Summary
The indolocarbazole staurosporine is a potent inhibitor of a variety of protein kinases. It contains a sugar moiety attached through C-N linkages to both indole nitrogen atoms of the indolocarbazole core. Staurosporine biosynthesis was reconstituted in vivo in a heterologous host Streptomyces albus by using two different plasmids: the ‘aglycone vector’ expressing a set of genes involved in indolocarbazole biosynthesis together with staG (encoding a glycosyltransferase) and/or staN (coding for a P450 oxygenase), and the ‘sugar vector’ expressing a set of genes responsible for the biosynthesis of the sugar moiety. Attachment of the sugar to the two indole nitrogens of the indolocarbazole core was dependent on the combined action of StaG and StaN. When StaN was absent, the sugar was attached only to one of the nitrogen atoms, through an N-glycosidic linkage, as in the indolocarbazole rebeccamycin. The StaG glycosyltransferase showed flexibility with respect to the sugar donor. When the ‘sugar vector’ was substituted by constructs directing the biosynthesis of L-rhamnose, L-digitoxose, L-olivose and D-olivose, respectively, StaG and StaN were able to transfer and attach all of these sugars to the indolocarbazole aglycone.
Introduction
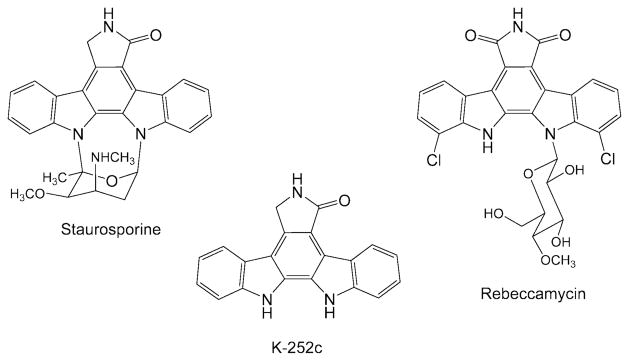
The indolocarbazole family of natural products constitutes a new class of antitumour drugs divided into two major groups depending on their mechanisms of action and structural features (Gribble and Berthel, 1993). One group includes inhibitors of DNA topoisomerase I, such as rebeccamycin (Fig. 1). Most members of this group contain a sugar moiety attached by a β-glycosidic linkage to one of the indole nitrogen atoms of the aglycone. The second group includes protein kinase inhibitors, such as staurosporine (Fig. 1). Usually, they contain a sugar moiety linked to both indole nitrogen atoms of the indolocarbazole core. The presence of the sugars appears to be essential for the biological activity of the compounds (Bailly et al., 1999).
Fig. 1.
Chemical structures of indolocarbazoles.
Staurosporine is a very potent inhibitor of protein kinase C (Tamaoki et al., 1986), but lacks the selectivity required for pharmaceutical applications involving the very specific inhibition of individual protein kinases (Gescher, 2000). Its discovery stimulated the development of medicinal chemistry programs aimed to generate novel derivatives with higher inhibitory specificity for protein kinase C. Some of these novel compounds (UCN-01, CGP 41251, CEP-751) have entered clinical trials for their use as antitumour agents (Akinaga et al., 2000). In addition, several staurosporine derivatives have been isolated from culture broths of different producer actinomycetes (Cai et al., 1995; 1996), from blocked mutants (Goeke et al., 1995) and from an actinomycete isolated from a marine environment (Williams et al., 1999).
From a biosynthetic point of view, the main building blocks of staurosporine are two tryptophan molecules, one glucose, and two methyl groups derived from methionine (Yang and Cordell, 1997a,b,c; Yang et al., 1999). By classical mutagenesis of staurosporine-producing Streptomyces longisporoflavus, blocked mutants were obtained that accumulated the aglycone K-252c (Goeke et al., 1995) and O-demethyl-staurosporine (Hoehn et al., 1995; Weidner et al., 1998) respectively. In recent years an increasing amount of information has become available on the molecular genetics of indolocarbazole biosynthesis. The gene cluster for the biosynthesis of rebeccamycin was isolated and characterized from the producer organism Lechevalieria aerocolonigenes (Sánchez et al., 2002; Hyun et al., 2003; Onaka et al., 2003). Heterologous expression of the gene cluster and subsequent production of rebeccamycin have been achieved in Streptomyces albus, at high level (Sánchez et al., 2002), and in Escherichia coli, at low level (Hyun et al., 2003). In addition, a number of rebeccamycin derivatives have been generated by combinatorial biosynthesis (Sánchez et al., 2005). In the case of staurosporine, the biosynthetic gene cluster has also been isolated and expressed in Streptomyces lividans (Onaka et al., 2002).
Here we report the functional characterization of two genes from the staurosporine cluster: staG coding for an N-glycosyltransferase and staN coding for a P450 oxygenase. We demonstrate that StaG is required to establish the N-glycosidic bond and that StaN subsequently forms the second C-N linkage between the sugar and the indolocarbazole aglycone. We also show that StaG is a ‘sugar flexible’ glycosyltransferase with the capability of transferring various deoxysugars.
Results
Isolation of the staG and staN genes
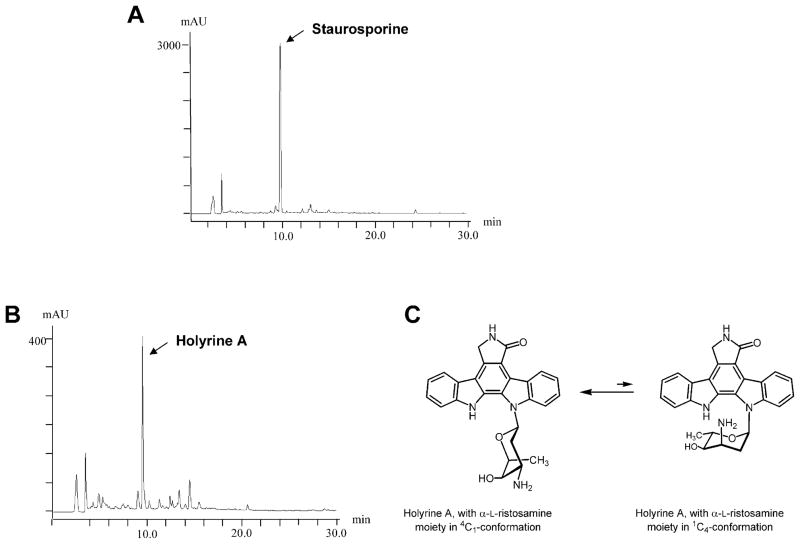
A cosmid library of chromosomal DNA from the staurosporine producer S. longisporoflavus DSM10189 was constructed in the bifunctional (Streptomyces-Escherichia coli) vector pKC505. To carry out in situ colony hybridization of this library, we generated a PCR product by amplification of a sequence previously noticed by us to be highly similar to rebD, a gene involved in rebeccamycin biosynthesis in L. aerocolonigenes (Sánchez et al., 2002). Information on this sequence was taken from a DNA sequence deposited in the databases under patent application WO97/08323 and reported to be involved in staurosporine biosynthesis (Schupp et al., 1997). Nine positively hybridizing clones were isolated. Restriction analysis of these cosmids showed that they were overlapping and covered a chromosomal region of about 45 kb. These cosmids were introduced into S. albus by protoplast transformation and transformants, selected for apramycin resistance, were tested for their ability to produce staurosporine. HPLC-MS analyses of cultures of the different transformants showed, in one case (cosmid cos32D1), a peak with retention time and m/z ion 467 consistent with the molecular weight of staurosporine (Fig. 2A). To further characterize the insert in this cosmid, isolated cosmid DNA was digested with PstI and hybridized against a PCR amplified product derived from the rebG gene that codes for an N-glycosyltransferase in the rebeccamycin biosynthetic cluster (Sánchez et al., 2002). The hybridization signal was found in a 4.5 kb PstI fragment. This DNA fragment was sequenced and analysed for the presence of open reading frames (orf). Two complete and two incomplete orfs were identified, and the sequence was deposited at the EMBL Nucleotide Sequence Database with Accession number AJ520100. At the time we were running these experiments, the cloning and sequencing of the staurosporine gene cluster was reported by another laboratory (Onaka et al., 2002). The genes we identified in our PstI fragment exactly corresponded to the staA (incomplete), staN, staG and staO (incomplete) genes described by these authors. Although different strains were used, namely Streptomyces sp. TP-A0274 by Onaka et al. and S. longisporoflavus for our work, the identity of the different gene products was higher than 95% for all four genes compared. Comparison of the deduced products of staG and staN showed that the StaG protein showed similarities in databases with RebG and other glycosyltransferases, while the StaN protein belonged to the family of P-450 oxygenases. Partial sequencing of the nine cosmids confirmed the existence of a sta gene cluster with a genetic organization identical to that reported for Streptomyces sp. TP-A0274.
Fig. 2.
HPLC analysis of cultures of S. albus cos32D (A), and of S. albus 39GT(pETAS) (B) and chemical structure of holyrine A (only found with sugar in 4C1-conformation) (C).
In vivo assay for the StaG glycosyltransferase
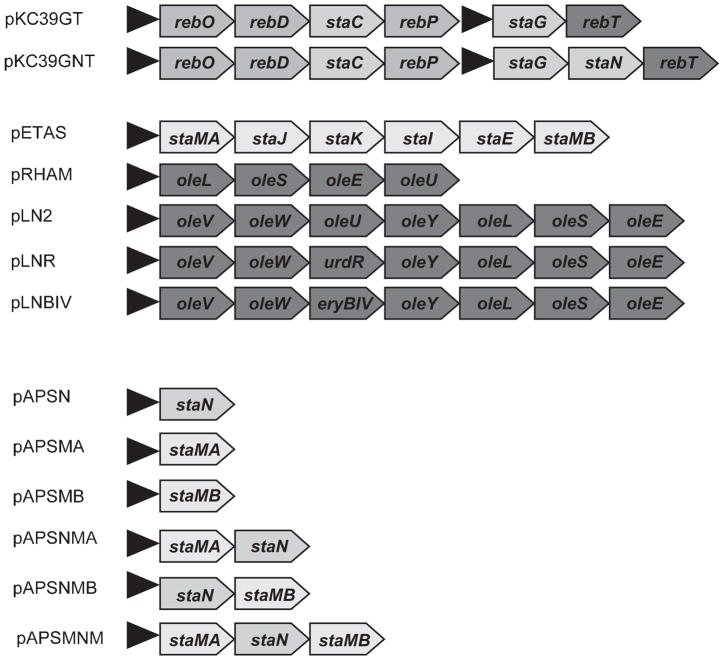
In order to establish an in vivo assay for testing the activity of the StaG glycosyltransferase, we constructed a S. albus recombinant strain containing two different plasmids: the ‘aglycone plasmid’ and the ‘sugar plasmid’. The ‘aglycone plasmid’ was based on an integrative vector (which can be selected for resistance to apramycin), in which we cloned the genes required for the biosynthesis of the staurosporine aglycone. As has been recently shown, expression of the rebODP genes from the rebeccamycin cluster together with the staC gene from the staurosporine cluster leads to the formation of the staurosporine aglycone (also named K252c; Fig. 1) (Sánchez et al., 2005). In the ‘aglycone plasmid’ we also cloned the staG gene, coding for the glycosyltransferase. The genes were cloned under the control of the erythromycin resistance promoter, ermE*p. The resultant construct pKC39GT (Fig. 3) was introduced into S. albus. HPLC-MS analysis of cultures of the resultant recombinant strain 39GT confirmed the production of the staurosporine aglycone: a peak was observed showing the same mobility as pure K252c and a m/z ion of 312, consistent with the expected mass for this compound. In addition, two other peaks were also observed with the characteristic absorption spectrum of staurosporine; these two products were not detected when the staG was omitted. Mass analyses of these peaks showed m/z ions of 440 and 458. These data are consistent with the possible attachment of a ketodideoxy-hexose and a monodeoxyhexose, respectively, to the staurosporine aglycone. Because no sugar biosynthetic genes were cloned into this construct, it was assumed that formation of these sugars was dependent on host chromosomal genes. These compounds were not further characterized. When strains carrying ‘sugar plasmids’ were analysed (see below), the mentioned compounds were very minor products.
Fig. 3.
Plasmid constructs used in this work. Constructs pRHAM (Rodriguez et al., 2000), pLN2 and pLNR (Rodriguez et al., 2002) and pLNBIV (Fischer et al., 2002) have been already described. They contain different sets of genes involved in the biosynthesis of L-olivose in Streptomyces antibioticus (ole genes), L-mycarose in Saccha-ropolyspora erythraea (eryB gene) or L-rhodinose in Streptomyces fradiae (urdR gene). The black triangle indicate the position of the erythromycin resistance promoter ermE*p.
The second plasmid, the ‘sugar plasmid’, was based on a replicative vector (which can be selected for resistance to thiostrepton) in which we cloned the genes required for the biosynthesis of the staurosporine sugar moiety. A 6.9 kb NruI fragment from the chromosome of S. longisporoflavus was cloned into pEM4 under the control of the ermE*p promoter, generating plasmid pETAS (Fig. 3). This DNA fragment contains the staMA (N-methyltransferase), staJ (2,3-dehydratase), staK (4-ketoreductase), staI (3-aminotransferase), staE (3,5-epimerase) and staMB (4-O-methyltransferase) genes from the staurosporine gene cluster (Schupp et al., 1997; Onaka et al., 2002). It was suggested that all these genes (located in a contiguous DNA fragment) were involved in the biosynthesis of the aminosugar moiety of staurosporine. Two other genes putatively involved in the biosynthesis of this aminosugar, staA (NDP-D-glucose synthase) and staB (NDP-D-glucose-4,6-dehydratase) are located at the other end of the cluster and were not included in these constructs because we assumed (as was verified later on) that probably the S. albus host could provide the corresponding enzymatic activities.
Strain 39GT (containing the ‘aglycone plasmid’) was then transformed with pETAS and cultures of the resultant strain was analysed by HPLC-MS (Fig. 2B). A single peak was observed with the characteristic indolocarbazole absorption spectrum. The compound in this peak showed a molecular ion of m/z 441, consistent with a glycosylated indolocarbazole lacking the N-methyl and O-methyl groups at C-3 and C-4 of the sugar moiety respectively. Its presumptive chemical structure was confirmed after purification of the compound by preparative HPLC and the elucidation of its chemical structure by NMR and MS (Fig. 2C). The compound turned out to be identical with a previously isolated minor product of cultures of a marine actinomycete, holyrine A (Williams et al., 1999). As already described by Williams et al. the sugar moiety of holyrine A, L-ristosamine, was found to exist predominantly in a 4C1 conformation (unusual for an L-sugar), which places the α-glycosidic bond in an equatorial position (Fig. 2C). This is evident from the 1H-NMR data, and is most likely a consequence of the mode of action of the glycosyltransferase StaG, as will be discussed below.
Several conclusions can be drawn from these experiments. First, StaG is the glycosyltransferase responsible for attaching the sugar to the aglycone through an N-glycosidic linkage. Second, another enzymatic activity is required for establishing the additional, non-glycosidic, C-N linkage between the sugar and the aglycone. Third, the absence of the N- and O-methyl groups in the sugar in holyrine A suggests that probably these two methylation steps occur after the sugar has been linked to the aglycone by the two C-N bonds. Fourth, the S. albus host can provide the synthase and 4,6-dehydratase activities required for catalysing the first two enzymatic steps in deoxysugar biosynthesis.
In vivo assay for the StaN monooxygenase
A possible candidate for establishing the second C-N bond could be the StaN P450 oxygenase (Onaka et al., 2002). To test this hypothesis, we incorporated the staN gene into pKC39GT, generating pKC39GNT. This construct was then introduced in S. albus harbouring pETAS and cultures of the resultant transformants were analysed by HPLC-MS. A single peak was observed in strain 39GNT (pETAS) that corresponded in HPLC mobility, absorption spectrum and mass analysis (molecular ion m/ z 467) to staurosporine (data not shown). This experiment demonstrated that StaN is responsible for establishing the second C-N bond between the sugar and the aglycone and strengthened the earlier hypothesis that the two methylation steps occurred after the sugar had been attached through two C-N linkages. The following experiments confirmed these conclusions. The staN, staMA (coding for a putative N-methyltransferase), and staMB (encoding a putative 4-O-methyltransferase) genes were expressed in S. albus, either individually or combined, under the control of the ermE*p promoter to yield the following recombinant strains: APSN (staN), APSMA (staMA), APSMB (staMB), APSNMA (staN plus staMA), APSNMB (staN plus staMB) and APSMNM (staN plus staMA plus staMB) (Fig. 3). All of these strains were grown in the presence of 20 μg ml−1 holyrine A and, after incubation for 5 days, the products of the bioconversion were analysed by HPLC-MS. Strain APSN (staN) showed a peak with a m/z ion of 439 consistent with the formation of O-demethyl-N-dem-ethyl-staurosporine, again confirming the role of the StaN oxygenase in the second C-N linkage. Holyrine A was not methylated when fed to strains APSMA and APSMB, indicating that the O- and N-methyltransferases could not act on this substrate. In contrast, feeding holyrine A to strains APSNMA, APSNMB and APSMNM lead to the formation of holyrine A derivatives showing molecular ions consistent with the generation of O-demethyl-staurosporine (m/z 453), N-demethyl-staurosporine (m/z 453) and staurosporine (m/z 467) respectively. Several conclusions can be drawn from these experiments: (i) O- and N-methylation steps occur once the sugar has been attached to the aglycone through two C-N bonds and (ii) each one of the methylation steps can occur in the absence of the other methyltransferase.
Sugar substrate flexibility of the StaG glycosyltransferase
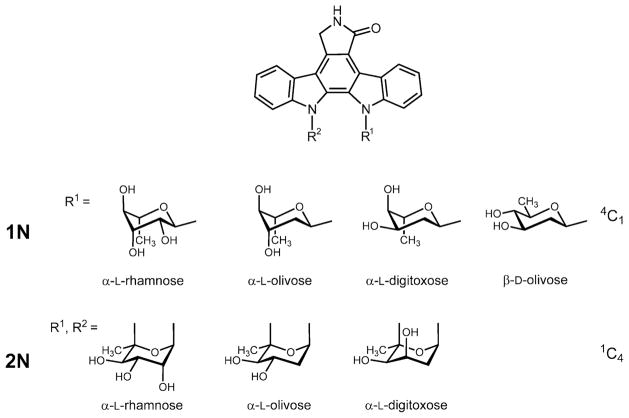
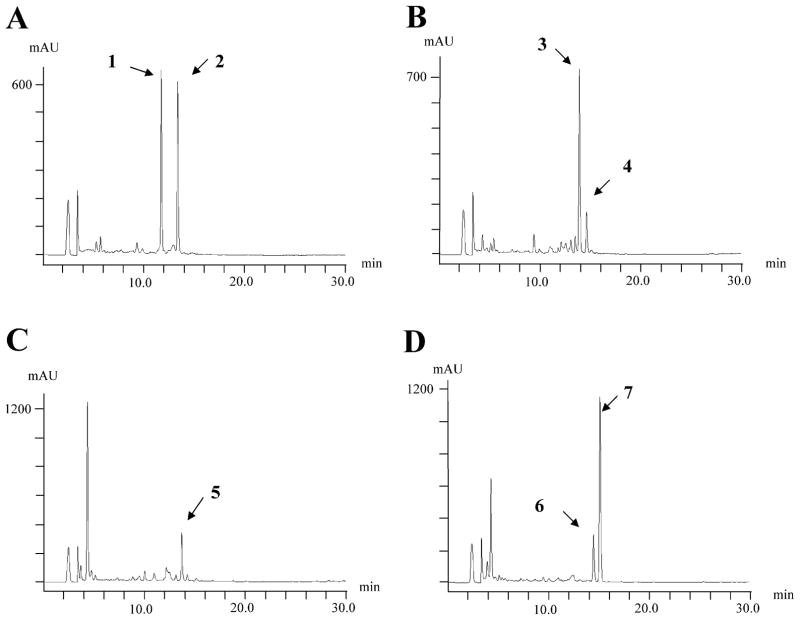
We tested the ability of the StaG glycosyltransferase to transfer different deoxysugars to the aglycone. We used as host S. albus 39GNT (harbouring staG and staN genes) and we independently transformed protoplasts of this strain with plasmids pRHAM, pLN2, pLNR and pLN-BIV (Fig. 3). These plasmids have been shown to direct the biosynthesis of L-rhamnose (pRHAM) (Rodríguez et al., 2000), L-olivose (pLN2) and D-olivose (pLNR) (Rodríguez et al., 2002) and L-digitoxose (pLNBIV) (Fischer et al., 2002). Transformants containing these plasmids, selected for thiostrepton resistance, were then incubated and the possible presence of glycosylated derivatives analysed by HPLC-MS (Fig. 5). Strain 39GNT(pRHAM) showed two major peaks with molecular ions of m/z 458 and 456 respectively (Fig. 4A). These are consistent with the presence of a L-rhamnose moiety attached either to one or both nitrogen atoms of the indolocarbazole ring. Strain 39GNT (pLN2) also showed two peaks with the characteristic spectrum of indolocarbazoles: the major one with m/z ion at 440 and the minor one at 442 (Fig. 4B). These are the masses expected for the incorporation of an L-olivose moiety to the aglycone through two or one C-N linkages respectively. When using pLNR (Fig. 4C), only a single peak was observed with a molecular ion of m/z 442, consistent with the attachment of a D-olivose moiety to one of the nitrogens of the indolocarbazole ring. Finally, strain 39GNT(pLNBIV) showed two peaks with molecular ions of m/z 442 and 440, respectively, consistent with the attachment of an L-digitoxose moiety through one and two C-N bonds respectively (Fig. 4D). Three of these compounds were produced in sufficient amounts that their chemical structures could be confirmed by NMR and MS (Fig. 5).
Fig. 5.
Chemical structures of the novel glycosylated staurosporine derivatives. The sugar residues were found exclusively in 4C1-conformation, when singly attached (1 N compounds), but were found in 1C4-conformation, when doubly attached (2 N compounds)
Fig. 4.
HPLC analysis of cultures of recombinant strains expressing different sugar plasmids.
A. S. albus 39GNT(pRHAM).
B. S. albus 39GNT(pLN2).
C. S. albus 39GNT(pLNR).
D. S. albus 39GNT(pLNBIV).
1: L-rhamnosyl-2N-K252c; 2: L-rhamnosyl-1N-K252c; 3: L-olivosyl-2N-K252c; 4: L-olivosyl-1N-K252c; 5: D-olivosyl-1N-K252c; 6: L-digitoxosyl-1N-K252c; 7: L-digitoxosyl-2N-K252c.
Structural elucidation of the novel compounds
The chemical structures of three compounds (2, 3 and 7 as shown in Fig. 4) containing different sugar moieties attached to the indolocarbazole core were elucidated by NMR spectroscopy in combination with mass spectrometry and UV spectroscopy. All three compounds showed similar UV spectra to that of K252c, suggesting the same indolocarbazole chromophore. One of these, compound 2, was identified by TLC, UV, MS and NMR to be known, with an L-rhamnose attached through one C-N linkage to the indolocarbazole frame. 1H and 13C NMR data (see Tables S1 and S2 in Supplementary material) of 2 were consistent with those reported for K252d (= L-rhamnosyl-1N-K252c, Yasuzawa et al., 1986). As in holyrine A (see above), the NMR data (see Tables S1 and S2 in Supplementary material) indicate that the L-rhamnose moiety is in 4C1-conformation and linked through an equatorial glycosidic bond instead of the typical axial glycosidic bond found in L-sugars with 1C4-conformation. The large coupling constant (9 Hz) found for the anomeric 1′-H, the small e,e-couplings between 4′-H and 5′-H as well as between 4′-H and 3′-H, and also the less downfield shifted C-1′ (δ 77.1, due to an increased distance from the aromatic ring current) are evidence for this 1C4-conformation. Although both the 3′-OH and the 4′-OH residues are forced into disadvantageous axial positions in this conformation, 2′-OH and the glycosidic bond are both equatorial, which helps to stabilize the 4C1-conformation. The other two compounds were identified as L-olivosyl-2N-K252c (3) and as L-digitoxosyl-2N-K252c (7) respectively. Both are new compounds, displaying the same molecular ion of m/z 440 in a positive MS analysis mode. The 1H and 13C NMR spectra of these two compounds were similar as found for the known compound 2 considering the signals for the aglycone, except for one big difference, namely the absence of one NH signal in the lower field of the 1H NMR spectra: only one NH-proton (δ 8.62 for 3; δ 8.51 for 7) NMR instead of two NH protons at δ 11.69 and 8.57 found in 2. Moreover, no. 5′-H was observed in the sugar moieties of compounds 3 and 7 as opposed to the sugar moiety in 2. Taken together, it was concluded that the sugar moieties in compounds 3 and 7 were linked by two covalent bonds to the indolocarbazole core, and that these linkages were two C-N bonds, the glycosidic bond (between C-1′ and N-13) and the second bond established between C-5′ and N-12. The observed downfield shift for C-5′ (this carbon resonates at δ 76.4 in compound 2) to δ 96.2 in 3 and to δ 92.3 in 7 was also consistent with the deduced structures. The 1H NMR spectra for compounds 3 and 7 were very similar, except for the signals of the sugar moieties. Both of them showed the anomeric 1′-H with a smaller coupling constant (~5 Hz, typical for an e,a-coupling), which is consistent with an axial α-glycosidic linkage typical for an L-sugar moiety in the preferred 1C4-conformation, a conclusion which was also consistent with the rest of the NMR data. The most significant difference between the 1H NMR data of compounds 3 and 7 was found for the coupling constant resulting from the coupling between 3′-H and 4′-H: the large transaxial coupling constant of 10 Hz found between 3′-H and 4′-H in compound 3 indicates that both 3′-OH and 4′-OH are in an equatorial position. In contrast, the much smaller coupling constant found for the coupling between 3′-H and 4′-H in compound 7 (J = 3.2 Hz) indicates that the 3′-OH residue is in an axial position. Thus, this coupling between 3′-H and 4′-H revealed the key difference between these two compounds, namely that the sugar moiety in 3 is an olivose, while the one in 7 is a digitoxose (see Fig. S1 in Supplementary material). Otherwise, both compounds showed the same mass peak in the HPLC-MS (m/z 440 in a positive APCI mode) and identical UV absorptions. Further 2D-NMR data were generated for compound 3, in agreement with the deduced structure (see Table S3 in Supplementary material). The 13C-NMR data along with the 2D NMR spectra (H,H-COSY, HSQC and HMBC) helped to unequivocally distinguish between sugar moieties and aglycon resonances and allowed for the detailed assignments given in Tables S1 and S2 in Supplementary material.
Discussion
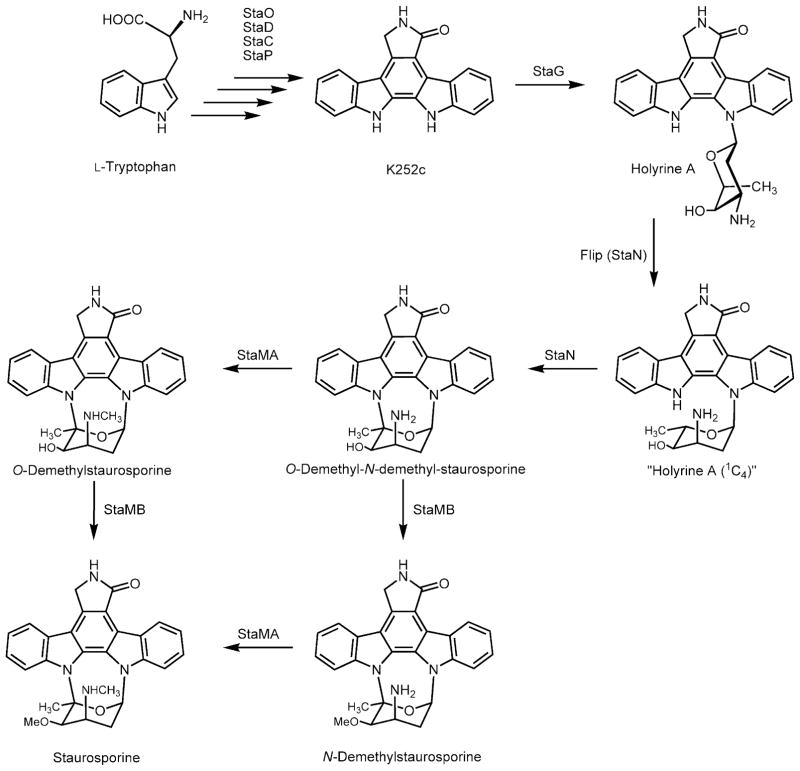
Most of the studies on the biosynthesis of the indolocarbazole staurosporine have been carried out by determining the pattern of incorporation of different precursors into the molecule. The isolation of the staurosporine gene cluster has opened up the possibility of further characterization of staurosporine biosynthesis by taking advantage of recombinant DNA technology. In the present work, we have dissected and reconstituted staurosporine biosynthesis using two independent plasmids (the ‘aglycone’ and the ‘sugar’ plasmids). By coexpressing both plasmids in the heterologous host S. albus, staurosporine production was achieved. On this basis and using these plasmids as starting material, we carried out experiments aimed to decipher late steps in staurosporine biosynthesis by combinatorial biosynthesis. The staurosporine aglycone (and that of its closely structurally related rebeccamycin) is synthesized through the condensation of L-tryptophan moieties after an oxidative deamination and several oxidation and decarboxylation steps (Fig. 6). Both aglycones, K252c (for staurosporine) and arcyriaflavin (the non-chlorinated aglycone for rebeccamycin) are then glycosylated by the action of the StaG and RebG glycosyltransferases respectively. This glycosylation event is very important because biological activity of indolocarbazoles requires the presence of the sugar. A major structural difference between rebeccamycin and staurosporine is the way in which the sugar is attached to the indolocarbazole structure: either through a single nitrogen (in the case of rebeccamycin) or through two nitrogens (in the case of staurosporine). Expression of the StaG glycosyltransferase in the presence of the ‘sugar’ plasmid produced holyrine A, previously isolated as a minor product from cultures of a marine actinomycete (Williams et al., 1999). Holyrine A seems to be a staurosporine biosynthetic intermediate because it was converted into staurosporine when fed to a S. albus strain expressing the staN, staMA and staMB genes, which catalyse the final steps in staurosporine biosynthesis. Interestingly, holyrine A is not methylated by StaMA or by StaMB, suggesting that these methyltransferases act once the sugar has been attached to the indolocarbazole through the two C-N linkages (Fig. 6). The StaN P450 oxygenase is responsible for the establishment of this second linkage. The result of the action of the StaN enzyme on holyrine A is the formation of the O-demethyl-N-demethyl-staurosporine intermediate. Both StaMA and StaMB methyltransferases can now act on this intermediate in a successive way leading to the formation of staurosporine (Fig. 6). However, it could not be unequivocally established which is the preferred order of action for these methylation events. In the case of the O-methylation event, there was previous experimental evidence supporting this view, through experiments in which O-demethyl-staurosporine was converted in vitro into staurosporine by using cell-free extracts of S. longisporoflavus (Weidner et al., 1998). It is worth mentioning that the sugar N-methyltransferases so far characterized and involved in antibiotic biosynthesis act before the sugar has been transferred to the aglycone (Chen et al., 1998; 2002; Chang et al., 2000). In our knowledge, the StaMA N-methyltransferase is the first reported example of a sugar N-methyltransferase acting after the linkage of the sugar moiety to the aglycone.
Fig. 6.
Proposed pathway for the late staurosporine biosynthesis.
The glycosyltransferases involved in antibiotic biosynthesis are increasingly being reported to be flexible with respect to their nucleotide-diphosphate (NDP)-sugar donor substrate. It has been shown that ElmGT, the elloramycin glycosyltransferase, has the ability to transfer at least 10 different deoxysugars to its corresponding aglycone (Wohlert et al., 1998; Blanco et al., 2001; Fischer et al., 2002; Rodriguez et al., 2002; Lombó et al., 2004; Pérez et al., 2005). Tandem action of the vancomycin pathway glycosyltransferases GtfE and GtfD has also allowed the formation of analogues of the glycopeptide antibiotics vancomycin and teicoplanin (Losey et al., 2002). In contrast, the NovM noviosyl glycosyltransferase has been reported to possess surprisingly stringent substrate specificity when tested against a panel of more than 40 different NDP-sugars (Albermann et al., 2003). The staurosporine StaG glycosyltransferase appears to possess at least some flexibility towards its NDP-sugar donor substrate. By endowing the recombinant strains with the capability of synthesizing different deoxysugars we showed here that StaG is able of transferring L-olivose, L-rhamnose, L-digitoxose and D-olivose with a good efficiency. Interestingly, StaG appears to establish an equatorial N-glycosidic bond, thereby forcing the L-sugars into an unusual 4C1-conformation, as discussed above in context with the NMR data of holyrine A and K252d (= L-rhamnosyl-1N-K252c, 2). Additionally, the StaN P450 oxygenase seems also able to act on these different deoxysugars, with the exception of D-olivose, establishing the C-N linkage between the second nitrogen of the indolocarbazole frame and C-5′ of the sugar. To establish the second linkage between C-5′ of the sugar moiety and N-13 of the indolocarbazole system, the sugar moiety needs to be ‘flipped’ from its 4C1- into a 1C4-conformation, which is typical for L-sugars. D-sugars prefer the 4C1-conformation and therefore perfectly fit the StaG enzyme pocket. Thus StaG, when confronted with NDP-D-olivose as sugar donor substrate, can easily establish an equatorial glycosidic link. However, P-450 enzyme StaN needs to abstract both 5′-H and the H-atom bound to N-13 to form the second C-N-link. The 5′-H abstraction seems to be facilitated when L-sugars are flipped into a 1C4-conformation, but is probably sterically hindered by the 5′-methyl residue of D-olivose. Changing this sugar moiety into a 1C4-conformation would render all residues (both OH groups, the methyl group and the glycosidic bond) into thermodynamically very unfavourable axial positions. Thus, probably both thermodynamic and steric reasons hinder the formation of a second C-N bond by StaN, when D-sugars are present. For these reasons we believe that doubly N-linked sugar moieties of the staurosporin-type can only be established in indolocarbazole anticancer antibiotics with L-sugars, in which such a conformational flip leads to an arrangement, from which an abstraction of both 5′-H and 13-H is facilitated.
Experimental procedures
Bacterial strains, culture conditions and vectors
Streptomyces longisporoflavus DSM 10189, S. albus J1074 (Chater and Wilde, 1980), E. coli DH10B (Invitrogen) and E. coli ED8767 (Sambrook et al., 1989) were used in this work. Plasmids pKC505 (Richardson et al., 1987), pKC796 (Kuhs-toss et al., 1991), pEM4 (Quirós et al., 1998), LITMUS 28 (New England BioLabs) and pKC039 (Sánchez et al., 2005) were used in this work. For sporulation, S. longisporoflavus and S. albus were routinely grown for 7 days at 30°C on agar plates containing either R2YE or Bennett’s agar (Kieser et al., 2000). When antibiotic selection of transformants was needed, we used 100 μg ml−1 ampicillin (for pEM4 and LITMUS 28 derivatives in E. coli), 25 μg ml−1 thiostrepton (5 μg ml−1 in liquid media, for pEM4 derivatives in S. albus), and 25 μg ml−1 apramycin or 20 μg ml−1 tobramycin (for pKC505, pKC796 and pKC039 derivatives in S. albus or E. coli respectively). For metabolite production and detection, S. albus strains were cultured and analysed by HPLC-MS as described before (Sánchez et al., 2005).
DNA manipulation
Total DNA isolation, plasmid DNA preparations, restriction endonuclease digestions, ligations and other DNA manipulations were performed according to standard procedures for E. coli (Sambrook et al., 1989) and for Streptomyces (Kieser et al., 2000). A genomic library of S. longisporoflavus was constructed in pKC505 as previously described (Richardson et al., 1987), and screened with a probe made from a rebD-homologue orf (Sánchez et al., 2002), later known as staD. This probe was obtained by polymerase chain reaction (PCR) according to standard procedures. For this purpose, we used total DNA from S. longisporoflavus and oligoprimers CS035 (5′-ATATAAGCTTGATGGCCCAGCACTTCGG-3′, altered sequence in italics, HindIII underlined) and CS036 (5′-TATCTAGACGGCGGGCGGAAGCGGTC-3′, altered sequence in italics, XbaI underlined), designed to amplify a DNA fragment encompassing nucleotides 1–323 from the sequence with Accession number A60304 (Schupp et al., 1997). This amplification product was cloned as a HindIII-XbaI fragment into LITMUS 28, and the resulting plasmid was sequenced to confirm the fidelity of the PCR reaction. Another probe, made from the rebG gene, was used for Southern analysis of selected cosmids. This probe consisted of a complete rebG obtained by PCR as described before (Sánchez et al., 2005). Digoxigenin labelling of DNA probes, Southern analysis, hybridization and detection were performed according to literature procedures and manufacturer recommendations (Boehringer Mannheim). Transformation of S. albus protoplasts followed procedures routinely used for Streptomyces (Kieser et al., 2000). DNA sequencing was performed as described before (Sánchez et al., 2002).
Construction of plasmids
pKC39GT
This plasmid resulted from the addition of the staG and rebT genes to plasmid pKC039, which already contained rebO, rebD, staC and rebP. The rebT gene confers rebeccamycin resistance to the otherwise sensitive S. albus host (Sánchez et al., 2002), and was included in the ‘aglycone plasmids’ in order to prevent potential toxic effects due to production of novel inhibitory indolocarbazole derivatives. The staG gene was obtained by PCR using total DNA from S. longisporoflavus and oligoprimers CS043 (5′-TATATTACTAGTCGCGGAGGCGACGTTGAC-3′, altered sequence in italics, SpeI underlined) and CS044 (5′-TATCTAGATTCAAC GGGTGATACCTCCG-3′, altered sequence in italics, XbaI underlined). The rebT gene was also obtained by PCR, as described before (Sánchez et al., 2005). Then, the ermE*p promoter (excised from pEM4 as a HindIII-BamHI fragment), staG and rebT were cloned together, in tandem, in LITMUS 28 to yield plasmid pLe5152GTF. Finally, the whole insert from pLe5152GTF was transferred as an XbaI fragment into the same site of pKC039, to yield pKC39GT.
pKC39GNT
This plasmid resulted from the addition of staG, staN and rebT to plasmid pKC039. A fragment including staG and staN was obtained by PCR using total DNA from S. longisporoflavus and oligoprimers CS043 (see above) and STAN2 (5′-TATCTAGAGTCAGTTCAGTACGGCGGGC-3′, altered sequence in italics, XbaI underlined), and cloned as an SpeI-XbaI fragment in the same sites of LITMUS 28 to yield pLGTFstaN. Then, the ermE*p promoter, the insert from pLGTFstaN and rebT were cloned together, in tandem, in the XbaI site of pKC039, to yield pKC39GNT.
pETAS
A 6.9 kb NruI fragment from cos32D1 containing the staMA, staJ, staK, staI, staE and staMB genes from the staurosporine cluster was subcloned into the EcoRV site of LITMUS 28, yielding plasmid pLAZSTA. From this construct, the genes were now recovered as an XbaI-EcoRI fragment and subcloned into the same sites of pEM4. In the final construct (pETAS) the genes are now under the control of the erythromycin resistance promoter ermE*p.
pAPSN
The staN gene was excised from plasmid pLGTF-staN as an NcoI-XbaI fragment and cloned into pEM4, under the control of the ermE*p promoter, to yield pAPSN.
pAPSMA
The staMA gene was excised from plasmid pLAZSTA as a MscI-NsiI fragment and cloned into pEM4, under the control of the ermE*p promoter, to yield pAPSMA.
pAPSMB
The staMB gene was obtained by PCR using total DNA from S. longisporoflavus and oligoprimers MB1 (5′-TATATTACTAGTCGAAAGGCCGTTCCCATGAC-3′, altered sequence in italics, SpeI underlined) and MB2 (5′-TATCTAGA GTCAGGGGCGCTCGGCGGT-3′, altered sequence in italics, XbaI underlined), and cloned into LITMUS 28 to yield pLstaMB. From this plasmid, staMB was cloned as an SpeI-XbaI fragment into pEM4, under the control of the ermE*p promoter, to yield pAPSMB.
pAPSNMA
The staN gene was excised from plasmid pLGTFstaN as an NcoI-XbaI fragment and cloned, downstream of staMA, into pAPSMA to yield pAPSNMA.
pAPSNMB
The staMB gene was excised from plasmid pLstaMB as an SpeI-XbaI fragment and cloned, downstream of staN, into pAPSN to yield pAPSNMB.
pAPSMNM
The staMB gene was excised from plasmid pLstaMB as an SpeI-XbaI fragment and cloned, downstream of staN, into pAPSNMA to yield pAPSMNM.
Isolation of staurosporine derivatives
For the isolation of compounds, strains were inoculated in TSB medium (Merck) and incubated for 24 h at 30°C and 250 r.p.m. Each seed culture was used to inoculate (at 2.5% v/v) several 2 l Erlenmeyer flasks (see below) each containing 400 ml of R5A medium. After incubation for 4–5 days in the above conditions, the cultures were centrifuged (12 000 r.p.m., 30 min) and the supernatants were discarded (in all cases most of the product was found in the cellular pellets). The pellets were extracted with acetone, shaking for about 2 h, after which the suspensions were centrifuged and the organic extract was evaporated in vacuo. For purification, the material extracted from pellets of each strain was redissolved in a small volume of a mixture of acetone and DMSO (50:50).
For the isolation of holyrine A, 9.6 l of culture of strain 39GT (pETAS) (from three independent experiments) were prepared. The extracts were first chromatographed in a μBonda-pak C18 radial compression cartridge (PrepPak Cartridge, 25 × 100 mm, Waters), eluting isocratically with acetonitrile and 0.1% trifluoroacetic acid (TFA) in water (37:63), at 10 ml min−1. The collected material was repurified in a semi-preparative column (Symmetry C18, 7.8 × 300 mm, Waters), with the same mobile phase, but at a flow rate of 3 ml min−1. Digitoxosyl derivatives were isolated from 3.2 l of culture of strain 39GNT (pLNBIV). The extract was chromatographed in the semipreparative column mentioned above with acetonitrile and 0.1% TFA in water (50:50), at 3 ml min−1. The two compounds collected were repurified in the same column with methanol and 0.1% TFA in water (70/30). L-olivosyl derivatives were isolated from 6.8 l of culture of strain 39GNT (pLN2) (in two independent experiments). The extracts were first chromatographed in the preparative cartridge with acetonitrile and 0.1% TFA in water (50:50), at 10 ml min−1, followed by a second purification in the semipreparative column with acetonitrile and 0.1% TFA in water (45:55), at 3 ml min−1. Rhamnosyl derivatives were isolated from extracts of 6.8 l of culture of strain 39GNT (pRHAM) (in two independent experiments) in a two step purification: first with acetonitrile and 0.1% TFA in water (45:55) and then with methanol and 0.1% TFA in water (70:30), both in the semipreparative column at 3 ml min−1. The D-olivosyl derivative was purified from an extract obtained from 3.2 l of culture of strain 39GNT (pLNR). The mobile phase used in this case was acetonitrile and 0.1% TFA in water (42:58) in a first step, and methanol and 0.1% TFA in water (75:25) in a second step, both at 3 ml min−1 in the semipreparative column. In all cases, after every purification step, the collected compounds were diluted fourfold with water and were desalted and concentrated by solid phase extraction, being finally lyophilized.
MS and NMR methods
The structures of the novel staurosporine derivatives were elucidated by NMR spectroscopy and mass spectrometry. HPLC-atmospheric pressure chemical ionization (APCI)-MS analysis was performed on a Waters Alliance 2695 instrument coupled with a Waters 2996 photodiode array detector and a Waters/Micromass ZQ 2000 mass spectrometer. All NMR data were recorded using either a Varian Marcury-300 or a Varian Inova-400 instrument at magnetic field strengths of B0 7.05 T and B0 9.4 T respectively. The NMR data for compounds 3 and 7, recorded in CD3OD, were listed in Tables S1 and S2 in Supplementary material. Additional 1H-NMR data for both compounds, recorded in DMSO-d6, are given below, because this solvent allows also the observation of the NH and OH signals. All NMR assignments are based on chemical shift rules and on comparison with previously elucidated indolocarbazoles compounds. All assignments were furthermore confirmed with homo- and heteronuclear 2D-experiments, particularly H,H – COSY, HSQC and HMBC.
1H NMR (DMSO-d6, 400 MHz) of compound 3 (l-olivosyl-2N-K252c)
δ 9.32 (d, J = 8.0 Hz, 1H), 8.62 (s, 1H), 8.07 (d, J = 8.4 Hz, 1H), 8.00 (d, J = 8.0 Hz, 1H), 7.66 (d, J = 8.4 Hz, 1H), 7.50 (ddd, J = 8.4, 8.0, 1.2 Hz, 1H), 7.44 (ddd, J = 8.4, 8.0, 1.2 Hz, 1H), 7.33 (t, 1H), 7.30 (t, 1H), 6.91 (dd, J = 4.4, 1.2 Hz, 1H), 6.04 (d, J = 5.6 Hz, OH), 4.99 (s, 1H), 4.98 (s, 1H), 4.94 (d, J = 4.4 Hz, OH), 3.87 (dd, J = 10, 5.2 Hz, 1H), 3.44 (m, 1H), 2.40 (m, 1H), 2.36 (s, 3H), 2.33 (m, 1H).
1H NMR (DMSO-d6, 400 MHz) of compound 7 (l-digitoxosyl-2N-K252c)
δ 9.28 (d, J = 8.0 Hz, 1H), 8.51 (s, 1H), 8.08 (d, J = 8.4 Hz, 1H), 7.96 (d, J = 8.0 Hz, 1H), 7.59 (d, J = 8.4 Hz, 1H), 7.46 (ddd, J = 8.4, 8.0, 1.2 Hz, 1H), 7.41 (ddd, J = 8.4, 8.0, 1.2 Hz, 1H), 7.28 (t, 1H), 7.26 (t, 1H), 6.76 (dd, J = 5.6, 1.6 Hz, 1H), 5.48 (br s, OH), 4.95 (s, 1H), 4.93 (s, 1H), 4.10 (br t, 1H + OH), 3.97 (d, J = 3.2 Hz, 1H), 2.64 (m, 1H), 2.39 (m, 1H), 2.29 (s, 3H).
Supplementary Material
Supplementary material.
Supplementary material is available for this article online.
Acknowledgments
This research was supported by grants from the Spanish Ministry of Education and Science (BIO2000-00474 and BMC2003-00478), of the Plan Regional de I+D+I del Principado de Asturias (GE-MED01-05) to J.A.S., and by the US National Institutes of Health (Grant CA 102102 to J.R.) We thank Obra Social Cajastur for financial support to C.S. and also the support of the Red Temática de Investigación Cooperativa de Centros de Cáncer (Ministerio de Sanidad y Consumo, Spain).
References
- Akinaga S, Sugiyama K, Akiyama T. UCN-01 (7-hydroxystaurosporine) and other indolocarbazole compounds: a new generation of anticancer agents for the new century? Anticancer Drug Des. 2000;15:43–52. [PubMed] [Google Scholar]
- Albermann C, Soriano A, Jiang J, Vollmer H, Biggins JB, Barton WA, et al. Substrate specificity of NovM: implications for novobiocin biosynthesis and glycorandomization. Org Lett. 2003;5:933–936. doi: 10.1021/ol0341086. [DOI] [PubMed] [Google Scholar]
- Bailly C, Qu X, Graves DE, Prudhomme M, Chaires JB. Calories from carbohydrates: energetic contribution of the carbohydrate moiety of rebeccamycin to DNA binding and the effect of its orientation on topoisomerase I inhibition. Chem Biol. 1999;6:277–286. doi: 10.1016/S1074-5521(99)80073-8. [DOI] [PubMed] [Google Scholar]
- Blanco G, Patallo EP, Braña AF, Trefzer A, Bechthold A, Rohr J, et al. Identification of a sugar flexible glycosyltransferase from Streptomyces olivaceus, the producer of the antitumor polyketide elloramycin. Chem Biol. 2001;8:253–263. doi: 10.1016/s1074-5521(01)00010-2. [DOI] [PubMed] [Google Scholar]
- Cai Y, Fredenhagen A, Hug P, Peter HH. A nitro analogue of staurosporine and other minor metabolites produced by a Streptomyces longisporoflavus strain. J Antibiot. 1995;48:143–148. doi: 10.7164/antibiotics.48.143. [DOI] [PubMed] [Google Scholar]
- Cai Y, Fredenhagen A, Hug P, Meyer T, Peter HH. Further minor metabolites of staurosporine produced by a Streptomyces longisporoflavus strain. J Antibiot. 1996;49:519–526. doi: 10.7164/antibiotics.49.519. [DOI] [PubMed] [Google Scholar]
- Chang CW, Zhao L, Yamase H, Liu HW. DesVI: a new member of the sugar N,N-dimethyltransferase family involved in the biosynthesis of desosamine. Angew Chem Int Ed. 2000;39:2160–2163. doi: 10.1002/1521-3773(20000616)39:12<2160::aid-anie2160>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Chater KF, Wilde LC. Streptomyces albus G mutants defective in the SalGI restriction-modification system. J Gen Microbiol. 1980;116:323–334. doi: 10.1099/00221287-116-2-323. [DOI] [PubMed] [Google Scholar]
- Chen H, Guo Z, Liu HW. Expression, purification, and characterization of TylM1, an N,N-dimethyltransferase involved in the biosynthesis of mycaminose. J Am Chem Soc. 1998;120:9951–9952. [Google Scholar]
- Chen H, Yamase H, Murakami K, Chang CW, Zhao L, Zhao Z, Liu HW. Expression, purification, and characterization of two N,N-dimethyltransferases, tylM1 and desVI, involved in the biosynthesis of mycaminose and desosamine. Biochemistry. 2002;41:9165–9183. doi: 10.1021/bi020245j. [DOI] [PubMed] [Google Scholar]
- Fischer C, Rodríguez L, Patallo EP, Lipata F, Braña AF, Méndez C, et al. Digitoxosyltetracenomycin C and glucosyltetracenomycin C, two novel elloramycin analogues obtained by exploring the sugar donor substrate specificity of glycosyltransferase ElmGT. J Nat Prod. 2002;65:1685–1689. doi: 10.1021/np020112z. [DOI] [PubMed] [Google Scholar]
- Gescher A. Staurosporine analogues – pharmacological toys or useful antitumour agents? Crit Rev Oncol Hematol. 2000;34:127–135. doi: 10.1016/s1040-8428(00)00058-5. [DOI] [PubMed] [Google Scholar]
- Goeke K, Hoehn P, Ghisalba O. Production of the staurosporine aglycon K-252c with a blocked mutant of the staurosporine producer strain Streptomyces longisporoflavus and by biotransformation of staurosporine with Streptomyces mediocidicus ATCC 13279. J Antibiot. 1995;48:428–430. doi: 10.7164/antibiotics.48.428. [DOI] [PubMed] [Google Scholar]
- Gribble GW, Berthel SJ. A survey of indolo[2,3-á]carbazoles and related natural products. In: Rahman A-U, editor. Studies in Natural Products Chemistry. Vol. 12. Amsterdam: Elsevier Science Publishers; 1993. pp. 365–409. [Google Scholar]
- Hoehn P, Ghisalba O, Moerker T, Peter HH. 3′-Demethoxy-3′-hydroxystaurosporine, a novel staurosporine analogue produced by a blocked mutant. J Antibiot. 1995;48:300–305. doi: 10.7164/antibiotics.48.300. [DOI] [PubMed] [Google Scholar]
- Hyun CG, Bililign T, Liao J, Thorson JS. The biosynthesis of indolocarbazoles in a heterologous Escherichia coli host. Chembiochem. 2003;4:114–117. doi: 10.1002/cbic.200390004. [DOI] [PubMed] [Google Scholar]
- Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hop-wood DA. Practical Streptomyces Genetics. Norwich: The John Innes Foundation; 2000. [Google Scholar]
- Kuhstoss S, Richardson MA, Rao RN. Plasmid cloning vectors that integrate site-specifically in Streptomyces spp. Gene. 1991;97:143–146. doi: 10.1016/0378-1119(91)90022-4. [DOI] [PubMed] [Google Scholar]
- Lombó F, Gibson M, Greenwell L, Braña AF, Rohr J, Salas JA, Méndez C. Engineering biosynthetic pathways for deoxysugars: branched-chain sugar pathways and derivatives from the antitumor tetracenomycin. Chem Biol. 2004;11:1709–1718. doi: 10.1016/j.chembiol.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Losey HC, Jiang J, Biggins JB, Oberthur M, Ye XY, Dong SD, Kahne D, et al. Incorporation of glucose analogs by GtfE and GtfD from the vancomycin biosynthetic pathway to generate variant glycopeptides. Chem Biol. 2002;9:1305–1314. doi: 10.1016/s1074-5521(02)00270-3. [DOI] [PubMed] [Google Scholar]
- Onaka H, Taniguchi S, Igarashi Y, Furumai T. Cloning of the staurosporine biosynthetic gene cluster from Streptomyces sp. TP-A0274 and its heterologous expression in Streptomyces lividans. J Antibiot. 2002;55:1063–1071. doi: 10.7164/antibiotics.55.1063. [DOI] [PubMed] [Google Scholar]
- Onaka H, Taniguchi S, Igarashi Y, Furumai T. Characterization of the biosynthetic gene cluster of rebeccamycin from Lechevalieria aerocolonigenes ATCC 39243. Biosci Biotechnol Biochem. 2003;67:127–138. doi: 10.1271/bbb.67.127. [DOI] [PubMed] [Google Scholar]
- Pérez M, Lombó F, Zhu L, Gibson M, Braña AF, Rohr J, et al. Combining sugar biosynthesis genes for the generation of l- and d-amicetose and formation of two novel antitumor tetracenomycins. Chem Commun (Camb) 2005;21:1604–1606. doi: 10.1039/b417815g. [DOI] [PubMed] [Google Scholar]
- Quirós LM, Aguirrezabalaga I, Olano C, Méndez C, Salas JA. Two glycosyltransferases and a glycosidase are involved in oleandomycin modification during its biosynthesis by Streptomyces antibioticus. Mol Microbiol. 1998;28:1177–1185. doi: 10.1046/j.1365-2958.1998.00880.x. [DOI] [PubMed] [Google Scholar]
- Richardson MA, Kuhstoss S, Solenberg P, Schauss NA, Nagaraja RN. A new shuttle cosmid vector, pKC505, for streptomycetes: its use in the cloning of three different spiramycin-resistance genes from a Streptomyces ambofaciens library. Gene. 1987;61:231–241. doi: 10.1016/0378-1119(87)90187-9. [DOI] [PubMed] [Google Scholar]
- Rodríguez L, Oelkers C, Aguirrezabalaga I, Braña AF, Rohr J, Méndez C, Salas JA. Generation of hybrid elloramycin analogs by combinatorial biosynthesis using genes from anthracycline-type and macrolide biosynthetic pathways. J Mol Microbiol Biotechnol. 2000;2:271–276. [PubMed] [Google Scholar]
- Rodríguez L, Aguirrezabalaga I, Allende N, Braña AF, Méndez C, Salas JA. Engineering deoxysugar biosynthetic pathways from antibiotic-producing microorganisms. A tool to produce novel glycosylated bioactive compounds. Chem Biol. 2002;9:721–729. doi: 10.1016/s1074-5521(02)00154-0. [DOI] [PubMed] [Google Scholar]
- Sambrook S, Fritsch EF, Maniatis T. Molecular Cloning: a Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sánchez C, Butovich IA, Braña AF, Rohr J, Méndez C, Salas JA. The biosynthetic gene cluster for the antitumor rebeccamycin: characterization and generation of indolocarbazole derivatives. Chem Biol. 2002;9:519–531. doi: 10.1016/s1074-5521(02)00126-6. [DOI] [PubMed] [Google Scholar]
- Sánchez C, Zhu L, Braña AF, Salas AP, Rohr J, Méndez C, Salas JA. Combinatorial biosynthesis of antitumor indolocarbazole compounds. Proc Natl Acad Sci USA. 2005;102:461–466. doi: 10.1073/pnas.0407809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupp T, Engel N, Bietenhader J, Toupet C, Pospiech A. Staurosporine Biosynthesis Gene Clusters. WO9708323 WIPO. 1997
- Tamaoki T, Nomoto H, Takahashi I, Kato Y, Morimoto M, Harada S. Staurosporine, a potent inhibitor of phospholipid/calcium dependent protein kinase. Biochem Biophys Res Commun. 1986;135:397–402. doi: 10.1016/0006-291x(86)90008-2. [DOI] [PubMed] [Google Scholar]
- Weidner S, Kittelmann M, Goeke K, Ghisalba O, Zahner H. 3′-Demethoxy-3′-hydroxystaurosporine-O-methyltransferase from Streptomyces longisporoflavus catalyzing the last step in the biosynthesis of staurosporine. J Antibiot. 1998;51:679–682. doi: 10.7164/antibiotics.51.679. [DOI] [PubMed] [Google Scholar]
- Williams DE, Bernan SE, Ritacco FV, Maiese WM, Greenstein M, Andersen RJ. Holyrines A and B, possible intermediates in staurosporine biosynthesis produced in culture by a marine actinomycete obtained from the North Atlantic Ocean. Tetrahedron Lett. 1999;40:7171–7174. [Google Scholar]
- Wohlert SE, Blanco G, Lombó F, Fernández E, Braña AF, Reich S, et al. Novel hybrid tetracenomycins through combinatorial biosynthesis using a glycosyltransferase encoded by the elm genes in cosmid 16f4 and which shows a broad sugar substrate specificity. J Am Chem Soc. 1998;120:10596–10601. [Google Scholar]
- Yang SW, Cordell GA. Metabolism studies of indole derivatives using a staurosporine producer, Streptomyces staurosporeus. J Nat Prod. 1997a;60:44–48. doi: 10.1021/np960566u. [DOI] [PubMed] [Google Scholar]
- Yang SW, Cordell GA. Further metabolic studies of indole and sugar derivatives using the staurosporine producer Streptomyces staurosporeus. J Nat Prod. 1997b;60:230–235. doi: 10.1021/np960674g. [DOI] [PubMed] [Google Scholar]
- Yang SW, Cordell GA. Origin of nitrogen in the indolocarbazole unit of staurosporine. J Nat Prod. 1997c;60:788–790. [Google Scholar]
- Yang SW, Lin LJ, Cordell GA, Wang P, Corley DG. O- and N-Methylation in the biosynthesis of staurosporine. J Nat Prod. 1999;62:1551–1553. doi: 10.1021/np990261q. [DOI] [PubMed] [Google Scholar]
- Yasuzawa T, Lida T, Yoshida M, Hirayama N, Takahashi M, Shirahata K, Sano H. The Structures of The Novel Protein Kinase C Inhibitors K-252a, b, c and d. J Antibiotics. 1986;39:1072–1078. doi: 10.7164/antibiotics.39.1072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Supplementary material is available for this article online.