Abstract
This paper presents methods to build histo-anatomically detailed individualised cardiac models. The models are based on high-resolution 3D anatomical and/or diffusion tensor magnetic resonance images, combined with serial histological sectioning data, and are used to investigate individualised cardiac function. The current state-of-the-art is reviewed, and its limitations are discussed. We assess the challenges associated with the generation of histo-anatomically representative individualised in-silico models of the heart. The entire processing pipeline including image acquisition, image processing, mesh generation, model set-up and execution of computer simulations, and the underlying methods are described. The multi-faceted challenges associated with these goals are highlighted, suitable solutions are proposed, and an important application of developed high-resolution structure-function models in elucidating the effect of individual structural heterogeneity upon wavefront dynamics is demonstrated.
Keywords: in-silico heart model, multi-scale modelling, cardiac electrical activity, monodomain, bidomain, parallel computing, individualised medicine
1. Introduction
Why do we need computer models?
Biophysically-detailed cell modelling was introduced as early as 1952, through the seminal work of Hodgkin and Huxley (Hodkin & Huxley, 1952), as an approach to advance our understanding of bioelectric phenomena. For quite some time, however, all endeavours in this regard remained limited to models of a single neuron. Pioneered by Noble (1960) in the sixties, the first models of cardiac cell activity were proposed, but it was not before the mid-eighties that computer models for studying bioelectrical activity at the tissue level emerged (Spach 1982, Spach & Kootsey 1985; Barr & Plonsey 1984). Among the motivations for development of modelling techniques was the desire to quantitatively integrate experimental insight, and to overcome the limitations associated with “wet” research techniques. These include the difficulty of selectively changing a single parameter in a well-controlled manner, as well as observation of only a restricted set of parameters, often with limited spatio-temporal resolution, that can be insufficient to draw sufficiently accurate conclusions. As a consequence, models have helped to build-up a mechanistic framework for integration of data, through which observed phenomena may be assessed, interpreted, contextualised, and to formulate hypotheses that can be submitted to targeted experimental validation.
In the context of cardiac electrophysiology research, a major breakthrough in experimental techniques was the introduction of optical mapping of voltage-sensitive dyes to tissue level studies (Cohen & Salzberg 1978, Dillon & Morad 1981, Windisch et al. 1986, Fast & Kléber 1993, Efimov et al. 1994). This offered two key advantages over traditional electrical mapping techniques: i) the recorded signal is proportional to the main quantity of interest, the transmembrane voltage, Vm; ii) optical maps can be recorded immediately before, during and after the application of interventions such as electrical shocks, with no lag-time and without the overwhelming artefacts omnipresent in electrical recordings due to electrode polarisation induced by strong electric fields. In addition, it was initially believed that optically recorded signals provided highly localised data, less affected by volume effects than their directly-recorded electrical counterparts, where current flow in the volume conductor (if the heart is immersed in a torso or a fluid bath), driven by remote sources, contributes substantially to the recorded extracellular potential, ɸe (Plonsey 1982). Although experimental techniques have been devised to improve locality of measurements of ɸe by suppressing far-field effects (Plank & Hofer 2000, Lemay et al. 2007) an unambiguous relationship of extracellular ɸe with local intrinsic membrane parameters, mainly the transmembrane voltage, Vm, and the transmembrane current, Im, is not always possible, particularly in the presence of fractionated electrograms. However, based on recent theoretical evidence (Bishop et al. 2007, Bishop et al. 2008), it would appear that optical sensors pick up signals from a three-dimensional (3D) volume of tissue, due to photon scattering, which poses some limitations for optical mapping techniques that have yet to be assessed quantitatively.
Today, computer models play increasingly important roles for integration of experimental findings into a comprehensive, quantitative and mechanistic description of biological phenomena. They enable us to bridge gaps where experimental techniques cannot provide data at all, or at an insufficient spatio-temporal resolution. In turn, experiments remain the key to corroborate or disprove conclusions drawn from computer simulations. Thus, iteration between experimental work and computer simulations is required to arrive at a sufficiently complete picture. The advantages of modelling lie in the fact that virtual experimentation allows the observation of all quantities of interest at almost any desired spatio-temporal resolution, simultaneously or consecutively, over the entire domain of interest. For instance, with mapping techniques a trade-off has to be made regarding the choice of spatio-temporal resolution and observation domain where signals are recorded either from a small domain such as a small patch at a cardiac surface at a fairly high spatio-temporal resolution, or from a larger domain at a low spatio-temporal resolution (Sharifov et al. 1994). Further, although optical mapping sensors pick up signals from the tissue depth as well, they are mainly constrained to the observation of electrical activity occurring close to the cardiac surfaces. Efforts are under way to overcome this limitation by using optrodes (Byars et al. 2003, Caldwell et al. 2005, Kong et al. 2007) or inverse recovery techniques (Hillman et al. 2007). However, these approaches are still in their infancy and may not allow, within the foreseeable future, to be developed towards a full 3D mapping technique that would permit construction of high-resolution transmural potential maps.
Furthermore, modelling approaches allow one to test hypotheses in a more straightforward manner than in any experimental context. Experimentally, it is difficult to manipulate a single parameter only, since manipulations tend to have undesired side effects on several other parameters, while other parameters cannot be modified at all, as the technology required is not available, or as sufficiently selective substances have not been developed yet. “Explorative modelling” has proven to be a viable approach for the development of new hypotheses and has aided the prediction of previously undiscovered mechanisms. For instance, assuming that the conductivity ratios in the intracellular domain and the interstitial domain are different, computer simulations predicted the formation of “virtual electrodes” during the application of strong electric fields, such as for electrical defibrillation of cardiac muscle (Sepulveda et al. 1989). Several years later these findings were confirmed experimentally by numerous groups (Knisley et al. 1994, Neunlist & Tung 1995, Wikswo et al. 1995).
Often it is not possible to conduct experiments without modifying the substrate. For instance, the dyes required for optical mapping are known to be cytotoxic, and most current optical mapping techniques require suppression of mechanical activity by administration of electro-mechanical uncouplers. Experimental results from such studies have to be interpreted with care, as potentially important regulatory mechanisms such as mechano-electric feedback (Kohl & Ravens 2003) are excluded, giving rise to possibly false-negative observations. At the same time, apparent findings may be modulated, or fully accounted for, by measurement artefacts, contributing to false-positive interpretations. Computer simulations can be invaluable for scrutinising such experimental evidence. To give an example, using computer simulations it was suggested that the so-called “dual humped” action potentials, often seen with optical mapping techniques, are a result of photon scattering effects. As such they would have to be classed as an artefact of the physical measurement principle and not as an electrophysiological finding per se (Efimov et al. 1999, Bishop et al. 2007). Similarly, in another study (Plank et al., 2008) it has been reported that the finding of exclusively negative shock-induced polarisations of the cardiac membranes under strong electrical shocks (Fast et al. 2002) is likely to be a measurement artefact, as such behaviour would be in contrast to the principle of charge conservation, which predicts shock-induced changes in transmembrane voltage of both polarities. These insights stimulated further experimental research, aiding the discovery of novel biological insight.
In conclusion, it is of paramount importance that computational models and experimental research are conducted in close iteration, and that conclusions drawn from both approaches are qualitatively and quantitatively in agreement with bio-physical principles. Quantitative computer models are exquisitely suited to scrutinising whether this is actually the case, since - at least theoretically - they could account for all known mechanisms and their interactions present in a system. On the other hand, computer models require a continuous input from, and careful validation against, experimental data to match biological reality and simulation.
Review of the State of the Art in Modelling Tissue and Organ Electrophysiology
In the following, the current state-of-the-art in cardiac modelling will be outlined, and the need for more detailed models will be discussed. Recent progress in the generation of high-resolution histo-anatomically detailed and individualised whole cardiac models will be presented. This will encompass advances in both experimental and computational fields. The advances in experimental imaging technologies and protocols will focus chiefly on anatomical magnetic resonance imaging (MRI) and diffusion-tensor MRI (DT-MRI), as well as whole-heart histology, used to obtain high-resolution structural/functional information. In addition, recent developments in computational image analysis, co-registration and mesh generation algorithms for the extraction of relevant information for automated construction of 3D cardiac computational models will be discussed. Finally, computational issues related to the simulators on current high performance computing hardware will be addressed. Computational efficiency is a requirement for the use of cardiac simulation software to perform “in-silico” experiments, which typically involve tens to hundreds of simulations to sweep the parameter space of interest. The lack of efficiency has been among the most important limiting factors over the last decade that forced drastic model simplifications to reduce the computational burden. Recent improvements both in available computing power and in numerical techniques promise to lift some of these restrictions to allow unprecedentedly realistic simulations of the electrical and mechanical function without current trade-offs in terms of representation of structural or functional detail.
In the 1980s, when pioneering computer simulations of cardiac tissue function were executed to study initiation, maintenance and termination of impulse propagation, available computing resources were limited. Initially, the one-dimensional (1D) core-conductor model was state-of-the-art, detailed descriptions of the cellular dynamics were not available, and efficient numerical techniques had not been fully developed. These constraints prevented realistic simulations of the heart at the organ level. For more than a decade, geometrically simple structures such as 1D strands (Spach & Kootsey 1986; Leon & Roberge 1990; Qu et al. 2006; Thomas et al. 2003; Garny et al. 2003), two-dimensional (2D) sheets (Leon & Roberge 1990; Weiss et al. 2005; Kneller et al. 2002; Samie et al. 2001; Anderson et al. 2001; Skouibine et al. 1999; Beaumont et al. 1999), and 3D slabs (Plank et al. 2005; Cherry et al. 2003; Vigmond & Leon 1999) were used to represent the myocardium.
Geometrically simple models continue to play important roles in the armoury of bio-computational analysis (Garny et al 2005). The main advantage - simplicity - can facilitate the dissection of mechanisms. Therefore, for many applications in basic research the simple geometry models of the heart remain a valid and important, if perhaps intermediary, tool for mechanistic enquiry. At the same time, for the clinical setting, complex patient-specific and anatomically detailed models will be needed. The combined effects of structural complexity and the various functional heterogeneities in a realistic heart lead to complex behaviour that is hard to predict or investigate with overly simplified models. Besides computational restrictions, simple geometries were popular since electric circuitry analogs can be used to represent the tissue as a lattice of interconnected resistors or a set of transversely interconnected core-conductor models (Leon & Roberge 1990). Thus, well-established techniques for electric circuit analysis can be applied, and simple discretisation schemes based on the finite difference method (FDM) became a standard due to their ease of implementation. With the advent of continuum formulations, derived by homogenisation of discrete tissue components (Henriquez 1993), and the adoption of more advanced spatial discretisation techniques such as the finite element method (Rogers & Mc Culloch 1994, Vigmond et al. 2002;) and the finite volume method (Harrild & Henriquez 1997;Trew et al. 2005;), the stage was set for anatomically realistic models of the heart. It is important to note, in this context, that the FDM does not easily accommodate grids with complex boundaries. This, to a certain degree, was an impediment to the development of anatomically realistic grids. With the introduction of phase field methods (Fenton et al. 2005) this main limitation of the FDM was overcome and, indeed, the FDM is still widely used. Its main advantage - ease of implementation - is, however, lost. During the last few years, anatomically realistic computer simulations of electrical activity in ventricular slices (Hillebrenner et al. 2004; Meunier et al. 2002; Trayanova & Eason 2002), in the whole ventricles (Xie et al. 2004; Rodriguez et al. 2005, Ashihara et al. 2008; Potse et al. 2006; Ten Tusscher et al. 2007) and in the atria (Harrild & Henriquez 2000; Vigmond et al. 2002; Virag et al. 2002; Seemann et al. 2006) have become accessible.
Over the same period of time, models describing cellular dynamics underwent a dramatic increase in complexity. Early efforts employed either simple Hodgkin-Huxley-type models, such as the Ebihara-Johnson model (1980) which characterised the cellular state with three variables only, or non-physiological analytical membrane models (FitzHugh 1968). These were employed to study action potential propagation in 1- or 2D models (Rogers & McCulloch 1994). Nowadays, highly-complex models with up to fifty state variables can be used for studies with anatomically realistic geometries (Plank et al. 2008). Besides the increase in the number of state variables, mathematically described by a set of non-linear ordinary differential equations (ODEs), the stiffness, and thus the numerical and computational effort to solve them efficiently, has increased even more dramatically (MacLachlan et al. 2007, Plank et al. 2008). The increase in stiffness is particularly pronounced with the introduction of Markov state formulations (Clancy & Rudy 1999) that gradually substitute more traditional phenomenological descriptions based on the Hodgkin-Huxley formalism. While in most Hodgkin-Huxley based models the fastest processes, typically the activation of the sodium channel, are governed by time constants at the order of some tens of microseconds, this is decreased to a fraction of a microsecond in Markov state models such as when describing the positive feedback loop between ryanodine receptor and L-type Ca2+ current which governs Ca2+ regulation in the ventricular myocyte (Jafri et al. 1998). A detailed review on computational aspects related to the increased stiffness of recent ionic models when used within tissue and organ simulations can be found elsewhere (Plank et al. 2008).
Despite the impressive development of numerical methods and available computing power, and notwithstanding the technical feasibility of macro-anatomically realistic organ simulations that use the most advanced membrane models available, many limitations continue to exist, and several potentially important physiological mechanisms remain unaccounted for. Even when simulations are executed using the most powerful supercomputers available today, the computational burden is significant, and simplifications have to be made to keep simulations tractable. The computational burden can be characterised by
| (1.1) |
where LCPU is the computational workload, Nv the number of vertices required to discretise the heart and NΔt the number of time steps to execute. Attempts to minimise LCPU aim at reducing either Nv or NΔt. Neither of the two can be freely chosen, due to physical and numerical constraints. Attempts to increase the maximum Δt can lead to a reduction in the number of time steps NΔt required. However, there are upper limits above which numerical schemes become unstable (Courant 1928), or where the solution shows artificial transients and/or becomes inaccurate (Whiteley 2006). The required NΔt increased substantially over the last decade. While initially the focus of tissue level simulations was on the observation of activation patterns over a few hundreds of milliseconds, observation periods of minutes are considered desirable, for instance, to study the temporal evolution of slow metabolic processes (Zhou et al. 2008). On the other hand, Nv can be constrained by choosing coarser mesh discretisations. However, this impedes the representation of finer anatomical detail, such as endocardial trabeculations or papillary muscles, and discontinuities such as vascularisation, cleavage planes, or patches of fat and collagen within the myocardial volume. In general, the myocardium has been treated as a continuum in all organ level studies published so far. Further, due to the characteristics of the biophysical problem that has to be solved on these grids, the mesh discretisation cannot be chosen arbitrarily coarse. The fast upstroke of cellular depolarisation, for example, gives rise to a steep wavefront, which covers a spatial extent of only 1 mm, or less. The space constant, λ, can be defined as
| (1.2) |
where σi and σe are the intracellular and extracellular conductivities with ζ designating the eigendirection of the conductivity tensor and β being the surface to volume ratio of the intracellular matrix. This is typically used as a measure to quantify the distance within which a local depolarising event affects the adjacent tissue. Spatial discretisation has to be chosen sufficiently small, relative to λ, to avoid spatial undersampling. Such undersampling can lead to a gradual deviation of conduction velocities from those observed in finer grids (Pollard et al. 1993), even giving rise to conduction block when the spatial discretisation becomes too coarse. The space constant is direction-dependent being largest along the axis of the cells, and smallest in the direction orthogonal to the myocardial laminae (Hooks et al. 2007). Further, pathological conditions that affect the conductivities of the substrate, such as downregulation of connexin expression during heart disease (Poelzing & Rosenbaum 2004, Akar et al. 2004), can lead to further changes in λ, requiring even finer discretisation steps. To avoid artefactual deviation of the conduction velocities as a function of grid granularity it is of major importance for computer simulation studies to tune the conductivity tensors for a given mesh discretisation. Due to the large variation in reported conductivities (Roth 1997, Hooks et al. 2007) there is a wide range of possible adjustments. Without proper tuning of the conductivity tensors, simulated depolarisation wavefronts may propagate too slowly or too quickly. This entails a scaling of the model’s wavelength, i.e. the product of action potential duration times conduction velocity. For instance, running a simulation with an artefactual increase in conduction velocity by 20% can be interpreted as reducing the size of the heart by 20%. Since the wavelength is the decisive factor that determines the presence and distribution of excitable gap and refractory tissue, wavelength matching is of key importance when comparing theoretical and experimental observations. This is particularly important for whole heart studies where conduction velocities cannot be estimated as easily as in simple models.
Current state-of-the-art models of the heart largely utilise stylized representations of cardiac anatomy, where finer structural detail is partially ignored. This is caused not only by the computational expenses associated with the large number of degrees of freedom in fine-grained meshes. It is also related to technical difficulties of image acquisition and processing of high-resolution data sets of the entire heart, and the generation of high-resolution grids that faithfully represent cardiac histo-anatomic detail at different spatial scales, from microscopic features (intercellular clefts, vascularisation, fibrosis, Purkinje system) through macroscopic structural organisation, such as fibre curvature and rotation of cell layers as well as trabeculations and papillary muscles, to the gross anatomy of the heart.
In addition to the simplifications made to represent cardiac structure, a further shortcoming of many current models is the omission (with notable exceptions such as Vigmond & Clements 2007; Ten Tusscher & Panfilov 2008; Berenfeld et al. 1998) of the specialised pacemaker and cardiac conduction system, i.e. the sinoatrial and atrioventricular nodes and the Purkinje network. In the absence of a realistic Purkinje system, for example, it is difficult to compute the activation sequence of the most important control case, a sinus beat, and it is impossible to elucidate the role played by Purkinje-related mechanisms during onset and maintenance of cardiac arrhythmias.
As a result of the limitations of currently used models, a new generation of high-resolution cardiac models, derived directly from MR data, is on the horizon. Studies (firstly by Burton et al. 2006, and more recently by Plotkowiak et al. 2007 and Vadakkumpadan et al. 2008) have demonstrated the feasibility of establishing a computational pipeline of technology which facilitates the automated generation of high-resolution finite element cardiac histo-anatomical models from MR voxel stacks and/or DT-MRI data, for use in computational simulations of whole-organ function. Building on this earlier work, the present paper provides a summary of underlying challenges, in particular of developing co-registration algorithms for incorporation of whole heart histological data.
(a) New Data and Concepts
The main focus of this article is to illustrate concept, implementation and validation strategies to construct “in-silico” organ models at an unprecedented level of anatomical fidelity. There are several major requirements that render the development of such models a challenging task. Key needs include:
Sufficiently accurate and detailed imaging data sources of intact 3D volumes that represent the cardiac gross anatomy, intra-myocardial histo-architectural heterogeneity, and structural anisotropy.
Image analysis tools, including segmentation of relevant structures and inter-modality or inter-patient registration algorithms to provide accurate anatomical information that is suited for generating comprehensive computational grids.
Tesselation of segmented anatomical image stacks into finite element meshes that preserve the relevant anatomical detail and represent the organ’s surface in a sufficiently smooth manner, allowing the selection of proper spatial discretisation that keeps the overall degrees of freedom within bounds that are compatible with computational tractability.
Transfer of anatomical information such as the classification of groups of voxels as left ventricular (LV), septal, or right ventricular (RV), obtained during image processing or on a per rule-base using a-priori knowledge (Streeter et al. 1969), for the purpose of “tagging” or annotating the finite element meshes to allow assignment of spatially varying electro-mechanical parameters.
Development and implementation of efficient parallel solver strategies which support simulation of cardiac electrophysiology over several seconds to minutes, where spatial discretisation may lead to up to 50 million degrees of freedom and temporal discretisation to several millions of time steps, ideally to be executed near real time.
In the following we present the development of an automated pipeline that outlines how to address these challenges. The test cases are a high-resolution rabbit heart MRI data set (~25 μm isotropic voxel dimension; Burton et al. 2006), and a rat data set including anatomical MRI (21.5 μm isotropic voxel dimension) and DT-MRI (100 μm isotropic resolution), both with with complete histological follow-up of the entire heart (0.5×0.5×10 μm voxel dimension). The extraction of significant structures from these data sets and the subsequent generation of finite element computational meshes at unprecedented resolution is demonstrated. The final model includes detailed structural information including blood vessels, papillary muscles, trabeculations and either rule-based or DT-MRI derived representation of the prevailing alignment of cardiac myocytes, also referred to as “fibre orientation”.
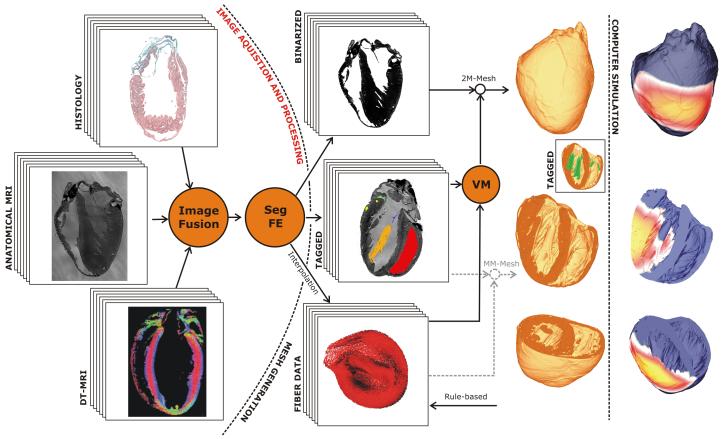
The computational challenge of solving the equations governing electrical dynamics over such a high-resolution whole-ventricular models (consisting of 4.3 million finite element node points, making-up 24 million tetrahedral elements) is met using the Cardiac Arrhythmia Research Package (CARP) (Vigmond et al. 2003; Plank et al. 2006), a highly optimised simulation environment for solving the cardiac bidomain equations in parallel (or the monodomain equations where current flow in the interstitial space and its feedback onto wavefront propagation is neglected). The overall workflow of the automated cardiac histo-anatomy processing pipeline for generation of high-resolution whole-ventricular models is shown in Fig. 1.
Figure 1.
Workflow of an automated cardiac histo-anatomy processing pipeline. Left panel: Image stacks obtained from different “wet” experimental modalities are fused into a single consistent 3D image stack. Subsequently, the image stack is segmented (Seg) and relevant features are extracted (FE). Middle Panel: A binarized input stack is fed into the mesh generator. A voxel mapper (VM) transfers “tags” that were assigned during the segmentation process, over to the final “tagged” mesh. Meshes may have an arbitrary number of tags, while the mesh itself is a “two-material” (2M) mesh only. Alternatively, the tagged image stack could be fed directly into the mesh generator to generate “multi–material”(MM) meshes, that is, the mesh generation strategy can be varied as a function of the tags assigned. Similarly, data on the tissue anisotropy (e.g. “fibre data”), either obtained from imaging data interpolated onto the mesh, or generated on a per rule-base, are mapped onto the final grid by the VM. In the inset, those regions of the mesh which were tagged as the endocardial surface of the RV are shown in red, the left atrio-ventricular valve in blue, a LV papillary muscle in orange, etc. Right panel: After assigning electrophysiological properties to the mesh and choosing electrode locations for stimulation, meshes are passed on to computer simulation. In the given example, the spread of electrical activation from an epicardial stimulation site is shown.
2. Methods
(a) Wet Lab Techniques
All experimental investigations conformed to the UK Home Offce guidance on the Operation of Animals (Scientific Procedures) Act of 1986. Hearts were excised from animals, killed according to schedule 1 of the Act, either by terminal anaesthesia (New Zealand white rabbits > 1 kg) using 100 mg×kg−1 sodium pentobarbitone (Rhône Merieux, Harlow UK), or by cervical dislocation subsequent to cerebral concussion (Wistar rats, 220-350 g). The isolated hearts were swiftly perfused in Langendorff mode, and then fixed, either in their slack (resting) state, during volume overload, or in contracture. Fixation was by coronary perfusion, using the fast-acting Karnovsky’s fixative (for detail see Burton et al. 2006).
Hearts were left overnight in Karnovsky’s fixative. For subsequent anatomical MRI, this contained 2 mM (in the case of rabbit) or 4 mM (in the case of rat) gadodiamide contrast agent (Omniscan 0.5 mmol/ml, Nycomed Imaging AS, UK). Hearts were then rinsed in cacodylate buffer, and embedded in bubble free 1% low melting temperature agar (Cambrex, Nusieve GTG agarose). For anatomical MRI, the agar contained 2 mM gadodiamide (unless otherwise stated) to improve the contrast between agar and tissue in the MRI scans (see Section c).
Subsequent histological follow-up of the hearts varied with species. In the case of rabbit hearts, longer dehydration periods in alcohol and longer infiltration times in xylene were required (see Methods in Burton et al. 2006). For rat hearts, dehydration involved steps of 2 h each in 20%/30%/50% alcohol (compared to 8 h each in 20%/30%/50% in rabbit), followed by an overnight exposure to 70% (same as in rabbit), 24 h in 90%, and 4 × 8 h in 100% alcohol (rabbit 48h in 90%, 4 × 6 h in 100% alcohol). Alcohol was replaced with xylene using increasing concentration steps (25%/50%/75%/100%) over 48 h each (similar to rabbit) and the tissue was then gradually infiltrated with wax at 25%/50% for 24 h, 75% for 48 h, and 100% for one week (for rabbit heart, 48 h 25%; 12h 50%; 12h 75%; 1-2 weeks in 100%).
(b) Sectioning, Histological Staining and Image Acquisition
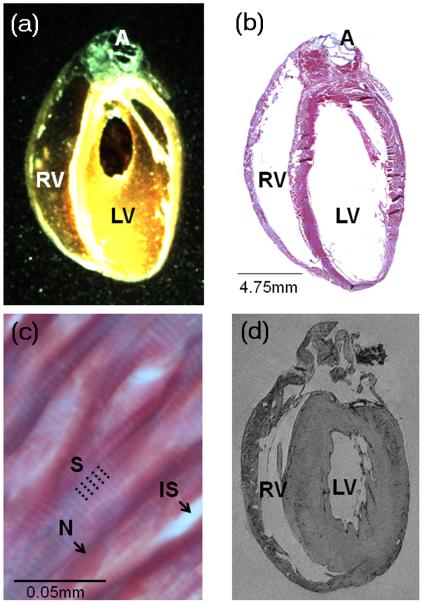
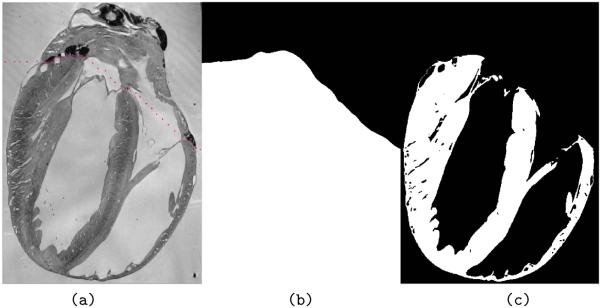
Whole hearts were embedded in black wax (Cat. No. 090740001, Fred Aldous, UK) to improve contrast between wax block and tissue surface (Fig. 2a). In order to achieve good quality whole-mount images from the tissue block before sectioning, a Leica MZ 95 stereomicroscope (Achromat objective, f = 300 mm) was equipped with a 62 mm circular polarising filter (Jessop Group Ltd., Leicester, UK), suspended from a custom-made holder to suit long working distance imaging. Specimen illumination from a cold light source (Microtec Fibre Optics, Micro Instruments, Long Hanborough, UK) was also passed through a polarising filter. Suitable rotational alignment of illumination and collection filters allowed significant reduction of unwanted light reflections from the wax surface, and weighing of signal acquisition towards superficial tissue layers. These overview images provide useful information on tissue shape and boundaries prior to sectioning, important for the subsequent correction of non-rigid deformations induced by blade-tissue interaction and variability in slice relaxation. This is also relevant for the alignment of tissue sections (Fig 2a-b) with anatomical MRI-derived sections of the previously imaged whole heart (Fig. 2d).
Figure 2.
Illustration of key “wet-lab” cardiac tissue processing stages. (a) Low resolution image of a black wax-embedded rat heart ready for sectioning, showing left ventricle [LV], right ventricle [RV] and aorta [A]. (b) Corresponding Trichrome stained montage image of an entire long-section, taken from the wax block shown in (a). (c) High-resolution fragment of the same image, obtained using ×63 objective, revealing sub-cellular detail such as sarcomeres [S], nuclei [N] and interstitial spaces [IS]. (d) Long-axis MRI section, from the same heart, before processing for co-alignment of histology and MRI data (see Section e, below).
Sections were cut at a thickness of 10 μm using a Leica SM2400 heavy duty, sledge-type microtome. Prior to mounting, the tissue was allowed to relax on a water bath (Leica Microsystems, HI 1210) at 39°C for 2 to 15 min, depending on the size of the tissue section. Slides were air dried in a Laminar Airflow Bench (overnight), followed by de-waxing in a 60°C oven (overnight) and Trichrome staining, using an automated stainer (Leica AutoStainer XL, ST50-10). The protocol used is as follows: heat (1 h at 60°C), leave to cool (30 min), expose to Xylene (2×15min), 100% alcohol (10 min), 70% alcohol (10 min), rinse in flowing tap water (10 min), stain with haemotoxylin (5 min), rinse in flowing tap water (4 min), stain with xylidine ponceau acid fuchsin (3 min), rinse in flowing tap water (1 min), expose to phosphomolybdic acid (3 min), expose to flowing tap water (30 seconds), stain with light green (1 min), rinse in flowing tap water (1×1 min; 1×30 seconds), to 70% alcohol (15 seconds), 100% alcohol (10 min) and finally in xylene (30 min).
After the last exposure to xylene, slides were mounted in DPX (Sigma, Poole, UK), taking care to avoid dry mounting (i.e. blotting the excess xylene with soft paper tissue before mounting, but not allowing the section to dry), and left overnight in a fume hood. The Trichrome stain labels connective tissue bluish-green, myocytes pinkish-red and nuclei blue-black (see Fig. 2c).
The stained and mounted sections were then imaged at high-resolution with Leica Application Suite, operating the LAS Power Mosaic module (which allows the entire specimen to be scanned at high speed and immediately combines the captured images to form a seamless mosaic image) integrated with an automated Leica DM4000B light microscope and a Märzhäuser IMI 10 × 1001 mm scanning stage (Part. No. 12723940), fitted with stepper motors on each axis which move the stage by means of a leadscrew when the motor is rotated. The distance moved by the stage is determined by a leadscrew pitch (here 1 mm). Motor rotation is set by an Oasis XY control board, using nano-stepping to advance in very small steps (typically 40,000 steps per rotation, yielding a step size of 25 nm). Reproducibility is determined by the mechanical accuracy of the leadscrews and the gearing that it uses. The operating software is an enhanced version of the Leica QWIN package, Leica LAS Power Mosaic, kindly adapted by Geoff Jenkinson (Leica, UK) to improve pixel alignment in neighbouring mosaic images, speed of scanning, removing image size restrictions, and offering an improved auto-alignment and calibration mode. With a ×10 objective and a Leica DFC 320 camera, pixel size of specimen images is (0.546 μm × 0.546 μm). The updated software also reduces the time required to save images, from previously 15-20 min (using Leica’s QWIN application) to now 4-5 min per section. The LAS Power Mosaic not only offers the benefits of acquiring and viewing massive mosaic images, but also supports image streaming (where mosaic image size is limited only by the free disk space). With this operating system, additional scans can be added to the initial scan to include all parts of a specimen. The overlapping of tiles allows smooth merging of joints. Power Mosaic automatically records camera rotation and stage skew, and allows for adjustment which is compensated for in the mosaic creation. The system was set-up and updated (with the help of Manuel Beynon and Tracey McKeown, Leica, UK) to the following specification: CPU: Pentium ®4 CPU 3 - Leica Q550 IW with 2GB of RAM and 3.40 GHz. Using a ×63 objective, sub-regions of the specimen have been imaged at significantly higher in-plane resolution (0.086 μm × 0.086 μm), (Fig. 2c). This data is suitable to compare, for example, sarcomere lengths in different regions of hearts.
The complete histological data set per heart, comprises a data volume of 46 GB for rat and 1.6 TB for rabbit heart. These were stored on a Network-Attached Storage (NAS) system (ReadyNAS NV+). While set-up to be accessed via the internet, on-line transfer even of individual sections (up to 1.85 GB for rabbit long-sections) at full resolution is slow, so back-up copies were positioned at two locations for on-site access and processing.
The sheer size of individual mosaic images makes it impossible to render them natively under 32 Bit Microsoft Windows. A bitmap viewer was therefore developed. The software can be freely downloaded from http://mef.physiol.ox.ac.uk/Software/BMPViewer.exe, and handles images of any size on any Microsoft Windows PC (we were able to visualise a 1.85 GB image on a machine with only 512 MB of memory). Using an explorer-like interface, the user can select an image. This automatically generates a thumbnail version of the image in a preview window, and renders part of the image at its native resolution in the main window. The content of the main window is automatically updated when a different area is selected in the preview window (and vice versa).
(c) MRI
Prior to the inherently destructive histological processing, MRI was used to non-invasively provide 3D data sets of all the ex-vivo hearts. Imaging was carried out on a 9.4 T (400 MHz) MR system (Varian Inc, Palo Alto, CA, USA), comprising of a horizontal magnet (bore size 210 mm), a VNMRS Direct Drive console, and a shielded gradient system (1 T/m, rise time 130 μs). A birdcage coil with an inner diameter of 28 mm (Rapid Biomedical, Würzburg, Germany) was used to transmit/receive the MR signals.
(i) Anatomical Imaging
Anatomical MRI scans were performed on all ex-vivo hearts, using the 3D gradient echo technique described by Schneider et al. (2004). The highest resolution data sets acquired were 21.5 μm isotropic resolution for rat (data reconstructed as described before in Schneider et al. 2004) and 26.4 μm × 26.4 μm in plane, 24.4 μm out-of-plane for rabbit (as described by Burton et al. 2006).
(ii) Diffusion Tensor Imaging
A 3D fast spin echo pulse sequence was developed to provide diffusion weighted MR images, and applied to ex-vivo rat hearts. During acquisition, a pair of unipolar trapezoidal diffusion gradients was applied either side of the first 180° pulse, and eight echoes were collected per excitation to reduce scan times. All diffusion and imaging gradients (including cross-terms, to account for the interactions between the two) were included in the calculation of the diffusion weighting factor (or b-value, typically 700-870 s/mm2). Diffusion gradients were applied in 6 non-co-linear directions, according to an optimised gradient scheme, based on the analogy of electrostatic repulsion described by Jones et al. (1999). Here, we collected diffusion weighted 3D data sets at an isotropic resolution of 101.6 μm in the ex-vivo rat heart.
Following data acquisition, the diffusion tensor was calculated on a voxel-by-voxel basis via a weighted linear least squares fit method (Kingsley et al. 2006), using in-house software developed in IDL (Interactive Data Language, ITT Corporation, Colorado, USA). The diffusion tensor was then decomposed to obtain its primary, secondary and tertiary eigenvectors.
There is very strong evidence, based on comparisons with histological data (Hsu et al. 1998; Scollan et al. 1998), that the primary eigenvector obtained using cardiac DT-MRI is locally aligned with the orientation of ventricular cardiac myocytes (usually, but potentially misleadingly, referred to as “fibre orientation”). Cardiomyocytes are additionally organised in layers (or sheets), that are 4-6 cells thick, resulting in a greater diffusion coefficient within the plane of the sheets, compared to the sheet normal (assumed tertiary eigenvector direction) (LeGrice et al. 1995). The secondary eigenvector is believed to be pointing in the plane of cell layers, but perpendicular to the main cell axis (Rohmer et al. 2007).
Various rotationally invariant parameters can also be derived from the diffusion tensor, such as the Fractional Anisotropy, which describes the level of anisotropy of the diffusion of water at a given voxel, and the Apparent Diffusion Coefficient, which is the mean of the three eigenvalues of the diffusion tensor, and describes the magnitude of local water diffusibility.
(d) Image Processing
Segmentation is the critical first stage in extracting geometrical information from the MR data sets to allow construction of a high-resolution anatomically detailed computational cardiac model. Initially, one must identify the voxels that represent “tissue” and differentiate them from others that represent “non-tissue”. This is achieved by creating a binary mask that allows discretisation between the myocardium and the surrounding background volume and cavities, as well as the identification of larger extracellular cleft space and vessels.
(i) Data Set Pre-Processing
The full resolution MR data set was first downsampled by a factor of 2 in each direction to produce a more computationally-managable volume containing 512 × 512 × 1024 voxels. Although such an approach reduces the definition of finer structures (such as the thin blood vessel walls on the epicardial surface, which in turn leads to problems in accurately delineating them via automated segmentation), it was necessary due to the large memory footprint produced by the high-resolution MR data set. Finally, the data set was further reduced in size to 451 × 416 × 714 voxels by removing those regions that contained no cardiac tissue. In addition, the original MR data was converted from 16-bit image data to 8-bit, and rescaled to occupy the full range of grey-scale intensities.
(ii) Segmentation Pipeline
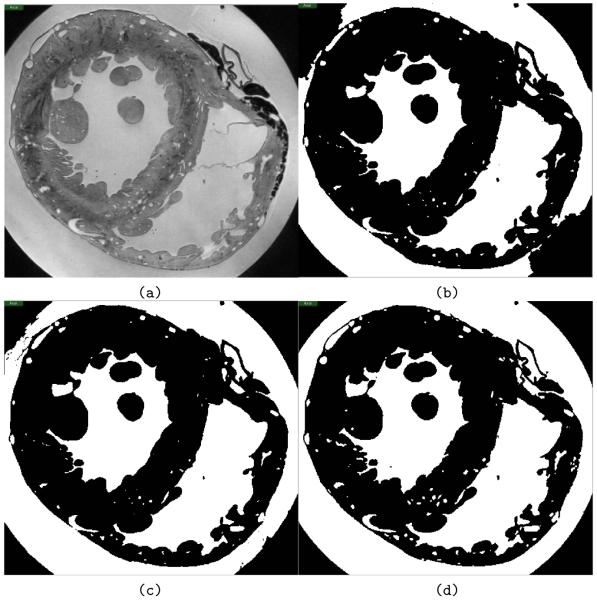
Spatial variations in coil sensitivity create a bias field, which causes partial overlap in image intensity of tissue and background voxels, rendering simple segmentation approaches, such as intensity thresholding, impractical. Bias field correction was attempted without success. Instead, a pipeline of three level-set segmentation filters was applied (described in more detail below), which allowed successful segmentation of the data set (Fig. 3).
Figure 3.
Automated segmentation results. (a) Transversal slice through the unsegmented original MR data set. (b) Output from the first step of the segmentation pipeline, the threshold level-set filter, which acts as a good initial approximation for segmentation. (c) Data after the geodesic active contour filtering. (d) Final result of the segmentation pipeline, following the Laplacian level-set filter.
Firstly, an initial approximation was performed using a level-set threshold filter based segmentation (Fig. 3a and b), following prior selection of seed-points throughout the volume of the data set, chosen based on intensity values to obtain a very conservative threshold (avoiding any effects of the bias field). Propagation of the evolving contour in the threshold level-set method depends on user-defined lower and upper threshold limits. By selecting a high intensity value (i.e. relatively bright) for the lower threshold, an approximate segmentation was achieved without the fronts “leaking” into regions of tissue with relatively high intensity values. However, due to spatial variations in coil sensitivity, this also left areas of the extra-cardiac background incorrectly defined. The result of the threshold level-set filter (Fig. 3b) was subsequently used as an initial level-set for a geodesic active contour level-set filter, which attracts the contour to object boundaries (Fig. 3c). The output of the geodesic filter was used as an initial level-set for a Laplacian level-set filter, which is useful for fine-tuning an initial segmentation (Fig. 3d).
(iii) Segmentation Post-Processing
Following the application of the segmentation pipeline, the binary mask was further improved by a series of morphological operations. Isolated segmentation artefacts were removed using a connected component algorithm. This is important, since all structures of the heart are physically connected. Thin-walled blood vessels on the outer-surface of the cardiac volume, for example, which were not always sufficiently well-defined during the segmentation process, were manually corrected. Finally, despite care in the sample preparation stage, a small number of air bubbles were inevitably present in the MR data set. These were also manually removed.
As the goal of this study was the generation of detailed ventricular models, atrial tissue was removed from the segmented data set. To achieve this, a series of points were manually selected along the line believed to separate ventricles and atria. Approximately 20 to 30 points were identified in every 5th slice along the longitudinal axis of the data set. B-splines were used to fit a smooth surface to the points which allowed dissection of the image volume. The smoothness of the surface was controlled by adjusting parameters in the B-splines algorithm. This 2D surface in the 3D object space defined a secondary binary mask that separates the ventricles from the atrial tissue.
The final stage of post-processing involved tagging of different structures in the heart, whose specific electrical, mechanical and/or functional properties may be important for simulations of heart function. Binary masks were created manually for the heart valves and the papillary muscles, and then convoluted with the segmented image stack to numerically tag each region with a number different from bulk myocardium (tagged as #1). For example, the valves were tagged as #2 and the papillary muscles as #3. These numerical tags are preserved at the meshing stage (see below), and thus can be used to assign different functional properties to these tissue regions prior to simulations.
The automatic segmentation algorithms were implemented using the Insight Toolkit (ITK, www.itk.org), which contains a range of techniques for 3D medical image segmentation, including several level set algorithms. Matlab (The Math-Works, Inc.) was employed for a varixety of tasks, including image downsampling. When manual interaction was required, Seg3D (Scientific Computing and Imaging Institute, University of Utah, USA) was used.
(e) Histology/MRI Co-Registration
Building upon work presented before (Mansoori et al. 2007), we have begun co-registration of histology and MRI data, to merge cell-type specific information with the new generation of high-resolution histo-anatomical models. Although histological images provide resolution and cell type discrimination that is superior to MRI, the process for acquiring histological slices is exceedingly time-consuming, and tissue processing introduces several rigid and non-rigid geometrical transformations, due to dehydration, embedding, cutting and rehydration of individual slices. This hinders the construction of valid 3D models based solely on stacked histological data. In order to overcome these problems, we have developed an algorithm for 3D co-registration of histology and MRI. The algorithm involves an iterative process combining 2D and 3D rigid registration in order to obtain MRI slices corresponding to the histological sections, and currently finishes with a 2D non-rigid registration between these corresponding slices, in the following way:
3D rigid registration between MRI and histological volumes ().
2D rigid registrations between each histological section and its corresponding slice in the MRI data set ().
If convergence has not been reached, go back to 1.
2D non-rigid registration between histological sections and their corresponding MRI slices.
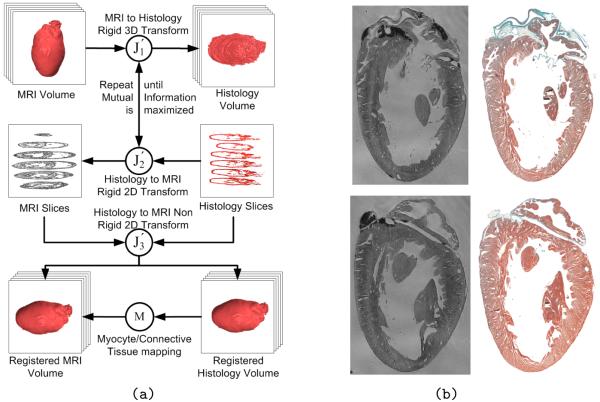
Details of the registration algorithm can be found in (Mansoori et al. 2007). Fig. 4 shows a schematic representation of the registration algorithm. It is important to note that, in comparison to algorithms that perform 3D reintegration based on a slice-to-slice registration of histological sections only, we obtain a realistic 3D geometry for the reconstructed histological data set, thanks to the prior use of non-invasively obtained MRI data as a reference data source.
Figure 4.
(a) Schematic depiction of the histology/MRI co-registration process (Mansoori et al. 2007), including the final step in which tissue segmentation from histology can be transformed into the MRI coordinate reference. (b) Result of the co-registration step, showing two selected histological sections next to their nearest corresponding MRI “slices”, which were found by non-rigid co-registrations.
The success of the registration algorithm described above depends largely upon the acquisition of “perfect” histological sections, without any nicks or folds which may occur during the cutting procedure (see regional damage of sections in papillary muscle area of Fig. 4b). Data “lost” in these areas is approximated by interpolation from neighbouring sections. In addition, a digital camera is used to take pre-cutting “in-the-wax” images of the tissue block (described in Section b), which will be incorporated into the registration algorithm to facilitate correction of non-rigid deformation of slices.
(f) Mesh Generation
Although unstructured mesh generation techniques were applied previously to develop anatomically realistic meshes of the ventricles (Trayanova et al. 2002, Plank et al. 2007, Vigmond & Clements 2007) and the atria (Harrild & Henriquez 2000), hearts in these studies were modelled in a stylized fashion. Anatomical details of potentially important structures, such as the papillary muscles and trabeculations, were ignored. Unstructured mesh generation techniques employed, although perfectly adequate for the sake of generating single high-quality meshes of a stylized heart, were not applied in this study for two reasons: firstly, the previously employed methods relied on highly interactive processing techniques, where a steep learning curve required substantial training. This means that generation of a single mesh of an anatomically detailed heart will normally require weeks. Secondly, traditional mesh generation approaches require a discrete description of the cardiac surfaces to serve as an input for subsequent mesh generation steps. This can be avoided when grid-based mesh generation techniques are used that operate directly from the segmented image stack to discretise the volume (Young et al. 2008, Prassl et al. 2008).
A fully automatic mesh generation technique would offer significant advantages for a high-throughput model generation pipeline, ideally requiring no user interaction at all. This is of particular practical relevance considering the volume of individual high-resolution imaging data sets, and the desire to process multiple data sets in order to feed into probabilistic atlas generation projects. Furthermore, due to the fine discretizations that are required to resolve histo-anatomical detail, the mesh generation software has to meet certain technical requirements to be able to deal with the memory demands and computational load associated with the generation of large-scale meshes.
To avoid complex mesh generation steps one could resort to a naïve technique by simply assuming that each voxel in the image stacks corresponds to a hexahedral finite element in the mesh. However, such an approach would be inadequate to meet the objectives of this study for two main reasons. Firstly, structured meshes suffer from limited applicability. The delivery of external currents to the heart, for instance, inevitably leads to spurious polarisations along the jagged surfaces of structured grids. This is the result of tip effects, as in this region the alignment of conductivity tensors is parallel to the anatomical surface of the heart, but not to the jagged geometric surface representation in a structured mesh. Furthermore, structured grids lead to substantial overestimation of the organ’s surface area, even with infinitely fine resolutions (Young et al. 2008). The effects of this artificially enlarged surface and, thus, extended tissue-bath interface, have not yet been studied in suffcient detail. Secondly, whenever a surrounding volume conductor is included into the model formulation, for instance to model the in-vivo setting where the heart is surrounded by a conductive torso volume, the computational load can be substantially lessened by reducing the spatial resolution of the mesh with increasing distance from the cardiac surfaces. This can be done without any loss in accuracy since the lowpass filtering effect of the torso means that the extracellular potential, ɸe, is a much smoother function that can be captured at a lower spatial resolution. However, inside the myocardial tissue, where steep depolarisation wavefronts need to be resolved, the mesh discretisation must be significantly finer. Thus, spatially adaptive unstructured mesh generation techniques are ideally suited for this purpose, allowing one to minimise the degrees of freedom required to represent the volume conductor, while preserving the conformity of the mesh.
In this study, the commercial mesh generation software Tarantula (www.meshing.at) was used. The underlying algorithms are similar to a recently published image-based unstructured mesh generation technique (Prassl et al. 2008). Briefly, the method uses a modified dual mesh of an Octree applied directly to segmented 3D image stacks. The method automatically produces conformal, boundary-fitted, hexahedra-dominant meshes without requirement for interactivity. Generated meshes are ac-curate volume-preserving representations of arbitrarily complex geometries, while conserving smooth surfaces.
(g) Accounting for Tissue Anisotropy
In order to realistically represent the anisotropic conduction within the heart, an anatomically-based model of cell alignment (“fibre architecture”) has to be incorporated into the cardiac mesh. Two strategies were considered. In those cases where both DT-MRI and anatomical MRI data are available for the same heart, DT-MRI tensor data are interpolated from the structured voxel grid onto the unstructured mesh. In absence of DT-MRI data, fibre information was incorporated on a per rule base using a-priori knowledge.
(i) Rule-based Approach using a-priori Knowledge
The method used was based on that of Potse et al. (2006). Before implementation of the fibre orientation algorithm, the three discrete surfaces (epicardium and the LV/RV endocardium) were defined. This was achieved by performing a secondary segmentation of the MR data, where seed-points were placed within the two ventricular cavities and outside the volume of the heart (i.e. excluding the blood vessels and the extracellular cleft spaces). Meshing of the resulting segmented data set produced elements with three different numerical tags representing the heart tissue volume, LV cavity, and the RV cavity. The epicardial and two endocardial surfaces could then be identified by examining the numerical tags of elements bordering each surface triangle. The spatial coordinates of the node points, defining the triangles on each of the surfaces, were then mapped onto the final ventricular mesh, as shown in the inset of the middle panel in Fig. 1 which shows the LV endocardium in red.
Subsequently, the minimum distance of each node point within the mesh from both the epicardium (depi) and the endocardium (dendo) was computed, and used to define a normalised transmural location parameter e
| (2.1) |
Note that e = 0 when dendo = 0 and e = 1 when depi = 0. In the septum, the depi was taken to be the minimum distance from the endocardial surface of the RV and dendo from the endocardial surface of the LV, replicating the apparently continuous cell alignment of LV and septum, described in previous studies (LeGrice et al. 1995, Scollan et al. 2000). The gradient of e within each tetrahedral element (▽e) then defined the transmural direction. Circumferential cell orientation was assigned to be orthogonal to ▽e. The actual direction of cardiac cell alignment was further determined by a helix angle
| (2.2) |
to account for their transmural rotation through the myocardial wall (Streeter et al. 1969). R was taken to be equal to π/3 for the LV and septum and π/4 for the RV. Cell orientation in the papillary muscles (tagged using the procedure detailed above) had a direction imposed to be along the axis of the papillary muscle. Finally, a smoothing operation was performed on fibre orientations in order to avoid discontinuities.
(ii) Interpolation of DT-MRI data
In order to use DT-MRI data to describe tissue anisotropy, DT-MRI tensor data are mapped to each voxel in the image stack. An interpolation scheme has to be devised that interpolates data, defined on the centroids of the voxels in the image stack, to the centroids of the elements spanning the unstructured grid. Since the segmented object in the image stack and its discrete representation by a mesh are defined in the same physical space, for each finite element the enclosing set of voxel centres can be easily identified. The selected voxel centres span a hexahedral element, where at each spanning node DT-MRI data are defined. Interpolation of vector data is then achieved by trilinear interpolation within the hexahedral element. Accuracy of this approach is limited along the surfaces of the heart, since a subset of nodes will be located outside the tissue (where water diffusibility is isotropic). In those cases, a nearest neighbour interpolation may reduce isotropic bias, introduced by non-myocardial voxels.
(h) Governing Equations
The bidomain equations (Plonsey et al. 1988), a continuum approximation of cardiac tissue that is based on the idea of a syncytium, state that currents that enter the intracellular or extracellular spaces by crossing the cell membrane represent the sources for the intracellular, ɸi, and extracellular potentials, ɸe
| (2.3) |
| (2.4) |
| (2.5) |
| (2.6) |
| (2.7) |
where and are the intracellular and extracellular conductivity tensors (respectively), β is the membrane surface to cell volume ratio, Im is the transmembrane current density, Itr is the current density of the transmembrane stimulus (used to initiate an action potential), Ie is the current density of the extracellular stimulus, Cm is the membrane capacitance per unit area, Vm is the transmembrane voltage, and Iion is the density of the total current flowing through the membrane ionic channels, pumps and exchangers, which in turn depends on the transmembrane voltage and on a set of state variables . At the tissue boundaries, electrical isolation is assumed, which is accounted for by imposing no-flux boundary conditions on ɸe and ɸi.
In those cases where cardiac tissue is surrounded by a conductive medium, such as blood in the ventricular cavities or a perfusing saline solution, an additional Poisson equation has to be solved
| (2.8) |
where σb is the isotropic conductivity of the conductive medium and Ie is stimulation current injected into the conductive medium. In this case, no-flux boundary conditions are assumed at the boundaries of the conductive medium, whereas continuity of the normal component of the extracellular current and continuity of ɸe are enforced at the tissue-bath interface. The no-flux boundary conditions for ɸi remain the same as in the case when the tissue is surrounded by a non-conductive medium, described above.
In this study, the bidomain equations were recast by applying the widely used transformation, proposed by Pollard et al. (1992), where Eqs. (2.3) and (2.4) are added and ɸi is replaced by Vm + ɸe. This yields
| (2.9) |
| (2.10) |
where Vm and ɸe, the quantities of primary concern, are retained as the independent variables. Note that Eq. 2.9 treats the bath as an an extension of the interstitial space.
(i) Spatiotemporal Discretisation
The bidomain equations were discretised based on Eqs. (2.9-2.10). An operator splitting technique was applied (Keener & Bogar 1998) to decouple the computing scheme into three components, an elliptic partial differential equation (PDE), a parabolic PDE, and a set of non-linear ODEs. Solutions are then found by leap-frogging between the decoupled components where either Vm in Eq.(2.9) or Φe in Eq.(2.10) are considered as constant. Discretising the decoupled bidomain equations leads to a three-step scheme, which involves a solution of the parabolic PDE, the elliptic PDE and the non-linear system of ODEs at each time step. In the simplest case, both parabolic PDE and the non-linear ODE systems can be solved with an explicit forward Euler scheme (Vigmond et al. 2002)
| (2.11) |
| (2.12) |
| (2.13) |
| (2.14) |
where Aξ is the discretised −▽ · (σ̅ξ▽)/(βCm) operator with ~ being either i or e; Δt is the time step; Vk, and are the temporal discretizations of Vm, ɸe and , respectively, at the time instant of kΔt.
In this study, due to the fine discretization of the mesh, it was advantageous to employ the (computationally more expensive, but larger integration time steps are possible) semi-implicit Crank-Nicolson scheme instead of the forward Euler to solve the parabolic PDE
| (2.15) |
which replaces Eq. (2.11).
Both monodomain and bidomain simulations were conducted. The numerical scheme in the monodomain case is essentially the same, with the difference that the step to solve the elliptic PDE and the term including ɸe in Eq. (2.10) are omitted, and the conductivity tensor σi in Eq. (2.10) is replaced by σm where
| (2.16) |
with ζ being one of three possible eigendirections of the conductivity tensor (Hooks et al. 2007).
(j) Numerical Setup and Benchmarking
The larger (rabbit) ventricular mesh consists of 4.3 million vertices and 24 million tetrahedral elements. To demonstrate the feasibility of using large high-resolution meshes for in-silico electrophysiological studies, benchmark runs were executed employing CARP. The numerical set-up was similar to a previous study (Plank et al. 2006), with the minor difference in the temporal discretization method for the parabolic PDE as described above. The parabolic problem was solved using a Block-Jacobi preconditioner together with an Incomplete Cholesky ICC(0) sub-block preconditioner with a zero fill-in level to preserve the sparsity pattern (Golub & Van Loan 1996). The solution was advanced using a fixed global time step of 25 μs. Membrane dynamics were modelled using a recent rabbit ventricular action potential model (Mahajan et al. 2007). The rabbit ventricular model was stimulated at the LV freewall using a transmembrane current pulse of 50×10−4 μA/cm3 and 1 ms duration. Electrical activity was simulated over 400 ms to×observe a full action potential cycle everywhere over the entire ventricular volume. The simulations were executed in parallel on the Oxford National Grid Service cluster, which is equipped with four Dual-Core AMD Opteron 885 Processors per node, varying the number of nodes from 1 to 32 (and, hence, of processors between 4 and 128). Execution times spent on solving the parabolic PDE and the set of ODEs were determined. In addition, the same set of runs was also executed using a bidomain formulation and the execution time required to solve the elliptic PDE was measured as well. For the benchmarks of the elliptic solver performance, only 10 ms of activity was simulated.
3. Results
(a) Image Acquisition
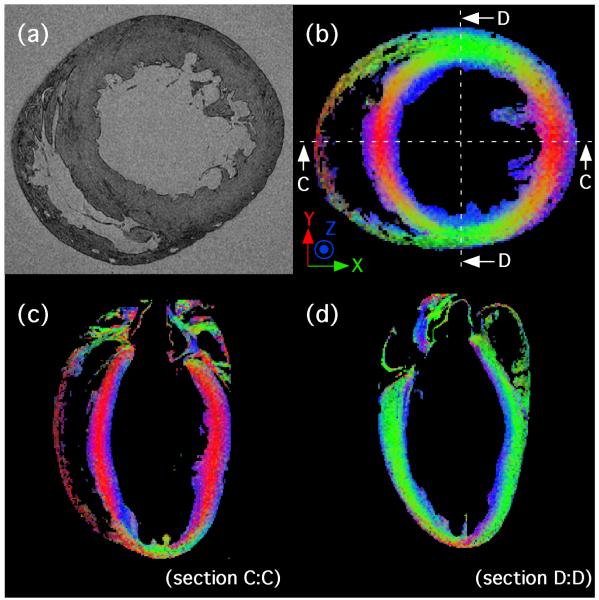
A short axis section from a 3D anatomical scan of an ex-vivo rat heart is shown in Fig. 5a, along with three colour-coded slices from a 3D DT-MRI data set from the same heart, showing primary eigenvector distribution. An XYZ → RGB colour scheme has been used to represent the direction of the primary eigenvector at each voxel (see Fig. 5b-d). The mean fractional anisotropy value in the entire LV volume is 0.29 ± 0.06, and is generally greatest in the mid-wall region, where the cardiomyocytes are orientated primarily in the short-axis plane of the heart.
Figure 5.
(a) Short axis section from a 3D anatomical scan of an ex-vivo rat heart. Voxel resolution is 21.5 μm isotropic. (b) Transverse view from a 3D, 101.6 μm isotropic resolution DT-MRI scan of the same heart. (c) Four chamber view (section C:C as illustrated in (b)) and (d) two chamber view (section (D:D). Colours are determined using an XYZ - GRB colour scheme according to the voxelwise primary eigenvector orientation in the transverse plane. Note the XYZ - GRB colour scheme applies to (b) only, as views C:C and D:D have been rotated.
(b) Image Processing
(i) Segmentation
Results of the segmentation process are shown in Fig. 3. Image voxels have been separated into two classes (tissue and non-tissue) with non-tissue including cavities, vessels and interstitial space. Separation between these can be achieved based on connectivity. The results of the subsequent segmentation between ventricles and atria are shown in Fig. 6.
Figure 6.
Removal of the atria to generate a ventricular mesh. (a) Longitudinal slice through the same rabbit MR data set as in Figure 3 with manually-placed points along the line believed to separate the atria and ventricles. (b) Binary mask produced to remove atrial tissue. (c) Final segmented image stack after removal of the atria.
As can be seen from Fig. 3d and Fig. 6c, the final result of the segmentation pipeline very clearly identifies the ventricular cavities, medium and large blood vessels, and extracellular cleft spaces from the MR data set. The detail contained in the final segmented image is significantly higher than in previous studies (Plotkowiak et al. 2007) where the use of a data set downsampled by an additional factor of 2 in each direction was combined with only one level-set segmentation filter (a Fast Marching Method filter), which reduced identification of detail (e.g. only the largest blood vessels within the myocardial wall).
(ii) Histology/MRI Co-Registration
In order to quantitatively validate the accuracy of the registration results, an expert selected corresponding landmarks on data sets previously downsampled to 53×53×49 μm resolution for computational tractability. The mean Euclidean distance between corresponding landmarks was found to be 0.24 mm (4.4 pixels), suggesting reasonable agreement between co-registered structures.
(c) Mesh Generation and Assignment of prevailing Cell Orientation
The segmented rabbit heart image stack was fed directly into the mesh generation software Tarantula. Using 2 Intel Xeon processors, clocked at 2.66 GHz, the required execution time was 39 minutes. The final mesh, consisting of 7.2 million nodes and 42 million tetrahedral elements, included the entire ventricles, the ventricular cavities and a surrounding fluid bath. Out of these, 4.3 million nodes and 24 million tetrahedral elements formed the myocardial grid. The minimum / mean / maximum edge length of elements within the myocardial grid was 11.8 / 125.0 / 228.8 μm. A visualisation of the ventricular surface and a cross section which exposes the endocardial structures is shown in the middle panel of Fig. 1. All segmentation tags used to label different regions in the heart were directly transferred over from the regular image stack to the unstructured grid, and can thus be used to define electrophysiological properties on a per region basis (see inset in middle panel of Fig. 1 where the elements tagged as papillary muscle are coloured in green).
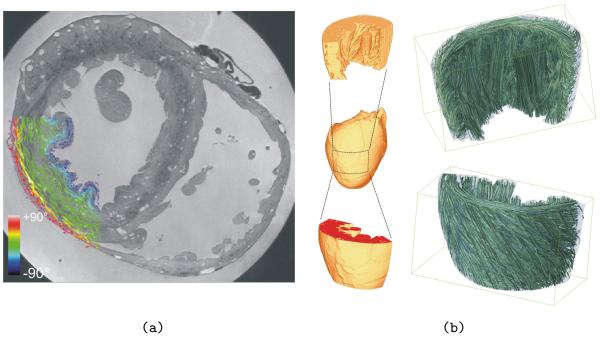
In the absence of DT-MRI data for this particular data set, the generated mesh was equipped with regionally prevailing cell orientation by applying the algorithm laid out in Methods (Section g). Fig. 7a shows this “fibre orientation” in a segment of the LV wall. Panel b shows streamline renderings in a LV wedge preparation generated from the primary eigenvectors which define the representation of “fibre orientation” in the model.
Figure 7.
(a) Vector representation of fibre directions (defined via the algorithm described in Section g) within a section of rabbit LV wall. The colour-bar represents the out-of-plane components of the fibre vectors. (b) Primary eigenvectors were used to generate streamline renderings in a LV wedge preparation (approximate region indicated by dashed rectangle) to allow a qualitative assessment of tissue anisotropy in the model. Top panel shows streamlines from an intra-cardiac viewpoint, bottom panel from an extra-cardial viewpoint. Note that streamlines here serve to illustrate 3D local orientation of myocardial cells only, and do not suggest the existence of myofibres in the heart.
(d) Simulations and Benchmark Results
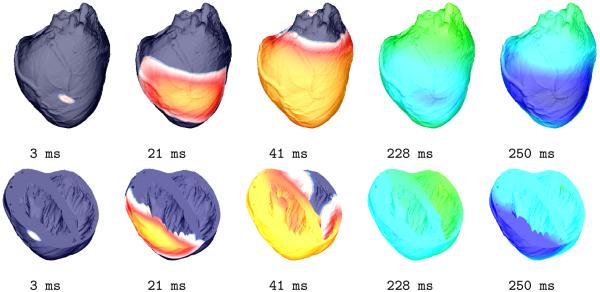
As described in Methods (Section j), a point-like stimulus was delivered to the central LV free wall, to initiate a propagating wavefront. Snapshots of the activation sequence at the epicardium, and over a cross-section through the ventricles, are shown in Fig. 8 at different phases of an entire action potential cycle, illustrating the feasibility of model application to electrophysiological in-silico research.
Figure 8.
Activation sequence used for proof-of-principle simulation. The distribution of Vm (dark blue corresponds to −90 mV, red to +40 mV) on the epicardial surface is shown for various time-points of an action potential cycle (top row). The bottom row shows the same sequence where a transversal clipping plane exposes transmural and endocardial activity.
Execution times, Texec, for the different portions of the the numerical scheme were measured as a function of the number of processors, Np. Benchmark results are summarised in Tab. 1. The simulation of 1 ms activity, using Np = 64 processors, lasted 6.7 s for monodomain and 70.6 s for bidomain formulation. Thus, with the hardware used, monodomain simulations are an order of magnitude faster than bidomain, but both are significantly slower (104 – 105) than real time.
Table 1.
Summary of average execution times (Texec, in seconds) required to simulate 1 ms of activity in a tissue model with one 4.3 million cells, running on the Oxford NGS cluster with varying numbers of processors
|
Np # |
Parabolic [s] |
ODE [s] |
Elliptic [s] |
Total Mono [s] |
Total Bidomain [s] |
|---|---|---|---|---|---|
| 4 | 73.03 | 7.46 | X* | 80.49 | X* |
| 8 | 42.50 | 3.72 | 401.82 | 46.22 | 448.04 |
| 16 | 21.84 | 1.86 | 214.34 | 23.70 | 238.04 |
| 32 | 13.23 | 0.94 | 114.33 | 14.17 | 128.50 |
| 64 | 6.32 | 0.39 | 63.89 | 6.70 | 70.59 |
| 128 | 4.03 | 0.17 | 61.01 | 4.22 | 65.23 |
Simulation could not be executed on a single node since the available memory was insufficient.
4. Discussion
(a) MRI
While MRI offers non-destructive 3D imaging in a fraction of the time required for serial sectioning-based histology, spatial resolution and cell type discrimination are inferior to that achieved with histological techniques. This is due to the physical limitations of the technique, such as signal-to-noise-ratio (SNR) and T2 decay. This problem is augmented further during diffusion tensor imaging due to the attenuation of the MR signal by the applied diffusion gradients, which provides an additional constraint on spatial resolution.
The collection of high-resolution (~100 μm) 3D DT-MRI data sets also creates significant demands on scan time, due to the necessary repetitions of the scan with diffusion gradients applied in at least 6 non-co-linear directions, in order to calculate the diffusion tensor. This necessitates the use of fast imaging techniques such as the fast spin-echo pulse sequence mentioned earlier. While this reduces scan times, typically by a “turbo” factor of between 4 and 16, it can create a fresh set of challenges for interrelating anatomical MRI and DT-MRI scan data. Thus, optimal sample preparation for anatomical MRI scans involves doping of the tissue with gadodiamide to enhance contrast between extracellular space and myocardium. The ideal preparation for DT-MRI, however, contains no paramagnetic contrast agent, as the resulting decrease in the T2 decay constant reduces the SNR and limits the maximum possible “turbo” factor. Observations such as this have helped us develop an efficient “pipeline” for the imaging of ex-vivo hearts, in which, for instance, DT-MRI is carried out immediately after excision, and before overnight addition of gadodiamide to the fixative to improve subsequent anatomical scans. Also, in particular for long-term studies, we have replaced agarose by gelatine as the embedding medium, as the former is gradually resolved, apparently by the fixative residue contained in the cardiac tissue.
Application of similar imaging techniques to the in-vivo setting is associated with new challenges. The high-resolution, achieved in anatomical scans ex-vivo, requires long scan times (e.g. ~12 hours for 213 μm3 resolution in the rabbit heart). This would be impractical for in-vivo work. Similar scan time limits affect the resolution attainable with DT-MRI, while cardiac and respiratory motion cause additional problems. Our current DT-MRI techniques probe the diffusion of water on a characteristic length scale of 10-100 μm, in order to examine architectural features associated with cardiac histo-architecture. They are therefore extremely sensitive to any bulk motion. If the fast spin echo pulse sequence, developed for ex-vivo DT-MRI, proves to be too slow for in-vivo studies, it will need to be adapted to single / multi-shot echo planar imaging. Whereas the echo planar technique is significantly faster than fast spin echo, the associated images are prone to artefacts caused by field inhomogeneities and eddy current distortions.
(b) Image Processing
Due to the size and complexity of the images, a pure manual segmentation approach is impractical. The algorithm we have developed and presented here automates the majority of the segmentation process without compromising the accuracy of the results. However, a certain amount of user interaction is still required. For example, walls of vessels located at the surface of the myocardium can sometimes have a width close to image resolution, and thus are not always fully detected by the segmentation. Though the extent of the undetected region is usually only of a few voxels, this introduces topological errors which could affect simulation results. At present, we correct these areas manually. Future work will include a vessel identification algorithm, which will eliminate this error source. Also, atria and ventricles are separated manually using a point cloud. Though the time spent on this is short compared to other stages in the process (such as sample preparation) a full automation is desirable and will be a topic of further development.
At present, images are downsampled before segmentation is done. This is due to the computational cost of applying the algorithms to full-resolution data sets. This introduces segmentation errors which, though local, may affect tissue topology, requiring manual inspection and correction. The downsampling conducted here was a factor of 2 in each direction less than in our previous study (Plotkowiak et al. 2007), and we are currently developing high-performance computing algorithms to avoid this step entirely.
A mathematical algorithm has been implemented to assign local cell orientation to the myocardium. Though this is superseded by DT-MRI images, there are cases when directly recorded data may not be available. For example, DT-MRI is currently not available with sufficiently high-resolution in-vivo. The accuracy of representation by the rule-based approach, when compared to DT-MRI, is another important aspect of further research and development.
We have demonstrated an algorithm to correct for geometrical deformations in histological images via co-registration with MRI. Quantitative validation provided an average distance between manually placed landmarks of 0.24 mm. Possible causes for this are a) the inaccuracy in manual landmark placement and b) the presence of 3D non-rigid deformations. To assess the effect of (a), a possibility is the automated identification of line or surface landmarks, e.g. vessel surfaces, which are easier to define than point landmarks. Regarding (b), 3D deformations introduced in the cardiac volume prior to sectioning by differences in embedding for MRI (agar or gelatine) and histology (wax) are not yet taken into account in the algorithm. Additional refinement will involve an attempt to add 3D non-rigid registration steps. However, this also has the potential of introducing deformation artefacts. Further assessment of the performance of the algorithm will also be achieved when 3D structures are reconstructed from the histological data sets, which is a topic of current work.
In a refinement step during the project’s progress, acquisition of an additional set of low-resolution images prior to slicing was introduced (Fig. 2a). The co-registration algorithm described here was developed before these images were available, and thus does not yet make use of them. Further development will include these in the co-registration pipeline, with the expected result of an increase of robustness.
(c) Computational Aspects
Ideally, “in-silico” experiments should match as closely as possible experiments conducted in the “wet” lab, or clinical observations. The employed computer models should include sufficient anatomical and functional details to yield an acceptable match, in order to allow predictive modelling. Due to the tremendous computational burden associated with highly realistic simulations of cardiac electrical activity, this remains challenging. This study is an initial attempt to bring cardiac modelling closer to the idea of a “virtual heart”. The simulations performed here used a model of an entire ventricle at an unprecedented level of anatomical detail. The ventricular volume was faithfully represented by a high-resolution mesh that matched rabbit and rat ventricular tissue structure with an uncertainty of less than 50 μm. The ionic model by Mahajan et al. (2007) used in this study is among the most detailed models published thus far.
Although the current performance lags significantly behind real time simulations (by a factor of 104-105, even using 64 or 128 processors), it is sufficient to address the requirements of many studies of potential interest, particularly when phenomena under investigation occur over a time scale of up to a few seconds.
Simulations involving the observation of processes over minutes or more are difficult, and further significant performance improvements are required. With the given rabbit ventricular model set-up and Np = 64, the simulation of 1 min of cardiac activity takes over 4 days (using monodomain formulations), which is too slow for efficient exploration of parameter spaces. However, these simulations were proof-of-principle work on a new hardware platform, i.e. without spending additional effort on optimisation. Simple measures such as using hardware optimised math libraries and a selection of compilers that give best performance on this platform can help to increase performance noticeably. For hearts of smaller species such as rats, however, the current performance suffices to observe activity over a minute with execution times of a few hours only.
To allow parameter sweeps with anatomically detailed models, in particular using hearts of larger species such as humans, or to observe phenomena over time scales of minutes and more, compiler optimisations will not suffice and other approaches have to be investigated to arrive at the desired performance increase. Although using 128 processors (the maximum assessed in this study) may seem an expensive simulation today, future computing platforms will employ similar processing units as a default.
While further increasing the number of cores may improve performance for larger hearts, for the given rabbit ventricular model the data presented in Tab. 1 suggest that 128 processors is an upper bound for good parallel scaling for the linear solver components, but not for solving the set of ODEs. Solving the ODEs is unlikely to level off with increased Np, since state variables do not diffuse between cells. Solution to the systems of ODEs governing the state variables within each “cell” can therefore be easily split-up and computed independently on different computational nodes. This renders the problem “embarrassingly parallel”. While the time spent on solving the ODEs can be scaled down linearly with Np this is not the case with the linear solver steps. When increasing Np from 64 to 128 almost no reduction of the elliptic solver time could be achieved and the parallel speedup of the parabolic solve was rather poor (≈59%). While the employed methods are likely to scale up to larger Np as the problem size increases, for instance, when human hearts are under investigation, the increase of Np will become inefficient at a certain maximum Np and execution times will remain unsatisfactory using the current approach, not allowing the exploration of parameters spaces. The current problem size with 4.3 million degrees of freedom is sufficiently large, with local parallel partition sizes of ~ 17 × 103 degrees of freedom, so that the ratio between computation and communication, theoretically, should support an increase of Np beyond 128 processors. Further investigations are required to identify and, if possible, eliminate the limiting factor that prevented efficient parallel scaling beyond 128 processors.
Even though further performance improvements can be achieved by optimising the techniques used here, and by executing simulations on cutting edge HPC hardware, an increase beyond roughly one order of magnitude cannot be expected when using current computer architectures. But even today, a first glance at novel computing paradigms demonstrates the impressive computing power that will become available with multi-processor PetaFlop computing hardware. Also, accelerator technologies, such as general purpose graphics processing units (GPUs), offer significant gains in processing power for many (but not all) applications, with cheap off-the-shelve standard graphics hardware. It has been demonstrated recently that the elliptic PDE, associated with a whole ventricular bidomain simulation, can be solved in double precision on a single Tesla GPU in the same time as using 32 nodes of a cluster with low-latency interconnect, such as used in this study (Liebmann et al 2008). Although it is unclear which hardware paradigm will prevail, a consequent optimisation of algorithms for the novel platforms, the use of more computational cores, and further improvements in low-latency interconnects are likely to provide a performance boost of at least 3 orders of magnitude. Such environments, with appropriately optimised codes, will enable whole heart simulations without trade- offs in terms of reduced anatomical or functional detail, and with near-real time performance, opening entirely new areas of application, for example, support of emergency decision making in the clinical setting.
(d) Limitations
Although the novel methods presented here allow one to generate structural models of the heart at an unprecedented level of detail, by definition these models remain simplifications of reality.
The presented model resolves cardiac geometry at an average resolution of about 100 μm. That is, a discrete length scale is equivalent to roughly one cell length, or about 5 cell diameters. Anatomical properties below this resolution cannot be directly accounted for.
The extent to which a study requires representation of structural detail is determined by the particular application. Clearly, structures such as the Purkinje network, endocardial trabeculations, cleavage planes, or vessels have pronounced effects on cardiac electro-mechanical function, and computational evaluation of their roles will benefit from the here presented detail.
Finally, this article is focused on developing a methodology and on demonstrating feasibility. Specific electrophysiological implications of using such highly detailed models remained largely unexplored. It will take serious additional research efforts over the next years to study the multitude of effects which can be explored at high structural detail. Based on preliminary evidence, we expect new insight from highly detailed models in areas such as investigation of mechanisms underlying the formation of arrhythmias, or the response of the heart to the application of strong electric fields.
5. Conclusion
The development of a new generation of cardiac organ models, based on imaging modalities with para- or sub-cellular resolution, has the potential of significantly advancing insight into basic cardiac function and arrhythmia mechanisms, and - in the more distant future - may serve as tools for diagnosis and patient-specific assessment of treatment modalities. This development requires a combination of advanced imaging and simulation techniques, and automated tools to enable seamless handshake between them. The pipeline presented in this paper contributes towards approaching this goal.
6. Acknowledgements
This project is subject of a BBSRC technology development grant (#E003443 to PK, JS & DG). GP was supported by a Marie Curie Fellowship (MC-OIF 040190) and holds an Austrian Science Fund grant (SFB F3210-N18), while VG is a Research Councils UK Fellow. The authors acknowledge the support provided by the EPSRC Integrative Biology project (#GR/S720223/01) and an EPSRC grant (EP/F011628/1), as well as the European Commission Virtual Physiological Human initiative. TM holds a stipend by IBM and BR an MRC Career Development Award.
References
- Akar FG, Spragg DD, Tunin RS, Kass DA, Tomaselli GF. Mechanisms underlying conduction slowing and arrhythmogenesis in nonischemic dilated cardiomyopathy. Circ Res. 2004;95:717–25. doi: 10.1161/01.RES.0000144125.61927.1c. [DOI] [PubMed] [Google Scholar]
- Akar FG, Aon MA, Tomaselli GF, O’Rourke B. The mitochondrial origin of postischemic arrhythmias. J. Clin. Invest. 2005;115:3527–3535. doi: 10.1172/JCI25371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr RC, Plonsey R. Propagation of excitation in idealized anisotropic two-dimensional tissue. Biophys. J. 1984;45(6):1191–202. doi: 10.1016/S0006-3495(84)84268-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenfeld O, Jalife J. Purkinje-muscle reentry as a mechanism of polymorphic ventricular arrhythmias in a 3-dimensional model of the ventricles. Circ. Res. 1998;82:1063–1077. doi: 10.1161/01.res.82.10.1063. [DOI] [PubMed] [Google Scholar]
- Bishop MJ, Rodriguez B, Qu F, Efimov IR, Gavaghan DJ, Trayanova NA. The role of photon scattering in optical signal distortion during arrhythmia and defibrillation. Biophys J. 2007;93(10):3714–26. doi: 10.1529/biophysj.107.110981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop MJ, Bub G, Garny A, Gavaghan DJ, Rodriguez BR. An investigation into the role of the optical detection set-up in the recording of cardiac optical mapping signals: a Monte Carlo simulation study. Physica D. 2008 in press. [Google Scholar]
- Burton RAB, Plank G, Schneider JE, et al. Three-dimensional models of individual histoanatomy: tools and challenges. Ann. N.Y. Acad. Sci. 2006;1080:301–319. doi: 10.1196/annals.1380.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byars JL, Smith WM, Ideker RE, Fast VG. Development of an optrode for intramural multisite optical recordings of Vm in the heart. J Cardiovasc Electrophysiol. 2003;14(11):1196–202. doi: 10.1046/j.1540-8167.2003.03203.x. [DOI] [PubMed] [Google Scholar]
- Caldwell BJ, Legrice IJ, Hooks DA, Tai DC, Pullan AJ, Smaill BH. In-tramural measurement of transmembrane potential in the isolated pig heart: validation of a novel technique. J Cardiovasc Electrophysiol. 2005;16(9):1001–10. doi: 10.1111/j.1540-8167.2005.40558.x. [DOI] [PubMed] [Google Scholar]
- Cherry EM, Greenside HS, Henriquez CS. Efficient simulation of three-dimensional anisotropic cardiac tissue using an adaptive mesh refinement method. Chaos. 2003;13:853–865. doi: 10.1063/1.1594685. [DOI] [PubMed] [Google Scholar]
- Clancy CE, Rudy Y. Linking a genetic defect to its cellular phenotype in a cardiac arrhythmia. Nature. 1999;400:566–569. doi: 10.1038/23034. [DOI] [PubMed] [Google Scholar]
- Cohen LB, Salzberg BM. Optical measurement of membrane potential. Rev Physiol Biochem Pharmacol. 1978;83:3588. doi: 10.1007/3-540-08907-1_2. [DOI] [PubMed] [Google Scholar]
- Courant R, Friedrichs K, Lewy H. Über die partiellen Differenzengleichungen der mathematischen Physik. Mathematische Annalen. 1928;100:32–74. [Google Scholar]
- Dillon S, Morad M. A new laser scanning system for measuring action potential propagation in the heart. Science. 1981;214(4519):453–6. doi: 10.1126/science.6974891. [DOI] [PubMed] [Google Scholar]
- Ebihara L, Johnson EA. Fast sodium current in cardiac muscle. A quantitative description. Biophys J. 1980;32(2):779–90. doi: 10.1016/S0006-3495(80)85016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov IR, Sidorov V, Cheng Y, Wollenzier B. Evidence of three-dimensional scroll waves with ribbon-shaped filaments as a mechanism of ventricular tachycardia in isolated rabbit heart. J. Cardiovasc. Electrophysiol. 1999;10:14511462. doi: 10.1111/j.1540-8167.1999.tb00204.x. [DOI] [PubMed] [Google Scholar]
- Efimov IR, Huang DT, Rendt JM, Salama G. Optical mapping of repolarization and refractoriness from intact hearts. Circulation. 1994;90(3):1469–80. doi: 10.1161/01.cir.90.3.1469. [DOI] [PubMed] [Google Scholar]
- Fast VG, Sharifov OF, Cheek ER, Newton JC, Ideker RE. Intramural virtual electrodes during defibrillation shocks in left ventricular wall assessed by optical mapping of membrane potential. Circulation. 2002;106(8):1007–14. doi: 10.1161/01.cir.0000027103.54792.9c. [DOI] [PubMed] [Google Scholar]
- Fast VG, Kléber AG. Microscopic conduction in cultured strands of neonatal rat heart cells measured with voltage-sensitive dyes. Circ Res. 1993;73(5):914–25. doi: 10.1161/01.res.73.5.914. [DOI] [PubMed] [Google Scholar]
- Fenton FH, Cherry EM, Karma A, Rappel WJ. Modeling wave propagation in realistic heart geometries using the phase-field method. Chaos. 2005;15(1):13502. doi: 10.1063/1.1840311. [DOI] [PubMed] [Google Scholar]
- Garny A, Kohl P, Hunter PJ, Boyett M, Noble D. One-dimensional rabbit sinoatrial node models: Benefits and limitations. J. Cardiovasc. Electrophysiol. 2003;14:S121–S132. doi: 10.1046/j.1540.8167.90301.x. [DOI] [PubMed] [Google Scholar]
- Golub GH, Van Loan CF. Matrix Computations. 3rd edn. Johns Hopkins Studies in Mathematical Sciences; 1996. [Google Scholar]
- Harrild D, Henriquez C. A computer model of normal conduction in the human atria. Circ. Res. 2000;87:E25–E36. doi: 10.1161/01.res.87.7.e25. [DOI] [PubMed] [Google Scholar]
- Henriquez CS. Simulating the electrical behavior of cardiac tissue using the bidomain model. Crit Rev Biomed Eng. 1993;21(1):1–77. [PubMed] [Google Scholar]
- Hillman EM, Bernus O, Pease E, Bouchard MB, Pertsov A. Depth-resolved optical imaging of transmural electrical propagation in perfused heart. Opt Express. 2007;15(26):17827–17841. doi: 10.1364/oe.15.017827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117(4):500–44. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks DA, Trew ML, Caldwell BJ, Sands GB, LeGrice IJ, Smaill BH. Laminar arrangement of ventricular myocytes influences electrical behavior of the heart. Circ. Res. 2007;101(10):e103–12. doi: 10.1161/CIRCRESAHA.107.161075. [DOI] [PubMed] [Google Scholar]
- Hsu EW, Muzikant AL, Matulevicius SA, et al. Magnetic resonance myocardial fiber-orientation mapping with direct histological correlation. American Journal of Physiology. Heart and Circulatory Physiology. 1998;274(5):1627–1634. doi: 10.1152/ajpheart.1998.274.5.H1627. [DOI] [PubMed] [Google Scholar]
- Jafri MS, Rice JJ, Winslow RL. Cardiac Ca2+ dynamics: the roles of ryanodine receptor adaptation and sarcoplasmic reticulum load. Biophys J. 1998;74:1149–68. doi: 10.1016/S0006-3495(98)77832-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DK, Horsfield MA, Simmons A. Optimal strategies for measuring diffusion in anisotropic systems by magnetic resonance imaging. Magnetic Resonance in Medicine. 1999;42:515–525. [PubMed] [Google Scholar]
- Keener J, Bogar K. A numerical method for the solution of the bidomain equations in cardiac tissue. Chaos. 1998;8:234–241. doi: 10.1063/1.166300. [DOI] [PubMed] [Google Scholar]
- Kingsley PB. Introduction to diffusion tensor imaging mathematics: Part III. Tensor calculation, noise, simulations and optimization. Concepts in Magnetic Resonance Part A. 2006;28A(2):155–179. [Google Scholar]
- Knisley SB, Hill BC, Ideker RE. Virtual electrode effects in myocardial fibers. Biophys J. 1994;66:719–28. doi: 10.1016/s0006-3495(94)80846-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl P, Bollensdor C, Garny A. Effects of mechanosensitive ion channels on ventricular electrophysiology: experimental and theoretical models. Experimental Physiology. 2006;91:307–321. doi: 10.1113/expphysiol.2005.031062. [DOI] [PubMed] [Google Scholar]
- Kohl P, Ravens U. Cardiac mechano-electric feedback: past, present, and prospect. Progress in Biophysics and Molecular Biology. 2003;82:3–9. doi: 10.1016/s0079-6107(03)00022-1. [DOI] [PubMed] [Google Scholar]
- Kong W, Fakhari N, Sharifov OF, Ideker RE, Smith WM, Fast VG. Optical measurements of intramural action potentials in isolated porcine hearts using optrodes. Heart Rhythm. 2007;4(11):1430–6. doi: 10.1016/j.hrthm.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeGrice IJ, Smaill BH, Chai LZ, Edgar SG, Gavin JB, Hunter PJ. Laminar structure of the heart: ventricular myocyte arrangement and connective tissue architecture in the dog. Am. J. Physiol. 1995;269:H571–H582. doi: 10.1152/ajpheart.1995.269.2.H571. [DOI] [PubMed] [Google Scholar]
- Lemay M, Vesin JM, van Oosterom A, Jacquemet V, Kappenberger L. Cancellation of ventricular activity in the ECG: evaluation of novel and existing methods. IEEE Trans Biomed Eng. 2007;54(3):542–6. doi: 10.1109/TBME.2006.888835. [DOI] [PubMed] [Google Scholar]
- Leon LJ, Roberge FA. Directional characteristics of action potential propagation in cardiac muscle. A model study. Circ Res. 1991;69(2):378–95. doi: 10.1161/01.res.69.2.378. [DOI] [PubMed] [Google Scholar]
- Leon LJ, Roberge FA. A new cable model formulation based on Green’s theorem. Ann Biomed Eng. 1990;18(1):1–17. doi: 10.1007/BF02368414. [DOI] [PubMed] [Google Scholar]
- Liebmann M, Plank G, Prassl AJ, Haase G. GPU accelerated algebraic multigrid methods for the virtual heart model; Nvision Conference; San Jose, USA. Aug. 25-27.2008. [Google Scholar]
- Maclachlan MC, Sundnes J, Spiteri RJ. A comparison of non-standard solvers for ODEs describing cellular reactions in the heart. Comput. Methods Biomech. Biomed. Eng. 2007;10(5):317–26. doi: 10.1080/10255840701259301. [DOI] [PubMed] [Google Scholar]
- Mahajan A, Shiferaw Y, Sato D, Baher A, Olcese R, Xie LH, Yang MJ, Chen PS, Restrepo JG, Karma A, Garfinkel A, Qu Z, Weiss JN. A rabbit ventricular action potential model replicating cardiac dynamics at rapid heart rates. Biophys. J. 2008;94:392–410. doi: 10.1529/biophysj.106.98160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansoori T, Plank G, Burton R, Schneider J, Kohl P, Gavaghan D, Grau V. Building Detailed Cardiac Models by Combination of Histoanatomical and High-Resolution MRI Images; IEEE International Symposium on Biomedical Imaging; 2007.pp. 572–575. [Google Scholar]
- Neunlist M, Tung L. Spatial distribution of cardiac transmembrane potentials around an extracellular electrode: dependence on fiber orientation. Biophys J. 1995;68(6):2310–22. doi: 10.1016/S0006-3495(95)80413-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble D. Cardiac action and pacemaker potentials based on the Hodgkin-Huxley equations. Nature. 1960;5(188):495–497. doi: 10.1038/188495b0. [DOI] [PubMed] [Google Scholar]
- Plank G, Prassl AJ, Hofer E, Trayanova NA. Evaluating intramural virtual electrodes in the myocardial wedge preparation: simulations of experimental conditions. Biophys J. 2008;94(5):1904–15. doi: 10.1529/biophysj.107.121343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plank G, Liebmann M, Weber dos Santos R, Vigmond EJ, Haase G. Algebraic multigrid preconditioner for the cardiac bidomain model. IEEE Trans. Biomed. Eng. 2007;54:585–596. doi: 10.1109/TBME.2006.889181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plank G, Leon LJ, Kimber S, Vigmond EJ. Defibrillation depends on conductivity fluctuations and the degree of disorganization in reentry patterns. J. Cardiovasc. Electrophysiol. 2005;16:205–216. doi: 10.1046/j.1540-8167.2005.40140.x. [DOI] [PubMed] [Google Scholar]
- Plank G, Hofer E. Model study of vector-loop morphology during electrical mapping of microscopic conduction in cardiac tissue. Ann Biomed Eng. 2000;28(10):1244–52. doi: 10.1114/1.1320841. [DOI] [PubMed] [Google Scholar]
- Plonsey R. Bioelectric sources arising in excitable fibers (ALZA lecture) Ann Biomed Eng. 1988;16(6):519–46. doi: 10.1007/BF02368014. [DOI] [PubMed] [Google Scholar]
- Plotkowiak M, Rodriguez B, Plank G, Schneider JE, Kohl P, Grau V. High performance computer simulations of cardiac electrical function based on high resolution MRI datasets. Lecture Notes in Computer Science. 2008;5101:571–580. [Google Scholar]
- Potse M, Dube B, Richer J, Vinet A, Gulrajani RM. A comparison of monodomain and bidomain reaction-diffusion models for action potential propagation in the human heart. IEEE Trans. Biomed. Eng. 2006;53:2425–2435. doi: 10.1109/TBME.2006.880875. [DOI] [PubMed] [Google Scholar]
- Prassl AJ, Kickinger F, Ahammer H, Grau v., Schneider JE, Hofer E, Vigmond EJ, Trayanova NA, Plank G. Automatically Generated, Anatomically Accurate Meshes for Cardiac Electrophysiology Problems. under review. 2008 doi: 10.1109/TBME.2009.2014243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JM, McCulloch AD. A collocation–Galerkin finite element model of cardiac action potential propagation. IEEE Trans. Biomed. Eng. 1994;41(8):743–57. doi: 10.1109/10.310090. [DOI] [PubMed] [Google Scholar]
- Rohmer D, Sitek A, Gullberg G. Reconstruction and visualization of fiber and sheet structure with regularized tensor diffusion MRI in the human heart. Invest. Radio. 2007;42(11):777–784. doi: 10.1097/RLI.0b013e3181238330. [DOI] [PubMed] [Google Scholar]
- Roth BJ. Electrical conductivity values used with the bidomain model of cardiac tissue. IEEE Trans. Biomed. Eng. 1997;44:326–328. doi: 10.1109/10.563303. [DOI] [PubMed] [Google Scholar]
- Poelzing S, Rosenbaum DS. Altered connexin43 expression produce arrhythmia substrate in heart failure. AJP Heart. 2004;287:1762–1770. doi: 10.1152/ajpheart.00346.2004. [DOI] [PubMed] [Google Scholar]
- Schneider JE, Bose J, Bamforth S, et al. Identification of cardiac malformations in mice lacking PTDSR using a novel high-throughput magnetic resonance imaging technique. Biomed. Central Develop. Biol. 2004;4:16. doi: 10.1186/1471-213X-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scollan F, Holmes F, Winslow R, Forder J. Histological validation of myocardial microstructure obtained from diffusion tensor magnetic resonance imaging. American Journal of Physiology. Heart and Circulatroy Physiology. 1998;275(6):2308–2318. doi: 10.1152/ajpheart.1998.275.6.H2308. [DOI] [PubMed] [Google Scholar]
- Scollan DF, Holmes A, Zhang J, Winslow RL. Reconstruction of cardiac ventricular geometry and fiber orientation using magnetic resonance imaging. Ann. Biomed. Eng. 2000;28:934–944. doi: 10.1114/1.1312188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifov OF, Ideker RE, Fast VG. High-resolution optical mapping of intramural virtual electrodes in porcine left ventricular wall. Cardiovasc Res. 2004;64(3):448–56. doi: 10.1016/j.cardiores.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Skouibine KB, Trayanova NA, Moore PK. Anode/cathode make and break phenomena in a model of defibrillation. IEEE Trans. Biomed. Eng. 1999;46:769–777. doi: 10.1109/10.771186. [DOI] [PubMed] [Google Scholar]
- Spach MS. The discontinuous nature of electrical propagation in cardiac muscle. Consideration of a quantitative model incorporating the membrane ionic properties and structural complexities. The ALZA distinguished lecture. Ann Biomed Eng. 1983;11(3-4):209–61. doi: 10.1007/BF02363287. [DOI] [PubMed] [Google Scholar]
- Spach MS, Kootsey M. Relating the sodium current and conductance to the shape of transmembrane and extracellular potentials by simulation: effects of propagation boundaries. IEEE Trans Biomed Eng. 1985;32(10):743–55. doi: 10.1109/TBME.1985.325489. [DOI] [PubMed] [Google Scholar]
- Streeter DD, Spontnitz HM, Patel DP, Ross J, Sonnenblick EH. Fiber orientation in the canine left ventricle during diastole and systole. Circ. Res. 1969;24:339–347. doi: 10.1161/01.res.24.3.339. [DOI] [PubMed] [Google Scholar]
- Ten Tusscher KH, Panfilov AV. Modelling of the ventricular conduction system. Prog. Biophys. Mol. Biol. 2008;96:152–170. doi: 10.1016/j.pbiomolbio.2007.07.026. [DOI] [PubMed] [Google Scholar]
- Ten Tusscher KH, Hren R, Panfilov AV. Organization of ventricular fibrillation in the human heart. Circ. Res. 2007;100:e87–e101. doi: 10.1161/CIRCRESAHA.107.150730. [DOI] [PubMed] [Google Scholar]
- Trayanova NA, Eason JC, Aguel F. Computer simulations of cardiac debrillation: a look inside the heart. Computing and Visualization in Science. 2002;4:259270. [Google Scholar]
- Trew M, Le Grice I, Smaill B, Pullan A. A finite volume method for modeling discontinuous electrical activation in cardiac tissue. Ann. Biomed. Eng. 2005;33:590–602. doi: 10.1007/s10439-005-1434-6. [DOI] [PubMed] [Google Scholar]
- Vadakkumpadan F, Arevalo H, Prassl AJ, Chen J, Kickinger F, Kohl P, Plank G, Trayanova N. IEEE Trans. Med. Eng. 2008 doi: 10.1002/wsbm.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigmond EJ, Clements C. Construction of a computer model to investigate sawtooth effects in the Purkinje system. IEEE Trans. Biomed. Eng. 2007;54:389–399. doi: 10.1109/TBME.2006.888817. [DOI] [PubMed] [Google Scholar]
- Vigmond EJ, Hughes M, Plank G, Leon LJ. Computational tools for modeling electrical activity in cardiac tissue. J. Electrocardiol. 2003;36(Suppl):69–74. doi: 10.1016/j.jelectrocard.2003.09.017. [DOI] [PubMed] [Google Scholar]
- Vigmond EJ, Aguel F, Trayanova NA. Computational techniques for solving the bidomain equations in three dimensions. IEEE Trans. Biomed. Eng. 2002;49(11):1260–9. doi: 10.1109/TBME.2002.804597. [DOI] [PubMed] [Google Scholar]
- Whiteley JP. An efficient numerical technique for the solution of the monodomain and bidomain equations. IEEE Trans. Biomed. Eng. 2006;53:2139–2147. doi: 10.1109/TBME.2006.879425. [DOI] [PubMed] [Google Scholar]
- Wikswo JP, Jr, Lin SF, Abbas RA. Virtual electrodes in cardiac tissue: a common mechanism for anodal and cathodal stimulation. Biophys J. 1995;69(6):2195–210. doi: 10.1016/S0006-3495(95)80115-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young PG, Beresford-West TB, Coward SR, Notarberardino B, Walker B, Abdul-Aziz A. An efficient approach to converting three-dimensional image data into highly accurate computational models. Philos Transact A Math Phys Eng Sci. 2008;366(1878):3155–73. doi: 10.1098/rsta.2008.0090. [DOI] [PubMed] [Google Scholar]
- Zhou L, Plank G, Cortossa S, Greenstein JL, Winslow RL, Tayanova NA, O’Rourke B. Mitochondrial Energetics Dysfunction Shortens the Action Potential and Forms a Metabolic Sink in the Disease Zone; 29th Annual Scientific Sessions of the Heart Rhythm Society; San Francisco, USA. May 14-17, 2008.2008. [Google Scholar]