Abstract
Wnts are secreted lipid-modified glycoproteins that carry out various signaling functions during development and in adult tissue. Wnt signaling is mediated by Fzd receptors at the cell surface and can be modulated by the secreted frizzled related proteins (SFRPs) and other molecular antagonists. Abnormal Wnt signaling has been implicated in several diseases. However, due to the complexity of the Wnt signal and lack of knowledge pertaining to the binding properties of different Wnt ligands, no therapeutic agents exist that target this pathway. Using a novel ELISA-based technique we were able to determine the first measurements of binding affinity for specific Wnt interactions. This study shows that purified Wnt3a, Wnt7a, and Wnt5a have different binding specificities for Fzds and SFRPs.
Keywords: Wnt, β-catenin, JNK, binding affinity, SFRP4, elisa, frizzled
The Wnt signaling pathway is highly conserved across many species and the investigation of this pathway is crucial for understanding development and disease [1;2]. However, several aspects of Wnt signal transduction and pharmacology are unclear. Wnts are secreted fatty-acid modified glycoproteins that regulate important cellular processes such as proliferation, differentiation, and cell fate specification [1;3]. To date, 19 wnt and 10 frizzled genes have been identified in the human and mouse genomes [4]. The binding of a Wnt ligand to its corresponding seven-transmembrane frizzled receptor (Fzd) on a target cell leads to the activation of at least one of the three possible signaling pathways: 1. the canonical/β-catenin pathway, 2. the non-canonical/JNK pathway, and 3. the Ca2+-mediated pathway [1;5]. Though there has been significant progress in the area of Wnt signaling, little is known about the specificity for the Wnt-Fzd interaction and which of the three pathways the individual Wnt-Fzd pairs activate.
Activation of Wnt signaling can be regulated by a host of diverse extracellular secreted proteins including members of the secreted frizzled-related protein (SFRP), Dickkopf, Cerberus, Sclerostin and Wnt inhibitory factor families [6;7]. The five SFRP family members share sequence homology with the cysteine-rich domain (CRD), or putative ligand binding region, of Fzd located in the N-terminal extracellular portion of the receptor [6]. SFRPs are thought to antagonize Wnt action by competing with Fzd for the binding of Wnt and preventing Wnt-Fzd interaction [6].
The biochemical and structural properties of Wnts and their binding affinities for the Fzd and SFRP proteins remain poorly defined. Post-translational modifications render the Wnts hydrophobic, poorly soluble, and difficult to isolate in large quantities [3;8]. A few studies have reported Wnt binding to Fzd CRDs through indirect means such as conditioned medium and transfection with affinities ranging from 9 to 70 nM [9;10]. Recently, nanomolar Kds for purified Wnt-SFRP binding were reported using surface plasmon resonance [11].
We have developed a novel bioassay to help elucidate Wnt-protein specificity and quantify relative affinities. This ELISA-based technique was previously employed and demonstrated that purified recombinant Wnt7a could bind to the CRD of Fzd5 with a Kd = 75.4 nM [12]. In the following report we quantitatively assess the potential for purified Wnt3a, Wnt5a, and Wnt7a to interact with multiple Fzds and SFRPs in a multi-well solid-state platform. These data combined with the analysis of pathway-specific gene reporter activity suggest that Wnt signal transduction is dependent on the diverse specificities and binding affinities of the various Wnt ligands. These findings may prove to be crucial to the future development of pharmacologic mediators of Wnt signaling.
Materials and Methods
Cell Culture and Transfection
Ishikawa endometrial carcinoma cell lines were kept in RPMI 1640 medium (Invitrogen-Gibco) and supplemented with 10% FBS, 0.01 M HEPES, and penicillin/streptomyosin. Cells were split two days prior to transient transfection, and grown to 60–70% confluency. Transient transfections of Ishikawa cells were conducted in 60 mm culture plates using FuGene 6 (Roche) according to manufacturer guidelines. Ishikawa control and Fzd5 V5/6xHis stable cell lines were generated as previously described [13].
Protein Pulldown and Immunoprecipitation Experiments
Recombinant rWnt7a and rSFRP4 V5/6xHis-tagged proteins were purified and quantitated as described [13]. To detect interactions between purified Wnt proteins, 500 ng/ml rWnt7a V5/6xHis, rSFRP4 V5/6xHis, or commercially obtained mWnt3a or mWnt5a (Millipore) were combined in TX-100 buffer (20 mM Hepes, 1.0% Triton X-100, 250 mM NaCl, 10% glycerol, 2 mM EDTA, 1 mM PMSF, 2.5 μg/ml aprotinin, 2.5 μg/ml leupeptin, 100 μM sodium orthovanadate, 50 mM sodium fluoride) with either 500 ng/ml hFzd5CRD-Fc IgG (R&D Systems), hFzd10CRD-Fc IgG (R&D Systems), or Fc IgG (Bethyl Laboratories). To demonstrate whether rSFRP4 bound mWnt3a, 500 ng/ml SFRP4 V5/6xHis was combined with 500 ng/ml mWnt3a and immunoprecipitated with 1 μg rat anti-mWnt3a antibody (R&D Systems).
Western Blot Analysis
Proteins were separated by electrophoresis on SDS-polyacrylamide gels and western analysis was performed using mouse anti-V5 (Invitrogen), rat anti-mWnt3a (R&D Systems), rabbit anti-mWnt5a (Cell Signaling), or rabbit anti-human IgG (Santa Cruz). Immunoreactive proteins were visualized using appropriate secondary antibodies and the enhanced chemiluminescence western blotting system (Amersham).
Luciferase Assays
Cells were co-transfected using 0.5 μg TopFlash firefly luciferase reporter vector and 0.1 μg of TK-Renilla reporter vector. For analysis of c-jun activity, cells were co-transfected using 0.5 μg pFR-Luc reporter vector and 0.05 μg pFA2-c-jun from the PathDetect c-Jun trans-Reporting System (Stratagene), 0.05 μg of the TK-Renilla reporter vector, and 0.5 μg Fzd10 pcDNA3.1 V5/6xHis or pcDNA3.0 empty vector (EV). Cells were reseeded into 12-well plates 24 hours post-transfection and treated with purified proteins as described 6 hours later. Luciferase activities were quantitated 24 hours post-treatment using the dual luciferase reporter assay system (Promega). Data are represented as ratios of Firefly to Renilla luciferase activity. Each experiment was repeated at least three times in duplicate.
Subcloning Wnt3a
The mWnt3a pIRESpuro3 expression vector was obtained from R. Moon, University of Washington. The sequence encoding mWnt3a was PCR amplified using primers (forward primer 5′-CACCATGGCCCCACTCGGATA- 3′ and reverse primer 5′-CTTGCAGGTGTGCACGTCA-3′) designed for ligation into the pcDNA3.1 directional V5/6xHis TOPO vector (Invitrogen) to incorporate a c-terminal V5/6xHis tag. The Wnt3a pcDNA3.1 V5/6xHis vector was verified by DNA sequencing and mWnt3a-V5/6xHis protein expression was confirmed by western analysis. Purification and subsequent quantitation of mWnt3a-V5/6xHis protein was performed as described [13].
ELISA-Based Protein Binding Technique
ELISA was performed for hFzd5CRD-Fc and hFzd10CRD-Fc binding experiments using methodology similar to that previously reported [12] and described in Table 1. For detection of SFRP4 binding, a 96-well plate was initially coated with an excess of SFRP4 antibody (4 μg/ml) (Santa Cruz) followed by immobilization of purified hSFRP4 (R & D Systems) (300 ng/50μl/well). Experiments were performed with using various concentrations of recombinant rWnt7a-V5/6xHis or mWnt3a-V5/6xHis. Absorbance at 450 nm was measured with a plate reader and OD values were converted to concentration based on a V5 standard curve. Non-linear regression analysis was performed using GraphPad Prism 5 Software.
Table 1.
Steps required for the ELISA-based protein binding assay.
| Step | Condition | Time (min) |
|---|---|---|
| Receptor immobilizationa | 2μg/ml FzdCRD-Fc or Fc in carbonate buffer pH 9.0, 4°C | Overnight |
| Wash | 3 × PBST (0.05% Tween-20, PBS pH 7.5), 25°C | 2 |
| Blocking | PBSTM (0.05% Tween-20, 5% dried milk, PBS pH 7.5), 25°C | 45 |
| Wash | 2 × PBST, 25°C | 2 |
| Ligandb | Wnt diluted at various concentrations, 25°C | 120 |
| Wash | 4 × PBST, 25°C | 2 |
| Primary antibody | 1 μg/ml mouse anti-V5 antibody diluted in PBSTM, 25°C | 90 |
| Wash | 4 × PBST, 25°C | 2 |
| Secondary antibody | HRP conjugated goat anti-mouse (1:500) in PBSTM, 25°C | 60 |
| Wash | 4 × PBST, 25°C | 2 |
| Detection | TMB Reagent | 30 |
| Stop reaction | 1M H2SO4 | N/A |
| Analysis | Plate reader, OD450 | N/A |
Modifications for SFRP protocol:
The receptor in this step is substituted with 4μg/ml anti-SFRP antibody.
An additional step is added to immobilize SFRP protein (300ng/well) for 120 minutes at 25°C, followed by a 4 × PBST wash step.
Results and Discussion
Purified Wnt3a binds Fzd5CRD and Activates β-catenin Signaling in Endometrial Cells
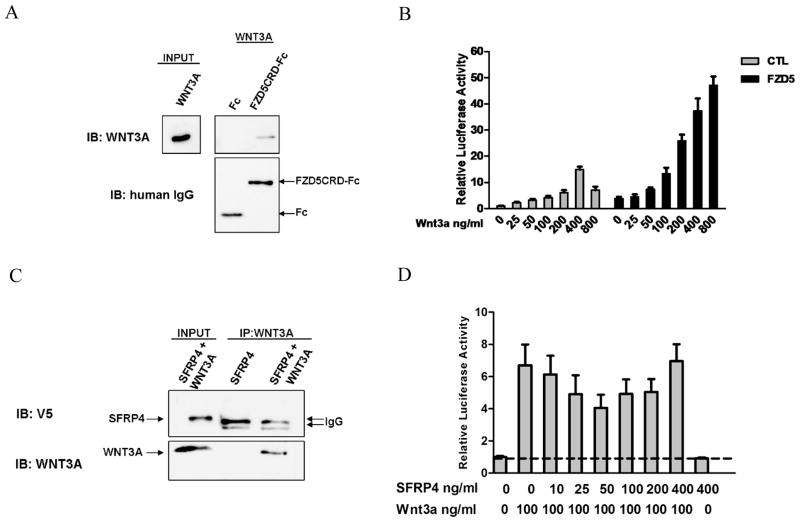
Reports have shown that Wnts bind to the extracellular cysteine-rich domains (CRD) of frizzled receptors as means to activate Wnt signaling [14]. We found that purified Wnt7a-V5/6xHis interacted with the Fzd5CRD in a pulldown experiment and activated β-catenin signaling in a dose-dependent manner [13]. Therefore, we tested whether Wnt3a and Wnt5a could directly bind the CRD of Fzd5 in a similar manner. Wnt3a was incubated with protein A/G agarose and a purified Fzd5CRD protein fused to the Fc portion of human IgG. As a negative control, Wnt3a was incubated with Fc IgG alone. Western analysis confirmed that Wnt3a specifically bound to the Fzd5CRD-Fc and not Fc (Figure 1A). Next, it was investigated whether purified Wnt3a was capable of transducing the canonical/β-catenin signal in the Ishikawa endometrial cancer cell line. The β-catenin-responsive TopFlash reporter was activated in response to treatment with purified Wnt3a and exhibited dose dependency (Figure 1B). Wnt3a protein produced a 15-fold induction of reporter activation in control (vector only) cells and 47-fold induction in Fzd5-overexpressing cells compared to control cell baseline. The EC50 value was calculated to be 240 ng/ml (6.0 nM) for control cells and 200 ng/ml (5.0 nM) for Fzd5-overexpressing cells. Parallel experiments were conducted using purified Wnt5a. Wnt5a failed to bind the Fzd5CRD and did not activate the TopFlash reporter activity in both control and Fzd5 cells (Supplementary Figure 1A and 1B). Wnt5b also had no observable effect on β-catenin signaling in Fzd5 cells (Supplementary Figure 2).
Figure 1. Interaction of purified Wnt3a with Fzd5CRD and activation of β-catenin signaling is not inhibited by SFRP4.
(A) Wnt3a protein was incubated with protein A/G agarose and a purified Fzd5CRD-Fc fusion or Fc alone. Western analysis was conducted with anti-Wnt3a, and rabbit anti-human IgG. (B) TopFlash dose response experiment indicates activation of β-catenin/canonical signaling by purified Wnt3a treatment of Ishikawa control cells (gray bars) and to a greater extent in stable Fzd5-overexpressing Ishikawa cells (black bars). (C) SFRP4 failed to co-immunoprecipitate with Wnt3a protein. Immunoprecipitation was performed using anti-Wnt3a antibody. Western analysis was conducted with anti-V5, anti-Wnt3a, and rabbit anti-human IgG. (D) Concurrent addition of SFRP4 protein had no significant effect on purified Wnt3a activation of the TopFlash reporter in Fzd5-overexpressing cells.
Based on our previous report, this shows that Wnt3a can activate canonical/β-catenin with a greater relative efficacy than Wnt7a [13] (Supplementary Figure 2). We considered that the inclusion of the V5/6xHis-tag may alter the biological activity of Wnt3a. Based on Topflash experiments comparing the activity of untagged Wnt3a to Wnt3a-V5/6xHis, this finding was determined not to be an artifact of the V5/6xHis-tag fused to Wnt7a (data not shown). Instead, this suggests that Wnt3a may promote the receptor active state for a longer time period, compared to Wnt7a. Alternatively, this observation may also be attributed to the recruitment of different intracellular Dsh proteins or other cytosolic proteins that exhibit diverse activation potentials for downstream Wnt signaling. This differential mediation of the canonical/β-catenin pathway by mammalian dishevelleds-1, -2, and -3 has been reported [15]. Each Wnt could also be binding other endogenous Fzds, secreted antagonists, or chaperone proteins that function to modulate signaling and, in turn, impact their efficacy. These findings establish biochemically, that like Wnt7a, Wnt3a can associate with the CRD region of the Fzd5 receptor and activate the canonical/β-catenin pathway.
SFRP4 Fails to Suppress Wnt3a Signaling
There are a number of secreted proteins that can perturb Wnt-Fzd interaction in order to modulate Wnt function, including members of the SFRP family [6]. Our previous studies have shown that SFRP4 can directly bind to Wnt7a and inhibit Wnt signaling in both an autocrine and paracrine fashion [13]. We tested whether SFRP4 could directly bind to Wnt3a in a similar fashion and subsequently modulate downstream signaling. Our pulldown experiment followed by western analysis confirmed no direct physical association between SFRP4 and Wnt3a (Figure 1C). Similarly, SFRP4 failed to bind Wnt5a (data not shown). Ishikawa cells overexpressing Fzd5 were concurrently treated with 100 ng/ml Wnt3a and increasing concentrations of purified SFRP4 protein (Figure 1D). SFRP4 had no significant dose-dependent inhibition on Wnt3a induced β-catenin signaling. Likewise, we observed that SFRP1 suppressed Wnt7a-mediated signaling but had no effect on signaling induced by Wnt3a (Supplementary Figure 2). Wawrzak et al reported that both SFRP1 and SFRP4 bind Wnt3a by surface plasmon resonance analysis [11]. This contradictory result may be due to different techniques employed for the binding studies. Nonetheless, similar to our findings, they reported that SFRP4 did not inhibit Wnt3a signaling [11]. Our data indicate SFRP4 specificity for Wnt7a and not Wnt3a or Wnt5a.
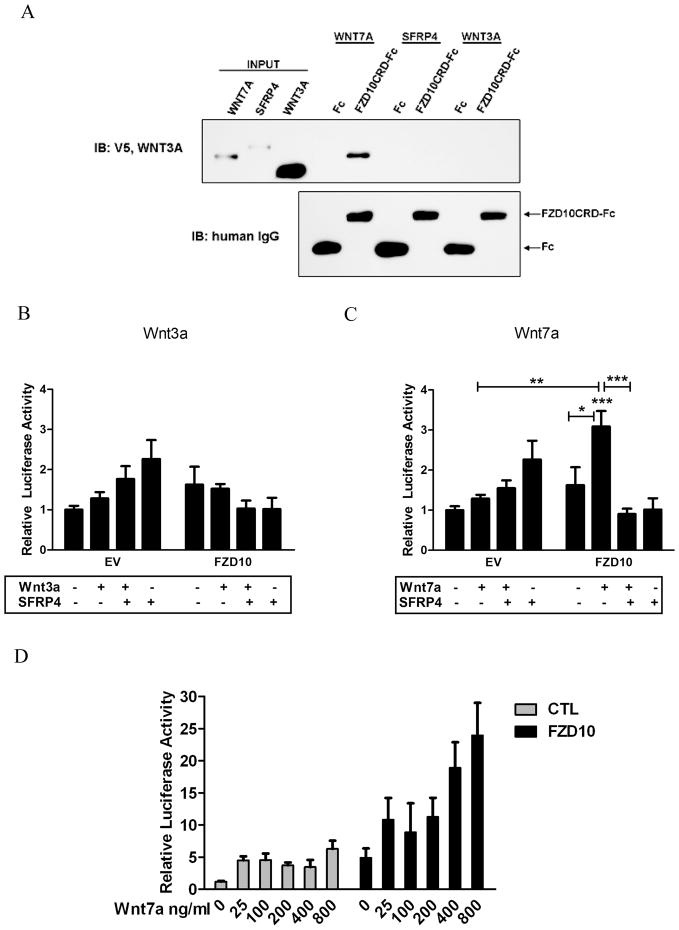
Purified Wnt7a and not Wnt3a binds Fzd10 CRD and Activates the Non-Canonical/JNK Pathway
A pulldown experiment was performed to explore whether Wnt7a, SFRP4, and Wnt3a directly bind to the Fzd10CRD. Western analysis confirmed that Wnt7a bound to the CRD of Fzd10 (Figure 2A). No direct association was detected between Fzd10CRD and SFRP4 or Wnt3a (Figure 2A) or Wnt5a (Supplementary Figure 1A).
Figure 2. Regulation of the non-canonical/JNK pathway by purified Wnt signaling proteins.
(A)Wnt7a, SFRP4, and Wnt3a proteins were incubated with protein A/G agarose and a purified Fzd10CRD-Fc fusion or Fc alone. Western analysis was performed using anti-V5, anti-Wnt3a, and rabbit anti-human IgG. (B), (C), and (D) c-jun luciferase reporter measurements of Ishikawa cell and Fzd10-overexpressing cells treated with (B) 300ng/ml purified Wnt3a and/or SFRP4, (C) 300ng/ml Wnt7a and/or SFRP4 or (D) increasing concentrations of purified Wnt7a as indicated. Luciferase activity is presented as relative light units Firefly/Renilla ± S.E. of three independent experiments runs in duplicate. Statistical significance is based on ANOVA and Bonferroni post-hoc for multiple comparisons. *, **, and *** indicate (p≤0.05), (p≤0.01), and (p≤0.001) respectively.
To detect potential activation of the non-canonical/JNK pathway, Ishikawa control cells or Fzd10-overexpressing cells were transfected with a phospho c-jun transactivation reporter and treated with various purified Wnt proteins. Wnt3a did not activate non-canonical signaling in either cell type (Figure 2B). Treatment with purified recombinant Wnt7a significantly induced non-canonical/JNK signaling in the presence of Fzd10 and exhibited dose-dependency with EC50 = 290 ng/ml (5.8 nM) (Figure 2C and 2D). SFRP4 suppressed Wnt7a activation of signaling mediated by Fzd10 (Figure 2C). Treatment with purified Wnt5a protein had no effect on modulating JNK activation in the absence or presence of Fzd10 overexpression in Ishikawa cells (Supplementary Figure 1C). However, Wnt5a has been shown to mediate non-canonical/JNK signaling in the context of other receptors, including ROR2, in different cell systems [16]. These data demonstrate that purified Wnt7a, and not Wnt3a or Wnt5a, can activate Fzd10 mediated JNK signaling in a dose-responsive manner and this signal can be inhibited by SFRP4.
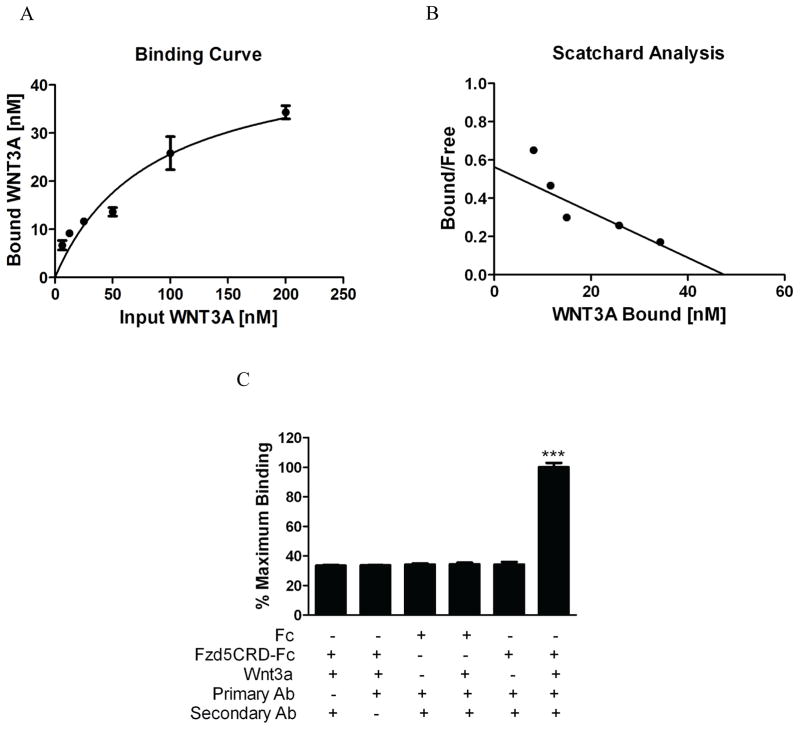
Relative Binding Affinities of Wnt Interactions
An ELISA-based technique was developed in order to determine the relative affinities of each observed Wnt-protein interaction. As shown by our binding curve (Figure 3A), when increasing amounts of Wnt3a were added to Fzd5CRD-Fc coated wells, an increase in binding was detected at concentrations up to 200 nM. Using non-linear regression analysis we constructed a Scatchard plot (Figure 3B) and calculated Bmax and Kd values of 47.1 nM and 83.7 nM, respectively. When compared to wells containing Fzd5CRD-Fc plus the maximum concentration of Wnt3a, very little background signal was measured from control wells that did not include primary or secondary detection antibodies (Figure 3C). Similarly, no significant signal was detected from wells coated with Fc or in wells not containing Wnt3a. Our results show that Wnt3a binds to Fzd5CRD with similar affinity and higher Bmax compared to Wnt7a, as we previously reported [12]. Equal affinities and different Bmax values may indicate different ligand-receptor stoichiometries. It has been speculated that more than one Wnt binding site may be present in the extracellular region of the Fzd receptor [9]. A higher Wnt3a-Fzd5CRD Bmax is also consistent with Wnt7a simply being a lower efficacy ligand for Fzd5, as was established by our TopFlash results. The fact that the Wnt3a and Wnt7a EC50 values were lower than their Kds may be an indication that the Fzd receptors are present in excess. This discrepancy could also be an artifact of recombinant protein heterogeneity during purification or the heterogeneous arrangement of immobilized FzdCRD proteins on the surface of the multi-well ELISA plate. Given that Wnt3a, Wnt7a, and Fzd5 are endogenously expressed in the Ishikawa cell line [13] and both Wnts have equal affinities for Fzd5 implies that Wnt3a and Wnt7a likely compete for binding to Fzd5. These data strongly indicate that Wnt3a binds to Fzd5CRD with affinity in the nanomolar range.
Figure 3. Quantitative binding data for Wnt3a-Fzd5CRD interaction using ELISA-based technique.
(A) Wnt3a-Fzd5CRD binding curve, (B) Scatchard plot, and (C) bar graph indicating negative controls for ELISA-based technique. The calculated Bmax and Kd were 47.1 nM and 83.7 nM, respectively. Binding data was determined using GraphPad Prism non-linear regression analysis. ***p≤0.001 compared to all controls using ANOVA and Bonferroni post-hoc analysis.
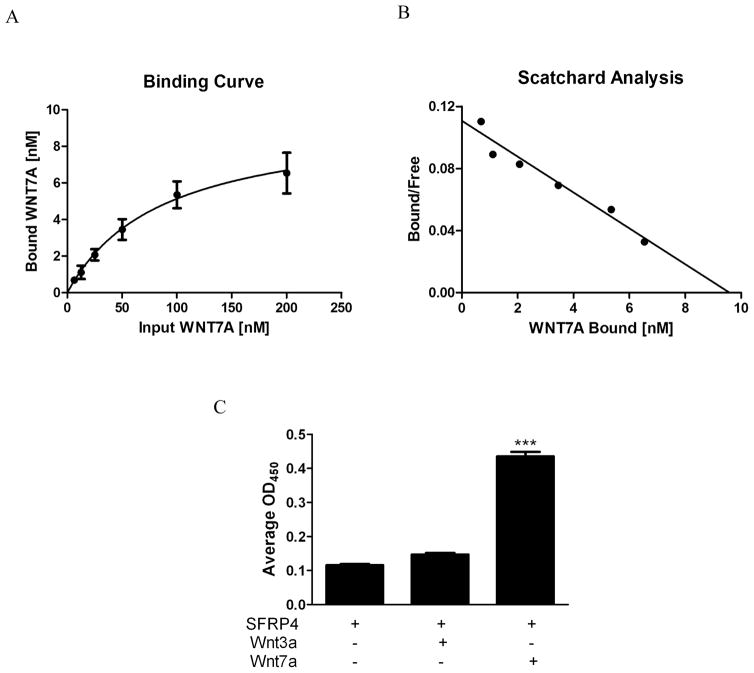
Our work revealed that SFRP4 binds to Wnt7a and inhibits Wnt7a-induced signaling. Therefore, we employed our ELISA-based technique to quantitatively determine the relative affinity of Wnt7a binding to SFRP4. When increasing amounts of Wnt7a were added to wells coated with SFRP4 protein, an increase in binding was detected at concentrations up to 200 nM (Figure 4A). Using non-linear regression analysis and Scatchard plot was obtained (Figure 4B) and the Bmax and Kd were calculated to be 9.6 nM and 86.5 nM, respectively. Minimal background signal was measured from control wells that did not have the primary or secondary detection antibodies added and no signal was detected from wells coated with anti-SFRP4 alone (data not shown). A comparison of the average measured OD450 values for no ligand, Wnt7a (200nM), and the non-binding Wnt3a ligand (200nM) is shown in Figure 4C. Unlike Wnt7a, when Wnt3a was added to the surface immobilized SFRP4 no significant change in the assay OD450 value was observed compared to baseline. This exhibits the specificity of the assay for different ligands. The finding that Wnt7a affinity for SFRP4 is similar to its affinity for the Fzd5CRD suggests that SFRP4 competes with Fzd5 to bind Wnt7a and the relative level of expression of the two proteins would determine if a Wnt signal is propagated.
Figure 4. Quantitative binding data for Wnt7a-SFRP4 interaction using ELISA-based technique.
(A) Wnt7a-SFRP4 binding curve and (B) Scatchard plot. (C) Comparison of average OD450 values obtained for no ligand baseline, Wnt3a, and Wnt7a. Wnt7a-SFRP4 binding curve and scatchard plot indicate Bmax = 9.6 nM and Kd = 86.5 nM. Binding data was determined using GraphPad Prism nonlinear regression analysis. ***p≤0.001 compared to all controls using ANOVA and Bonferroni post-hoc analysis.
The ELISA-based technique was employed for the remaining Wnt-protein pairs and the Kd and Bmax values for detectable binding are listed in Table 2. The results indicate that Wnt7a binds to the Fzd10CRD with lower affinity compared to the binding data collected from the Wnt7a-Fzd5CRD, Wnt3a-Fzd5CRD and Wnt7a-SFRP4 experiments. The low affinity of Wnt7a-Fzd10CRD binding may indicate that this interaction does not occur under physiological conditions in vivo. Alternatively, this observation may be attributed to the possibility that an alternative extracellular portion of the Fzd10 receptor must be present in order to mediate optimal binding of the Wnt7a ligand.
Table 2.
Quantitative binding parameters of Wnt signaling proteins
Conclusions
In conclusion, these findings indicate that purified Wnt3a, Wnt5a, and Wnt7a have different binding specificities for Fzd5, Fzd10, and SFRP4. We have developed a novel ELISA-based technique to quantitatively measure the relative affinity of different Wnt interactions. Nanomolar affinity and EC50s were obtained for each positive Wnt-protein complex as summarized in Table 2. These data uniquely demonstrate that the nature of the Wnt signal may be dependent upon the binding affinity of distinct Wnt ligands for the different Fzd receptors and SFRPs expressed in individual tissues. The determination of Wnt-receptor affinity is fundamental to elucidating protein function, the biochemical impact on cell signal transduction, and targeting the Wnt pathway for drug discovery. Additional studies need be conducted to address the binding specificities and affinities of other Wnt ligands for their cognate binding partners.
Supplementary Material
Acknowledgments
This work was supported by a training fellowship from the Keck Center for Interdisciplinary Bioscience Training of the Gulf Coast Consortia NIH Grant #5 T90 DK070109-03 and 5 R90 DK071505-03, the American Legion Auxiliary Fellowship, John S. Dunn Pilot Screening Grant, and the NIH Uterine Cancer Specialized Programs of Research Excellence Grant#P50-CA098258.
Abbreviations
- Fzd
Frizzled
- SFRP
Secreted frizzled-related protein
- JNK
jun N-terminal kinase
- ELISA
Enzyme-linked immunosorbent assay
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 2.Polakis P. Wnt signaling and cancer. Genes & Development. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 3.Willert K, Brown JD, Danenberg E, Duncan AW, Weissman IL, Reya T, Yates JR, Nusse R. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature. 2003;423:448–452. doi: 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
- 4.Miller JR. The Wnts. Genome Biol. 2002;3:REVIEWS3001. doi: 10.1186/gb-2001-3-1-reviews3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mlodzik M. Planar cell polarization: do the same mechanisms regulate Drosophila tissue polarity and vertebrate gastrulation? Trends Genet. 2002;18:564–571. doi: 10.1016/s0168-9525(02)02770-1. [DOI] [PubMed] [Google Scholar]
- 6.Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116:2627–2634. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- 7.Li X, Zhang Y, Kang H, Liu W, Liu P, Zhang J, Harris SE, Wu D. Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. J Biol Chem. 2005;280:19883–19887. doi: 10.1074/jbc.M413274200. [DOI] [PubMed] [Google Scholar]
- 8.Wainwright BJ, Scambler PJ, Stanier P, Watson EK, Bell G, Wicking C, Estivill X, Courtney M, Boue A, Pedersen PS, Williamson R, Farrall M. Isolation of A Human-Gene with Protein-Sequence Similarity to Human and Murine Int-1 and the Drosophila Segment Polarity Mutant Wingless. EMBO J. 1988;7:1743–1748. doi: 10.1002/j.1460-2075.1988.tb03003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsieh JC, Rattner A, Smallwood PM, Nathans J. Biochemical characterization of Wnt-Frizzled interactions using a soluble, biologically active vertebrate Wnt protein. Proc Natl Acad Sci USA. 1999;96:3546–3551. doi: 10.1073/pnas.96.7.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu CH, Nusse R. Ligand receptor interactions in the Wnt signaling pathway in Drosophila. J Biol Chem. 2002;277:41762–41769. doi: 10.1074/jbc.M207850200. [DOI] [PubMed] [Google Scholar]
- 11.Wawrzak D, Metioui M, Willems E, Hendrickx M, de Genst E, Leyns L. Wnt3a binds to several sFRPs in the nanomolar range. Biochemical and Biophysical Research Communications. 2007;357:1119–1123. doi: 10.1016/j.bbrc.2007.04.069. [DOI] [PubMed] [Google Scholar]
- 12.Carmon KS, Loose DS. Wnt7a interaction with Fzd5 and detection of signaling activation using a split eGFP. Biochem Biophys Res Commun. 2008;368:285–291. doi: 10.1016/j.bbrc.2008.01.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carmon KS, Loose DS. Secreted frizzled-related protein 4 regulates two Wnt7a signaling pathways and inhibits proliferation in endometrial cancer cells. Mol Cancer Res. 2008;6:1017–1028. doi: 10.1158/1541-7786.MCR-08-0039. [DOI] [PubMed] [Google Scholar]
- 14.Dann CE, Hsieh JC, Rattner A, Sharma D, Nathans J, Leahy DJ. Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains. Nature. 2001;412:86–90. doi: 10.1038/35083601. [DOI] [PubMed] [Google Scholar]
- 15.Lee YN, Gao Y, Wang HY. Differential mediation of the Wnt canonical pathway by mammalian Dishevelleds-1, -2, and -3. Cell Signal. 2008;20:443–452. doi: 10.1016/j.cellsig.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oishi I, Suzuki H, Onishi N, Takada R, Kani S, Ohkawara B, Koshida I, Suzuki K, Yamada G, Schwabe GC, Mundlos S, Shibuya H, Takada S, Minami Y. The receptor tyrosine kinase Ror2 is involved in non-canonical Wnt5a/JNK signalling pathway. Genes Cells. 2003;8:645–654. doi: 10.1046/j.1365-2443.2003.00662.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.