Abstract
Tuberculosis (TB) has substantial mortality worldwide with 5-10% of those exposed progressing to active TB disease. Studies in mice and humans indicate that the inducible nitric oxide synthase (iNOS) molecule plays an important role in immune response to TB. A mixed case-control association study of individuals with TB, relatives, or close contact controls was performed in 726 individuals (279 case and 166 control African-Americans; 198 case and 123 control Caucasians). Thirty-nine single nucleotide polymorphisms (SNPs) were selected from the NOS2A gene for single SNP, haplotype, and multilocus interaction analyses with other typed candidate genes using generalized estimating equations. In African-Americans, ten NOS2A SNPs were associated with TB. The strongest associations were observed at rs2274894 (odds ratio (OR) = 1.84, 95% confidence interval (CI) [1.23-2.77], p = 0.003) and rs7215373 (OR 1.67, 95% CI [1.17-2.37], p = 0.004), both of which passed a false discovery rate (FDR) correction for multiple comparisons (q*=0.20). The strongest gene-gene interactions were observed between NOS2A rs2248814 and IFNGR1 rs1327474 (p = 0.0004) and NOS2A rs944722 and IFNGR1 rs1327474 (p = 0.0006). Three other SNPs in NOS2A interacted with TLR4 rs5030729 and five other NOS2A SNPs interacted with IFNGR1 rs1327474. No significant associations were observed in Caucasians. These results suggest that NOS2A variants may contribute to TB susceptibility, particularly in individuals of African descent, and may act synergistically with SNPs in TLR4 and IFNGR1.
Keywords: tuberculosis, epistasis, complex disease, infectious disease, genetic epidemiology
Introduction
Approximately 30% of the population worldwide is infected with Mycobacterium tuberculosis (M.TB), with only a subset (5-10%) of those infected progressing to active TB disease (Flynn and Chan 2001). Despite this small percentage, in the United States (US), the progression to active TB among non-immunocompromized non-Hispanic African-Americans is eight times higher than the rate among non-Hispanic Caucasians. Environmental and socioeconomic factors only explain a small portion of this disparity (Center for Disease Control (CDC) 2004). M.TB organisms survive within macrophages during the latent period of infection and it remains unclear why some individuals successfully control M.TB infections and others do not.
The evidence from human and animal studies shows that M.TB clearance is under genetic regulation (Berrington and Hawn 2007). Twin studies of M.TB-infected individuals show that latently-infected monozygotic twins are more likely to develop pulmonary TB than latently infected dizygotic twins (Comstock 1978). Recently research is focusing on genes involved in macrophage responses to M.TB to better understand mechanisms for the increased susceptibility to TB.
Nitric oxide (NO) is vital for macrophage function and granuloma formation in the immune response to M.TB, and kills M.TB in vitro. NO synthase is produced with isoforms from three different genes: inducible nitric oxide synthase (iNOS) produced by NOS2A, neuronal nitric oxide synthase (nNOS) produced by NOS1, and endothelial NOS (eNOS) produced by NOS3. Among these genes, NOS2A has become an established candidate for TB (Gomez et al. 2007; Jamieson et al. 2004; Qu et al. 2007). In a mouse model, it has been shown that iNOS knockouts (iNOS −/−) infected with aerosolized M.TB have a higher mortality rate relative to iNOS (+/+) wild-type mice (Macmicking et al. 1997). Also, macrophages from iNOS-expressing mice infected with M.TB in vitro cannot fight infection if the macrophages are treated with iNOS inhibitors (Chan et al. 1992). These data suggest that iNOS has either an indirect or direct role in antimycobacterial response to M.TB in mice. Whether this translates to humans has not been determined.
The majority of association studies examining iNOS for association with TB have focused on promoter variants because the regulation of NOS2A is predominantly transcriptional (de Vera et al. 1996; Gomez et al. 2007; Kun et al. 2001; Linn et al. 1997; Marks-Konczalik et al. 1998; Qu et al. 2007). We comprehensively examined NOS2A for association with TB among African-Americans and Caucasians by testing 39 single nucleotide polymorphisms (SNPs) throughout the gene. We also tested gene-gene interactions with 11 other biologically plausible TB candidate genes to identify NOS2A variants that interact with genes, either upstream or downstream of NOS2A macrophage signaling for further study in independent samples.
Methods
Study Population
Participants for this study were ascertained through the North Carolina and South Carolina TB Control Programs, USA, or as patients at the outpatient clinic at F.J. Muñiz Hospital in Buenos Aires, Argentina, between 2002 and 2006. Criteria for inclusion as a TB case were (a) age 14 years or older and culture-confirmed pulmonary TB or (b) less than 14 years old and either culture-confirmed or clinically diagnosed pulmonary TB that included a positive tuberculin skin test plus an infiltrate or hilar adenopathy on chest X-ray. Individuals were eligible to participate in the study if their TB had been diagnosed in the past, or if they were currently receiving TB treatment. All TB cases remained eligible if they also had a diagnosis of extrapulmonary TB. Family members with a history of TB were enrolled as part of a multi-case family only if review of their records established that diagnosis of TB was culture-confirmed or clinically diagnosed pulmonary or extrapulmonary TB.
The diagnosis of a TB case was confirmed by review of medical records and laboratory reports, or by documentation through electronic surveillance databases used by each state’s TB program to document and report TB cases to the US Centers for Disease Control and Prevention [the TB Information Management System (TIMS)]. TIMS comprises data collected by local jurisdictions using the form called the Report of Verified Case of TB (RVCT). We attempted to document HIV status for all subjects but this was not required for participation, HIV status was measured directly through blood test results abstracted from the medical record.
Unaffected individuals who were in close contact with cases during the infectious phase of the disease (household contacts such as spouses and partners, and relatives such as parents and siblings) were enrolled as controls. Informed consent was obtained from all subjects or their legal representatives before participation in the study. Human experimentation guidelines of the US Department of Health and Human Services and those of participating research institutions were followed. The protocol was IRB approved at Duke University Medical Center, Centro de Educación Médica e Investigaciones Clínicas “Norberto Quirno” (CEMIC), F.J. Muñiz Hospital, Buenos Aires, Argentina, and the University of Miami Miller School of Medicine.
Genotyping and SNP Selection
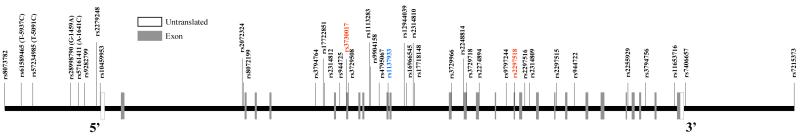
A total of 39 tag SNPs were screened in NOS2A. Tag SNPs were selected based on their ability to tag surrounding variants in the African (Yoruba) and Centre d’Etude du Polymorphisme Humain (CEPH) populations from the International HapMap project (http://www.hapmap.org), NCBI build 35 assembly HapMap phase II (The International HapMap Consortium 2003). Tagger software included in Haploview software (Barrett et al. 2005) was used to select tag SNPs. The criteria used in tag SNP selection included a tag SNP 2,000 base pairs (bp) 5′ upstream and 3′ downstream of NOS2A, a minor allele frequency (MAF) of 0.05, a pairwise correlation coefficient (r2) of 0.80, and SNPs that met the criteria for aggressive tagging using 2- and 3-marker haplotypes option on Haploview software. We also included potential functional variants reported in previous studies (Hobbs et al. 2002). A schematic representation of NOS2A and genotyped SNPs is provided in Figure 1. The same procedure was used in selection of tag SNPs for the non-NOS2A genes examined in interaction analyses: solute carrier family 11 (SLC11A1), toll-like receptor 9 isoform B (TLR9), toll-like receptor 2 (TLR2), interferon gamma receptor 1 (IFNGR1), tumor necrosis factor alpha (TNF-α), PARK2 co-regulated (PACRG), parkin isoform 1 (PARK2), toll-like receptor 4 (TLR4), interferon gamma (IFNG), vitamin D (1,25-dihydroxyvitamin D3) receptor (VDR), and ubiquitin protein ligase E3A (UBE3A). These SNPs and their genes are listed in Supplemental Table 1. The selection of PACRG and PARK2 (adjacent genes on chromosome 6), two less commonly examined TB candidate genes, is based on work by Mira et al. (Mira et al. 2004) identifying association between SNPs in the shared promoter region of these two genes and risk of leprosy (caused by Mycobacterium leprae).
Fig. 1. NOS2A SNP density map.
27 exons are labeled on the gene map of NOS2A. Untranslated regions are labeled in white and exons are labeled in gray. The map is oriented 5′ to 3′; the 5′ and 3′ ends are of the gene are labeled on the map. SNPs are indicated by vertical lines with SNP Rs number or relative position designated. Among these SNPs there were two nonsynonymous SNPS (rs3730017 and rs2297518, color coded in red in the online version) and one synonymous (rs1137933, color coded in blue in the online version) SNPS. The remaining SNPs are noncoding SNPs (color coded in black in the online version).
DNA was isolated for 445 African-American individuals from the USA (233 families; 279 cases and 166 controls) and 321 Caucasian individuals (163 families; 198 cases and 123 controls, 64 cases and 41 controls from Argentina) using the Autopure automated system (Gentra Systems, Minneapolis, MN). The TaqMan genotyping platform was used to genotype tag SNPs using the TaqMan assay on an ABI 7900 (Applied Biosystems, Foster City, CA). All SNPs used in this study had genotyping call rates of 95% or better (mean call rate 98%) and quality control sample match rates of 100%. DNA samples from CEPH families were duplicated between and across plates for use as a quality control, and laboratory personnel were blinded to the affection status of the individuals being genotyped. A list of the TaqMan probes and primers examined for NOS2A are included in Supplemental Table 2.
Statistical Analysis
All statistical analyses were performed stratified by race. Genetic Data Analysis software (GDA) was used to test for deviations from Hardy-Weinberg equilibrium (HWE) (Weir and Cockerham 1996). In African-Americans, two SNPs, rs7406657 (p = 0.04) and rs2297516 (p = 0.02), had nominally significant deviations from HWE in controls, while there was only one deviation from HWE in Caucasian controls at rs7215373 (p = 0.04). Pairwise linkage disequilibrium (LD) using standard summary statistics D’ and r2 were calculated using Haploview statistical software separately for cases and controls (Barrett et al. 2005). Haplotype blocks were assigned using the D’ confidence interval (CI) algorithm created by Gabriel et al. (2002).
Genotypic tests of association were performed using generalized estimating equations (GEE) implemented in SAS (Proc GENMOD) statistical software version 9.1 (SAS Institute, Cary, NC). GEE with an independent correlation matrix has been shown to be a valid test of gene x gene and gene x environment interactions in mixed case-control and family data (Hancock et al. 2007). Additive genotypic models were performed modeling the number of minor alleles as the risk category (0 vs. 1 vs. 2) adjusting for potential confounders’ age and gender for all analyses. In Caucasians, we tested for genetic heterogeneity between study sites by incorporating ascertainment site and a genotype-ascertainment site interaction term in all models.
Multilocus analysis between statistically significant (p < 0.05) NOS2A SNPs and SNPs in other TB candidate genes (only SNPs with MAF ≥ 0.05) was performed in GEE using additive risk models adjusting for potential confounders’ age and gender for all analyses. These analyses were only performed between genes-98 SNPs in 11 non NOS2A genes that met the criteria for analysis of interaction with 9 NOS2A SNPs. Models included both SNPs individually with an interaction term for the interactions between risk alleles of both SNPs. We followed up interactions in genes with the most significant associations (p ≤ 0.001 for the interaction term) with GEE analyses of their multilocus diplotypes. Odds ratios (OR) and 95% CI were calculated for both single SNP and stratified GEE analyses.
A false discovery rate (FDR) correction was performed to adjust for multiple comparisons using a q* of 0.20 (Benjamini and Hochberg 1995; Juenger et al. 2006; Smith et al. 2007). The q* indicates the expected proportion of results that are identified as interesting that are actually false. This is in contrast to α (typically set to 0.05) which indicates the probability of obtaining a false-positive result among all tests performed. FDR is used to measure global error, i.e., the expected number of false rejections of the null hypothesis among the total number of rejections. The critical significance level is calculated by ranking the results by decreasing magnitude of p then multiplying this rank by q* divided by the total number of tests.
Results
A description of the 396 families analyzed in this study and a summary of the demographic characteristics are presented in Table 1. The majority of families had only one case per family (87% in African-Americans and 83% in Caucasians). The average TB case age at enrollment was 44.9 ± 17.9 years in African-American cases and 52.56 ± 21.50 for African-American controls (p = 0.0001) and 42.3 ± 20.3 years for Caucasian cases and 46.94 ± 50.57 for Caucasian controls, (p = 0.05). HIV status was documented in 72% of cases (74% of African-American, 67% of Caucasian) collected. Among all cases, 11% of African-Americans and 4% of Caucasians were seropositive and these results do not differ when cases and controls are stratified by HIV status (data not shown). As a result analyses were not stratified by HIV status. The majority of cases were male (67% of African-Americans and 55% of Caucasians) but controls were mostly female (82% of African-Americans and 67% of Caucasians). The proportion of males and females was statistically different between cases and controls in both African-Americans (p < 0.0001) and Caucasians (p < 0.0001). The majority of the sites of infection were pulmonary or pulmonary with extrapulmonary for both African-Americans (91%) and Caucasians (97%). A small portion of our study subjects enrolled as part of a multi-case-family had extrapulmonary TB only.
Table 1.
Description of study population and demographic characteristics
| (a) | ||||
|---|---|---|---|---|
| Family Type | African-American Families (Na = 233) |
Caucasian families (Na = 163) |
||
| N | % | N | % | |
| 1 Case + Parents or Children | 20 | 0.09 | 17 | 0.10 |
| 1 Case + 1 Parent | 55 | 0.24 | 41 | 0.25 |
| 1 Case + Siblings, No Parents | 28 | 0.12 | 22 | 0.13 |
| 1 Case Only | 100 | 0.43 | 56 | 0.34 |
| Multiplex (2 + Cases) | 30 | 0.13 | 27 | 0.17 |
| (b) | ||||||
|---|---|---|---|---|---|---|
| Variable | African-American Cases (N = 279) |
Controls (N = 166) | p | Caucasian Cases (N = 198) |
Controls (N = 123) | p |
| Mean (SD) Age Enrollment (years) | 44.88 (17.73) | 52.56 (21.50) | 0.0001 | 42.31 (20.30) | 46.94 (20.57) | 0.05 |
| Gender | ||||||
| Proportion Male | 0.67 | 0.18 | <0.0001 | 0.55 | 0.33 | <0.0001 |
| Proportion Female | 0.33 | 0.82 | 0.49 | 0.67 | ||
| Proportion HIV Positive | 0.11 | – | 0.04 | – | ||
| Site of Infection | ||||||
| Extrapulmonary | 0.09 | – | 0.03 | – | ||
| Pulmonary | 0.89 | – | 0.94 | – | ||
| Extrapulmonary and Pulmonary | 0.02 | – | 0.03 | – | ||
SD standard deviation
445 individuals represent the 233 African-American families and 321 individuals represent the 163 Caucasian families
Single Locus Association Analyses
No significant single SNP or multilocus associations in NOS2A were observed among Caucasians (Supplemental Table 3). In African-Americans genotype tests of association using GEE (Table 2) identified nine nominally significant associations: rs7215373 (OR = 0.1.67, 95% CI [1.17-2.37], p = 0.004), rs2255929 (OR = 1.45, 95% CI [1.06-1.99], p = 0.02), rs944722 (OR = 1.63, 95% CI [1.14-2.33], p = 0.01), rs2314809 (OR = 1.40, 95% CI [1.03-1.92], p = 0.03), rs2274894 (OR = 1.84, 95% CI [1.23-2.77], p = 0.003), rs3729718 (OR = 0.48, 95% CI [0.28-0.85], p = 0.01), rs2248814 (OR = 1.68, 95% CI [1.13-2.50], p = 0.01), rs3729508 (OR = 1.76, 95% CI [1.16-2.67], p = 0.01), and rs7234985 (T-5091C) (OR = 0.44, 95% CI [0.22-0.91], p = 0.03). Marker rs2314809 was in moderate LD (Supplemental Figure A1, LD plots are only presented for controls, similar patterns were observed in cases) with surrounding markers, and in stronger LD with rs2255929 (r2=0.78 in control samples). The allelic association at rs2314809 and all of the genotypic associations remained statistically significant after an FDR correction for multiple comparisons.
Table 2.
NOS2A single marker association in African-Americans
| Rs # | SNP Typea | Allele Frequency (Allele) |
OR | 95% CI | Uncorrected p valueb |
|
|---|---|---|---|---|---|---|
| rs8073782 | Promoter | 0.80(C) / 0.20(T) | 0.91 | 0.65 | 1.27 | 0.57 |
| rs61589465 (T-5937C) | Promoter | 0.96(A) / 0.04(G) | 2.15 | 0.87 | 5.30 | 0.10 |
| rs57234985 (T-5091C) | Promoter | 0.96(A) / 0.04(G) | 0.44 | 0.22 | 0.91 | 0.03 |
| rs28998790 (G-1459A) | Promoter | 0.93(G) / 0.07(A) | 1.16 | 0.53 | 2.52 | 0.72 |
| rs57161411 (A-1641C) | Promoter | 0.99(A) / 0.01(C) | 0.67 | 0.20 | 2.18 | 0.50 |
| rs9282799 | Promoter | 0.94(G) / 0.06(A) | 0.96 | 0.48 | 1.93 | 0.91 |
| rs2779248 | Promoter | 0.53(T) / 0.47(C) | 1.03 | 0.78 | 1.37 | 0.83 |
| rs10459953 | 5′UTR | 0.70 (G) / 0.30(C) | 0.91 | 0.66 | 1.27 | 0.58 |
| rs2072324 | Intron | 0.88(C) / 0.12(A) | 0.93 | 0.56 | 1.55 | 0.79 |
| rs8072199 | Intron | 0.82(C) / 0.18(T) | 1.29 | 0.88 | 1.89 | 0.20 |
| rs3794764 | Intron | 0.71(G) / 0.29(A) | 0.74 | 0.52 | 1.05 | 0.09 |
| rs17722851 | Intron | 0.96(T) / 0.04(A) | 1.34 | 0.45 | 3.99 | 0.60 |
| rs2314812 | Intron | 0.84(C) / 0.16(T) | 1.00 | 0.67 | 1.50 | 0.99 |
| rs944725 | Intron | 0.51(C) / 0.49(T) | 0.78 | 0.57 | 1.08 | 0.13 |
| rs3730017 | Coding Exon (R/W 221) | 0.76(G) / 0.24(A) | 1.20 | 0.84 | 1.70 | 0.32 |
| rs3729508 | Intron | 0.83(C) / 0.17(T) | 1.76 | 1.16 | 2.67 | 0.01 |
| rs1113283 | Intron | 0.84(C) / 0.16(T) | 0.99 | 0.66 | 1.48 | 0.95 |
| rs9904158 | Intron | 0.94(G) / 0.06(C) | 1.23 | 0.60 | 2.54 | 0.57 |
| rs4795067 | Intron | 0.82(A) / 0.18(G) | 1.14 | 0.76 | 1.71 | 0.52 |
| rs1137933 | Coding Exon (D/D 346) | 0.85(G) / 0.15(A) | 1.00 | 0.66 | 1.52 | 1.00 |
| rs12944039 | Intron | 0.76(G) / 0.24(A) | 0.75 | 0.53 | 1.08 | 0.12 |
| rs16966545 | Intron | 0.83(T) / 0.17(A) | 0.84 | 0.55 | 1.31 | 0.45 |
| rs2314810 | Intron | 0.99(G) / 0.01(C) | 5.71 | 0.81 | 40.27 | 0.08 |
| rs17718148 | Intron | 0.88(C) / 0.12(T) | 1.03 | 0.64 | 1.68 | 0.89 |
| rs3729966 | Intron (boundary) | 0.86(C) / 0.14(T) | 0.75 | 0.48 | 1.18 | 0.22 |
| rs2248814 | Intron (boundary) | 0.83(G) / 0.17(A) | 1.68 | 1.13 | 2.50 | 0.01 |
| rs3729718 | Intron (boundary) | 0.94(T) / 0.06(G) | 0.48 | 0.28 | 0.85 | 0.01 |
| rs2274894 | Intron | 0.83(G) / 0.17(T) | 1.84 | 1.23 | 2.77 | 0.003 |
| rs9797244 | Intron | 0.83(T) / 0.17(C) | 0.92 | 0.63 | 1.36 | 0.68 |
| rs2297518 | Coding Exon (S/L 569) | 0.89(G) / 0.11(A) | 1.17 | 0.72 | 1.91 | 0.51 |
| rs2297516 | Intron | 0.66(A) / 0.34(C) | 0.77 | 0.54 | 1.09 | 0.15 |
| rs2314809 | Intron | 0.57(T) / 0.43(C) | 1.40 | 1.03 | 1.92 | 0.03 |
| rs2297515 | Intron | 0.97(A) / 0.03(C) | 1.80 | 0.64 | 5.05 | 0.27 |
| rs944722 | Intron | 0.79(T) / 0.21(C) | 1.63 | 1.14 | 2.33 | 0.01 |
| rs2255929 | Intron | 0.57(A) / 0.43(T) | 1.45 | 1.06 | 1.99 | 0.02 |
| rs3794756 | Intron | 0.91(C) / 0.09(T) | 0.71 | 0.42 | 1.18 | 0.19 |
| rs11653716 | Intron | 0.66(C) / 0.34(G) | 1.01 | 0.73 | 1.42 | 0.93 |
| rs7406657 | Downstream | 0.82(G) / 0.18(C) | 0.90 | 0.61 | 1.32 | 0.59 |
| rs7215373 | Downstream | 0.76(C) / 0.24(T) | 1.67 | 1.17 | 2.37 | 0.004 |
Statistically significant p values (p<0.05) are in bold; an allele frequencies are for founders; and all analyses were adjusted for age (years) and gender
SNPs type was determined using SNPper software available from http://snpper.chip.org
These p values are uncorrected for multiple comparisons
Multilocus Association Analyses
The SNPs with nominally significant single SNP associations and with a minor allele frequency ≥ 0.05 in NOS2A were tested for interactions with SNPs in other TB candidate genes. We limited our interaction analyses to two SNP interactions due to sample size limitations, resulting in a total of 784 tests. We observed multiple strong interactions (p < 0.001) between SNPs in NOS2A and IFNGR1 and TLR4 (Table 3a). In IFNGR1, one SNP (rs1327474) in the promoter region of IFNGR1 had multiple statistically significant associations with eight of the nine NOS2A SNPs we examined for interaction analyses: rs7215373, rs2255929, rs944722, rs2314809, rs2274894, rs3729718, rs2248814, and rs3729508. The p values for the interactions ranged from 0.02 to 0.0004. One other SNP in TLR4 (rs5030729) interacted with three SNPs in NOS2A: rs7215373, rs2274894, and rs2248814. The p values for the interaction term for these analyses ranged from 0.005 to 0.002. TLR4 SNPs rs1927910 and rs5030729 were in moderate LD (r2 ~0.33) in both case and control samples. Among these interactions none remained statistically significant after FDR correction for multiple testing; however, NOS2A rs2248814 and IFNGR1 rs1327474 and NOS2A rs944722 and IFNGR1 rs1327474 were very close to the threshold for significance (critical value for rs2248814 × rs1327474 is 0.0003 and critical value for rs944722 × rs1327474 is 0.0005). The SNPs from TLR4 and IFNGR1 involved in the interactions did not have statistically significant main effects.
Table 3.
Gene-gene interactions and diplotype analyses
| (a) | ||||
|---|---|---|---|---|
| Interacting Gene | SNP Typea | NOS2A SNP | Interacting SNP | Interaction p value |
| Interferon Gamma Receptor 1 (IFNGR1) | Promoter | rs3729508 | rs1327474 | 0.01 |
| Promoter | rs2274894 | rs1327474 | 0.003 | |
| Promoter | rs2314809 | rs1327474 | 0.02 | |
| Promoter | rs2248814 | rs1327474 | 0.0004 | |
| Promoter | rs944722 | rs1327474 | 0.0006 | |
| Promoter | rs3729718 | rs1327474 | 0.01 | |
| Promoter | rs7215373 | rs1327474 | 0.003 | |
|
| ||||
| Toll-like Receptor 4 (TLR4) | Intron | rs2248814 | rs5030729 | 0.004 |
| Intron | rs2274894 | rs5030729 | 0.002 | |
| Intron | rs7215373 | rs5030729 | 0.005 | |
| (b) | ||||||
|---|---|---|---|---|---|---|
| Interacting Gene | SNPs | Multilocus Diplotype |
OR | 95% CI |
p value |
|
| IFNGR1 | rs1327474-rs3729508 (minor alleles G,T) | AG,GG / CT,TT | 0.77 | 0.33 | 1.79 | 0.55 |
| AG,GG / CC | 1.19 | 0.64 | 2.20 | 0.58 | ||
| AA / CT,TT | 2.07 | 1.23 | 3.47 | 0.01 | ||
|
|
||||||
| rs1327474-rs2274894 (minor alleles G,T) | AG,GG / GT,TT | 0.66 | 0.27 | 1.64 | 0.37 | |
| AG,GG / GG | 1.48 | 0.77 | 2.83 | 0.24 | ||
| AA / GT,TT | 2.47 | 1.44 | 4.22 | 0.001 | ||
|
|
||||||
| rs1327474-rs2314809 (minor alleles G,C) | AG,GG / CT,CC | 0.89 | 0.43 | 1.18 | 0.74 | |
| AG,GG / TT | 1.82 | 0.86 | 3.85 | 0.12 | ||
| AA / CT,CC | 1.61 | 1.00 | 2.59 | 0.05 | ||
| rs1327474-rs2248814 (minor alleles G,A) | AG,GG / AG,AA | 1.74 | 1.07 | 2.84 | 0.03 | |
| AG,GG / GG | 2.01 | 1.23 | 3.31 | 0.01 | ||
| AA / AG,AA | 2.15 | 1.27 | 3.64 | 0.004 | ||
|
|
||||||
| rs1327474-rs944722 (minor alleles G,C) | AG,GG / CT,CC | 0.68 | 0.32 | 1.43 | 0.31 | |
| AG,GG / TT | 1.50 | 0.76 | 2.97 | 0.25 | ||
| AA / CT,CC | 2.01 | 1.29 | 3.14 | 0.002 | ||
|
|
||||||
| rs1327474-rs3729718 (minor alleles G,G ) | AG,GG / GT,GG | 1.94 | 0.37 | 10.27 | 0.44 | |
| AG,GG / TT | 0.68 | 0.38 | 1.22 | 0.20 | ||
| AA / GT,GG | 0.37 | 0.19 | 0.71 | 0.003 | ||
|
|
||||||
| rs1327474-rs7215373 (minor alleles G,T) | AG,GG / CT,TT | 0.84 | 0.38 | 1.85 | 0.66 | |
| AG,GG / CC | 1.51 | 0.76 | 3.00 | 0.24 | ||
| AA / CT,TT | 1.92 | 1.19 | 3.10 | 0.01 | ||
|
|
||||||
| TLR4 | rs5030729-rs2248814 (minor alleles G,A) | AG,GG / AG,AA | 2.05 | 0.83 | 5.08 | 0.12 |
| AG,GG / GG | 0.42 | 0.23 | 0.77 | 0.01 | ||
| AA / AG,AA | 1.04 | 0.57 | 1.91 | 0.89 | ||
|
|
||||||
| rs5030729-rs2274894 (minor alleles G,T) | AG,GG / GT,TT | 2.48 | 0.97 | 6.36 | 0.06 | |
| AG,GG / GG | 0.37 | 0.20 | 0.68 | 0.001 | ||
|
|
||||||
| AA / GT,TT | 1.13 | 0.62 | 2.06 | 0.68 | ||
| rs5030729-rs7215373 (minor alleles G,T) | AG,GG / CT,TT | 1.52 | 0.70 | 3.29 | 0.29 | |
| AG,GG / CC | 0.41 | 0.22 | 0.78 | 0.01 | ||
| AA / CT,TT | 1.20 | 0.67 | 2.17 | 0.54 | ||
Markers with low minor allele frequency (< 0.05) were not included in the analysis. Statistically significant associations (p<0.05) are indicated by bold p-values
SNPs type was determined using SNPper software available from http://snpper.chip.org
We performed multilocus diplotype analyses examining the multilocus genotype of IFNGR1 rs1327474 and each of the interacting NOS2A SNPs using GEE (Table 3b). We combined the heterozygote and the homozygous minor genotypes for these analyses due to the low frequency of the homozygous minor genotype. These analyses showed that there was an elevated susceptibility to TB in the presence of both IFNGR1 rs1327474 AA and the minor allele of four of the NOS2A SNPs, with odds ratios ranging 1.61–2.47.
Multilocus diplotype analyses in TLR4 (Table 3b) showed that for the TLR4 SNP (rs5030729) and NOS2A SNPs there was a significant decrease in risk in the presence of the minor allele of rs5030729 and the homozygous major allele of the NOS2A SNPs. ORs ranged 0.37-0.42 for these models.
Discussion
Significant single SNP associations in NOS2A were found in African-Americans, providing further evidence of a role for NO synthesis in TB susceptibility. Although we did not find identical results among Caucasians the point estimates of the ORs for SNPs associated with TB in African-Americans are generally very close to 1 in Caucasians, indicating that the discordant results are not simply due to an underpowered Caucasian sample. Multilocus analyses also identified significant interactions between SNPs in NOS2A and SNPs in IFNGR1 and TLR4. These findings are particularly interesting, as these SNPs in IFNGR1 and TLR4 did not show significant single SNP effects and would not have been identified as associated with TB without the interaction with NOS2A. We do note that these genes were selected as candidates due to their previous associations with TB and other mycobacterial infections in other study populations and our findings only indicate that these genes did not associate with the tag SNPs we selected for our analyses (Cooke et al. 2006; Kamijo et al. 1995; Means et al. 1999; Tsuji et al. 2000). This study, unlike other studies of NOS2A, did not focus exclusively on functional variants of TB (Burgner et al. 2003; Gomez et al. 2007; Kun et al. 1998; Kun et al. 2001). Instead we comprehensively genotyped SNPs, tagging the common polymorphisms across the gene, providing better coverage and several key findings.
All of the interactions observed in African-Americans were not present in Caucasians. Such discrepancy can be explained by the allele frequency differences observed between African-Americans and Caucasians for most associated SNPs. The lack of association observed in Caucasians is potentially due to a selection process which has shifted the allele frequencies in the population, as a result of reduced evolutionary pressures to fight tuberculosis. African-Americans are a recently admixed population and selection or drift may not have removed the risk allele from this population. A large majority of African-Americans from the South and Southeast US have a majority West African ancestry (Salas et al. 2005). Studies by Souëf et al. (2000) examining the evolutionary adaptation of inflammatory responses among human populations have observed that risk alleles for inflammatory cytokines are more prevalent in populations, such as West Africa, with a tropical origin relative to those from temperate climates. Supporting this are the elevated rates of asthma and asthma-related allergic diseases among African-Americans (25% higher than Caucasians) and Puerto Ricans (Akinbami 2006). It may be that in African-Americans, not in Caucasians, there is a strong selective pressure predisposing to TB that may explain the elevated prevalence rates of TB in African-Americans.
The majority of the associations in NOS2A were in the 3′ end of the gene. However, we did observe a significant association at NOS2A promoter SNP rs7234985 (T-5091C), a SNP established as a TB candidate due to both its location in the promoter region of NOS2A and a previous association with severe malaria (Hobbs et al. 2002). Studies by Levesque et al. (2009) (in preparation), have also observed that rs7234985 (T-5091C) is associated with low exhaled NO levels in healthy African-American adults; these results suggest that this SNP may be a potential candidate for follow-up studies (Levesque et al. 2009). Interestingly, the 3′ end of NOS2A has not been previously recognized as a strong candidate for TB susceptibility. However, associations have been observed in the 3′ end of NOS2A and in intronic regions in other phenotypes including Parkinson disease (PD) (Hancock et al. 2008), spontaneous preterm birth (Gibson et al. 2007), and osteomyelitis (Asensi et al. 2007). Two of the associated TB SNPs, rs2255929 and rs2248814, were also previously associated with PD in a Caucasian population (Hancock et al. 2008). The fact that we are seeing similar associations in African-Americans with TB suggests that these SNPs are in LD with functionally relevant variants.
One possible reason we are seeing associations between SNPs in the 3′ end of NOS2A and TB is due to the importance of the 3′ end of the gene to mRNA stability (Kleinert et al. 2004; Schroder et al. 2006). iNOS production is regulated at both the transcriptional and post-transcriptional level (Kleinert et al. 2003, 2004). iNOS mRNA stability is mediated by the 3′ untranslated region (UTR) that contains conserved AUUUA sequence motifs, where different regulatory elements bind such as RNA binding protein HuR (Caput et al. 1986). Complex arrays of various proteins bind to this 3′UTR region providing stabilization and destabilization in response to various stimuli. Studies by Vodovotz et. al. (1993), e.g., have demonstrated that in mouse peritoneal macrophages, binding of transforming growth factor β1 (TGFβs) decreases iNOS mRNA stability and iNOS expression by increasing iNOS degradation (Vodovotz et al., 1993). We posit that the associations we described in the 3′ end of the iNOS gene may be due to our SNPs tagging of a variant in the 3′UTR region of NOS2A (perhaps in one of the conserved AUUUA sequence motifs) that prevent the iNOS mRNA stabilization necessary for efficient NO expression.
IFNGR1 and TLR4 both have important roles in immune response to M.TB. IFNG plays a significant role in induction of macrophage-mediated microbicidal immune response and in prevention of M.TB infection progression (Cooper et al. 1993; Flynn et al. 1993). The IFNG cell surface receptor consists of two heterodimeric subunits, IFNGR1 (α ligand binding receptor and signal transduction subunit expressed in hematopoietic cells) and IFNG receptor 2 (IFNGR2) (β ligand binding receptor expressed in nonhematopoietic cells). Mutations in these receptors have been associated with increased susceptibility to TB (Dorman et al. 2004; Hussain et al. 1999). Animal model studies have also found that IFNGR1 −/− mice infected with Mycobacterium bovis cannot successfully generate NO suggesting that IFNGR1 plays an important role in NO synthesis and immune response (Kamijo et al. 1995). TLR4 plays a significant role in both response to M.TB and NO production. iNOS is a target molecule for TLR4 signaling stimulated by microbial LPS. TLR4 initiates the immune response to M.TB by interacting with heat labile soluble mycobacterial factor and whole viable M.TB (Means et al. 1999; Tsuji et al. 2000). TLR4 (−/−) mice develop lung infections that resemble human exposure to aerosolized M.TB; these infections associate with M.TB growth and reduced macrophage response in lung tissue (Abel et al. 2002). Given the strong relationship between IFNGR1, TLR4, and iNOS and TB susceptibility, it may not be surprising that we observed statistical associations between these genes.
Although encouraging, our study has limitations. First, we observed some baseline differences for the ratio of males to females and mean age at exam in cases and controls. As a result, we adjusted for these covariates in our models. We acknowledge that multiple tests being performed may result in chance findings of statistical significance. We have employed an FDR correction for multiple comparisons, and several of our main effects passed a correction for multiple testing, while our interactions between NOS2A-TLR4 and NOS2A-IFNGR1 did not but were very close to their adjusted critical value for significance. This, coupled with similar associations previously observed in the 3′ end of NOS2A in other diseases (Hancock et al. 2008; Johannesen et al., 2001), leads us to conclude that our results are less likely to represent false-positive findings as NOS2A is important to several biological processes and a functional variant in such a pleiotropic gene might influence development of several diseases.. The tagging approach we used in our SNP selection also poses a limitation; it is unclear whether the associations we are observing are due to the variants we examined or SNPs in LD with the functional variant; further studies are required to identify the functionally relevant variant. We also acknowledge that including the HIV-infected individuals within our analysis might confound results; however, results did not substantially differ when excluding HIV-positive individuals from analysis. We were also underpowered to find associations with risk alleles with low frequency; as a result we did not observe associations with rare SNPs which would require a larger sample size. We did not select ancestry informative markers for our analyses but rather only relied on self-reported race and ethnicity and as a result our study is susceptibility issues related to population stratification. However, studies by Tang et al. (2005) have demonstrated that self-reported race is reliable for controlling confounding by ancestry and we are therefore confident that our analysis is not strongly confounded by residual stratification by ancestry (Tang et al. 2005). Finally, we also did not have access to a complete history of smoking use and therefore could not look at the potential interactions between smoking and NOS2A as previously observed in PD. (Hancock et al. 2008). Despite these limitations, the current results are promising and merit further examination.
A review by Schroder et. al., proposed a synergistic model whereby IFNG and TLR4 molecules engage in cross-talk that results in coordinated regulation of iNOS gene expression (Schroder et al., 2006). This model would support that polymorphisms in IFNGR1 and TLR4 may interfere with binding to the NOS2A promoter, and that polymorphisms in the 3′UTR of NOS2A may prevent post-transcriptional NOS2A mRNA stabilization by preventing binding of post-transcriptional elements, resulting in inefficient NO production and TB progressing to active disease. Our findings suggest that NOS2A genetic defects in both transcriptional and post-transcriptional regulatory functions may contribute to the susceptibility to TB progression in African-Americans. Several of our single SNP associations passed a correction for multiple testing further supporting an important role of NOS2A in the susceptibility to TB. While none of the interactions survived, a correction for multiple testing were a few that were very close to their new adjusted threshold for significance suggesting that these results merit further investigation. Further examination of NOS2A and the relationship with IFNGR1 and TLR4 is warranted to more clearly understand the biological interactions between these genes.
Supplementary Material
Acknowledgements
The work in this manuscript was supported by grant number R01 HL068534 from the National Heart, Lung and Blood Institute, National Institutes of Health. Dr. Hamilton acknowledges support from NIH K24-AI001833. We thank the study participants, without whom this study would have been impossible, the North Carolina TB Control Nurse Consultants (Myra Allen, Dee Foster, Julie Luffman and Elizabeth Zeringue) and county TB nurses who referred subjects to the study. We would also like to thank Martha Fletcher, Elizabeth Levine, Earline Little, and Carol Poszik for assistance in recruiting participants in South Carolina, and Courtney Linton, Regina Carney, and Ann Mosher for recruiting participants in North Carolina.
Footnotes
Published as:
Velez DR, Hulme WF, Myers JL, Weinberg JB, Levesque MC, Stryjewski ME, Abbate E, Estevan R, Patillo SG, Gilbert JR, Hamilton CD, Scott WK. Hum Genet. NOS2A, TLR4, and IFNGR1 interactions influence pulmonary tuberculosis susceptibility in African-Americans. Hum Genet. 2009 Nov;126(5):643-53.
The final publication is available at www.springerlink.com: (http://www.springerlink.com/content/wk832573j444j415/)
References
- Abel B, Thieblemont N, Quesniaux VJ, Brown N, Mpagi J, Miyake K, Bihl F, Ryffel B. Toll-like receptor 4 expression is required to control chronic Mycobacterium tuberculosis infection in mice. J Immunol. 2002;169:3155–3162. doi: 10.4049/jimmunol.169.6.3155. [DOI] [PubMed] [Google Scholar]
- Akinbami . Asthma prevalence, health care use and mortality: United States, 2003-05. 2006. [PubMed] [Google Scholar]
- Asensi V, Montes AH, Valle E, Ocana MG, Astudillo A, Alvarez V, Lopez-Anglada E, Solis A, Coto E, Meana A, Gonzalez P, Carton JA, Paz J, Fierer J, Celada A. The NOS3 (27-bp repeat, intron 4) polymorphism is associated with susceptibility to osteomyelitis. Nitric Oxide. 2007;16:44–53. doi: 10.1016/j.niox.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Berrington WR, Hawn TR. Mycobacterium tuberculosis, macrophages, and the innate immune response: does common variation matter? Immunol Rev. 2007;219:167–186. doi: 10.1111/j.1600-065X.2007.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soci Ser B (Methodological) 1995;57:289–300. [Google Scholar]
- Burgner D, Usen S, Rockett K, Jallow M, Ackerman H, Cervino A, Pinder M, Kwiatkowski DP. Nucleotide and haplotypic diversity of the NOS2A promoter region and its relationship to cerebral malaria. Hum Genet. 2003;112:379–386. doi: 10.1007/s00439-002-0882-4. [DOI] [PubMed] [Google Scholar]
- Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci U S A. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center for Disease Control (CDC) Racial disparities in tuberculosis--selected southeastern states, 1991-2002. MMWR Morb Mortal Wkly Rep. 2004;53:556–559. [PubMed] [Google Scholar]
- Chan J, Xing Y, Magliozzo RS, Bloom BR. Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. J Exp Med. 1992;175:1111–1122. doi: 10.1084/jem.175.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comstock G. Tuberculosis in twins: a re-analysis of the Prophit survey. Am Rev Respir Dis. 1978;117:621–624. doi: 10.1164/arrd.1978.117.4.621. [DOI] [PubMed] [Google Scholar]
- Cooke GS, Campbell SJ, Sillah J, Gustafson P, Bah B, Sirugo G, Bennett S, McAdam KP, Sow O, Lienhardt C, Hill AV. Polymorphism within the interferon-gamma/receptor complex is associated with pulmonary tuberculosis. Am J Respir Crit Care Med. 2006;174:339–343. doi: 10.1164/rccm.200601-088OC. [DOI] [PubMed] [Google Scholar]
- Cooper AM, Dalton DK, Stewart TA, Griffin JP, Russell DG, Orme IM. Disseminated tuberculosis in interferon gamma gene-disrupted mice. J Exp Med. 1993;178:2243–2247. doi: 10.1084/jem.178.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vera ME, Shapiro RA, Nussler AK, Mudgett JS, Simmons RL, Morris SM, Jr., Billiar TR, Geller DA. Transcriptional regulation of human inducible nitric oxide synthase (NOS2) gene by cytokines: initial analysis of the human NOS2 promoter. Proc Natl Acad Sci U S A. 1996;93:1054–1059. doi: 10.1073/pnas.93.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman SE, Picard C, Lammas D, Heyne K, van Dissel JT, Baretto R, Rosenzweig SD, Newport M, Levin M, Roesler J, Kumararatne D, Casanova JL, Holland SM. Clinical features of dominant and recessive interferon gamma receptor 1 deficiencies. Lancet. 2004;364:2113–2121. doi: 10.1016/S0140-6736(04)17552-1. [DOI] [PubMed] [Google Scholar]
- Flynn JL, Chan J. Immunology of tuberculosis. Annu Rev Immunol. 2001;19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, Bloom BR. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J Exp Med. 1993;178:2249–2254. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Gibson CS, MacLennan AH, Dekker GA, Goldwater PN, Dambrosia JM, Munroe DJ, Tsang S, Stewart C, Nelson KB. Genetic polymorphisms and spontaneous preterm birth. Obstet Gynecol. 2007;109:384–391. doi: 10.1097/01.AOG.0000252712.62241.1a. [DOI] [PubMed] [Google Scholar]
- Gomez LM, Anaya JM, Vilchez JR, Cadena J, Hinojosa R, Velez L, Lopez-Nevot MA, Martin J. A polymorphism in the inducible nitric oxide synthase gene is associated with tuberculosis. Tuberculosis (Edinb) 2007;87:288–294. doi: 10.1016/j.tube.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Hancock DB, Martin ER, Vance JM, Scott WK. Nitric oxide synthase genes and their interactions with environmental factors in Parkinson’s disease. Neurogenetics. 2008;9:249–262. doi: 10.1007/s10048-008-0137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock DB, Martin ER, Li YJ, Scott WK. Methods for interaction analyses using family-based case-control data: conditional logistic regression versus generalized estimating equations. Genet Epidemiol. 2007;31:883–893. doi: 10.1002/gepi.20249. [DOI] [PubMed] [Google Scholar]
- Hobbs MR, Udhayakumar V, Levesque MC, Booth J, Roberts JM, Tkachuk AN, Pole A, Coon H, Kariuki S, Nahlen BL, Mwaikambo ED, Lal AL, Granger DL, Anstey NM, Weinberg JB. A new NOS2 promoter polymorphism associated with increased nitric oxide production and protection from severe malaria in Tanzanian and Kenyan children. Lancet. 2002;360:1468–1475. doi: 10.1016/S0140-6736(02)11474-7. [DOI] [PubMed] [Google Scholar]
- Hussain S, Zwilling BS, Lafuse WP. Mycobacterium avium infection of mouse macrophages inhibits IFN-gamma Janus kinase-STAT signaling and gene induction by down-regulation of the IFN-gamma receptor. J Immunol. 1999;163:2041–2048. [PubMed] [Google Scholar]
- Jamieson SE, Miller EN, Black GF, Peacock CS, Cordell HJ, Howson JM, Shaw MA, Burgner D, Xu W, Lins-Lainson Z, Shaw JJ, Ramos F, Silveira F, Blackwell JM. Evidence for a cluster of genes on chromosome 17q11-q21 controlling susceptibility to tuberculosis and leprosy in Brazilians. Genes Immun. 2004;5:46–57. doi: 10.1038/sj.gene.6364029. [DOI] [PubMed] [Google Scholar]
- Johannesen J, Pie A, Pociot F, Kristiansen OP, Karlsen AE, Nerup J. Linkage of the human inducible nitric oxide synthase gene to type 1 diabetes. J Clin Endocrinol Metab. 2001;86:2792–2796. doi: 10.1210/jcem.86.6.7559. [DOI] [PubMed] [Google Scholar]
- Juenger TE, Wayne T, Boles S, Symonds VV, McKay J, Coughlan SJ. Natural genetic variation in whole-genome expression in Arabidopsis thaliana: the impact of physiological QTL introgression. Mol Ecol. 2006;15:1351–1365. doi: 10.1111/j.1365-294X.2006.02774.x. [DOI] [PubMed] [Google Scholar]
- Kamijo R, Gerecitano J, Shapiro D, Green SJ, Aguet M, Le J, Vilcek J. Generation of nitric oxide and clearance of interferon-gamma after BCG infection are impaired in mice that lack the interferon-gamma receptor. J Inflamm. 1995;46:23–31. [PubMed] [Google Scholar]
- Kleinert H, Pautz A, Linker K, Schwarz PM. Regulation of the expression of inducible nitric oxide synthase. Eur J Pharmacol. 2004;500:255–266. doi: 10.1016/j.ejphar.2004.07.030. [DOI] [PubMed] [Google Scholar]
- Kleinert H, Schwarz PM, Forstermann U. Regulation of the expression of inducible nitric oxide synthase. Biol Chem. 2003;384:1343–1364. doi: 10.1515/BC.2003.152. [DOI] [PubMed] [Google Scholar]
- Kun JF, Mordmuller B, Lell B, Lehman LG, Luckner D, Kremsner PG. Polymorphism in promoter region of inducible nitric oxide synthase gene and protection against malaria. Lancet. 1998;351:265–266. doi: 10.1016/S0140-6736(05)78273-8. [DOI] [PubMed] [Google Scholar]
- Kun JF, Mordmuller B, Perkins DJ, May J, Mercereau-Puijalon O, Alpers M, Weinberg JB, Kremsner PG. Nitric oxide synthase 2(Lambarene) (G-954C), increased nitric oxide production, and protection against malaria. J Infect Dis. 2001;184:330–336. doi: 10.1086/322037. [DOI] [PubMed] [Google Scholar]
- Le Souef PN, Goldblatt J, Lynch NR. Evolutionary adaptation of inflammatory immune responses in human beings. Lancet. 2000;356:242–244. doi: 10.1016/s0140-6736(00)02491-0. [DOI] [PubMed] [Google Scholar]
- Levesque, Allen A, Hauswirth D, Mervin-Blake S, Fernandez C, Patch K, Alexander K, Allgood S, McNair P, Sundy J. An a priori study of nitric oxide synthase type 2 (NOS2A) promoter polymorphisms and exhaled nitric oxide levels in African Americans. In preparation. [Google Scholar]
- Linn SC, Morelli PJ, Edry I, Cottongim SE, Szabo C, Salzman AL. Transcriptional regulation of human inducible nitric oxide synthase gene in an intestinal epithelial cell line. Am J Physiol. 1997;272:G1499–G1508. doi: 10.1152/ajpgi.1997.272.6.G1499. [DOI] [PubMed] [Google Scholar]
- Macmicking JD, North RJ, LaCourse R, Mudgett JS, Shah SK, Nathan CF. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc Natl Acad Sci U S A. 1997;94:5243–5248. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks-Konczalik J, Chu SC, Moss J. Cytokine-mediated transcriptional induction of the human inducible nitric oxide synthase gene requires both activator protein 1 and nuclear factor kappaB-binding sites. J Biol Chem. 1998;273:22201–22208. doi: 10.1074/jbc.273.35.22201. [DOI] [PubMed] [Google Scholar]
- Means TK, Wang S, Lien E, Yoshimura A, Golenbock DT, Fenton MJ. Human toll-like receptors mediate cellular activation by Mycobacterium tuberculosis. J Immunol. 1999;163:3920–3927. [PubMed] [Google Scholar]
- Mira MT, Alcais A, Nguyen VT, Moraes MO, Di FC, Vu HT, Mai CP, Nguyen TH, Nguyen NB, Pham XK, Sarno EN, Alter A, Montpetit A, Moraes ME, Moraes JR, Dore C, Gallant CJ, Lepage P, Verner A, Van D V, Hudson TJ, Abel L, Schurr E. Susceptibility to leprosy is associated with PARK2 and PACRG. Nature. 2004;427:636–640. doi: 10.1038/nature02326. [DOI] [PubMed] [Google Scholar]
- Qu Y, Tang Y, Cao D, Wu F, Liu J, Lu G, Zhang Z, Xia Z. Genetic polymorphisms in alveolar macrophage response-related genes, and risk of silicosis and pulmonary tuberculosis in Chinese iron miners. Int J Hyg Environ Health. 2007;210:679–689. doi: 10.1016/j.ijheh.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Salas A, Carracedo A, Richards M, Macaulay V. Charting the ancestry of African Americans. Am J Hum Genet. 2005;77:676–680. doi: 10.1086/491675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K, Sweet MJ, Hume DA. Signal integration between IFNgamma and TLR signalling pathways in macrophages. Immunobiology. 2006;211:511–524. doi: 10.1016/j.imbio.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Smith NL, Hindorff LA, Heckbert SR, Lemaitre RN, Marciante KD, Rice K, Lumley T, Bis JC, Wiggins KL, Rosendaal FR, Psaty BM. Association of genetic variations with nonfatal venous thrombosis in postmenopausal women. JAMA. 2007;297:489–498. doi: 10.1001/jama.297.5.489. [DOI] [PubMed] [Google Scholar]
- Tang H, Quertermous T, Rodriguez B, Kardia SL, Zhu X, Brown A, Pankow JS, Province MA, Hunt SC, Boerwinkle E, Schork NJ, Risch NJ. Genetic structure, self-identified race/ethnicity, and confounding in case-control association studies. Am J Hum Genet. 2005;76:268–275. doi: 10.1086/427888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Tsuji S, Matsumoto M, Takeuchi O, Akira S, Azuma I, Hayashi A, Toyoshima K, Seya T. Maturation of human dendritic cells by cell wall skeleton of Mycobacterium bovis bacillus Calmette-Guerin: involvement of toll-like receptors. Infect Immun. 2000;68:6883–6890. doi: 10.1128/iai.68.12.6883-6890.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodovotz Y, Bogdan C, Paik J, Xie QW, Nathan C. Mechanisms of suppression of macrophage nitric oxide release by transforming growth factor beta. J Exp Med. 1993;178:605–613. doi: 10.1084/jem.178.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Genetic Data Analysis. Sinauer; Sunderland, Massachusetts: 1996. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.