Abstract
Few studies have examined the association of SNPs in the adiponectin (ADIPOQ) and adiponectin receptor 1 and 2 (ADIPOR1, ADIPOR2) genes with the euglycemic clamp, i.e. the gold standard measure of insulin sensitivity. The association of comprehensive tag SNPs in these genes with insulin sensitivity was examined in a cohort of adolescents and their parents. Probands and siblings (n = 441, mean age = 17.9 years) were recruited along with their parents (n = 262, mean age = 47.9 years).Typed SNPs included 21 SNPs in ADIPOQ, 7 SNPs in ADIPOR1, and 13 SNPs in ADIPOR2. Mixed model linear regression was used to test the association of SNPs with euglycemic-clamp derived insulin sensitivity. All analyses were stratified by race. After corrections to account for multiple testing and the linkage disequilibrium structure of the genes, one SNP in the ADIPOQ gene (rs822393) was significantly associated with insulin sensitivity in white subjects. In whites, six SNPs in ADIPOQ, one SNP in ADIPOR1 and one SNP in ADIPOR2 were associated with insulin sensitivity at the p < .05 level. In African Americans, two SNPs in ADIPOR1 were associated with insulin sensitivity at the p < .05 level. These results suggest that a variant in the ADIPOQ gene influences levels of insulin sensitivity and age may modify the effects of this variant. There are several other variants in ADIPOQ, ADIPOR1, and ADIPOR2 that may influence insulin sensitivity and these variants should be further investigated in other populations.
Keywords: Insulin sensitivity, adiponectin, euglycemic clamp, SNP
Adiponectin, an adipokine, is inversely related to both adiposity and many chronic disease risk factors, including insulin resistance. Adiponectin has been associated, cross-sectionally, with measures of adiposity (Shand et al. 2003; Weyer et al. 2001) and both cross-sectionally and longitudinally with euglycemic-clamp derived insulin sensitivity (Rasmussen-Torvik et al. In press; Stefan et al. 2002). Because of these relations, adiponectin may play a causal role in the association of obesity and insulin resistance.
Previous studies have identified SNPs in the adiponectin gene (ADIPOQ) that appear to alter the transcription or activity of adiponectin (Menzaghi et al. 2007).Additional studies have identified SNPs in two receptors for adiponectin (ADIPOR1 and ADIPOR2) that may also affect the activity of adiponectin (Damcott et al. 2005; Stefan et al. 2005).Few studies have examined the association of SNPs in these genes with the moderately heritable (Rasmussen-Torvik et al. 2007) gold standard measure of insulin sensitivity, the euglycemic clamp (Buzzetti et al. 2007; Kantartzis et al. 2006; Salmenniemi et al. 2005; Stefan et al. 2005; Ukkola et al. 2005; Vozarova de Courten et al. 2005) and only two other studies attempted to use tag SNPs to capture variation throughout the entire gene (Hara et al. 2005; Vozarova de Courten et al. 2005).Additionally, most previous studies have been conducted in middle-aged or older populations. In this study, the association of tag SNPs (selected using the International Hap Map website (Thorisson et al. 2005)) in ADIPOQ, ADIPOR1, and ADIPOR2 with clamp-derived insulin sensitivity was determined in a population of adolescents and their parents.
Research Design and Methods
Subjects
Subjects were drawn from a longitudinal study of cardiovascular risk factors in adolescents. Details of the recruitment have been published previously (Sinaiko et al. 2001).Briefly, in 1996, Minneapolis school children were randomly selected with stratification according to sex, race (African American and white), and systolic blood pressure. Informed consent was obtained for 401 children (probands) and their parents. Probands returned for a subsequent study visit at mean age 19 at which time parents and siblings of the probands also were recruited into the study and underwent many of the same measurements as the probands. Individuals who self-reported a diagnosis of diabetes or use of diabetes medication did not have a euglycemic clamp measurement of insulin sensitivity performed. All genotyped individuals (probands, parents, and siblings) with insulin sensitivity measurements from this visit were included in this analysis. Individuals with extreme values of insulin sensitivity (n = 9) were excluded from this analysis. This resulted in a sample of 584 whites and 119 African Americans.
Phenotypic measurements
Overall adiposity was estimated by BMI (kg/m2).Weight was measured using a balance beam scale and height was measured using a stadiometer. Tanner stage was assessed in children during a physical exam by a pediatrician. Fasting insulin and glucose were collected as previously described (Sinaiko et al. 2001) and HOMA-IR was calculated according to the report by Matthews et al. with the formula HOMA-IR = (fasting insulin in µU/ml × fasting glucose in mg/dl )/405 (Matthews et al. 1985). Euglycemic clamp studies were conducted in the University of Minnesota Clinical Research Center after a 12 hour fast as previously described (Sinaiko et al. 2001). Plasma glucose was measured at baseline and every five minutes during the clamp. The insulin infusion was started at time 0 and continued at 1 mU/kg/min for 3 hours. An infusion of 20% glucose was started at time 0 and adjusted, based on plasma glucose levels, to maintain plasma glucose at 100 mg/dl. Insulin sensitivity was determined from the amount of glucose administered over the final 40 minutes of the euglycemic clamp, and expressed as glucose utilization/ kg lean body mass/ minute. Percentage of body fat and lean body mass (LBM), or fat-free mass were calculated by DEXA.
Genotyping
SNPs were selected using an algorithm provided on the International HapMap website (Thorisson et al. 2005).The algorithm was set to select tag SNPs that would cover all HapMap SNPs with a minor allele frequency of 0.05 or greater with an r2 of 0.8 or greater in a Caucasian (CEU) population. Additional candidate SNPs were selected after reviewing the literature. In total 15 tag SNPs and 6 additional SNPs were typed within ADIPOQ, 5 tag SNPs and 2 additional SNPs were typed in ADIPOR1, and 12 tag and 1 additional SNP were typed in ADIPOR2.
Multiplexed PCR reactions were run at the University of Minnesota BioMedical Genomics Center using primers selected using Sequenom® software. SNP typing was conducted using the Sequenom MassARRAY® mass spectrometry system at the BioMedical Genomics Center. A total of 27 blind duplicates were run to assess genotyping quality. All SNPs had 100% agreement between the duplicates, except one SNP in ADIPOR2 (rs2286384) that had 95% agreement between the duplicates.
Statistical Methods
All genetic association analyses were stratified by self-identified race (white vs. African American) to avoid spurious associations due to population stratification (Cardon and Palmer. 2003).Linkage disequilibrium (r2) and Hardy-Weinberg equilibrium were calculated separately by race using the computer program Haploview (Barrett et al. 2005).
Individual SNP associations and genotype means (adjusted for age and sex) of insulin sensitivity were calculated using linear regression as implemented in a mixed model (SAS v.9.1, Cary, NC).To account for the phenotypic correlation in insulin sensitivity that would be expected within families, a compound symmetry correlation structure between members of families was specified in the mixed model, and a sandwich estimator used to calculate the variance. SNP genotypes were included individually in regression models as 2-df class variables. There was some instability in the estimation of p-values using the sandwich estimator when there were small numbers for the homozygous minor allele genotype. Therefore, in cases where there were fewer than 10 individuals with the homozygous minor allele genotype, a 1 d.f. dominant model association test (with the homozygous minor allele and heterozygous genotypes combined) was used and the corresponding p-value reported in this paper. T-tests were performed in SAS. Estimates of the proportion of variance accounted for by SNPs and other covariates were performed in SOLAR v. 2.1.5 (Almasy and Blangero. 1998).
All regression models were adjusted for age and sex. Further adjustment for BMI did not materially alter the effect size of the β estimates for any SNPs, so models adjusted for BMI were not reported in the paper. Additional regression models were run stratified by generation (parents vs. siblings and probands) only in whites because of limited sample size in the African Americans. Because the associations of multiple SNPs in each gene were tested with insulin sensitivity, a multiple testing correction was implemented in the program SNPSpD (Nyholt 2004) using the Li and Ji estimate of effective number of SNPs (Li and Ji. 2005). This program takes into account both the number of SNPs tested in each gene, and the LD structure among the SNPs. Using this program with genotype information for the white participants in the study, the threshold for significance for SNP associations in the ADIPOQ gene was 0.0037, the threshold for significance for SNP associations in the ADIPOR1 gene was 0.0085, and the threshold for significance for SNP associations in the ADIPOR2 gene was 0.0063.Because of limited power in this study population, genotype means for all associations with p < .05 were reported in this paper.
Results
Cohort Characteristics
Table 1 lists characteristics of the individuals included in the study by self-identified race and generation. Parents had significantly greater mean BMI (p < 0.0001) than their offspring but mean insulin sensitivity was similar (p = 0.22).The majority of offspring were Tanner stage 5 or greater, but 22.3% of white offspring and 25 % of African American offspring had not yet reached Tanner stage 5. The majority of parents were female while the majority of offspring, particularly African American offspring, were male. Five white parents and one African American offspring did not report a diagnosis of diabetes but had fasting glucose measurements (range 127–137 mg/dl) that met the ADA definition of diabetes. Exclusion of these individuals from the study population did not materially change any results presented in this analysis.
Table 1.
Characteristics of study population, by self-identified race and generation
| White parents (n=235) |
African American parents (n=27) |
White offspring (n=349) |
African American offspring (n=92) |
|
|---|---|---|---|---|
| Age (years) | 48.4 (4.9) | 44.0 (7.3) | 17.9 (3.0) | 18.0 (4.3) |
| Waist (cm) | 91.7 (15.1) | 97.8 (17.9) | 82.6 (15.2) | 80.4 (14.4) |
| BMI (kg/m2) | 28.1 (5.4) | 31.8 (8.0) | 25.1 (6.2) | 25.2 (6.2) |
| Insulin sensitivity (mg/kg/min) |
11.2 (3.9) | 10.8 (4.2) | 10.9 (3.6) | 10.4 (3.3) |
| Fasting glucose (mg/dl) |
92.7 (10.8) | 91.0 (12.5) | 87.4 (8.3) | 86.8 (8.7) |
| Fasting insulin (uU/ml) |
8.8 (4.7) | 9.3 (4.9) | 9.6 (8.0) | 10.2 (9.1) |
| Lean body mass (kg) | 52.1 (11.8) | 51.9 (11.4) | 51.5 (12.8) | 53.3 (11.6) |
| Fat body mass (kg) | 29.8 (12.8) | 37.8 (16.9) | 22.8 (14.4) | 19.5 (16.0) |
| Percent body fat | 35.6 (10.1) | 40.9 (10.5) | 29.2 (12.2) | 24.5 (14.9) |
| Male (%) | 41.7 | 25.1 | 54.4 | 68.5 |
| Tanner stage 5 (%) | Not applicable | Not applicable | 77.7 | 75.0 |
Mean (SD)
Genotypes
Table 2 lists the rs numbers (SNP ID), chromosome positions, and previously used labels for the SNPs included in this analysis. Few SNPs encoded amino acid changes. Because ADIPOQ, ADIPOR1, and ADIPOR2 have been studied extensively, nearly all the SNPs had been reported in the literature and thus had labels from previous studies.
Table 2.
SNPs genotyped in study sample
| GENE | SNP ID | Chromosome position (bp) |
SNP label— previous literature |
Major/ Minor allele† |
|---|---|---|---|---|
| ADIPOQ | rs4632532 | 184987915 | −19169c/t | T/C |
| ADIPOQ | rs860291 | 184994175 | −12891/−12823 | C/T |
| ADIPOQ | rs16861194* | 184995644 | A/G | |
| ADIPOQ | rs17300539* | 184995679 | −11391 g/a | G/A |
| ADIPOQ | rs266729* | 184995693 | −11365/−11377 | C/G |
| ADIPOQ | rs182052* | 184997001 | −10066/−10069 | G/A |
| ADIPOQ | rs822391* | 185000021 | T/C | |
| ADIPOQ | rs822393 | 185002548 | −4522 | C/T |
| ADIPOQ | rs16861210* | 185002720 | G/A | |
| ADIPOQ | rs822396* | 185003099 | −3964 a/g | A/G |
| ADIPOQ | rs12495941* | 185004402 | G/T | |
| ADIPOQ | rs7649121* | 185005007 | A/T | |
| ADIPOQ | rs2036373 | 185006413 | −657 | T/G |
| ADIPOQ | rs9882205 | 185006620 | −450 | G/A |
| ADIPOQ | rs17366568* | 185006675 | G/A | |
| ADIPOQ | rs2241766 | 185007114 | 45 t/g | T/G |
| ADIPOQ | rs1501299* | 185007345 | 276 g/t | C/A |
| ADIPOQ | rs2241767* | 185007418 | 349 a/g | A/G |
| ADIPOQ | rs3821799* | 185007708 | C/T | |
| ADIPOQ | rs3774261* | 185007781 | 712 g/a | G/A |
| ADIPOQ | rs17366743* | 185008311 | Y111H | T/C |
| ADIPOR1 | rs10753929* | 176052424 | C/T | |
| ADIPOR1 | rs10494839* | 176051440 | T/C | |
| ADIPOR1 | rs12733285* | 176051286 | C/T | |
| ADIPOR1 | rs12045862* | 176046053 | C/T | |
| ADIPOR1 | rs1342387* | 176043603 | 5843 | G/A |
| ADIPOR1 | rs2275736 | 176040732 | 8714 | T/A |
| ADIPOR1 | rs7539542 | 176039221 | 10,225 (3’UTR) | C/G |
| ADIPOR2 | rs1029629* | 3407188 | −64,241 | A/C |
| ADIPOR2 | rs2058033* | 3412800 | −58,628 | A/C |
| ADIPOR2 | rs7975600* | 3423176 | A/T | |
| ADIPOR2 | rs12826079* | 3434472 | C/T | |
| ADIPOR2 | rs11061946* | 3436443 | C/T | |
| ADIPOR2 | rs10773983* | 3439160 | G/A | |
| ADIPOR2 | rs11612383* | 3439269 | G/A | |
| ADIPOR2 | rs4140992 | 3444724 | −26,696 | T/C |
| ADIPOR2 | rs1058322* | 3444891 | −26,529 | C/T |
| ADIPOR2 | rs929434* | 3469980 | −1,419 | C/T |
| ADIPOR2 | rs16928751* | 3497933 | 26,690 (Gln/Gln) | G/A |
| ADIPOR2 | rs2286384* | 3498777 | 27,534 | C/G |
| ADIPOR2 | rs1044471* | 3504679 | 33,447 (3’ UTR) | C/T |
indicates tag SNP
in white probands
Because of the large number of SNPs typed, a cut-off of p < 0.01 was set to define SNPs in Hardy-Weinberg disequilibrium. Based on this cut-off, only 1 SNP (rs7975600 in ADIPOR2) was in Hardy-Weinberg disequilibrium in both races. This SNP was not included in association analyses. Supplementary Figures 1 – 6 show the pair-wise linkage disequilibrium (LD) patterns as expressed by r2 across ADIPOQ, ADIPOR1, and ADIPOR2 in African Americans and whites. Because only the minimum number of SNPs required to tag the gene (in whites) were selected, there is relatively low-LD between the SNPs in each gene. As expected, the LD between SNPs is also lower in the African American population as compared to the white population.
Single SNP Associations
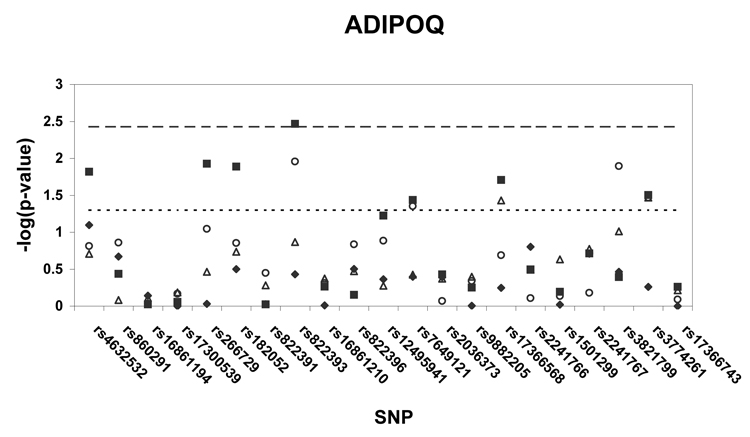
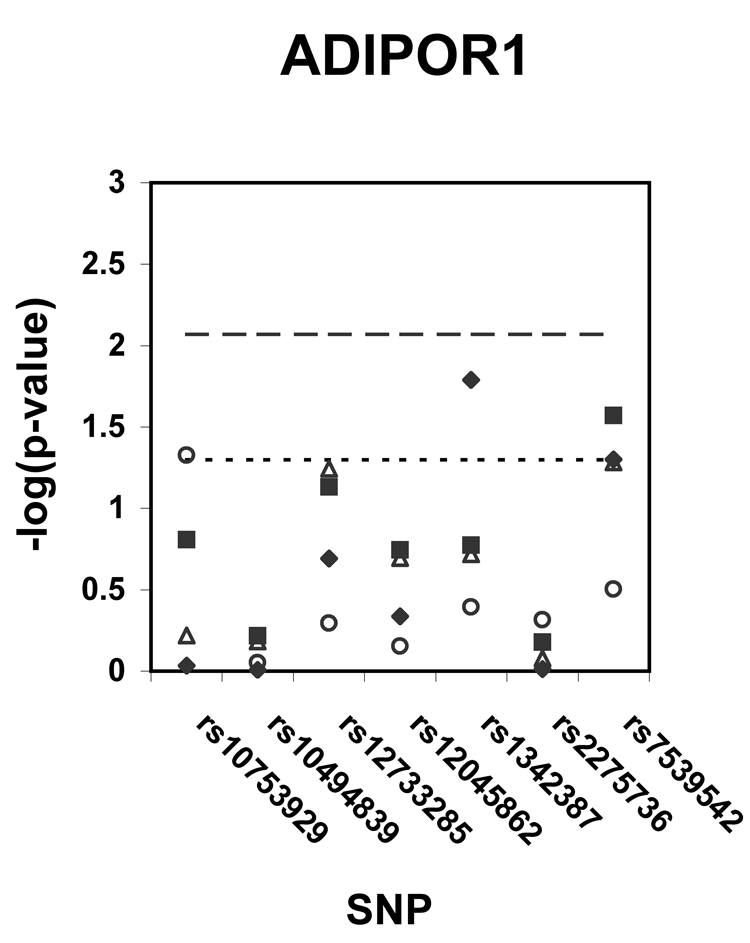
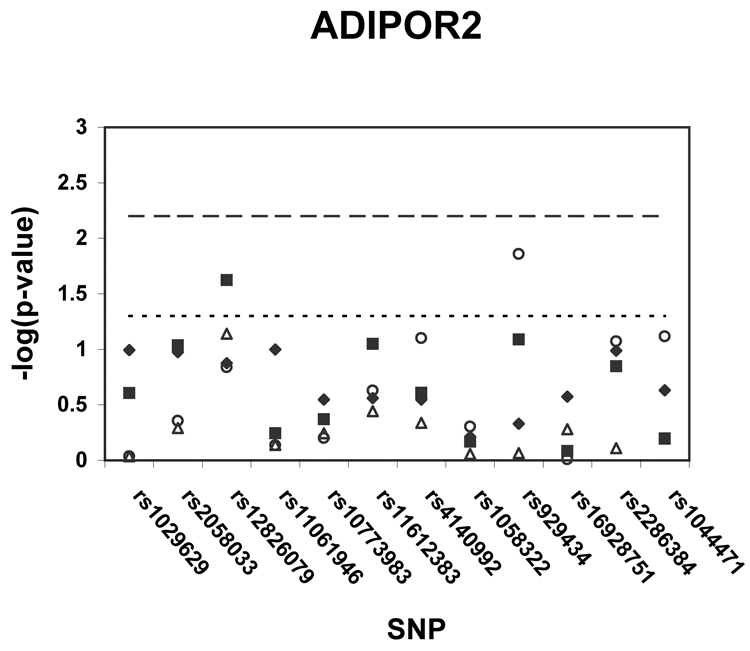
Figure 1 shows p-values (on a –log scale) for the single SNP association analyses with insulin sensitivity in African Americans, whites, and white generational subgroups. Using the pre-specified thresholds for significance (p < .0037 for ADIPOQ, p < .0085 for ADIPOR1, p < .0063 for ADIPOR2), one ADIPOQ SNP (rs822393) was significantly associated (p = .0034) with insulin sensitivity in whites. After adjustment for age and sex, the SNP rs822393 accounted for 1.9 % of the variance in clamp-derived insulin sensitivity. In whites, several additional SNPs in ADIPOQ, one SNP in ADIPOR1, and one SNP in ADIPOR2 were associated with insulin sensitivity at the p < .05 level. In African Americans, two SNPs in ADIPOR1 were associated at the p < .05 level. The association of SNPs in ADIPOQ, ADIPOR1, and ADIPOR2 were also tested with HOMA-IR, a surrogate measure of insulin resistance; no associations of this trait with HOMA-IR met the pre-specified levels of significance.
Figure 1. a–c. Association of tag SNPs in ADIPOQ, ADIPOR1, ADIPOR2 with insulin sensitivity.
All p-values are for regressions of insulin sensitivity on SNP genotype adjusted for age and sex. Black diamonds = African Americans, black squares = whites, white triangles = white probands and siblings, white circles = white parents. Points above the dashed line indicate p-values below the pre-specified significance threshold defined by SNPSpD (p < .0037 for ADIPOQ, p < .0085 for ADIPOR1, p < .0063 for ADIPOR2).Points above the dotted line indicate p-values below the threshold of p = .05. SNPs are listed in order of their appearance in the gene.
The association of rs822393 with insulin sensitivity was more highly significant (based on p-values) in the parents (p = .01), as compared to the offspring (p = .14), although there was not sufficient power to conduct a formal test of interaction for this association with age. There was no evidence of significant interaction of this association with BMI and the significance of the association was similar in those with BMI ≤ 25 (p = .0255) and those with BMI over 25 (p = .0174).Additionally, there was no evidence of significant interaction with BMI for any of the suggestive (p < .05) associations reported in this paper.
Table 3 lists adjusted means by genotype for all SNPs associated with insulin sensitivity at the p < .05 level. For five ADIPOQ SNPs (rs4632532, rs266729, rs182052, rs7649121, and the single significant SNP, rs822393) the pattern of genotype means was extremely similar. When two of these SNPs were included together in a regression model, the β estimates for SNP rs822393 were attenuated slightly. Generational subgroup (parents and offspring) genotype means were calculated for SNPs rs822393. The pattern of genotype means was similar in both subgroups, although the differences between the means were more pronounced in the parents (Supplementary Table 1).
Table 3.
Adjusted* means (mg/kg/min) and 95% confidence intervals (C.I.) of insulin sensitivity by SNP Genotype
| SNP | Gene | population | † Genotype Means | p-value†† | ||
|---|---|---|---|---|---|---|
| rs4632532 | ADIPOQ | Whites | TT= 10.7 (10.2 , 11.2) |
CT= 11.0 (10.5, 11.4) |
CC= 12.2 (11.3, 13.1) |
.015 |
| rs266729 | ADIPOQ | Whites | CC= 10.7 (10.2, 11.1) |
CG= 11.2 (10.6, 11.7) |
GG= 12.2 (11.2, 13.2) |
.012 |
| rs182052 | ADIPOQ | Whites | GG= 10.7 (10.1, 11.2) |
GA= 11.0 (10.5, 11.4) |
AA= 12.2 (11.3, 13.0) |
.013 |
| rs822393 | ADIPOQ | Whites | CC= 10.6 (10.2 , 11.0) |
CT= 11.1 (10.6, 11.6) |
TT= 12.7 (11.6, 13.9) |
.0034 |
| rs7649121 | ADIPOQ | Whites | AA= 10.7 (10.3, 11.2) |
AT= 11.3 (10.7, 12.0) |
TT= 12.7 (11.0, 14.4) |
.037 |
| rs17366568 | ADIPOQ | Whites | GG= 10.8 (10.4, 11.2) |
GA/AA‡ = 11.7 (11.1, 12.4) |
.020‡ | |
| rs3774261 | ADIPOQ | Whites | GG= 11.6 (10.9, 12.1) |
GA= 10.6 (10.2, 11.1) |
AA= 10.8 (10.1, 11.6) |
.031 |
| rs7539542 | ADIPOR1 | Whites | CC= 11.4 (11.0, 11.9) |
CG = 10.6 (10.1, 11.1) |
GG= 10.8 (9.8, 11.9) |
.027 |
| rs12826079 | ADIPOR2 | Whites | CC= 10.8 (10.4, 11.2) |
CT/TT‡ = 11.8 (11.0, 12.6) |
.024‡ | |
| rs1342387 | ADIPOR1 | African Americans |
GG= 11.7 (10.6, 12.7) |
GA= 9.6 (8.9, 10.4) |
AA= 11.0 (9.6, 12.5) |
.016 |
| rs7539542 | ADIPOR1 | African Americans |
GG= 11.2 (10.3, 12.1) |
GC/CC‡ = 9.9 (9.1, 10.8) |
.05‡ | |
All regression analyses adjusted for age and sex
Major allele
for a 2 d.f. test, or a 1 degree of freedom dominant model test
when genotypes were combined for analysis due to small number of homozygous recessives
Discussion
In this study, one SNP in the ADIPOQ gene (rs822393) was significantly associated with insulin sensitivity in whites. There were several other suggestive (p < .05) associations of SNPs in ADIPOQ, ADIPOR1, and ADIPOR2. In whites, four of the suggestive ADIPOQ SNPs (rs4632532, rs266729, rs182052, rs7649121) were in moderate (r2 = .33–.67) LD with the significant SNP, rs822393. Given these findings, and the similarity between the genotype means for these SNPs, all of these associations may be driven by the association of rs822393 with insulin sensitivity or by the association of an untyped variant in high LD with rs822393. As the size of the β estimates for SNP rs822393 were attenuated slightly when another of the correlated SNPs was included in the model, these associations may be driven by an untyped variant. None of the other suggestive SNPs in ADIPOQ (in whites) or in ADIPOR1 (in African Americans) can be explained due to high LD between SNPs and thus these associations may represent the effects of multiple genetic variants on the expression or activity of the gene.
With one exception (rs7539542), the SNPs found to be associated with insulin sensitivity in the African Americans were different from those associated with white subjects in this study (and rs7539542 showed differing genotype specific means across the races, with different alleles being associated with increased insulin sensitivity).These findings may be due to the different patterns of linkage disequilibrium present in the two racial groups. The differing results between the races also may be the result of different sample sizes between the white and African American study populations, or of different genetic variants acting in the two racial groups.
In whites, the association of rs822393 and insulin sensitivity is stronger in the parents than the offspring. This could indicate that the influence of variants in adiponectin on insulin sensitivity may take many years to manifest (essentially, that many years of exposure to such variants are necessary in order for an effect to be seen).If this is the case, the effect of these variants would not yet be evident in adolescents, explaining the less significant association seen in the siblings and probands than in the parents.
Although several previous studies have examined the association of clamp-derived insulin sensitivity with SNPs in ADIPOQ (Buzzetti et al. 2007; Salmenniemi et al. 2005; Stumvoll et al. 2002; Ukkola et al. 2005; Vozarova de Courten et al. 2005), only one previous study used tag SNPs (Vozarova de Courten et al. 2005), so there is little overlap between the results of this study and previous studies in the literature. Only one of the suggestive ADIPOQ SNPs has been examined previously in relation to insulin sensitivity;rs266729 was found to be significantly (p < .05) associated with clamp-derived insulin sensitivity in obese Italians (Buzzetti et al. 2007) but was not significantly associated with the same phenotype in Pima Indians (Vozarova de Courten et al. 2005). Only two previous studies have examined the association of ADIPOR1 and ADIPOR2 SNPs and clamp-derived insulin sensitivity (Kantartzis et al. 2006; Stefan et al. 2005) with neither using a tag SNP approach; one of these studies included the SNP rs16928751 and concluded this SNP was not significantly associated with insulin sensitivity in a Caucasian cohort, mirroring the results of this study.
Previous studies have examined the association of SNPs in ADIPOQ with HOMA-IR and found significant associations (Hivert et al. 2008; Menzaghi et al. 2007). However, the results of this study suggest that SNPs associated with clamp-derived insulin sensitivity are not associated with HOMA-IR, possibly due to differences in the traits. The lack of replication of significant HOMA-IR / ADIPOQ SNP associations in this cohort can be explained by a relatively small sample size (compared to studies measuring insulin sensitivity surrogates) and a higher threshold for significance. The results from the present study support previous results in this study population suggesting that the genetic determinants of clamp-derived insulin sensitivity differ from those of surrogate measures of insulin resistance such as fasting insulin or HOMA-IR (Rasmussen-Torvik et al. 2007)).
As with any genetic association study, it will be necessary to replicate these results in unrelated study populations. Given the results of this study, it is important to conduct the replication studies using clamp-derived measures of insulin sensitivity rather than surrogates, and the replication cohort should include individuals from multiple generations. Of course, the use of the clamp (an invasive and time-consuming procedure) will limit the size of these studies and so expectations about the level of statistical significance must be adjusted appropriately. Studies will also be needed to test the association of tag SNPs (particularly rs822393) in these three genes with insulin sensitivity in other ethnic groups. The results in African Americans in this study must be viewed with caution; the number of African Americans was very small, tag SNPs were selected for a Caucasian population (and thus likely did not adequately tag the genes for the African American population), significance thresholds were calculated for Caucasian populations (and this may have been too generous for African American populations), and no genomic control markers were typed to address confounding by varying levels of admixture in this population. However the moderate association (p < .05) of two SNPs in ADIPOR1, despite the small sample size in this study, should encourage others to more fully investigate these associations in a larger African American cohort.
In summary, this study tested the association of comprehensive tag SNPs in ADIPOQ, ADIPOR1, and ADIPOR2 with clamp-derived insulin sensitivity. One SNP, rs822393, was significantly associated with insulin sensitivity in whites. There were several additional SNPs with suggestive associations to insulin sensitivity in both whites and African Americans. These genetic results lend support to epidemiologic studies that have found associations between circulating adiponectin and insulin sensitivity (Chen et al. 2005; Tschritter et al. 2003; Weyer et al. 2001) and to bench studies showing that adiponectin (administered intravenously or through genetic up-regulation) increases insulin sensitivity in rodents (Yamauchi et al. 2001; Yamauchi et al. 2003) and strengthens the argument that changes in adiponectin level or activity cause insulin resistance.
Supplementary Material
Acknowledgments
This study was supported by grants HL52851 and M01RR00400 from the National Institutes of Health. LJR-T is supported by NHLBI training grant T32 HL07779.
References
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am.J.Hum.Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Buzzetti R, Petrone A, Zavarella S, Zampetti S, Spoletini M, Potenziani S, Leto G, Osborn J, Leonetti F. The glucose clamp reveals an association between adiponectin gene polymorphisms and insulin sensitivity in obese subjects. Int.J.Obes.(Lond) 2007;31:424–428. doi: 10.1038/sj.ijo.0803419. [DOI] [PubMed] [Google Scholar]
- Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- Chen L, Xiang KS, Jia WP, Lu JX, Bao YQ, Lu HJ. Serum adiponectin concentration in relation to body fat distribution and tissue insulin sensitivity in the glucose clamp study. Zhonghua Yi Xue Za Zhi. 2005;85:1456–1459. [PubMed] [Google Scholar]
- Damcott CM, Ott SH, Pollin TI, Reinhart LJ, Wang J, O'connell JR, Mitchell BD, Shuldiner AR. Genetic variation in adiponectin receptor 1 and adiponectin receptor 2 is associated with type 2 diabetes in the old order amish. Diabetes. 2005;54:2245–2250. doi: 10.2337/diabetes.54.7.2245. [DOI] [PubMed] [Google Scholar]
- Hara K, Horikoshi M, Kitazato H, Yamauchi T, Ito C, Noda M, Ohashi J, Froguel P, Tokunaga K, Nagai R, Kadowaki T. Absence of an association between the polymorphisms in the genes encoding adiponectin receptors and type 2 diabetes. Diabetologia. 2005;48:1307–1314. doi: 10.1007/s00125-005-1806-3. [DOI] [PubMed] [Google Scholar]
- Hivert MF, Manning AK, McAteer JB, Florez JC, Dupuis J, Fox CS, O'Donnell CJ, Cupples LA, Meigs JB. Common variants in the adiponectin gene (ADIPOQ) associated with plasma adiponectin levels, type 2 diabetes, and diabetes-related quantitative traits: The framingham offspring study. Diabetes. 2008 doi: 10.2337/db08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantartzis K, Fritsche A, Machicao F, Haring HU, Stefan N. The −8503 G/A polymorphism of the adiponectin receptor 1 gene is associated with insulin sensitivity dependent on adiposity. Diabetes Care. 2006;29:464. doi: 10.2337/diacare.29.02.06.dc05-2020. [DOI] [PubMed] [Google Scholar]
- Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: Insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- Menzaghi C, Trischitta V, Doria A. Genetic influences of adiponectin on insulin resistance, type 2 diabetes, and cardiovascular disease. Diabetes. 2007;56:1198–1209. doi: 10.2337/db06-0506. [DOI] [PubMed] [Google Scholar]
- Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am.J.Hum.Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen-Torvik LJ, Pankow JS, Jacobs DR, Steffen LM, Moran AM, Steinberger J, Sinaiko AR. Heritability and genetic correlations of insulin sensitivity measured by the euglycaemic clamp. Diabet.Med. 2007;24:1286–1289. doi: 10.1111/j.1464-5491.2007.02271.x. [DOI] [PubMed] [Google Scholar]
- Rasmussen-Torvik LJ, Pankow JS, Jacobs DR, Jr, Steinberger J, Moran A, Sinaiko AR. Influence of waist on adiponectin and insulin sensitivity in adolescence. Obesity. doi: 10.1038/oby.2008.482. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmenniemi U, Zacharova J, Ruotsalainen E, Vauhkonen I, Pihlajamaki J, Kainulainen S, Punnonen K, Laakso M. Association of adiponectin level and variants in the adiponectin gene with glucose metabolism, energy expenditure, and cytokines in offspring of type 2 diabetic patients. J.Clin.Endocrinol.Metab. 2005;90:4216–4223. doi: 10.1210/jc.2004-2289. [DOI] [PubMed] [Google Scholar]
- Shand BI, Scott RS, Elder PA, George PM. Plasma adiponectin in overweight, nondiabetic individuals with or without insulin resistance. Diabetes Obes.Metab. 2003;5:349–353. doi: 10.1046/j.1463-1326.2003.00279.x. [DOI] [PubMed] [Google Scholar]
- Sinaiko AR, Jacobs DR, Jr, Steinberger J, Moran A, Luepker R, Rocchini AP, Prineas RJ. Insulin resistance syndrome in childhood: Associations of the euglycemic insulin clamp and fasting insulin with fatness and other risk factors. J Pediatr. 2001;139:700–707. doi: 10.1067/mpd.2001.118535. [DOI] [PubMed] [Google Scholar]
- Stefan N, Vozarova B, Funahashi T, Matsuzawa Y, Weyer C, Lindsay RS, Youngren JF, Havel PJ, Pratley RE, Bogardus C, Tataranni PA. Plasma adiponectin concentration is associated with skeletal muscle insulin receptor tyrosine phosphorylation, and low plasma concentration precedes a decrease in whole-body insulin sensitivity in humans. Diabetes. 2002;51:1884–1888. doi: 10.2337/diabetes.51.6.1884. [DOI] [PubMed] [Google Scholar]
- Stefan N, Machicao F, Staiger H, Machann J, Schick F, Tschritter O, Spieth C, Weigert C, Fritsche A, Stumvoll M, Haring HU. Polymorphisms in the gene encoding adiponectin receptor 1 are associated with insulin resistance and high liver fat. Diabetologia. 2005;48:2282–2291. doi: 10.1007/s00125-005-1948-3. [DOI] [PubMed] [Google Scholar]
- Stumvoll M, Tschritter O, Fritsche A, Staiger H, Renn W, Weisser M, Machicao F, Haring H. Association of the T-G polymorphism in adiponectin (exon 2) with obesity and insulin sensitivity: Interaction with family history of type 2 diabetes. Diabetes. 2002;51:37–41. doi: 10.2337/diabetes.51.1.37. [DOI] [PubMed] [Google Scholar]
- Thorisson GA, Smith AV, Krishnan L, Stein LD. The international HapMap project web site. Genome Res. 2005;15:1592–1593. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschritter O, Fritsche A, Thamer C, Haap M, Shirkavand F, Rahe S, Staiger H, Maerker E, Haring H, Stumvoll M. Plasma adiponectin concentrations predict insulin sensitivity of both glucose and lipid metabolism. Diabetes. 2003;52:239–243. doi: 10.2337/diabetes.52.2.239. [DOI] [PubMed] [Google Scholar]
- Ukkola O, Santaniemi M, Rankinen T, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bergman R, Kesaniemi YA, Bouchard C. Adiponectin polymorphisms, adiposity and insulin metabolism: HERITAGE family study and oulu diabetic study. Ann.Med. 2005;37:141–150. doi: 10.1080/07853890510007241. [DOI] [PubMed] [Google Scholar]
- Vozarova de Courten B, Hanson RL, Funahashi T, Lindsay RS, Matsuzawa Y, Tanaka S, Thameem F, Gruber JD, Froguel P, Wolford JK. Common polymorphisms in the adiponectin gene ACDC are not associated with diabetes in pima indians. Diabetes. 2005;54:284–289. doi: 10.2337/diabetes.54.1.284. [DOI] [PubMed] [Google Scholar]
- Weyer C, Funahashi T, Tanaka S, Hotta K, Matsuzawa Y, Pratley RE, Tataranni PA. Hypoadiponectinemia in obesity and type 2 diabetes: Close association with insulin resistance and hyperinsulinemia. J.Clin.Endocrinol.Metab. 2001;86:1930–1935. doi: 10.1210/jcem.86.5.7463. [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Kamon J, Waki H, Terauchi Y, Kubota N, Hara K, Mori Y, Ide T, Murakami K, Tsuboyama-Kasaoka N, Ezaki O, Akanuma Y, Gavrilova O, Vinson C, Reitman ML, Kagechika H, Shudo K, Yoda M, Nakano Y, Tobe K, Nagai R, Kimura S, Tomita M, Froguel P, Kadowaki T. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat.Med. 2001;7:941–946. doi: 10.1038/90984. [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Kamon J, Waki H, Imai Y, Shimozawa N, Hioki K, Uchida S, Ito Y, Takakuwa K, Matsui J, Takata M, Eto K, Terauchi Y, Komeda K, Tsunoda M, Murakami K, Ohnishi Y, Naitoh T, Yamamura K, Ueyama Y, Froguel P, Kimura S, Nagai R, Kadowaki T. Globular adiponectin protected ob/ob mice from diabetes and ApoE-deficient mice from atherosclerosis. J.Biol.Chem. 2003;278:2461–2468. doi: 10.1074/jbc.M209033200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.