SUMMARY
Autophagy is a homeostatic process involved in the bulk degradation of cytoplasmic components including damaged organelles and proteins. In both genetic and dietary models of obesity, we observed a severe downregulation of autophagy, particularly in Atg7 expression levels in liver. Suppression of Atg7 both in vitro and in vivo resulted in defective insulin signaling and elevated ER stress. In contrast, restoration of the Atg7 expression in liver resulted in dampened ER stress, enhanced hepatic insulin action and systemic glucose tolerance in obese mice. The beneficial action of Atg7 restoration in obese mice could be completely prevented by blocking a downstream mediator, Atg5, supporting its dependence on autophagy in regulating insulin action. Our data demonstrate that autophagy is an important regulator of organelle function and insulin signaling and loss of autophagy is a critical component of defective insulin action seen in obesity.
INTRODUCTION
Autophagy is a highly regulated process involved in the turnover of long-lived proteins, cytosolic components or damaged organelles (Yorimitsu and Klionsky, 2005). As a putative adaptive catabolic process, autophagy could generate energy for cells under nutrient-poor conditions or during starvation and help maintaining cellular homeostasis in nutrient-rich environments through its constitutive activity (Lum et al., 2005; Yorimitsu and Klionsky, 2005). In fact, impairment of this process has been implicated in a number of human diseases (Levine and Kroemer, 2008).
Obesity represents a condition where the proper sensing and management of nutrients and energy status presents significant challenges at the cellular and organismic levels. The organism cannot adapt and maintain homeostasis under continuous energy and nutrient exposure and the consequent emergence of metabolic and oxidative stress leads to inflammation responses and organelle dysfunction (Hotamisligil, 2006, 2010). Chronic exposure to high energy and nutrient intake increases the demand on the cellular synthetic and degradation machinery in tissues such as liver, adipose tissue, and pancreas, all of which are central to systemic metabolic homeostasis. Several observations led us to explore a link between obesity and alterations in autophagy. First, obesity is a condition of continuous and excess energy and nutrient flow into the system that would challenge this adaptive response. Second, ineffective macromolecule turnover, such as lipids or glycogen, could compromise hepatic metabolic function and promote defective insulin action. Third, there are suggested links between autophagy and immune response (Levine and Deretic, 2007) which exhibits abnormalities in obesity. Fourth, autophagy might be beneficial for cells to dispose of damaged cell structures caused by these stresses, and defective autophagy may lead to the failure of restoration of cellular homeostasis, including organelle function, thus exacerbating insulin resistance and possibly other metabolic pathologies associated with obesity. This may be particularly relevant to the endoplasmic reticulum (ER) function, since obesity is characterized by ER stress (Gregor and Hotamisligil, 2007). It has been suggested that ER may be a source of the membranes during the formation of autophagic vesicles (Axe et al., 2008; Razi et al., 2009) and experimental ER stress can induce autophagy in mammalian cells (Ogata et al., 2006), with several canonical UPR (unfolded protein response) pathways implicated in this interaction (Kouroku et al., 2007; Ogata et al., 2006). It has also been postulated that ER stress-induced autophagy may have evolved as a mechanism to dispose misfolded proteins that cannot be degraded by ER-associated proteosomal degradation (ERAD) and consequently assist ER homeostasis (Ding and Yin, 2008). Interestingly, two recent studies (Ebato et al., 2008; Jung et al., 2008) independently demonstrated the importance of autophagy in the preservation of pancreatic β cell function. Since beta cell function is also influenced by ER stress and defective insulin secretion is critical in obesity-induced insulin resistance and diabetes (Lipson et al., 2006; Scheuner and Kaufman, 2008), these findings also support a role for autophagy in metabolic homeostasis. Finally, autophagy is regulated by the integrated actions of insulin and mTOR (Ellington et al., 2006; Mammucari et al., 2007; Xie and Klionsky, 2007), both of which are altered in obesity. Hence, dysregulation of autophagy could be a critical component of obesity and contribute to metabolic dysfunction triggered by this condition including the emergence of ER stress responses. In this study, we set out to examine the autophagic response in obesity and whether this process is involved in the emergence of insulin resistance and type 2 diabetes associated with positive energy balance.
RESULTS
Downregulation of Hepatic Autophagy in Obesity
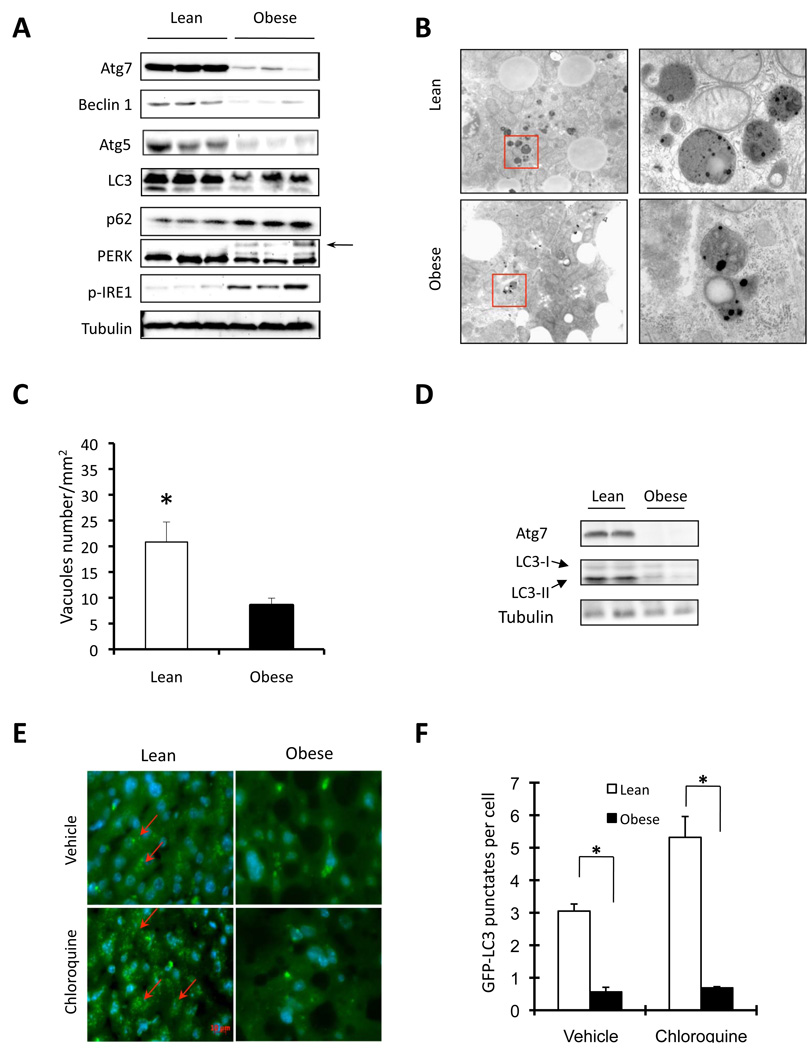
To study the regulation of autophagy in obesity, we first examined the expression patterns of several molecular indicators of autophagy in both genetic (ob/ob) and dietary (high-fat diet–induced, HFD) models of murine obesity. Surprisingly, we found that obesity resulted in markedly decreased autophagy indicators in liver of both genetic and dietary models, as evidenced by downregulation of LC3, Beclin 1 (also called Atg6), Atg5 and in particular, Atg7 protein levels (Fig. 1A and Suppl Fig. S1A). In contrast, p62 (also called SQSTM1, which is involved in aggresome formation and degraded through autophagy) (Bjorkoy et al., 2005) is elevated in the liver tissue of ob/ob mice compared to lean controls (Fig. 1A). The liver of ob/ob mice also exhibited elevated ER stress as evident by an induction of phosphorylated PERK (PKR-like endoplasmic reticulum kinase) and IRE1 (inositol requiring 1) proteins. Electron microscopic examination of the liver tissue demonstrated significant reduction in autophagosome/autolysosome formation in obese mice supporting the biochemical alterations in key autophagy molecules (Fig. 1B–C). We also examined whether the ob/ob mice are responsive to food withdrawal by induction of autophagy in the liver. As shown in figure 1D, food withdrawal failed to activate autophagy in the liver tissue of obese animals, further supporting the abnormal nature of autophagic response in obesity. Considering the possibility that the low levels of autophagy markers such as LC3-II seen in obese liver tissue may reflect a more rapid turnover rather than deficiency, we treated the ob/ob mice expressing GFP-LC3 in the liver with chloroquine, a lysosomal protease inhibitor, then monitored the formation of autophagosome. After an overnight fast, very few GFP-LC3 positive punctate structures typical of enhanced autophagy was detectable in the liver tissue of the ob/ob mice compared to the strong signals observed in lean controls (Fig. 1E–F). In addition to significantly higher amounts of punctate structures decorated with GFP-LC3, there was also enhanced distribution of LC3 in the lean liver tissue following lysosomal enzyme inhibition (Fig. 1E–F). Taken together, these observations confirmed that defective biochemical markers in obese liver are not reflecting rapid autophagic efflux but indicative of defects in the activity of this pathway.
Figure 1. Regulation of autophagy in obesity.
A. Autophagy and ER stress indicators were examined in liver tissue of obese (ob/ob) mice and age-and sex-matched lean controls by western blot analysis. Autophagy was evaluated by the conversion of LC3-I to LC3-II, expression of Beclin 1, Atg5, Atg7, and p62 proteins as markers. ER stress was examined by the phosphorylation of IRE1 and PERK (the arrow indicates phosphorylated PERK). B. Representative electron micrographs (9690×) of livers of lean and obese mice. The pictures shown on the right are high magnification (57800×) of the field marked with a red rectangle on the left panel. C. Quantification of autophagolysosome-like vacuoles per field in the EM images. D. Examination of autophagic markers in the liver of obese mice and its age-and sex-matched lean control after food withdrawal. E. Representative pictures of GFP-LC3 punctate structures in livers of lean and obese mice expressing GFP-LC3, in the presence or absence of chloroquine. The red arrows indicate the GFP-LC3 punctate structure; the nuclei were stained with DAPI shown in blue. F. Quantification of GFP-LC3 punctate structure per cell. Data are shown as mean ± SEM. Asterisk indicates statistical significance determined by student’s t test (*p<0.05). All mice are male and at the age of 12–16 weeks.
Regulation of Atg7 in Obesity
We next explored potential mechanisms that may underlie the defective Atg7 expression and/or autophagy in obese liver tissue. First, we examined the time course of the changes in Atg7 expression in mouse liver during the development of high fat diet-induced (HFD) obesity. As shown in figure S1B, the decreased hepatic Atg7 expression was evident at 16-weeks and essentially completely abolished at 22 weeks of HFD feeding. As hyperinsulinemia occurs with insulin resistance and autophagy is negatively regulated by the activity of insulin-mTOR axis, we explored whether deficiency of autophagy in obesity is the result of hyperinsulinemia. For this, we administered streptozotocin into the ob/ob mice, which leads to destruction of beta cells and insulinopenia, followed by examination of Atg7 expression. As shown in figure S2A, STZ treatment results in a 5-fold decrease in serum insulin levels in ob/ob mice. However, this decline in insulin level did not restore the Atg7 expression level in liver. We also examined hepatic autophagy in the liver tissue of db/db mice, which has been demonstrated to suffer from severe depletion of β-cells with age and develop severe diabetes in the KsJ genetic background. As shown in figure S2B, db/db mice also exhibit defective hepatic Atg7 expression compared to lean controls (which is consistent with our results in the ob/ob and HFD models) and reduction in insulin level (occurring at 15 w) did not restore Atg7 expression in the liver tissue. These results indicate that hyperinsulinemia seen in obese mice is unlikely the primary cause for the downregulation of Atg7.
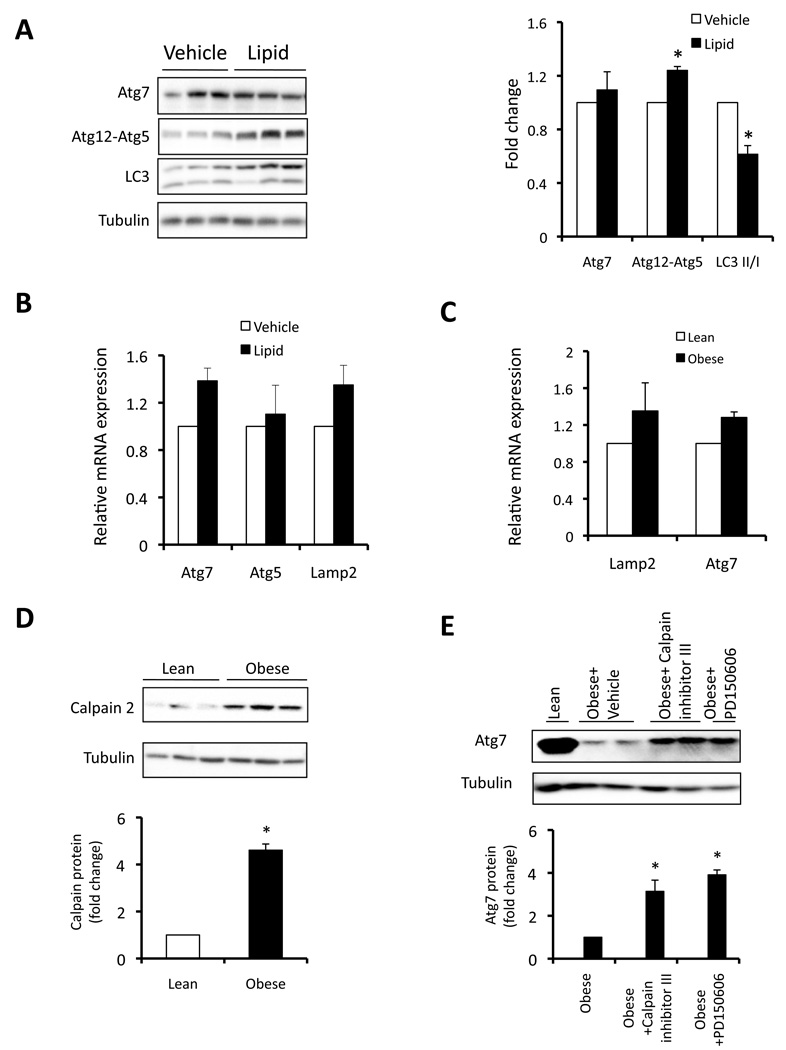
Excess fatty acid accumulation in non-adipose tissues is a hallmark of metabolic disease and recent studies suggested that an abnormal increase in intracellular lipids might impair autophagic clearance (Singh et al., 2009). Hence, we investigated whether lipid exposure will lead to defective autophagy in liver. As shown in figure 2A, there was no alteration in Atg7 protein expression in the liver tissue of mice after a short-term lipid infusion compared to controls. Although there was a mild increase in the Atg12-Atg5 conjugation, the LC3 conversion was decreased by exposure to lipids and no significant changes were evident in the expression of Atg7, Atg5 as well as Lamp2 mRNAs (Fig. 2B). Furthermore, although Atg7 protein level was dramatically decreased in the liver of obese mice (Fig. 1A), we did not observe a corresponding decrease in Atg7 mRNA expression (Fig. 2C) indicating that alterative mechanisms may be involved in regulation of autophagy observed in obesity. Earlier studies have also shown that Atg7, Atg5, and Beclin 1 could be cleaved and degraded by the calcium-dependent protease Calpain 2 (Kim et al., 2008; Yousefi et al., 2006). To explore the possibility of Calpain-mediated Atg7 depletion in obesity, we next examined Calpain 2 expression in the liver tissue of ob/ob mice. As shown in figure 2D, there was a marked (5-fold) increase in Calpain 2 expression in liver of obese mice compared to lean controls. Furthermore, acute inhibition of Calpain by two different specific Calpain inhibitors (peptide and small molecule inhibitors) was able to restore Atg7 expression (3–4 fold of obese levels) in the obese liver tissue (Fig. 2E). These results indicate that obesity-induced increase in Calpain 2 may be an important mechanism leading to downregulation of Atg7 then defective autophagy.
Figure 2. Regulation of Atg7 in liver of lean and obese mice.
A. Autophagy was examined in livers of wild type lean mice (10 week-old) treated with lipid infusion or vehicle for 5 hrs, using Atg7, conjugated Atg5 and LC3 conversion as autophagy markers. Quantification is shown on the right side panel. B. mRNAs coding for Atg7, Atg5 and Lamp2 were examined by quantitative RT-PCR in livers of lean mice treated with vehicle (n=3) or lipid infusion (n=5) for 5 hrs. Results are presented as gene expression levels in lipid infusion group normalized to vehicle controls. C. mRNAs coding for Atg7, Lamp2 were examined by quantitative RT-PCR in livers of lean mice (n=3) or ob/ob mice. Results are presented as gene expression levels in obese group normalized to lean controls. D. Representative western blot of Calpain 2 expression in the livers of ob/ob mice (n=8) and lean mice (n=7) control. Quantification of the blots is shown on the bottom. E. ob/ob mice were injected with Calpain inhibitor III (10mg/kg), PD150606 (10mg/kg), and liver Atg7 expression was examined by western blot assay. Quantification of the blots is shown on bottom, with 4 mice in each treatment. All data are shown as mean±SEM. Asterisk indicates statistical significance determined by student’s t test (*p<0.05).
Suppression of Autophagy Results in Insulin Resistance and ER stress
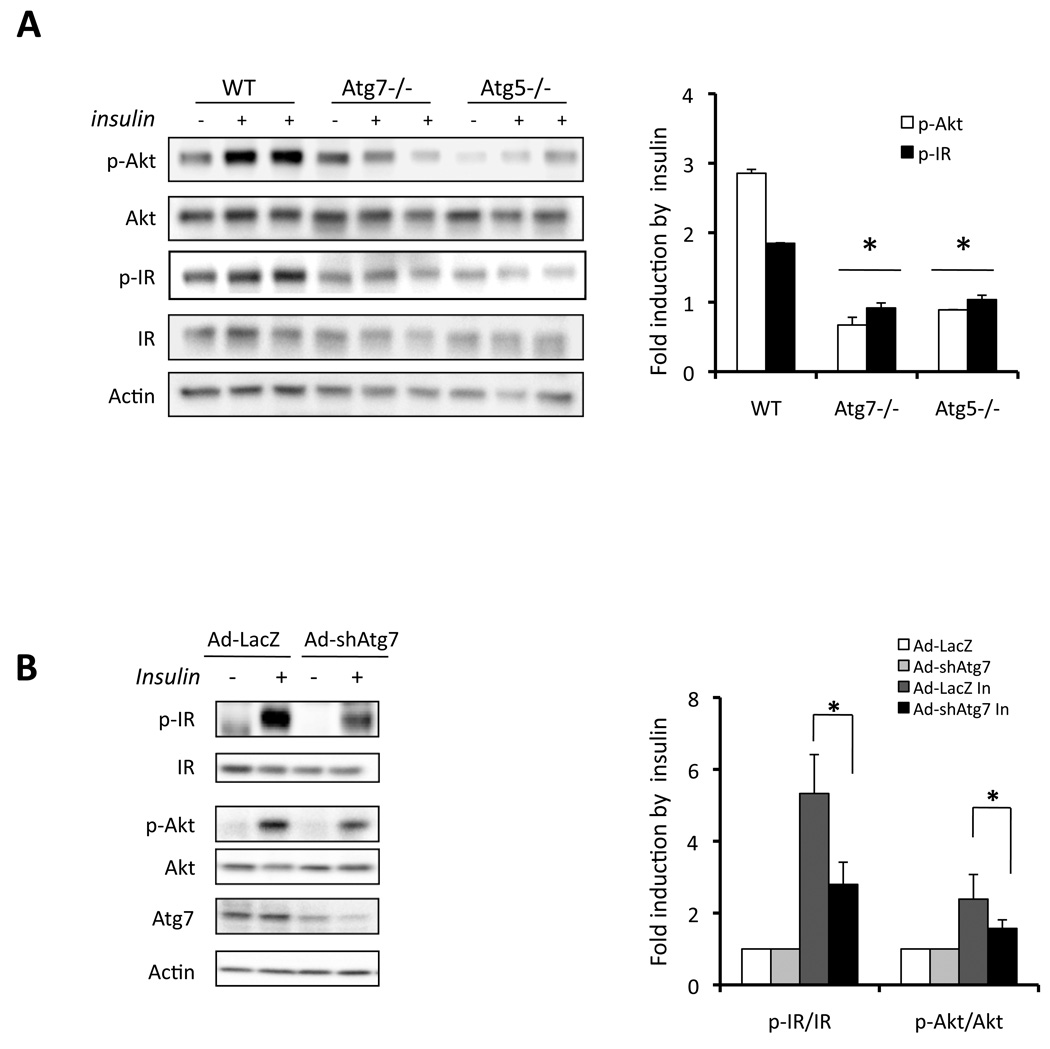
To investigate the impact of autophagy on metabolic regulation, particularly in the context of insulin responsiveness, we employed several approaches to interfere with autophagy and examined insulin action in cultured cells. To determine the regulation of insulin action during impaired autophagy, we examined insulin receptor signaling in two cellular models genetically deficient in Atg7 or Atg5 (Fig. 3A). Consistent with published reports, defective autophagy was also verified in Atg7-deficient cells (Suppl. Fig. S3A–B). In both of these cell types, there was a significant reduction in insulin-stimulated tyrosine 1162/1163 phosphorylation of insulin receptor β subunit (IRβ) and serine 473 phosphorylation of Akt, demonstrating severe insulin resistance (Fig. 3A). Interestingly, as shown in figure S3A, deficiency of Atg7 not only resulted in decreased level of Atg12-Atg5 conjugation but also defective expression of Atg5, suggesting that Atg7 is required for the maintenance of Atg5 and plays a central regulatory role in autophagy by controlling several downstream mediators. Considering the potential limitations of the mouse embryonic fibroblasts used in these experiments in reflecting physiological relevance, Atg7 expression was also suppressed in murine hepatoma cells, Hepa1–6, using an adenovirus-mediated shRNAi approach. In this system, suppression of Atg7 expression (∼70%) resulted in decreased insulin-stimulated Akt Ser-473 (∼50%) and IR beta subunit (∼60%) phosphorylation (Fig. 3B). These experiments indicated that genetic, or molecular suppression of autophagy in multiple types of cells was detrimental for insulin action.
Figure 3. Suppression of autophagy results in impaired insulin signaling.
A. Wild type (WT), Atg7−/− or Atg5−/− MEF cells were stimulated with 50nM insulin. Insulin receptor signaling was detected by western blot analysis of phosphorylation of IR tyrosine 1162/1163 (p-IR) and Akt serine 473 (p-Akt), with quantification shown on the right side panel. Results represent fold induction by insulin compared to basal levels in each group. B. Hepa1-6 cells were infected with adenovirus-mediated control shRNAi to LacZ (Ad-LacZ) or shRNAi to Atg7 (Ad-shAtg7) at a titer of 2×109 vp/ml per well in 12-well plate. Forty-eight hours later, cells were stimulated with 10nM insulin (In) for 3 mins and the insulin receptor signaling was examined by western blot analysis. Quantification is shown on the right side panel. Results represent fold induction by insulin compared to basal levels in each group. Asterisk indicates statistical significance determined by student’s-t test (*p<0.05).
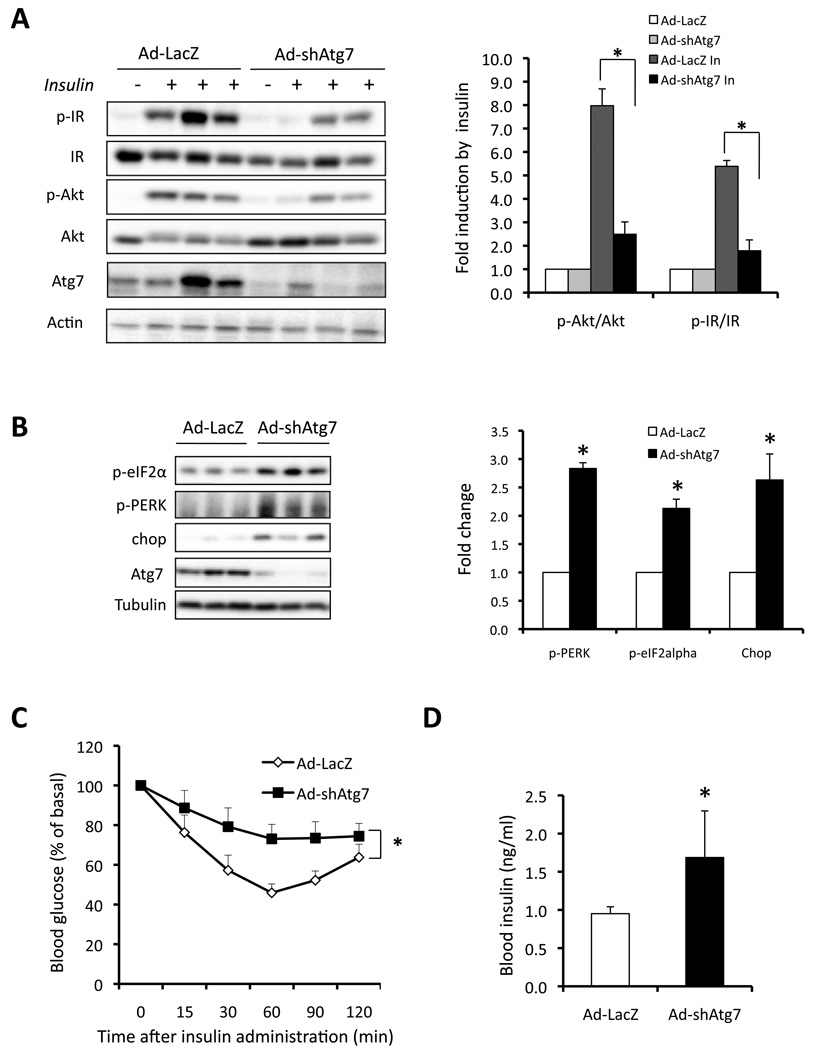
However, it is essential to determine the physiological relevance of such a relationship between autophagy and insulin action in an in vivo setting. To establish this system, we suppressed Atg7 expression in the liver tissue of lean mice with Atg7 or control shRNAs using an adenovirus-mediated approach in vivo. The shRNA treatment resulted in 80% reduction of Atg7 protein in these animals (Fig 4A–B). As shown in figure 4A, there was a marked reduction in the ability of insulin to stimulate Akt (∼80%) and IR beta subunit (∼70%) phosphorylation in liver of intact lean mice injected with adeno-Atg7 shRNAi, demonstrating severe insulin resistance upon suppression of Atg7. Hence, experiments in both cellular systems and whole animals demonstrate the importance of the autophagy process in insulin action.
Figure 4. Defective autophagy results in insulin resistance.
A. Hepatic insulin action in lean mice following in vivo Atg7 knock down with shRNAi. Lean mice (male, 14-week old, C57BL/6J) were injected with Ad-LacZ or Ad-shAtg7. Insulin signaling was examined in intact mice following hepatic insulin administration. Quantification of the data is shown on right panel with the averaged results of 4 mice in each treatment group. Results represent fold induction by insulin compared to basal levels in each group. Data are shown as mean±SEM. Asterisk indicates statistical significance determined by student’s-t test (*p<0.05). B. Examination of ER stress in livers of lean mice following Atg7 knock down in vivo. Chop, PERK and eIF2α phsophorylation were detected by western blot analysis and Atg7 and tubulin proteins are shown as controls. Quantification of the data shown on right panel. Results represent fold change in each molecule by Ad-shAtg7 to Ad-LacZ. C. Insulin tolerance test was performed in lean mice (n=8) following Atg7 knockdown (n=8). Results represent blood glucose concentrations relative to the starting value. All data are presented as mean±SEM, with statistical analysis performed by repeated measures two-way ANOVA followed by post-test (* indicates p<0.05). D. Serum insulin level was measured in lean mice after 6 hrs fasting following Atg7 knockdown.
We have previously demonstrated that the metabolic and inflammatory stresses of obesity disrupt the homeostasis of the ER which in turn contributes to insulin resistance (Ozcan et al., 2004). Recent studies also indicate that ER stress may be linked to autophagy potentially through the degradation of unfolded proteins and in the removal of superfluous ER membranes (Bernales et al., 2007). Inhibition of autophagy during ER stress increases apoptosis in many cellular settings, suggesting an adaptive role for autophagy during the unfolded protein response (Ogata et al., 2006). Interestingly, we observed that deficiency of autophagy induced by suppression of Atg7 also resulted in ER stress in the liver tissue of lean mice (Fig. 4B), as evident by the induction of eIF2α (eukaryotic initiation factor 2 alpha) and PERK phosphorylation, and induction of Chop (C/EBP homologus protein), three well-known ER stress indicators. This pattern is reminiscent of the observations made in obese liver and suggests that autophagy may be integrated to ER homeostasis in impacting insulin action. Consistent with the biochemical alterations, insulin resistance was evident in lean mice in insulin tolerance tests (ITT) upon Atg7 suppression (Fig. 4C). Serum insulin level was significantly increased, however glucose levels remained slightly lower in mice after the suppression of Atg7 (Fig. 4D, and Suppl. Fig. S4E). This decreased basal glucose level is likely caused by impairment of lysosome and autophagy-mediated glycogen breakdown (Kotoulas et al., 2004), which is supported by the evidence of increased hepatic glycogen content in lean mice after suppression of liver Atg7 (Suppl. Fig. S4F–G). In addition, expression of mRNAs coding for glycogen phosphorylase and α-glucosidase, two key glycogen breakdown enzymes, were also significantly decreased in lean mice following Atg7 suppression (Suppl. Fig. S4H). In these mice, despite indications for a mild hepatomegaly (Suppl. Fig. S4A), we did not observe a significant change in liver lipid accumulation and no alterations were evident in body weight, serum triglyceride or free fatty acid levels (Suppl. Fig. S4B–D). Taken together, these data link autophagy to insulin action and ER stress in the liver and demonstrate the systemic metabolic impact of hepatic autophagy-deficiency in vivo.
Restoration of Hepatic Autophagy in Obese Mice Results in Enhanced Systematic Glucose Homeostasis, Insulin Action and Ameliorated ER Stress
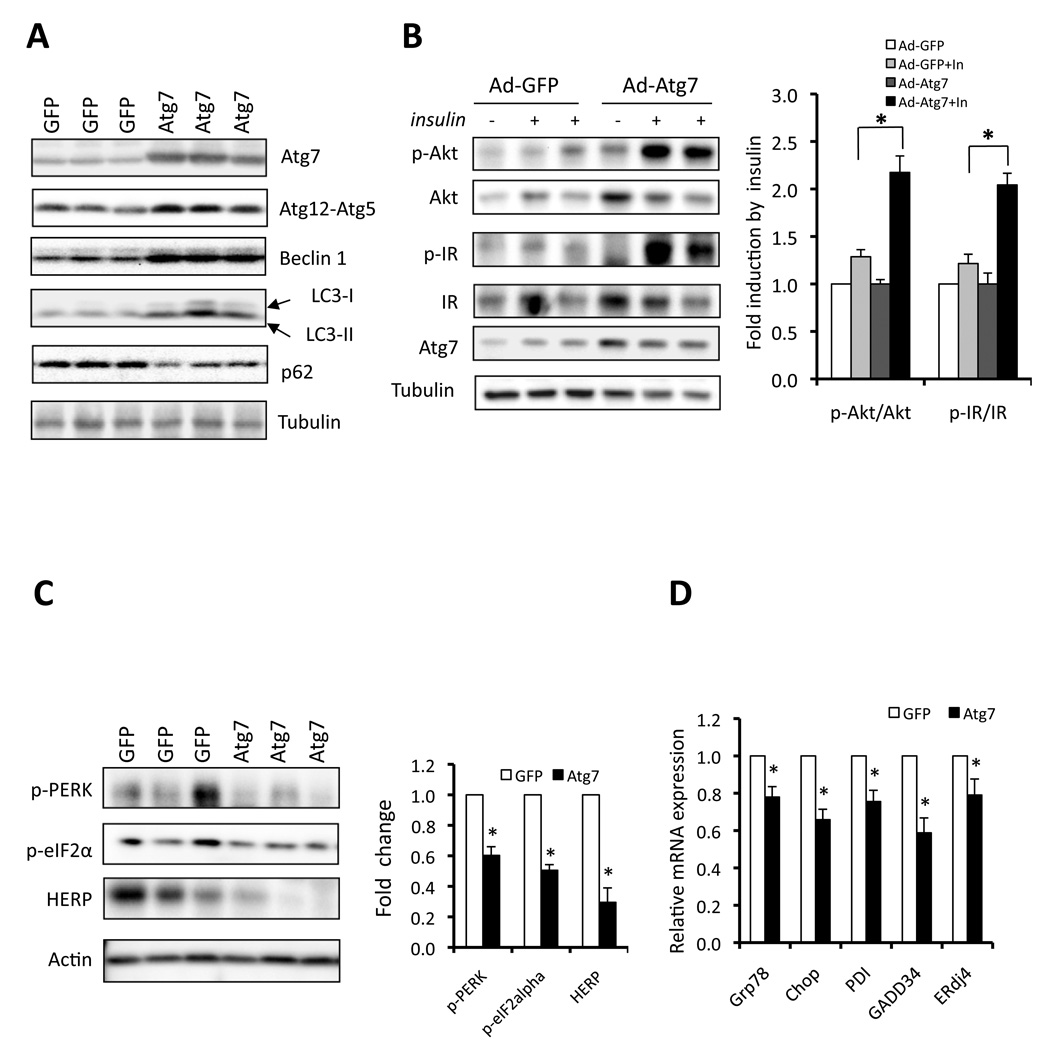
While disrupting the autophagic response in cells and liver tissue appeared causal to insulin resistance, these experiments do not provide evidence linking defective autophagy to abnormal insulin action seen in obesity. We reasoned that since autophagy response is already defective in obesity, a definitive link could be established if autophagy is reconstituted in the obese animals and this intervention results in enhanced insulin action. Among the many Atg (autophagy-related) genes that regulate autophagy, Atg7, which encodes a ubiquitin-activating enzyme (E1)-like enzyme, is central for autophagosome formation (Tanida et al., 1999) responsible for both Atg12-Atg5 conjugation and LC3 conversion (Suppl. Fig. S3A–B). Since Atg7 is dramatically down-regulated in the liver of ob/ob mice (>90%, Fig. 1A), we reasoned that reconstitution of Atg7 expression would likely to be an effective way to reestablish autophagy, at least in part, in the livers of obese mice. To test the possibility that the defects in hepatic autophagy and particularly in Atg7 seen in obesity might be causal for insulin resistance in vivo, we expressed Atg7 using an adenoviral system in the liver tissue of ob/ob animals to examine its metabolic impact. After adenoviral delivery, we verified that expression of Atg7 was significantly elevated (∼5-fold) in the liver tissue of obese mice, compared to vector controls (Fig. 5A), bringing its levels closer to what has been detected in lean controls. This modest increase in the expression of Atg7 in obese mice led to increased autophagy as evident by increased conjugated Atg5 expression, increased Beclin 1 expression, a higher LC3 conversion, and a decrease in p62 expression level in liver tissue compared to control mice, indicating that the strategy employed in this setting was, at least in part, successful in reconstituting Atg7 and autophagic activity in vivo. We also examined whether our adenoviral strategy primarily targeted liver tissue, by measuring both GFP and exogenous Atg7 expression in other insulin sensitive tissues/organs of the ob/ob mice. As shown in figure S6A–B, both GFP and Atg7 expression was readily detectable in the liver tissue of ob/ob mice injected with Ad-GFP or Ad-Atg7 but not detectable in muscle and adipose tissue.
Figure 5. Restoration of autophagy and improvement of insulin action by reconstitution of Atg7 in obese mice.
A. Autophagy in ob/ob mice following adenoviral expression of Atg7 or control vector (GFP). Atg7, Atg12-Atg5 conjugation, Beclin 1 expression, LC3 conversion and p62 levels were examined as autophagy markers. B. Insulin-stimulated IR tyrosine 1162/1163 (p-IR) and Akt serine 473 (p-Akt) phosphorylationin the livers of ob/ob mice expressing Atg7 or control vector (GFP). Atg7 and tubulin are used as controls. Quantification of the data is shown in right panel. Results represent fold induction by insulin compared to basal levels in each group. C. Phosphorylation of PERK, eIF2α and expression of HERP proteins in the livers of ob/ob mice expressing Atg7 or control (GFP) proteins by adenoviral delivery. Actin is shown as a control and quantification of the data is shown in right panel. Results represent fold change in each molecule by Ad-Atg7 to Ad-GFP. D. mRNAs coding for Grp78, Chop, PDI, Gadd34 and ERdi4 were examined by quantitative RT-PCR in livers of ob/ob mice reconstituted with Atg7. Results are presented as gene expression levels in Atg7 group normalized to controls (GFP). All data are shown as mean±SEM. Asterisk indicates statistical significance determined by student’s t test (*p<0.05).
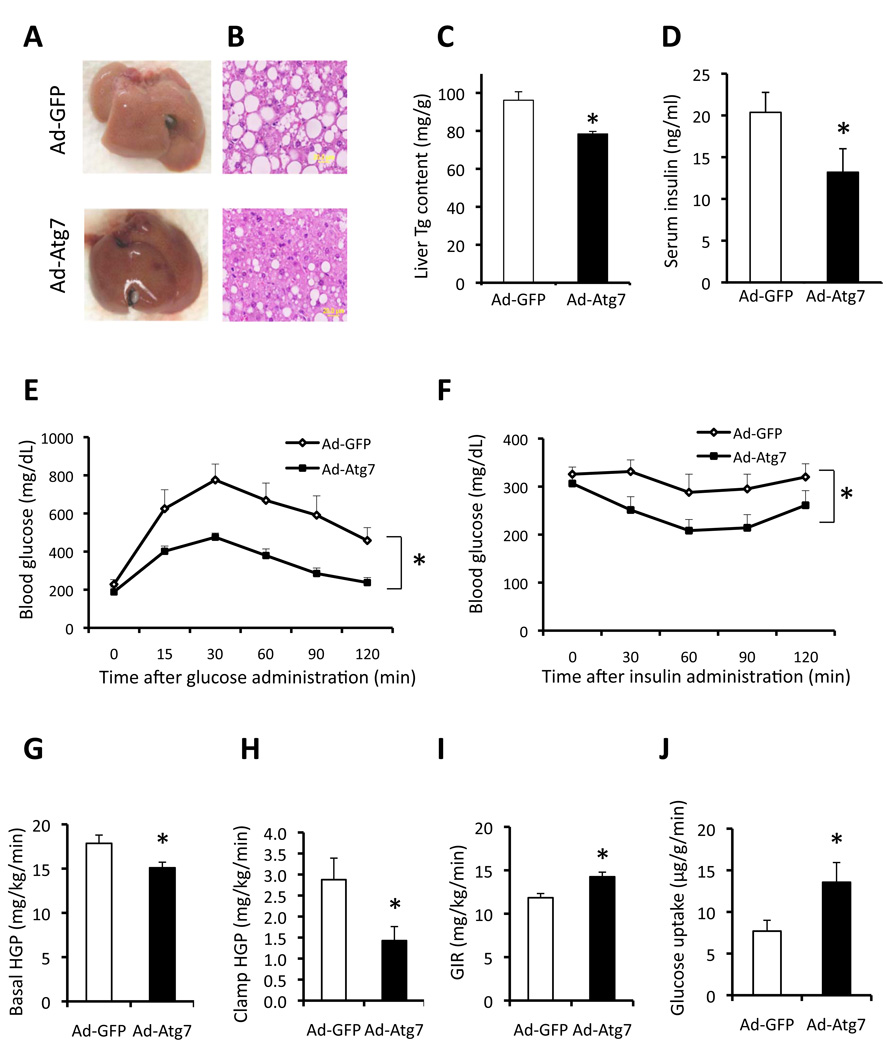
We next asked whether expression of Atg7 in the ob/ob liver tissue, which is severely defective in both Atg7 expression and insulin action, could rescue the defects in insulin receptor signaling. Upon insulin stimulation, Atg7-expressing obese animals exhibited a markedly enhanced tyrosine 1162/1163 phosphorylation of insulin receptor β subunit (IRβ) and serine 473 phosphorylation of Akt in liver tissue (Fig. 5B) compared to controls. Moreover, restoration of Atg7 expression resulted in significant reduction in obesity-induced ER stress in the liver of ob/ob mice, as evidenced by decreased levels of phosphorylated PERK, eIF2α and HERP (homocysteine-inducible ER stress protein) proteins as well as ER stress inducible mRNAs coding for Grp78 (glucose regulate protein 78), Chop, PDI (protein disulfide isomerase), Gadd34 (Growth arrest and DNA damage-inducible gene 34) and ERdj4 (endoplasmic reticulum-localized DnaJ homologue) compared to control mice (Fig. 5C–D). Examination of total liver tissue and liver sections demonstrated that Atg7 complementation resulted in reduced hepatic fatty acid infiltration (Fig. 6A–B), liver triglyceride content (Fig. 6C), and serum insulin level (Fig. 6D). There was a modest increase in serum triglyceride but not free fatty acid levels in mice expressing exogenous Atg7 (Suppl. Fig. S6C–D). It is possible that enhanced autophagy reduced lipid accumulation through an increase in lipid metabolism, as proposed by Singh et al. (Singh et al., 2009). Interestingly, in this system, a significant decrease was also apparent in the expression of FAS and SCD1 transcripts (Suppl. Fig. S7E) which are important mediators of hepatic lipogenesis. A similar trend was also seen for SREBP1 and ACC1 but these changes did not reach statistical significance (p=0.059 and 0.067, respectively). Hence, it is possible that in this setting, a reduction in lipid synthesis also contributed to improved hepatosteatosis. In agreement with enhanced hepatic insulin action, we also observed that expression of genes involved in gluconeogenesis were also attenuated by overexpression of Atg7 in liver of ob/ob mice (Suppl. Fig. S7E).
Figure 6. Metabolic effects of liver Atg7 restoration in obese mice.
A. Gross anatomical views of representative livers of ob/ob mice expressing Atg7 (Ad-Atg7) or control vector (Ad-GFP). B. Representative images of H&E staining (40×) in ob/ob mice liver expressing Atg7 or control vector. Triglyceride (Tg) content of the liver (C), serum insulin level (D) were measured in ob/ob mice expressing Atg7 (n=8) or control vector (n=8). Data are shown as mean±SEM. Asterisk indicates statistical significance determined by student’s-t test (*p<0.05). E. Glucose tolerance test (GTT) performed in ob/ob mice following injection of Adeno-GFP (Ad-GFP, n=8) or Adeno-Atg7 (Ad-Atg7, n=8). F. Insulin tolerance test in ob/ob mice following injection of Ad-GFP or Ad-Atg7. All data are presented as mean±SEM, with statistical analysis performed by repeated measures two-way ANOVA (* indicates p<0.05). Hyperinsulinaemic–euglycaemic clamp studies were performed in ob/ob mice transduced with Ad-GFP (n = 7) or Ad-Atg7 (n = 6). Basal and clamp hepatic glucose production (HGP) (G–H), glucose infusion rate (GIR) (I) and glucose uptake in gastrocnemius muscle (J) were analyzed. Data are shown as the mean±SEM, *P < 0.05.
We next examined insulin action and glucose metabolism in these animals. Glucose and insulin tolerance tests (GTT and ITT) demonstrated significantly improved glucose tolerance and insulin sensitivity in ob/ob mice expressing Atg7 compared with mice injected with the control virus (Fig. 6E–F). We also explored the effect of liver Atg7 complementation in a dietary model of obesity. Adenoviral delivery of Atg7 improved glucose tolerance in the high fat diet-induced model of obesity without any changes in body weight (Suppl. Fig. S1C–D). These results demonstrate enhanced systemic insulin sensitivity upon reconstitution of Atg7 expression in livers of mice with genetic or dietary obesity, and that reconstitution of Atg7 expression in the liver tissue of obese mice resulted in improved liver and systemic insulin sensitivity and glucose homeostasis.
To analyze the action of the autophagy on systemic glucose fluxes and insulin action in more detail, we performed hyperinsulinemic-euglycemic clamp studies in ob/ob mice upon reconstitution of hepatic Atg7. As shown in figure 6G–H, restoration of Atg7 expression resulted in a significant reduction in hepatic glucose production (HGP) both at baseline and during the clamp studies. Glucose infusion rates (GIRs) to maintain euglycemia were also higher in the ob/ob mice expressing Atg7 compared to controls (Fig. 6I). Consistent with this result, glucose uptake in muscle tissue was also increased in Atg7-expressing ob/ob mice compared with controls (Fig. 6J). These data demonstrate that the restoration of Atg7 in the liver tissue of ob/ob mice improves whole-body insulin sensitivity through the suppression of hepatic glucose production and enhancement of insulin-stimulated glucose disposal in the periphery.
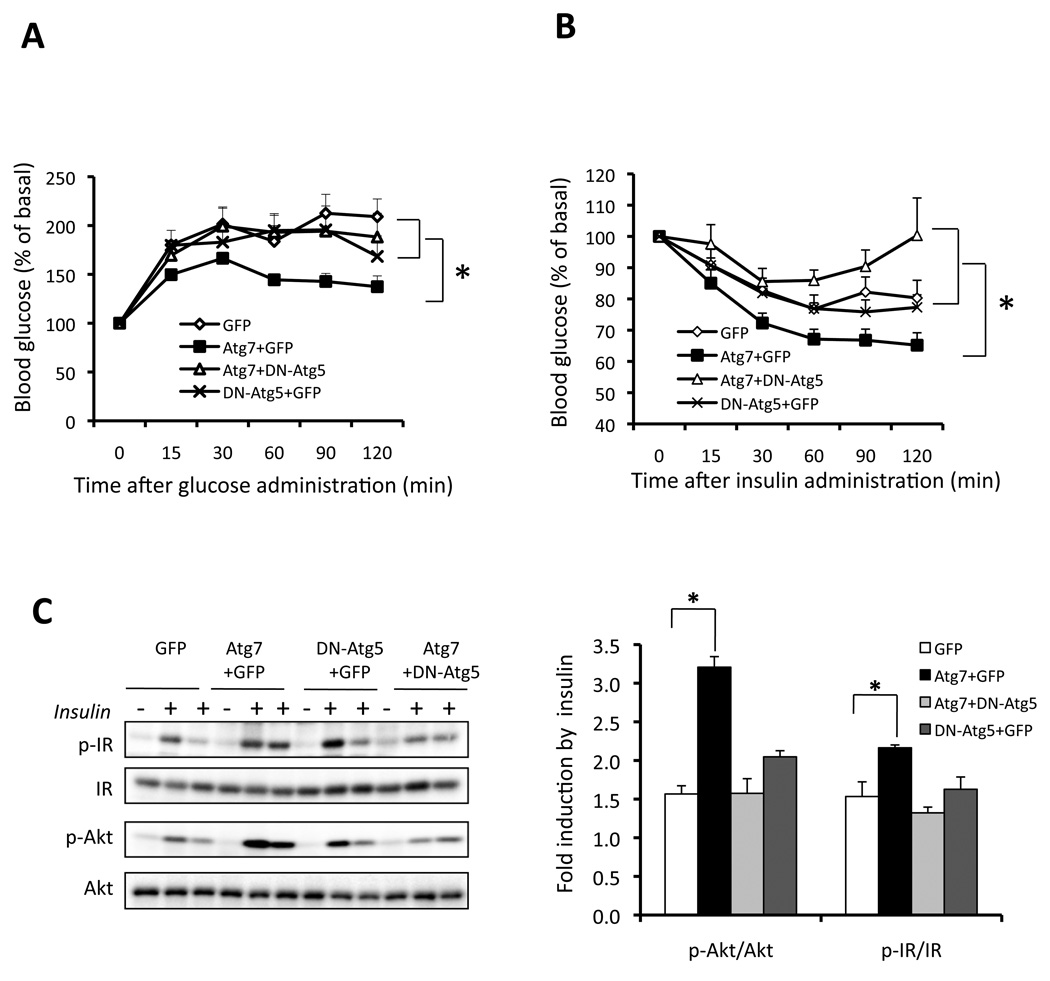
While our in vitro and in vivo experiments strongly link autophagy to insulin action, it is still possible that, particularly in light of the data produced by the expression of Atg7 in liver, the impact of Atg7 in enhancing insulin action may involve mechanisms that are independent of the autophagy process. To address this question in a relevant in vivo setting, we set out to introduce a block in autophagy using a dominant-negative form of Atg5 (DN-Atg5) (Hamacher-Brady et al., 2007) in ob/ob mice where Atg7 expression is also reconstituted in the liver. The action of the DN-Atg5 was first confirmed in a hepatocyte cell line where inhibition of autophagy was verified by formation of GFP-LC3 punctate structures (Suppl. Fig. S7). As shown in figure 7A–B, control ob/ob mice have impaired glucose tolerance and insulin tolerance, and restoration of Atg7 significantly improved the systemic glucose disposal and insulin sensitivity. Expression of DN-Atg5 did not have any effect on these parameters in obese mice. This is an expected result since autophagy is already severely defective in the ob/ob liver. However, co-expression of DN-Atg5 completely abolished the positive effects of Atg7 on systemic insulin sensitivity in the ob/ob animals. In the presence of DN-Atg5, Atg7 expression could not rescue obesity-induced defects in insulin action. Furthermore, we examined insulin signaling in the liver tissue of these obese mice. As shown in figure 7C–D, restoration of Atg7 improved insulin receptor signaling in the liver of ob/ob mice and this effect was completely negated by the co-expression of DN-Atg5. These experiments showed that the effects of Atg7 on enhancing insulin signaling and insulin sensitivity involve, at least in part, its regulation of autophagy and were blocked by interfering with this response through a downstream effector molecule in vivo.
Figure 7. Autophagy-dependent regulation of systemic insulin action by Atg7 in obese mice.
Glucose (A) and insulin (B) tolerance tests in ob/ob mice expressing dominant-negative Atg5 (DN-Atg5+GFP), Atg7 (Atg7+GFP), or combination (Atg7+Dn-Atg5). GFP virus alone is the control. Results represent blood glucose concentrations relative to the starting value. All data are presented as mean±SEM, with statistical analysis performed by repeated measures ANOVA followed by post test (* indicates p<0.05). Each group contained 8 mice at age of 9 weeks. C. Insulin-stimulated phosphorylation of IR tyrosine 1162/1163 (p-IR) and Akt serine 473 (p-Akt) in the livers of ob/ob mice expressing GFP, Atg7+GFP, DN-Atg5+GFP, or Atg7+DN-Atg5. Quantification of each molecule shown in panel C is on the right. Results represent fold induction by insulin compared to basal levels in each group of mice. Data are shown as mean±SEM. Asterisk indicates statistical significance determined by student’s-t test (*p<0.05).
DISCUSSION
In this study we demonstrate that defective autophagy is causal to impaired hepatic insulin sensitivity and glucose homeostasis in obesity. Combined with the potential reported role in islet function and survival (Ebato et al., 2008; Jung et al., 2008), the data presented here indicate that autophagy may be a relevant mechanism for the two major pathological arms of type 2 diabetes: impaired insulin secretion and insulin sensitivity. Our findings raise several interesting possibilities. In obesity, significant autophagy defects are observed in the liver tissue (Fig. 1 & Suppl. Fig. S1), a site which also presents the most pronounced ER stress. It has been shown that nutrient and energy surplus and inflammatory milieu associated with obesity compromise the function of the ER in major metabolic cell types and lead to insulin resistance (Hotamisligil, 2010). Here we show that impaired autophagy could also contribute to ER stress in lean mice and that restoration of autophagy dampens obesity-induced liver ER stress in vivo. Hence, it would be plausible that one function of autophagy is to facilitate elimination or repair of damaged/distressed organelles, and/or assist their adaptive responses to restore metabolic homeostasis. In fact, ER stress can induce autophagy to participate in the degradation of unfolded proteins and in the removal of superfluous ER membranes (Bernales et al., 2007). Failure to achieve this may compromise ER function in the face of continuous energy and nutrient stress, engage inhibitory signals for autophagic and ER stress responses, further organelle dysfunction, and create a vicious cycle resulting in metabolic deterioration, insulin resistance and type 2 diabetes. Future studies should address whether other mechanisms are also involved in integrating autophagy or critical molecules in autophagy to insulin action or other aspects of metabolism; for example the reported autophagy mediated clearance of misfolded insulin receptor induced by ER stress (Zhou et al., 2009). There are many parallels between age-related impairment in metabolism and pathologies associated with obesity both of which benefits from caloric restriction. Intriguingly, caloric restriction is also an effective strategy to enhance autophagy (Wohlgemuth et al., 2007). Hence, metabolic regulation by autophagy may have implications beyond obesity-induced insulin resistance and type 2 diabetes.
Although we observed severe deficiency of autophagy in the liver of obese mice, the cause of this defect and particularly the mechanisms leading to down-regulation of Atg7 could be diverse. Since obesity is characterized with enhanced lipid accumulation in the liver, it is plausible that chronic lipid overloading or sustained lipogenesis might be one of the triggers, although we did not observe significant alteration of autophagic markers in the liver tissue of lean mice treated with a short-term lipid infusion (Fig. 2A–B). Recently, Singh et al. (Singh et al., 2009) demonstrated that autophagy plays an important role in lipid metabolism and inhibition of autophagy increases hepatic lipid storage during starvation. We did not observe hepatic steatosis or changes in serum triglyceride and fatty acid levels in lean mice following shRNA-mediated suppression of Atg7 (Suppl. Fig. S4A–D). Although we did not investigate lipid metabolism in our model under prolonged starvation conditions, it is possible that the role of autophagy in lipid metabolism might be much more complicated than expected and may depend on the developmental stage of the interventions or the age of the experimental animals. It is also likely that severe defects induced by genetic deletion of major autophagy components disrupt major homeostatic pathways and therefore, the impact of such experimental manipulations could vary depending on the timing, severity of the effects, and the metabolic demands of the target tissues. In fact, it is reported that starvation-induced lipid droplet formation is suppressed in the liver of Atg7-deficient mice at a younger age (Shibata et al., 2009). Furthermore, ER stress could also act to regulate lipogenesis and decrease apoB secretion (Kammoun et al., 2009; Su et al., 2009), both of which could alter hepatic lipid accumulation. Thus, ER stress induced by impaired hepatic autophagy as observed in our model may further accelerate lipid accumulation in the liver under defined conditions such as starvation.
Moreover, it has been reported that insulin can suppress autophagy through the mTOR or Akt/FOXO3 pathways (Mammucari et al., 2007; Xie and Klionsky, 2007). Most recently, it has been proposed that insulin may also contribute to downregulation of some Atg genes, although this direct link was examined in lean mice (Liu et al., 2009). However, it is likely that impaired autophagy is also an upstream regulator of insulin resistance as demonstrated here. Furthermore, reduction of insulin level in obese mice (ob/ob and db/db mouse models) fail to recover Atg7 deficiency (Suppl. Fig. S2) but suppression of Atg7 directly results in systemic and hepatic insulin resistance in lean mice. It is nevertheless possible that during the early stages of obesity, hyperinsulinemia may contribute to the dampening of the initiation of autophagic machinery, which would further the organelle dysfunction, disrupt metabolic homeostasis, and promote the emergence of disease. Lastly, as an E1-like enzyme, Atg7 plays a major role for autophagosome formation and LC3 activation (Tanida et al., 1999). We showed that Atg7 is severely downregulated at the protein level in the liver of obese mice (Fig. 1A and Suppl. Fig. S1A–B). However, there was no decrease in the levels of Atg7 mRNA in the liver tissue of obese mice, a proposed mechanism of insulin action on Atg7 expression (Liu et al., 2009), compared to lean controls (Fig. 2C). Thus, a distinct mechanism(s) is likely to account for the downregulation of Atg7. Interestingly, we identified a dramatic increase in Calpain 2 protein expression in liver of ob/ob mice compared to lean controls (Fig. 2D) and inhibition of Calpain activity in vivo by two independent chemical Calpain inhibitors resulted in enhanced Atg7 protein expression level (Fig. 2E). It has also been reported that Calpain 2 may play an important role in Beclin 1 and Atg7 depletion in anoxic rat hepatocytes (Kim et al., 2008). Hence, we suggest that the Calpain-mediated depletion of Atg7 may be a critical contributing mechanism to obesity-related defects in Atg7 protein levels and related autophagic responses.
Regardless, defective autophagy and insulin resistance are highly integrated in mice and impose a major effect on systemic metabolism. In addition to insulin sensitivity, suppression of autophagy also results in alterations of glycogen accumulation in the liver of lean mice (Suppl. Fig. S4F–H), which is increased in obesity. Since autophagy is involved in gluconeogenesis and lysosome plays an important role in glycogen breakdown (Kotoulas et al., 2004) our results are consistent with a modest reduction in blood glucose despite insulin resistance upon suppression of Atg7 in vivo (Suppl. Fig. S4E). In fact, suppression of autophagy also results in a significant decrease in lyosomal acid α-glucosidase, which is produced in ER and routed through Golgi complex to the lysosomes. Given the enhanced ER stress in liver of lean mice upon suppression of Atg7, it is plausible that the defective glycogen breakdown in this model may be a direct consequence of disturbed ER homeostasis. While we have demonstrated that the impact of Atg7 on insulin sensitivity is, at least in significant part, dependent on autophagy, the possibility remains that some part of Atg7 action on liver metabolism may still involve yet unknown and autophagy-independent mechanisms. Future studies are warranted to explore such possibilities to fully understand the metabolic impact of this adaptive response and how insulin action is influenced by different autophagy mediators. Taken together, our observations underscore the importance of the delicate and coordinated regulation of autophagic responses during metabolic homeostasis. It remains an intriguing possibility that small molecules modulators of autophagy or central molecules controlling this process could facilitate the exploitation of this adaptive pathway for therapeutic interventions against obesity and type 2 diabetes.
EXPERIMENTAL PROCEDURES
Cell culture and reagents
Hepa1-6 (murine hepatoma cell line), HEK293A, and MEF cells were cultured in DMEM (Invitrogen) with 10% fetal bovine serum. Twenty µM E64d (Merck) was used to inhibit lysosomal proteases when indicated. For insulin signaling, Hepa1–6 cells were stimulated with 10nM insulin for 3 minutes; MEF cells were stimulated with 50nM insulin for 10 minutes.
Adenovirus transduction
The shRNAi for Atg7 was designed against mouse Atg7 (sense: 5’-ATGAGATCTGGGAAGCCAT-3’) using pSilencer design tool (Ambion). The hairpin template oligonucleotides were synthesized by Integrated DNA Technologies followed by annealing and inserting into adenovirus shuttle vector by using pSilencer system (Ambion) according to manufacturers’s protocol. The adenovirus carrying Atg7 is a generous gift from Dr. Kim (University of Florida). Adenoviruses were amplified in HEK293A cells and purified by CsCl gradient centrifugation. The viruses were tittered and transduced into the cells as described (Cao et al., 2008).
Western blot analysis
Proteins were extracted from tissues, subjected to SDS– polyacrylamide gel electrophoresis, as previously described (Furuhashi et al., 2007). Membranes were incubated with anti-LC3 (Novus), anti-Beclin 1 (Cell Signaling), anti-Atg7 (Abgent), anti-Atg5 (Abgent), anti-conjugated Atg12-Atg5 (Novus), anti-p62 (Abgent), anti-Actin (Santa Cruz), anti-tubulin (Santa Cruz), anti-p-Akt (Santa Cruz), anti-p-IR (Calbiochem), anti-Akt (Santa Cruz), anti-IR (Santa Cruz) , anti-p-PERK (Cell signaling), anti-PERK (home-made), anti-HERP (kind gift from Dr. Yasuhiko Hirabayashi of Tohoku University, Japan), anti-p-IRE1 (Novus), anti-p-eIF2α (Invitrogen), anti-Chop (Santa Cruz) or anti-calpain 2 (Cell Signaling) antibodies overnight at 4°C. The membranes were incubated with the secondary antibody conjugated with horseradish peroxidase (Amersham Biosciences), and visualized using the enhanced chemiluminescence system (Roche Diagnostics). The densitometric analysis of western blot images were done by using Quantity One Software (Bio-Rad).
Quantitative real time RT-PCR
Total RNA was isolated using Trizol reagent (Invitrogen). and converted into cDNA using a cDNA synthesis kit (Applied Biosystems). Quantitative real-time PCR analysis was performed using SYBR Green in a real-time PCR apparatus (Applied Biosystems). The primers were synthesized and purchased from Intergrated DNA Technologies, and the sequences are presented in Supporting Material section.
Electron microscopy
Mice were anesthetized with tribromoethanol followed by sequential portal vein perfusion of 10 ml NaCl (0.9%), dilute fixative and concentrated fixative. For EM analysis, tissues were treated for 1 h with 1% osmium tetroxide and 1.5% potassium ferrocyanide, and then 30 min with 0.5% uranyl acetate in 50mM maleate buffer, pH 5.15. After dehydration in ethanol, tissues were treated for 1 hr in propylenoxide and embedded in Epon/Araldite resin. Ultrathin sections were collected on EM grids and observed by using a JEOL 1200EX transmission electron microscope. For quantification of autophagolysosome-like vacuoles, the numbers of autophagolysosomal-like vacuoles were counted in each field and normalized by the surface area.
Mouse models, Calpain inhibitiont, and administration of the adenoviruses
Male ob/ob mice at 6 weeks of age were purchased from Jackson Labs, and kept on a 12-hrs light cycle and fed with regular diet for 4–8 weeks. Mice used in the diet-induced obesity model were male C57BL/6J purchased from Jackson Labs, which were placed on HFD (35.5% fat, 20% protein, 32.7% carbohydrates, Research Diets) immediately after weaning. Adenovirus carrying Atg7, GFP, DN-Atg5, LacZ-shRNA, or Atg7-shRNA was delivered into the ob/ob (or HFD) mice or lean mice via orbital venous plexus at a titer of 3×1011 vp/mice. In the case of co-expression experiments, two types of viruses were first mixed with equal titers amount, and each virus was delivered to mice at a titer of 1.5×1011 vp/mice. After 7–10 days, glucose and insulin tolerance tests and hepatic portal vein insulin injections were performed as described below. For Calpain inhibition experiments, male ob/ob mice (10 weeks) were injected with vehicle (DMSO), or Calpain inhibitor III (Calbiochem) or PD150606 (Calbiochem) at 10mg/kg through intraperitoneal injection. Six hour after injection, mice were sacrificed and tissues were removed and frozen in liquid nitrogen and kept at −80°C until processing.
In vivo analysis of GFP-LC3 punctuated structures
GFP-LC3 expressing adenovirus was delivered into lean or ob/ob mice via orbital venous plexus at a titer of 2×1011 vp/mice. After 7 days, mice were received either vehicle or chloroquine (10mg/kg/day) (Yuan et al., 2009) by intraperitoneal injection for two days followed by an overnight fast. Livers were then harvested and processed for frozen section. Thawed tissue slides were fixed with 4% paraformaldehyde in PBS for 10min at room temperature and sealed with mounting solution with DAPI (Vector Labs). Punctuated GFP-LC3 and nuclei in each view were quantified by using particle analysis tool of ImageJ software (NIH), GFP-LC3 punctuates per cell was calculated based on the number of nuclei.
Plasma and liver triglycerides measurements
Plasma insulin was measured in mice after a 6 hrs food withdrawal with a commercially available ultra sensitive ELISA assay (Crystal chemicals). Liver triglycerides were determined with colorimetric assay systems (Sigma-Aldrich) adapted for microtitre plate format (Furuhashi et al., 2007).
Glucose and insulin tolerance tests and insulin infusions
Glucose tolerance tests were performed by intraperitoneal glucose injection (1 g kg−1) after an overnight fast, and insulin tolerance tests were performed by intraperitoneal insulin injection (1.0 IU kg−1) after a 6 hrs fast (Furuhashi et al., 2007). Following 6 hrs of food withdrawal, ob/ob mice were anaesthetized with an intraperitoneal injection of tribromoethanol (250 mg kg−1), and insulin (1 IU kg−1) or phosphate buffered saline (PBS) in 200µl volume was infused into the portal vein. Three minutes after infusion, tissues were removed and frozen in liquid nitrogen and kept at −80°C until processing.
Hyperinsulinemic-euglycemic clamp studies
ob/ob mice were injected with Ad-Atg7 or Ad-GFP adenoviruses. Five days after the injections, surgery was performed to catheterize the jugular vein. Clamp experiments were performed at day 4 of post-surgery as previously described (Furuhashi et al., 2007). To determine [3H]-glucose and 2-[14C]-DG concentrations, plasma samples were deproteinized with ZnSO4 and Ba(OH)2, dried, resuspended in water, and counted in scintillation fluid for detection of 3H and 14C. Tissue 2-[14C]-DG-6-phosphate (2-DG-6-P) content was determined in homogenized samples that were subjected to an ion-exchange column to separate 2-DG-6-P from 2-[14C]-DG.
Lipid infusion experiments
Mice were anaesthetized, and the right jugular vein was catheterized. After a 3-day recovery, mice were fasted for overnight followed by an infusion of lipid (5ml/kg/h; Intralipid; Baxter Healthcare Corporation) or saline for 5 hrs (Nakamura et al., 2010).
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Hotamisligil laboratory for their contributions and discussions. Especially, we are grateful to Jason Fan, Haiming Cao, Takahisa Nakamura, Gürol Tuncman, Abdullah Yalcin, Cem Gorgun and Dr. Furuhashi (Sapporo Medical University School of Medicine, Japan) for their assistance with the hyperinsulinemic-euglycemic clamp studies. We thank Dr. Yuan (University of Texas Health Science Center, San Antonio) for providing the GFP-LC3 construct, Dr. Hirabayashi (Tohoku University, Japan) for providing anti-HERP antibody, Dr. Hu (University of New Mexico School of Medicine), Dr. Mizushima (Tokyo Medical and Dental University) and Dr. Komatsu (Tokyo Metropolitan Institute of Medical Science) for providing the Atg5−/− and Atg7−/− MEF lines and Dr. Kim (University of Florida) for providing the Adeno-Atg7. This work is supported in part by a grant from the National Institutes of Health to GSH. LY is supported by a Mentor-based Postdoctoral Fellowship from the American Diabetes Foundation (7-08-MN-26 ADA). PL is supported by a grant from Syndexa Pharmaceuticals. SF and EC are supported by training grants from the National Institutes of Health (T32 ES 007155 and T32 CA 009078).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, Schuck S, Walter P. ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy. 2007;3:285–287. doi: 10.4161/auto.3930. [DOI] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171. 2005:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Gerhold K, Mayers JR, Wiest MM, Watkins SM, Hotamisligil GS. Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell. 2008;134:933–944. doi: 10.1016/j.cell.2008.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding WX, Yin XM. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy. 2008;4:141–150. doi: 10.4161/auto.5190. [DOI] [PubMed] [Google Scholar]
- Ebato C, Uchida T, Arakawa M, Komatsu M, Ueno T, Komiya K, Azuma K, Hirose T, Tanaka K, Kominami E, Kawamori R, Fujitani Y, Watada H. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab. 2008;8:325–332. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Ellington AA, Berhow MA, Singletary KW. Inhibition of Akt signaling and enhanced ERK1/2 activity are involved in induction of macroautophagy by triterpenoid B-group soyasaponins in colon cancer cells. Carcinogenesis. 2006;27:298–306. doi: 10.1093/carcin/bgi214. [DOI] [PubMed] [Google Scholar]
- Furuhashi M, Tuncman G, Gorgun CZ, Makowski L, Atsumi G, Vaillancourt E, Kono K, Babaev VR, Fazio S, Linton MF, Sulsky R, Robl JA, Parker RA, Hotamisligil GS. Treatment of diabetes and atherosclerosis by inhibiting fatty-acid-binding protein aP2. Nature. 2007;447:959–965. doi: 10.1038/nature05844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor MG, Hotamisligil GS. Adipocyte stress: The endoplasmic reticulum and metabolic disease. J Lipid Res. 2007 doi: 10.1194/jlr.R700007-JLR200. [DOI] [PubMed] [Google Scholar]
- Hamacher-Brady A, Brady NR, Logue SE, Sayen MR, Jinno M, Kirshenbaum LA, Gottlieb RA, Gustafsson AB. Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ. 2007;14:146–157. doi: 10.1038/sj.cdd.4401936. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS. Endoplasmic Reticulum Stress and the Inflammatory Basis of Metabolic Disease. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HS, Chung KW, Won Kim J, Kim J, Komatsu M, Tanaka K, Nguyen YH, Kang TM, Yoon KH, Kim JW, Jeong YT, Han MS, Lee MK, Kim KW, Shin J, Lee MS. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8:318–324. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Kammoun HL, Chabanon H, Hainault I, Luquet S, Magnan C, Koike T, Ferre P, Foufelle F. GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J Clin Invest. 2009;119:1201–1215. doi: 10.1172/JCI37007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Nitta T, Mohuczy D, O’Malley KA, Moldawer LL, Dunn WA, Jr, Behrns KE. Impaired autophagy: A mechanism of mitochondrial dysfunction in anoxic rat hepatocytes. Hepatology. 2008;47:1725–1736. doi: 10.1002/hep.22187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy. Microsc Res Tech. 2004;64:10–20. doi: 10.1002/jemt.20046. [DOI] [PubMed] [Google Scholar]
- Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson KL, Fonseca SG, Ishigaki S, Nguyen LX, Foss E, Bortell R, Rossini AA, Urano F. Regulation of insulin biosynthesis in pancreatic beta cells by an endoplasmic reticulum-resident protein kinase IRE1. Cell Metab. 2006;4:245–254. doi: 10.1016/j.cmet.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Liu HY, Han J, Cao SY, Hong T, Zhuo D, Shi J, Liu Z, Cao W. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem. 2009 doi: 10.1074/jbc.M109.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Furuhashi M, Li P, Cao H, Tuncman G, Sonenberg N, Gorgun CZ, Hotamisligil GS. Double-stranded RNA-dependent protein kinase links pathogen sensing with stress and metabolic homeostasis. Cell. 2010;140:338–348. doi: 10.1016/j.cell.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, Shiosaka S, Hammarback JA, Urano F, Imaizumi K. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Gorgun C, Glimcher LH, Hotamisligil GS. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- Razi M, Chan EY, Tooze SA. Early endosomes and endosomal coatomer are required for autophagy. J Cell Biol. 2009;185:305–321. doi: 10.1083/jcb.200810098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuner D, Kaufman RJ. The unfolded protein response: a pathway that links insulin demand with beta-cell failure and diabetes. Endocr Rev. 2008;29:317–333. doi: 10.1210/er.2007-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Yoshimura K, Furuya N, Koike M, Ueno T, Komatsu M, Arai H, Tanaka K, Kominami E, Uchiyama Y. The MAP1-LC3 conjugation system is involved in lipid droplet formation. Biochem Biophys Res Commun. 2009;382:419–423. doi: 10.1016/j.bbrc.2009.03.039. [DOI] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q, Tsai J, Xu E, Qiu W, Bereczki E, Santha M, Adeli K. Apolipoprotein B100 acts as a molecular link between lipid-induced endoplasmic reticulum stress and hepatic insulin resistance. Hepatology. 2009;50:77–84. doi: 10.1002/hep.22960. [DOI] [PubMed] [Google Scholar]
- Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, Kominami E. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol Biol Cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth SE, Julian D, Akin DE, Fried J, Toscano K, Leeuwenburgh C, Dunn WA., Jr Autophagy in the heart and liver during normal aging and calorie restriction. Rejuvenation Res. 2007;10:281–292. doi: 10.1089/rej.2006.0535. [DOI] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12 Suppl 2:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon HU. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- Yuan H, Perry CN, Huang C, Iwai-Kanai E, Carreira RS, Glembotski CC, Gottlieb RA. LPS-induced autophagy is mediated by oxidative signaling in cardiomyocytes and is associated with cytoprotection. Am J Physiol Heart Circ Physiol. 2009;296:H470–H479. doi: 10.1152/ajpheart.01051.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou nL, Zhang J, Fang Q, Liu M, Liu X, Jia W, Dong LQ, Liu F. Autophagy-mediated insulin receptor down-regulation contributes to endoplasmic reticulum stress-induced insulin resistance. Mol Pharmacol. 2009;76:596–603. doi: 10.1124/mol.109.057067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.