstract
We describe micro-western arrays, which enable quantitative, sensitive and high-throughput assessment of protein abundance and modifications following electrophoretic separation of micro-arrayed cell lysates. This method allowed us to measure 91 phosphosites on 67 proteins at six time points following stimulation with five EGF concentrations in A431 human carcinoma cells. We inferred the connectivities among 15 phosphorylation sites across 10 receptor tyrosine kinases (RTK) and 2 sites from Src kinase using Bayesian network modeling and two mutual information-based methods; the three inference methods yielded significant agreement on the network topology. These results imply multiple distinct RTK coactivation mechanisms and support the notion that small amounts of experimental data collected from phenotypically diverse network states may enable network inference.
Systems-level understanding of protein functions in biological processes remains a challenge. The western1 blot is a powerful protein analysis method because the electrophoretic separation step allows for reduction in sample complexity and the antibody detection step then results in signal amplitude proportional to the abundance of the immobilized antigen at a physical location on the detection membrane that can be related to molecular size standards. Because western blots require a relatively large amount of sample and a great deal of human labor, they have been of limited utility in large-scale protein studies. Reverse phase lysate arrays (RPAs), performed by arraying lysates directly on nitrocellulose-coated slides and probing them with antibodies, are useful for quantifying large numbers of proteins from limited amounts of material such as in biomarker discovery2,3. In contrast to western blots, however, RPAs lack confirmatory data for signal veracity; in a side-by-side comparison of measurements from RPAs and western blots, only 4 of 34 phospho-specific antibodies examined generated equivalent information with the two approaches4. Antibody cross-reactivity was concluded to contribute substantial noise to RPAs, confounding true protein measurements. Many antibodies have been validated for use with the Luminex xMAP bead-sorting system, although about 1,000-fold greater cell material is required per protein analysis than RPAs and the cost of detection reagents per protein about 30-fold higher. Flow cytometry permits a (relatively small) cohort of proteins to be examined simultaneously in individual cells; this multiplexing feature has been exploited with Bayesian network modeling to predict new signaling network causalities5.
In contrast to antibody-based methods, mass spectrometry (MS) can identify novel proteins. Using MS, thousands of peptides were assessed in lung cancers to identify commonly activated receptor tyrosine kinases and downstream signaling pathways6. Relative abundances can be examined quantitatively using isotopic labels across time points, cell types, or perturbations as in examination of phosphorylation dynamics of Hela7 and mammary epithelial cells8 following EGF or heregulin treatment. However, the large sample amount required by MS can limit the number of conditions that can be analyzed; ~108 cells are typically required for an MS experiment5 versus ~105 cells for an immunoblot or ~103 cells for RPAs9.
Here we describe micro-western arrays (MWA), which combine the scalability of RPAs while retaining vital attributes of western blots for highly multiplexed proteomic measurements: reduction of sample complexity and signals that can be related to protein size standards. In combination with suitable pan- and modification-specific antibodies, dynamics of protein abundance and modification may be simultaneously monitored across many samples. We demonstrate that MWA in combination with computational modeling techniques can yield useful systems-level biological insights for EGF receptor signaling dynamics.
RESULTS
Fabrication of micro-western arrays
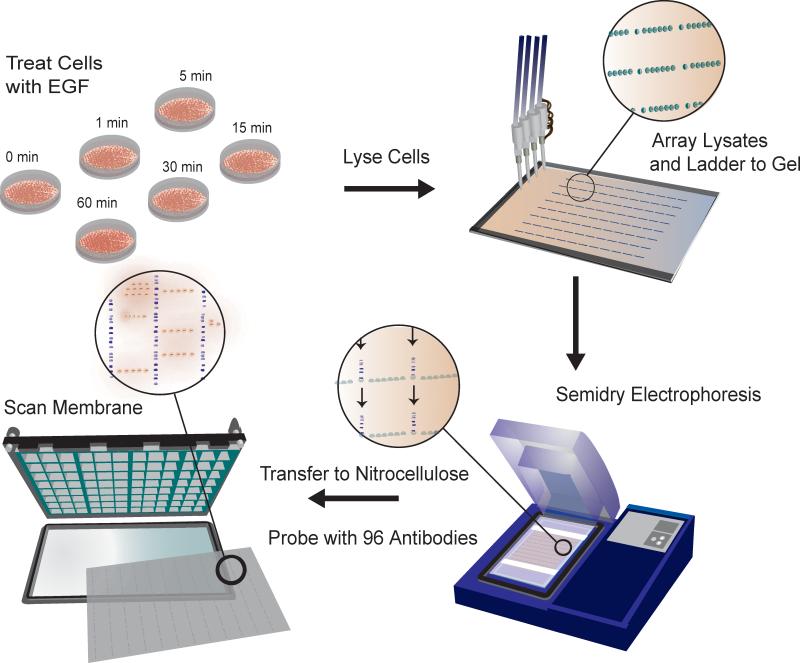
Our strategy allows us to compare protein abundances and differences in post-translational modifications for cells stimulated under different conditions (Fig. 1). To interface the microscopic western blots with micro-titer-based liquid handling methods, cell lysates are printed via a non-contact microarrayer on gels in a series of 96 identical blocks with dimensions of a 96-well plate10 (Fig. 1). Using these dimensions 6 different lysates may be examined with 96 different antibodies or 24 different lysates may be examined with 24 different antibodies. An acetate running buffer is used to increase the migration rate of large proteins and slow the rate of smaller ones, obviating the need for a stacking gel. For each spot, 6 nl of sample was arrayed over the same gel position ten times, allowing for greater spotting density and signal than micro-depositing the entire 60 nl in a single dispense. One spot of size standard and six spots of experimental sample were arrayed at one millimeter pitch at the top edge of each block. After printing, the samples are subject to semidry electrophoresis for 12 min and then transferred to a nitrocellulose membrane. The membrane is placed in a 96-well gasket to isolate each set of 6 separated lysates; each block is then incubated with a different antibody. Following incubation with dye-labeled secondary antibody, the blot is scanned with an infrared fluorescence scanner. This format allows interrogation of 192 antibodies against 6 samples when two antibodies from different hosts (e.g. rabbit and mouse) are utilized. A total of 1,152 antibody-sample readouts is therefore possible per MWA device. Each measurement requires ~1,000 cells (equivalent to 250 ng of protein) and 16 ng of detection antibody and allows for the analysis of ~4,000 protein abundances.
Fig. 1. Micro-western array (MWA) procedure.
Procedural schematic for the micro-western array method.
Validation of micro-western array method
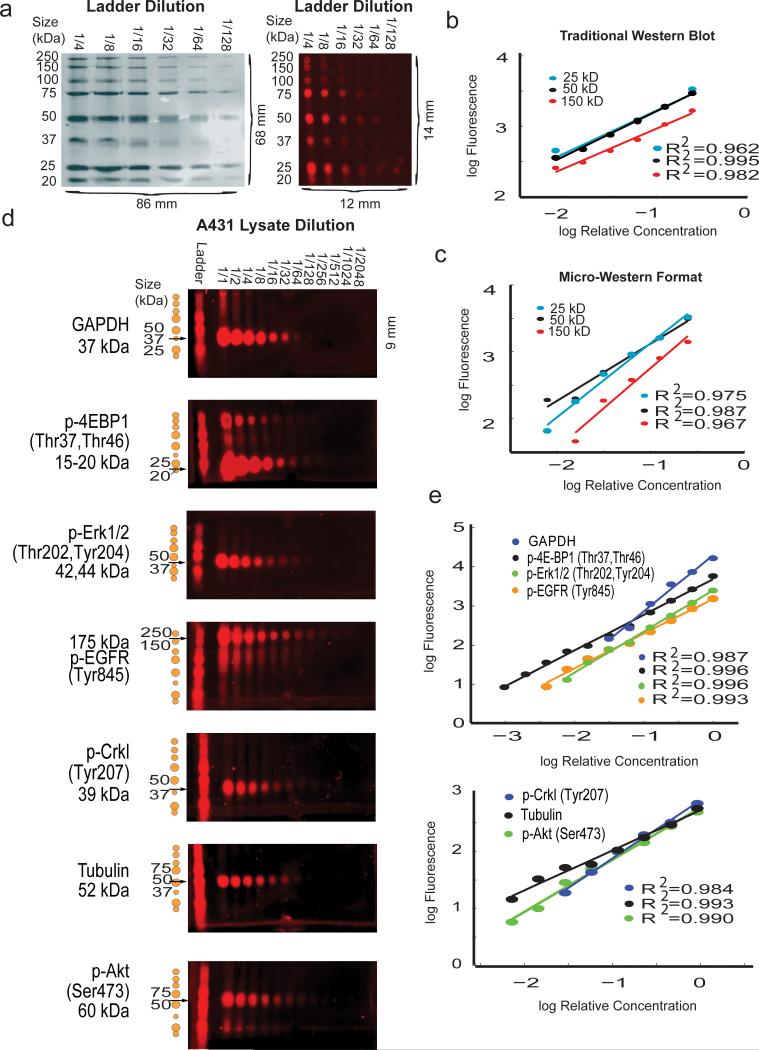
We compared the resolution and linearity in signal of MWAs with macroscopic gels using a labeled molecular weight standard (LI-COR 928-40000) (Fig. 2a). For proteins of 150, 50, and 25 kDa, the intensity of each ladder spot was proportional to the fold dilution over a range of two orders of magnitude for both methods (Fig. 2b,c). The coefficient-of-variation from arraying, rehydration, and transfer of a single band of the LI-COR ladder across the area of the membrane was < 9%.
Fig. 2. MWA Validation of Linear Response.
(a) (left) 5 μL of a series of 1/2 dilutions of LI-COR protein ladder were electrophoresed using traditional 10% SDS-PAGE. (right) 60 nl of the identical samples were run in the micro-western format. The difference in scale is noted below the figure. (b) The median net signal intensity was quantified for three bands (150 kDa, 50 kDa, and 25 kDa) of the LI-COR protein ladder in the traditional western blot as shown in part (a). The intensity vs. concentration of proteins displayed a linear relationship for all protein bands quantified. (c) The median net signal intensities quantified from micro-western dilution series. The intensity vs. concentration displayed a similar linear relationship as that from the traditional western. (d) 1/2 dilutions of lysates from A431 cells stimulated for 5 min with 200 ng ml-1 of EGF probed with seven rabbit primary antibodies directed at proteins spanning a range of molecular weights (175 kDa to 15 kDa) and detected with IR700-labeled secondary antibody. (e) The median net signal intensity for each band vs. relative concentration as shown in part (d). The graphs show a linear relationship between net fluorescence and concentration for all antibodies tested.
We then tested the linearity of signal response in quantifying proteins from A431 human carcinoma cell lysates using a two-stage fluorescent immuno-detection system (Fig. 2d,e). We used five phospho- and two pan-specific antibodies to analyze proteins from 15-175 kDa in EGF-stimulated A431 cells lysates. All MWAs showed a linear relationship between relative antigen concentration and signal intensity over their detectable range (from 100- to 1,000-fold). Assuming an expression level of 1.2 × 106 receptors per A431 cell11 EGFR was detectable down to one cell equivalent (~2 attomoles; ~340 femtograms). We assumed linearity for all further analysis.
Comparison of macro-western blots and micro-western arrays
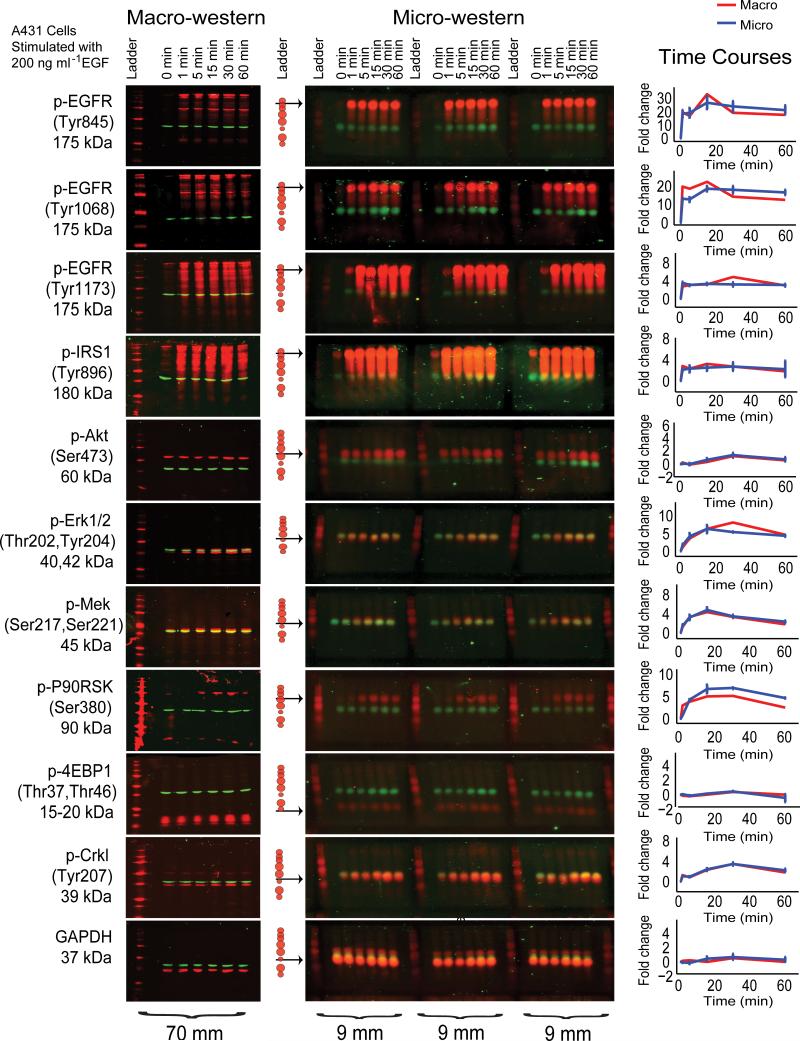
To compare performance of MWAs with macro-western blots for monitoring phosphorylation dynamics, we selected a representative test-set of 11 antibodies. Four were previously shown to generate equivalent quantitative data by RPAs and western blots4; another four were shown to result in substantial compression of dynamic range by RPAs due to antibody cross-reactivity4.
Measurements obtained by MWAs were similar to macro-western blots for all antibodies (Fig. 3) and did not display the dynamic range compression observed for RPAs. For many protein phosphosites, including EGFR, IRS1, and AKT, we observed bands at the predicted size as well as at additional sizes that could obscure quantitative measurements by RPAs. The precision in estimating sizes of proteins > 100 kDa by MWAs was ~10 kDa, and for smaller proteins ~5 kDa. While protein sizes could be determined with precision approaching that of a standard western blot, proteins were not completely resolved unless they differed by more than the following: 75 kDa for proteins greater than 200 kDa; 50 kDa for proteins between 100 and 200 kDa; 25kDa for proteins between 50 and 100 kDa; and 10 kDa for proteins less than 50 kDa (Fig. 2d, Fig. 3), corresponding to a migration distance of about 1.5 mm, double the diameter of spotted protein (Fig. 2d, Fig. 3). Resolution equal to a macro-western blot could be obtained by electrophoresing the samples for twice the distance (Fig. 2a).
Fig. 3. Comparison of MWA to traditional western blot.
A comparison of eleven traditional western blots is shown parallel to the identical samples run in triplicate in micro-western array format. Lysates from A431 cells stimulated with EGF (200 ng ml-1) and lysed at 0,1,5,15,30, and 60 minutes after stimulation along with the LI-COR protein ladder. β-actin monoclonal mouse primary antibody (detected with IR800 secondary antibody shown in green) was probed with each of the eleven rabbit primary antibodies (detected with IR700 secondary antibody shown in red) polyclonal antibodies to demonstrate equal loading of each sample. An arrow indicating the band quantified is indicated to the left of the blot along with the corresponding sizes of the LI-COR protein standard. Quantification of the fluorescence of the traditional western (red) is shown in comparison to the micro-western (blue). Error bars shown represent the standard error of the three technical replicates of the micro-westerns shown. Absolute sizes of the blots are indicated below to demonstrate the extent of the miniaturization of scale of the micro-western in comparison to the traditional western Blot.
Application of MWAs to analysis of EGFR signaling network
To examine EGF signaling dynamics using MWAs, we chose antibodies directed at a wide range of phosphosites to monitor many molecular biological processes (Supplementary Figure 1, Supplementary Table 1): 1) early positive growth factor response regulators; 2) negative signaling regulators; 3) downstream proliferation indicators; 4) nutrition sensing indicators; 5) adhesion and migration indicators; 6) phospho-lipid and calcium state indicators; 7) stress indicators; and 8) transcription and cell cycle indicators.
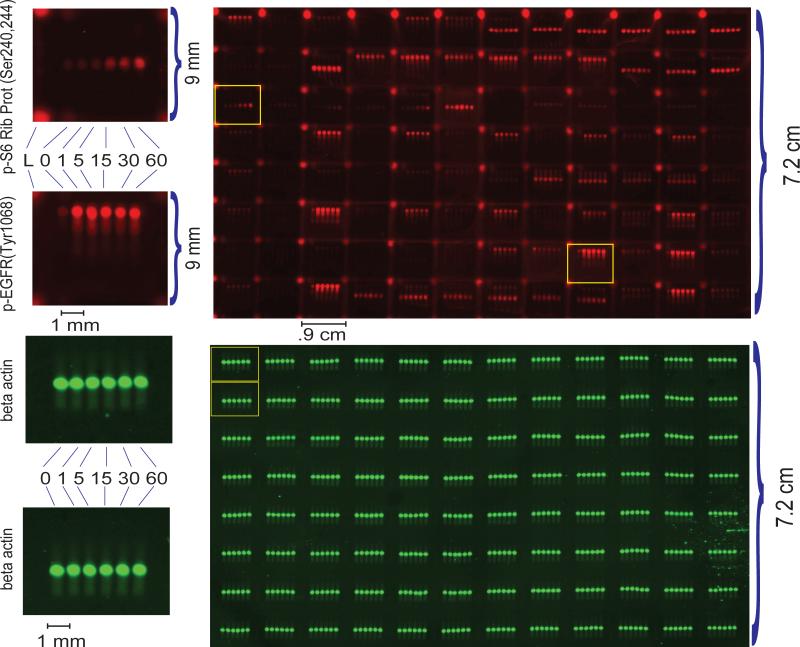
To observe signaling dynamics at doses approximating physiological levels12, we stimulated cells with 2, 50, 100, and 200 ng ml-1 EGF. A mock stimulation was performed to distinguish EGF-mediated signaling events from nutrition-related events. Figure 4 shows a MWA chip along with two magnified blocks demonstrating distinct temporal phosphorylation patterns of p-S6 Ribosomal Protein (Ser240,Ser244) and of p-EGFR (Tyr1068) (Supplementary Table 2). A mouse beta-actin antibody was used to probe all plate wells and demonstrates the reproducibility of the approach (Fig. 4, Supplementary Table 2). The coefficient-of-variation from arraying, rehydration, transfer, binding of primary antibody and secondary antibody was < 17%.
Fig. 4. An MWA containing 6 cell lysates probed with 192 antibodies.
The red channel (700 nm laser) shows the stimulation of A431 cells with 200 ng/ml EGF probed with a panel of rabbit anti-human polyclonal antibodies detected with IR700-labeled secondary antibodies. The green channel (800 nm laser) reflects a scan of the samples probed with mouse monoclonal antihuman β-actin antibody detected with IR800-labeled secondary antibodies to demonstrate the consistency of printing across the area of the membrane. For antibody layout, see Supplementary Table 2.
We quantified 91 phosphosites from 67 proteins and 18 pan-specific protein abundances for a total of 75 proteins in technical triplicate resulting in ~9,800 signaling observations with sufficient residual lysates for further analyses using different antibody panels. Intensity and inferred sizes from spots detected with each antibody were recorded along with signal-to-background ratio (Supplementary Table 1, Supplementary Figures 2, 3). 17 of 91 phosphosites quantified here were previously quantified in one recent MS report using pan-phospho enrichment7 and 22 in another13 employing phosphotyrosine-specific enrichment (Supplementary Table 3). Many ubiquitous EGFR signaling proteins quantified by MWAs, including p-EGFR(Tyr845), p-SHP2(Tyr542), p-p70S6K(Ser371), p-Raf(c-)(Ser338), p90RSK(Ser380), and p-Stat3(Ser727), were not quantified in either MS study, suggesting that MS detects only a fraction of phosphorylation events elicited by EGF. 4 of 91 phosphosites quantified here were quantified by RPA and demonstrated to generate equivalent information as a western blot3.
Comparison of signaling network at different EGF input levels
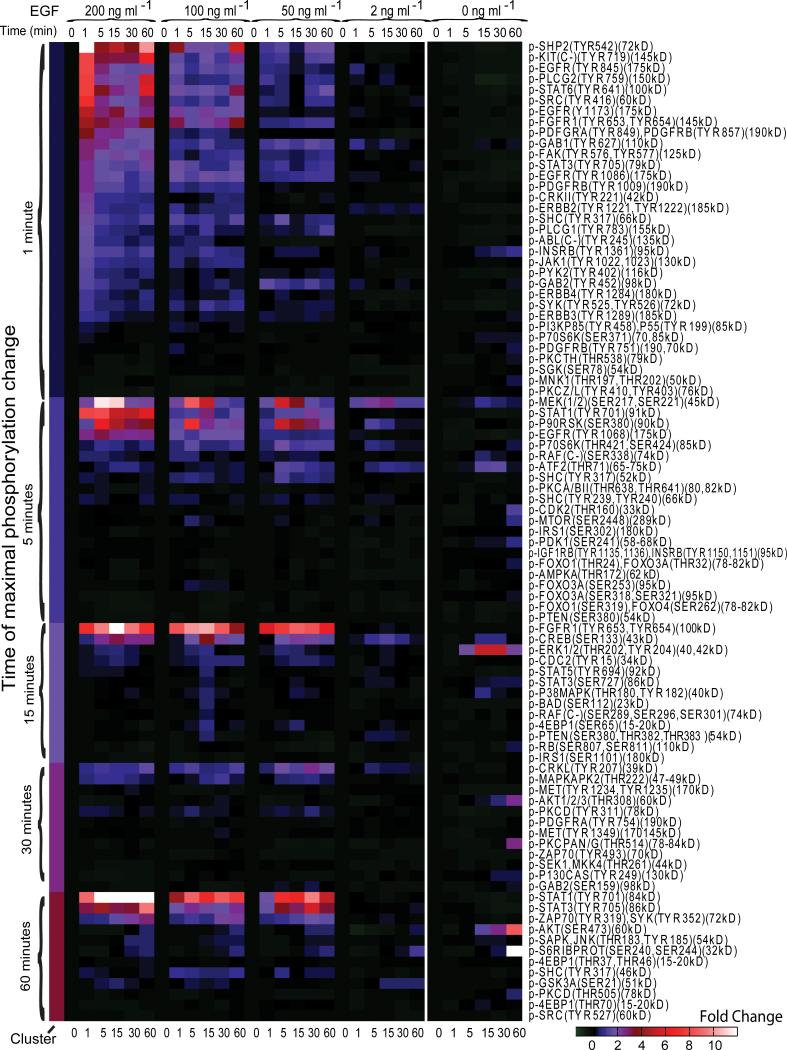
We next asked whether biological insights could be revealed using the MWA method. Five clusters of signaling profiles were organized based on time of maximal phosphorylation (Fig. 5). Phosphosites within clusters were rank ordered by fold-change. At the 2 ng ml-1 input level, we observed several phosphosites from EGFR, ErbB2, PLC gamma, Gab1, Mek, p90RSK, p70S6K, and Crkl that were absent in mock treatment (Fig. 5, Supplementary Figure 4, 5).
Fig. 5. A clustered heatmap profile of fold changes for antibody bands representing specific phosphorylation sites of proteins in A431 cells over six time points for four stimulation conditions and one 0 ng ml-1 control.
The net fold-change is color coded as indicated in the legend. Antibody bands are clustered into six clusters according to the time point (0, 1, 5, 15, 30, or 60 min) at which maximal fold change occurs. The antibodies are in descending order sorted in each cluster by the value of the fold change at the 200 ng ml-1 stimulation condition at the time point representative of that particular cluster. The antibody names are listed as given by the manufacturer followed in parentheses by an approximation of the size of each band. The EGF mock stimulation is shown on the right image block.
Conversely, many phosphosites related to phosphoinositide signaling displayed substantial fold-change in mock stimulation but not EGF treatment, including sites from PDK1 and its downstream targets AKT, PKC gamma, and PKC delta; downstream targets of AKT including mTOR, FOXO1; and mTor substrate p70S6K and its downstream target S6 Ribosomal Protein. We speculate that activation of PLC gamma following EGF stimulation leads to the hydrolysis of phosphatidylinositol 4,5-bisphosphate, causing down-regulation of PDK1 and AKT. Reduced AKT activity could produce the observed A431 cell cycle inhibition14 through decreased phosphorylation of cyclin dependent kinase (CDK) inhibitors, including p21CIP1(Thr145) and p27Kip1(Thr157). Consistent with this notion, Insulin-like Growth Factor (IGF), a potent PI3K stimulator, is also a potent mitogen for A431 cells12.
We then asked how the dynamic range and timing of the EGF signaling network were influenced by EGF input level. The first wave of phosphorylation peaking at the 1 minute point included 33 tyrosines from EGFR and other RTKs and membrane-localized proteins (Fig 5, Supplementary Figures 3-5, Supplementary Table 1). At 5 minutes, we observed serine and threonine sites from downstream kinases and transcription factors including Raf, MEK, p70S6 kinase, mTor, and ATF2. At 15 minutes, we observed phosphosites from Erk, P38 MAPK, and cell cycle-related kinases and substrates. Sites peaking at 30 minutes included those of the Crkl adapter protein and MAPKAPK2, a substrate of P38 MAPK. Proteins with sites peaking at 60 minutes included the PDK1 substrates AKT and PKCD, and the AKT substrate 4EBP1, among others. The timing of most phosphorylation events was not affected by EGF concentrations.
Bayesian network modeling of receptor layer connectivity
In order to try to elucidate the directional influences among phosphosites, we applied Bayesian network modeling approaches to the receptor layer of the EGF signaling network. This permitted us to verify known influences while identifying new directional relationships underlying receptor-level crosstalk. Bayesian networks are graphical representations of conditional independencies in a probability distribution over a set of variables15, and can potentially be inferred from experimental data such as that generated by MWAs. The network analyzed here comprised seventeen phosphosites: two from the Src kinase and fifteen from the ten RTKs for which fold-change measurements were observed specifically in all four EGF treatment input levels and for which antibodies were predicted to result in no cross-reactivity with each other (Fig. 6, Supplementary Table 4). Each time-point was considered as an independent sample of the EGF-stimulated network state, giving 20 samples for each phosphosite (4 conditions across 5 non-zero time points of one biological replicate). All data were normalized to the zero time point.
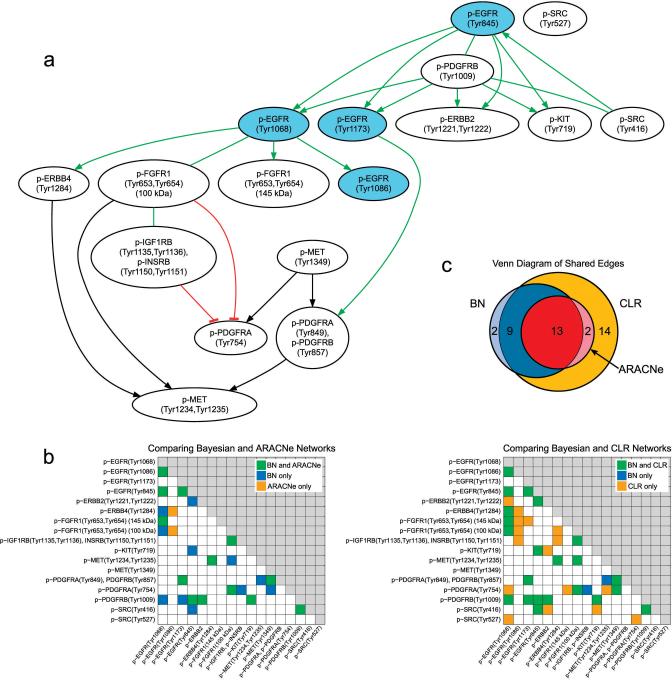
Fig. 6.
Consensus model of EGFR receptor level influences modeled by Bayesian network inference with comparison to ARACNe and CLR. (a) A consensus model of the EGF signaling network obtained by exact Bayesian model averaging following Bayesian network inference (Supplementary Note 1). Significant (p < 0.001) positive edges (green), significant (p < 0.05) negative edges (red blunt edges), and interactions with a non-significant correlation coefficient (black) are shown. Edges for which the directionality could not be determined using equivalence class analysis are shown as undirected. (b) Heatmaps show the undirected adjacency matrices comparing the Bayesian network to the ARACNe and CLR networks. An edge between node i and node j is represented by matrix value (i, j). Because the undirected networks are compared, the adjacency matrix is symmetric across the diagonal, and thus only the lower triangular matrix of the adjacency matrix is shown. Edge weight thresholds were set to > 0.3 for the Bayesian network and ARACNe (using ARACNe Data Processing Inequality parameter τ= 0.03) and to Z > 1.13 for CLR. Eight of 11 edges present only in the Bayesian network and not in the ARACNe network would induce three-node triplets in the ARACNe network, which is precisely what ARACNe is designed to prune out (see Supplementary Figure 8). (c) Venn diagram comparing edges across the three networks. The ARACNe network forms a complete subnetwork of the CLR network and a near complete subnetwork of the Bayesian network, which forms a near complete subnetwork of the CLR network.
Given typically limited amounts of data, a variety of graph structures can be generated by Bayesian inference modeling that describe the data reasonably well, so a consensus model is often sought rather than aiming to find a unique best-scoring graph15. Accordingly, we illustrate here (Fig. 6) a consensus model containing only edges with a score > 0.3, derived from exact Bayesian network model averaging over all directed acyclic graph (DAG) structures having at most three parents per node16,17. By considering only those DAG structures in the equivalence class of the consensus model with a directed edge from p-SRC(Tyr416) to p-EGFR(Tyr845), directionality of the remaining compelled edges was determined18 (Supplementary Note 1). Signs of directional influences (i.e. positive vs. negative) could also be discerned. EGFR(Tyr845) is a known Src kinase substrate which is not phosphorylated by the EGFR kinase19. This prior knowledge was only used to distinguish edge directionality in the equivalence class; no prior structural knowledge was used to derive the consensus model.
The three linked root nodes from which most downstream influences in the graph structure were derived included p-SRC(Tyr416), p-EGFR(Tyr845), and p-PDGFRB(Tyr1009). The model suggests that the EGFR and PDGFRA,B influence one another, with p-EGFR(Tyr1068), p-EGFR(Tyr1173), p-ERBB2(Tyr1221,Tyr1222), and p-KIT(C)(Tyr719) depicted directly downstream of both p-PDGFRB(Tyr1009) and p-EGFR(Tyr845). Notably, PDGFRB has previously been described to heterodimerize and transactivate the EGFR20 in response to PDGF, even in the presence of a PDGFR inhibitor. While others have previously suggested A431 cells lack PDGFR expression, we witnessed bands at the predicted molecular weights using several phospho- and pan-specific antibodies directed at the intracellular region of the receptor (Supplementary Figures 1, 6) as well as with RT-PCR using primers directed at the juxtamembrane region of PDGFR (data not shown).
Interestingly, p-PDGFRA(Tyr849),PDGFRB(Tyr857), the activation loop phosphosite of PDGFRA,B was depicted by the model to lie downstream of p-MET(Tyr1349), a root node, and p-EGFR(Tyr1173), which was downstream of the root nodes p-EGFR(Tyr845) and p-PDGFRB(Tyr1009). p-EGFR(Tyr1173) began to display robust phosphorylation at the 100 ng ml-1 EGF input level, the same level that the activation loop of PDGFRA,B began to display phosphorylation; at low EGF levels Src kinase may mediate the phosphorylation of a subset of PDGFR sites other than Tyr849,Tyr857, while at higher levels the PDGFR kinase itself becomes activated through a mechanism involving or concurrent with the phosphorylation of p-EGFR(Tyr1173).
p-EGFR(Tyr1068), modeled to be upstream of p-EGFR(Tyr1086), p-ERBB4(Tyr1284) and both p-FGFR1(Tyr653,Tyr654) activation loop isoforms, was distinct among EGFR sites in displaying maximal phosphorylation at 5 minutes and sustained phosphorylation amplitude for the duration of the time course. The edge directed from p-EGFR(Tyr1068) to p-FGFR1(Tyr653,Tyr654)(145 kDa) displayed a relatively high edge score (.80; see Supplementary Figure 7 for all consensus Bayesian network edge weights), similar to that displayed between p-EGFR(Tyr1068) and p-EGFR(Tyr1086) (which displayed an edge score of .89), suggesting that EGFR is able to mediate the activation of FGFR1. We speculate that the 145 kDa and 100 kDa forms of FGFR1 represent hyper- and hypoglycosylated forms of the receptor, respectively. Hyper-glycosylation of FGFR1 has been shown to inhibit its interaction with both FGF2 and heparin-derived oligosaccharides21, which was predicted to decrease its activity. Only the short form phosphosite was modeled to have subsequent downstream targets. The only site negatively regulated in the model was p-PDGFRA(Tyr754), which is known to recruit the SHP2 phosphatase22 resulting in dephosphorylation of RASGAP recruitment sites on PDGFRA and B and increased MAPK signaling. Therefore down-regulation of p-PDGFRA(Tyr754) would be predicted to decrease MAPK signaling. Consistent with previous reports, the model suggests that p-SRC(Tyr527), a known inhibitory site of Src kinase, is disconnected from the EGF network23.
To corroborate the Bayesian network results, we also inferred network connectivities using ‘Algorithm for the Reconstruction of Accurate Cellular Networks’ (ARACNe) and ‘Context Likelihood of Relatedness’ (CLR) algorithms24,25. 22 of 24 edges in the Bayesian network were also identified by ARACNe and/or CLR, though as undirected edges since these latter methods are based on mutual information notions (Fig. 6b,c, Supplementary Figures 8, 9). Consistency across network inference methods has been shown experimentally to characterize accurate edge inference26,27, and in our case here data permutation studies show that the topologies inferred by the Bayesian network, ARACNe, and CLR are significant (Supplementary Note 1 and Supplementary Figure 10). Because in the context of proteomic signaling networks it is problematic to make broad assumptions about edge directionality absent extensive prior knowledge (e.g., concerning particular kinase-substrate relationships), we believe that predicting edge directionality using methods such as Bayesian network modeling offers appealing advantage.
DISCUSSION
In contrast to RPAs, MWAs have the ability to reduce the complexity of lysates after arraying, minimizing effort in experimental scale-up. Most of the information of a traditional western blot can be obtained, using 200-fold less protein and antibody. MWAs should be useful for analysis of proteins from cell lines and tissues where lysates are sufficient to print hundreds of MWAs that could be distributed en masse in an analogous manner to spotted DNA microarrays for interrogation with the user's choice of antibodies. The only devices required following printing are commercially available 96-well gaskets and an imager. The ability to obtain information regarding hundreds of proteins with the MWA method should allow advances in our understanding of cell-context-specific networks underlying human disease when combined with appropriate computational modeling methods.
MWAs could also be very useful for large-scale, systematic validation of antibodies. Antibody collections could be systematically verified for selectivity by examining lysates from cells transfected with a cDNA or depleted for the cognate protein by RNAi. The amount of antibody obtained from a single rabbit immunization (~5 mg) would be sufficient for over 100,000 MWAs, thus minimizing lot-tolot variability of polyclonal antibodies. MWAs could be useful for current efforts to build a human protein atlas; samples from tissues used for in situ analyses could be examined with MWAs to verify that signals observed with each antibody resulted from proteins of the predicted molecular weight(s).
The ability to gather dynamic information regarding hundreds of proteins under many conditions poses new challenges for computational modeling. The Bayesian network described here represents direct and/or indirect effects of a given node on other nodes as indicated by high-probability connecting arcs, which are hypothesized to represent relationships of influence among the phosphoproteins in the network. Using prior knowledge to restrict edge directionality across a Bayesian network equivalence class, one can bolster the case for assigning directionality to these edges. To further support a case for interpreting network connections as causal, one could explicitly model the temporal data28 and/or use interventional data15,17 which will be the source of future inquiry.
The timing and amplitude of phosphorylation dynamics observed here coupled with the connectivities modeled in the Bayesian network suggest several candidate sources of RTK-coactivation, each of which may be important in specific cancer contexts: 1) direct dimerization and/or phosphorylation by EGFR or other downstream tyrosine kinases as suggested by the rapid phosphorylation kinetics of Src, ErbB2 and 4, coupled with their close proximity at the top of the network; 2) activation of proteases that activate precursor growth factors or latent RTKs as might be predicted from the delayed phosphorylation amplitudes of FGFR1(100kDa) and MET activation loop sites coupled with their distance from EGFR in the network and; 3) inactivation of tyrosine phosphatases through oxidation by reactive oxygen species29. Phosphorylation of Tyr542 of Shp2 phosphatase displayed the highest fold-change of any site in our analysis; this site has been suggested to relieve inhibition of phosphatase30 activity. The sustained phosphorylation of this and other tyrosine sites at EGF levels ≥ 50 ng/ml suggests that it (and other cysteine-based tyrosine phosphatases) may be inactivated thus unmasking many tyrosine kinase activities. Each of these mechanisms may play distinct roles in the context of cancers that have become resistant to single kinase inhibitors; systems level analysis of other tyrosine kinase-driven cancers may be helpful in revealing appropriate therapeutic targets.
Supplementary Material
EdSumm AOP
Micro-western arrays combine the advantages of scalability of reverse phase protein arrays and the information content of western blotting, for analyzing protein abundance and modification state with high sensitivity and throughput. The method is demonstrated for analyzing phosphorylation state changes in the EGF receptor signaling network using Bayesian network modeling.
EdSumm issue
Micro-western arrays combine the advantages of scalability of reverse phase protein arrays and the information content of western blotting, for analyzing protein abundance and modification state with high sensitivity and throughput. The method is demonstrated for analyzing phosphorylation state changes in the EGF receptor signaling network using Bayesian network modeling.
ACKNOWLEDGEMENTS
We thank C.Y. Chuang for technical assistance with MWAs. We thank M. R. Rosner and K. P. White, and W. L. McKeehan for helpful discussions during the course of this work. We thank C. May for the graphic design of Fig. 1. We thank J. Barkinge for operating support with micro-arraying. This work was supported, in part, by awards from the The University of Chicago Cancer Research Center, The American Cancer Society, The University of Chicago Breast Cancer Specialized Program of Research Excellence, The Cancer Research Foundation, and The Illinois Department of Health #86280156 to R.B.J., the National Institutes of General Medical Sciences P50-GM0686762 Cell Decision Processes Center grant and National Cancer Institute CA96504 to D.A.L., M.F.C. was supported by a National Institutes of Health Systems Biology of Oxygen Predoctoral Training Grant, J.P.W. was supported by a National Science Foundation Graduate Fellowship, and C.P.C. was supported by a National Institutes of Health Cancer Biology Postdoctoral Training Grant.
ONLINE METHODS
Cell stimulation and lysis
A431 cells were a gift from Dr. Shutsung Liao. Cells were passaged three times after thawing from frozen stocks. For the final passage, plates at 80% confluence were split 1:10 into fresh plates (6 plates per EGF concentration tested). 0.7 mL of Trypsin-EDTA was incubated with cells for seven minutes at 37 deg C. Newly plated cells were grown in 8% fetal bovine serum, 0 I.U. penicillin, and 50 μg/ml streptomycin, and the media was replaced with serum-free media when they reached 50% confluence. Cells were maintained in serum free DMEM with penicillin and streptomycin for 48 hrs. A stock solution of EGF was prepared at 200 μg/ml and was applied at the appropriate dilution in serum-free media after washing twice with PBS. At each time point the cells were again washed twice with PBS (see Supplementary Note 2 for all recipes). 1.0 ml of Lysis Buffer was added and the cells scraped into a 2 mL microcentrifuge tube. After lysis, the samples were boiled for ten minutes and then frozen at -80 deg C. The following day, the cells were sonicated at Power 5 on a Sonic Cell Disruptor for ten seconds, alternating one second on, one second off (total 20 seconds time). The samples were boiled for 2 minutes and 500 μL were immediately placed in a micro-concentration device with a 10 kDa molecular weight cutoff. The tubes were spun at 12000 G for approximately ten minutes until the volume was reduced by 5X. Each sample was then topped off to 100 μl with lysis buffer so that each sample was of equal volume. Samples were divided into 20 μl aliquots and frozen at -80 deg to avoid repeated freeze-thaw cycles until printing. Samples were boiled for 2 minutes immediately before printing. Product Numbers are provided in Supplementary Table 5.
Gel fabrication
Glass casting plates (one measuring 14 × 27 cm, the other measuring 14 × 28 cm) were sprayed with BlueSlick and wiped thoroughly with a paper towel to provide an even layer of solution over the entire side of the glass plate. Rubber spacers were placed on three sides of the inner coated sides of the glass plate (two long sides and one short side) making sure there were no gaps between the spacers on the corner (as this would be a potential source of acrylamide leakage). One rectangle of Netfix was placed on the glass plate on top of the spacers. The second glass plate was placed on top with the coated surface facing down, leaving a small lip (about 1 cm) to inject the unpolymerized gel. 12 clamps were placed around the three gasketed edges of the sandwich.
Gel reagents (Supplementary Note 2) were gently mixed to avoid bubble formation in a 50 ml conical tube. One corner of the sandwiched plates was placed on an object about three inches high so that gel could run at a slant in two dimensions. A 30 ml syringe with a 19 gauge needle was used to inject the gel mixtures so that it ran down the length of one long side, then along the length of the bottom short side, and then up the second length in order to avoid any trapped air. After the sample was poured, the sandwich was laid horizontally during polymerization to avoid leakage. After one hour, the clamps were removed and the top plate was pried off with a plastic wedge. The gel was removed from the bottom glass plate by lifting manually from the plate and placed between two plastic Gelfix sheets.
A standard paper cutter was used to remove the Netfix border and then used to divide the gel into two parts (with each half measuring at least 11.5 cm wide in each dimension). The gels were saved in vacuum sealed bags and refrigerated at 4 deg C prior to use. The performance was not found to differ substantially following storage for several weeks.
Micro-arraying
The lysates were spotted using a non-contact microarrayer (Gesim Nanoplotter 2.1 E) with active humidification. The printing performance of each tip was validated with a stroboscope prior to beginning each microarray print. If printing was inconsistent, 200 μL of 50% methanol/50% HCl was loaded into the tip and dispensed three times using manual mode. The tips were washed for sixty seconds using the Wash/Dry cycle and rechecked with the stroboscope. The Gelfix sheet was removed from the gel and the gels prehydrated in hydration buffer (100 ml) containing 1 ml 1M sodium bisulfite and 1 ml 1M DTT) for five minutes. Excess fluid was blotted off with a piece of cardboard.
To keep the gel and samples hydrated, the relative humidity was maintained at 80%. Z-height measurements were taken prior to the print of each gel. Micro-western arrays were printed onto two gels per array run. Tip dispense height was held at 1.5 mm above the gel surface while printing.
Samples were placed with the ladder in well A1 of a 384 well plate and samples consecutively in A2-A7. Software NP2.15.46 was used. The TransferTipMultiSim04H9 (Gesim) was run using the transfer text and the workplate definition file provided in Supplementary Note 3.
LI-COR protein ladder was printed in lane 1 at a 1:2 dilution in lysis buffer. This ladder is printed at high concentration so that it may be visible on the nitrocellulose membrane after transfer to facilitate alignment of the gasket.
Following print completion, the gel was maintained for an additional ten minutes in the humidified environment to insure that all droplets were absorbed beneath the gel surface. The gel was subsequently rehydrated for five in the rehydration buffer described above with gentle agitation. Following rehydration, the gel was placed onto the Genephor for horizontal semidry electrophoresis.
Horizontal Semidry Electrophoresis
Samples were separated by size using a horizontal electrophoresis device pre-chilled to 10 deg C. Three drops of kerosene were added on to the surface of the Genephor and spread with a Kimwipe to provide a homogenous layer on the surface. A clean piece of Gelfix measuring 14×14 cm was added to the surface of the Genephor. The hydrated gel was placed on top of the plastic sheet covering the area of both electrode rectangles. Three filter wicks were cut in half and each half stacked three deep. Each stack was placed in electrode buffer (Supplementary Note 2) for several seconds and air was manually pressed out of them while submerged. The first (bottom) lid of the Genephor was placed over the gel making sure that there was no fluid making contact with the lid from the gel. The two stacks of hydrated wicks were placed in the center of each electrode slat on the gel. The second and third (upper) lids of the Genephor were then closed. The power supply was set at 350V, 30 watts, and unlimited amps. The lowest molecular weight ladder bands migrated about 9 mm (the length of one well of a 96-well plate) in 12 minutes.
Transfer
Following electrophoresis, the gel was placed protein side down onto nitrocellulose pre-moistened in transfer buffer (Supplementary Note 2). Filter paper was placed on either side of the nitrocellulose and gel and clamped in a transfer box cartridge. All bubbles were pressed out with a roller. The gels were transferred either at 0.8 amps for 60 min or 0.15 amps overnight at 4 deg in a Criterion transfer box with plate electrode. The box was pre-chilled to 4 deg and kept in a cold room with gentle agitation to maintain a constant temperature.
Blotting
Nitrocellulose was removed from the transfer apparatus and washed for five minutes in TBS (without Tween 20) to remove methanol. The blots were blocked for 1 hour in Odyssey Blocking Buffer. The blot was aligned on the gasket by placing the visible ladder on the vertical lines and centering the ladder between the horizontal lines. The gasket was clamped into the 96-well isolation device and primary antibodies were pipetted into the appropriate wells, making sure that the membrane stayed wet during the process. The primary antibody was diluted in pure Odyssey blocking buffer (without Tween 20) overnight. 150 μl of diluted antibody was added per well. After incubation, the wells were washed four times with 200 μl of TBST per well using a multichannel pipettor. Goat anti-rabbit Alexa 680 conjugated secondary, goat anti-rabbit, and goat anti-mouse IR800-conjugated secondary antibodies (1:5000) were diluted in 20% Odyssey blocking buffer, 80% TBS (without Tween 20). 150 μl of the diluted secondary was added to the appropriate well. After incubation for an hour, the blot was washed three times with 200 μl TBST while clamped in the gasketing device. The blot was then removed from the gasket, placed in a box top and washed for an additional five minutes in TBST. For the fifth wash, TBS without Tween 20 was used, washing for five minutes. The membrane was completely dried using pressurized air and scanned using the LI-COR Odyssey imager at 24 micron resolution and high quality (Laser Intensity 1.0 on the 700 channel, Laser Intensity 2.0 on the 800 channel) settings.
Analysis
Scanned images were saved for analysis as 16-bit tiff files. Genepix 8.0 was used to record the mean by drawing an equally-sized circle around the appropriately sized band for each sample. Appropriate size is defined as within 10 kD of the size as defined by the antibody product sheet as measured in comparison to the LI-COR ladder bands. All bands within this region that were visible were recorded. Bands outside this region were noted but the intensities were not recorded or analyzed. The background fluorescence was recorded by placing an equal sized circle in the blank space to the left of the first sample (not covering sample or ladder space) and the minimum value of this circle was recorded. Net intensity was calculated by subtracting each sample intensity from the background. To normalize sample concentration, the net intensities were divided by a simple mean of the net intensities for GAPDH, Tubulin, and Actin calculated separately for each array print. To calculate fold change, each normalized net intensity was divided by the value at the zero minute time point and subtracted by one. Graphs, heatmaps, and clustergrams were generated using MATLAB 2007b.
Signaling network inference modeling
Bayesian networks were modeled using a dynamic programming algorithm that computes the exact marginal posterior probability of edges in the Bayesian network derived from the data set16. The algorithm was implemented using a modified version of the open-source Bayesian Network Structure Learning toolbox in MATLAB17. Node conditional probability distributions were represented by multinomials using a uniform Dirichlet prior with equivalent sample size of one, and a prior over graph structures was calculated by accounting for the number of ways to choose parents sets in a graph, as previously described31,16. Networks were scored using the Bayesian Dirichlet likelihood equivalent uniform (BDeu) score32. The BDeu score accounts for both model fit and complexity, and thus avoids overfitting the data. Although this dynamic programming algorithm introduces a non-uniform prior over graph structures, it has been shown to perform better at structure learning tasks16,17 than local search methods that use a uniform prior over graph structures, such as Markov Chain Monte Carlo searches over directed acyclic graphs33, as well as MCMC searches over node orderings, which uses a non-uniform prior31.
All nodes were discretized using three-level k-means clustering to indicate low, medium, and high phosphoprotein levels (Supplementary Table 6). Clustering was done using the squared Euclidean distance metric and repeated 50 times for each node to find the optimal clustering assignments. It is believed that by using k-means clustering, we are better representing the physiological diversity in signaling states of the phosphoproteins in the network, compared to more arbitrary discretization schemes, like interval and quantile discretization, that do not try to explicitly capture clusters in the data.
CLR was implemented using MATLAB code provided by the original authors, with Z scores (edge weights) calculated as previously described25. ARACNe was implemented using the minet package in R34. To minimize the sources of variation between algorithms, the same discretized data that was used to learn the Bayesian network model was also used to learn the CLR and ARACNe models. The mutual information matrix for ARACNe was calculated using a simple histogram method in the minet package, and for CLR was calculated directly from the discretized data. The edge score thresholds for CLR and ARACNe were varied in an effort to maximize the similarity between the Bayesian network and ARACNE (or CLR), both given the > 0.3 edge weight threshold for the Bayesian network (Fig. 6) and when this constraint on the Bayesian network edge weight threshold was removed, though in both cases staying within edge weight thresholds that gave significant network results (see Supplementary Note 1 and Supplementary Figs. 8-10).
The sign of the influences between nodes in the Bayesian network were estimated using pairwise correlation coefficients. 17 of 24 pairwise interactions had a highly significant (p < 0.001) positive correlation coefficient. 2 of 24 had a significant (p < 0.05) negative correlation coefficient. The remaining 5 of 24 pairwise interactions had a non-significant (p > 0.05) correlation coefficient, but were edges in two- or three-parent interactions, suggesting a simple pairwise correlation coefficient was not sufficient to capture the parent-child behavior. Interestingly, both negative interactions were directed at p-PDGFRA(Tyr754). Five of the six two-parent interactions (including all four with p-EGFR(Tyr845) and p-PDGFRB(Tyr1009) as the parent set) were consistent with ‘AND gate’ behavior. The parent-child raw data from all one-, two-, and three-parent interactions are plotted versus one another in Supplementary Figure 11.
Considering up to three parents per node in the Bayesian network captured almost all higher-order interactions in the data set (see Supplementary Note 1 and Supplementary Figure 12). While additional higher-order interactions may be present but there are simply not enough data for the Bayesian network to infer them, it may also be that such higher-order interactions are indeed not present, regardless of how much data are available describing the network. ARACNe and CLR, which only consider undirected pairwise interactions, thus represent useful, but likely not complete, approximations of interactions in this data set.
Footnotes
Statement of Author Contributions
C.P.C., M.F.C., and R.B.J, designed the experiments. C.P.C. and M.F.C. performed the cell culture, and growth factor stimulations. M.F.C., and R.B.J. designed the micro-western array method, M.F.C. carried out micro-western experiments and organized the data into heat maps. J.P.W. and D.A.L. performed Bayesian network, CLR, and ARACNe analysis of the data. M.F.C., J.P.W., D.A.L, and R.B.J. wrote the manuscript. All authors read and revised the manuscript.
References
- 1.Burnette W. Western Blotting”: Electrophoretic transfer of proteins from sodium dodecyl sulfate polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Analytical Biochemistry. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- 2.Paweletz CP, Liotta LA, Petricoin EF. New technologies for biomarker analysis of prostate cancer progression: Laser capture microdissection and tissue proteomics. Urology. 2001;57:160–3. doi: 10.1016/s0090-4295(00)00964-x. [DOI] [PubMed] [Google Scholar]
- 3.Paweletz CP, et al. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene. 2001;20:1981–9. doi: 10.1038/sj.onc.1204265. [DOI] [PubMed] [Google Scholar]
- 4.Sevecka M, MacBeath G. State-based discovery: a multidimensional screen for small-molecule modulators of EGF signaling. Nature Methods. 2006;3:825–31. doi: 10.1038/NMETH931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sachs K, Perez O, Pe'er D, Lauffenburger DA, Nolan GP. Causal Protein-Signaling Networks Derived from Multiparameter Single-Cell Data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- 6.Rikova K, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 7.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–48. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 8.Wolf-Yadlin A, et al. Effects of HER2 overexpression on cell signaling networks governing proliferation and migration. Molecular systems biology. 2006;2:54. doi: 10.1038/msb4100094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tibes R, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Molecular Cancer Therapeutics. 2006;5:2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 10.Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168–74. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- 11.Sunada H, Magun BE, Mendelsohn J, MacLeod CL. Monoclonal antibody against epidermal growth factor receptor is internalized without stimulating receptor phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 1986;83:3825–3829. doi: 10.1073/pnas.83.11.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gill GN, Lazar CS. Increased phosphotyrosine content and inhibition of proliferation in EGF-treated A431 cells. Nature. 1981;293:305–307. doi: 10.1038/293305a0. [DOI] [PubMed] [Google Scholar]
- 13.Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:5860–5. doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang F, et al. Involvement of PI3K//Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: a target for cancer chemotherapy. Leukemia. 2003;17:590–603. doi: 10.1038/sj.leu.2402824. [DOI] [PubMed] [Google Scholar]
- 15.Pe'er D. Bayesian Network Analysis of Signaling Networks: A Primer. Sci. STKE. 2005;2005pl4 doi: 10.1126/stke.2812005pl4. [DOI] [PubMed] [Google Scholar]
- 16.Koivisto M, Sood K. Exact Bayesian Structure Discovery in Bayesian Networks. J. Mach. Learn. Res. 2004;5:549–573. [Google Scholar]
- 17.Eaton D, Murphy K. Exact Bayesian structure learning from uncertain interventions.. Proc. 12th Conf. on AI and Stats; 2007.pp. 107–114. [Google Scholar]
- 18.Chickering DM. Learning equivalence classes of bayesian-network structures. J. Mach. Learn. Res. 2002;2:445–498. [Google Scholar]
- 19.Downward J, Parker P, Waterfield MD. Autophosphorylation sites on the epidermal growth factor receptor. Nature. 1984;311:483–5. doi: 10.1038/311483a0. [DOI] [PubMed] [Google Scholar]
- 20.Saito Y, Haendeler J, Hojo Y, Yamamoto K, Berk BC. Receptor Heterodimerization: Essential Mechanism for Platelet-Derived Growth Factor-Induced Epidermal Growth Factor Receptor Transactivation. Mol. Cell. Biol. 2001;21:6387–6394. doi: 10.1128/MCB.21.19.6387-6394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duchesne L, Tissot B, Rudd TR, Dell A, Fernig DG. N-Glycosylation of Fibroblast Growth Factor Receptor 1 Regulates Ligand and Heparan Sulfate Co-receptor Binding. Journal of Biological Chemistry. 2006;281:27178–27189. doi: 10.1074/jbc.M601248200. [DOI] [PubMed] [Google Scholar]
- 22.Ekman S, Kallin A, Engström U, Heldin C, Rönnstrand L. SHP-2 is involved in heterodimer specific loss of phosphorylation of Tyr771 in the PDGF beta-receptor. Oncogene. 2002;21:1870–1875. doi: 10.1038/sj.onc.1205210. [DOI] [PubMed] [Google Scholar]
- 23.Gould KL, Hunter T. Platelet-derived growth factor induces multisite phosphorylation of pp60c-src and increases its protein-tyrosine kinase activity. Mol. Cell. Biol. 1988;8:3345–3356. doi: 10.1128/mcb.8.8.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Margolin AA, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faith JJ, et al. Large-Scale Mapping and Validation of Escherichia coli Transcriptional Regulation from a Compendium of Expression Profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor RC, Acquaah-Mensah G, Singhal M, Malhotra D, Biswal S. Network Inference Algorithms Elucidate Nrf2 Regulation of Mouse Lung Oxidative Stress. PLoS Comput Biol. 2008;4:e1000166. doi: 10.1371/journal.pcbi.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cantone I, et al. A Yeast Synthetic Network for In Vivo Assessment of Reverse-Engineering and Modeling Approaches. Cell. 2009;137:172–181. doi: 10.1016/j.cell.2009.01.055. [DOI] [PubMed] [Google Scholar]
- 28.Husmeier D. Sensitivity and specificity of inferring genetic regulatory interactions from microarray experiments with dynamic Bayesian networks. Bioinformatics. 2003;19:2271–2282. doi: 10.1093/bioinformatics/btg313. [DOI] [PubMed] [Google Scholar]
- 29.Lou YW, et al. Redox regulation of the protein tyrosine phosphatase PTP1B in cancer cells. The FEBS journal. 2008;275:69–88. doi: 10.1111/j.1742-4658.2007.06173.x. [DOI] [PubMed] [Google Scholar]
- 30.Lu W, Shen K, Cole PA. Chemical dissection of the effects of tyrosine phosphorylation of SHP-2. Biochemistry. 2003;42:5461–5468. doi: 10.1021/bi0340144. [DOI] [PubMed] [Google Scholar]
- 31.Friedman N, Koller D. Being Bayesian about Bayesian network structure: A Bayesian approach to structure discovery in Bayesian networks. Mach. Learn. 2003;50:95–125. [Google Scholar]
- 32.Heckerman D, Geiger D, Chickering DM. Learning Bayesian networks: The combination of knowledge and statistical data. Machine Learning. 1995;20:197–243. [Google Scholar]
- 33.Madigan D, York J. Bayesian graphical models for discrete data. Int. Stat. Rev. 1995;63:215–232. [Google Scholar]
- 34.Meyer P, Lafitte F, Bontempi G. minet: A R/Bioconductor Package for Inferring Large Transcriptional Networks Using Mutual Information. BMC Bioinformatics. 2008;9:461. doi: 10.1186/1471-2105-9-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
EdSumm AOP
Micro-western arrays combine the advantages of scalability of reverse phase protein arrays and the information content of western blotting, for analyzing protein abundance and modification state with high sensitivity and throughput. The method is demonstrated for analyzing phosphorylation state changes in the EGF receptor signaling network using Bayesian network modeling.
EdSumm issue
Micro-western arrays combine the advantages of scalability of reverse phase protein arrays and the information content of western blotting, for analyzing protein abundance and modification state with high sensitivity and throughput. The method is demonstrated for analyzing phosphorylation state changes in the EGF receptor signaling network using Bayesian network modeling.