Abstract
In a genome-wide screen of 684 cancer cell lines, we identified homozygous intragenic microdeletions involving genes encoding components of the apical-basal cell polarity complexes. Among these, PARD3 is disrupted in cell lines and primary tumors from squamous carcinomas and glioblastomas. Reconstituting PARD3 expression in both cell types restores tight junctions and retards contact-dependent proliferation. Searching specifically for small intragenic microdeletions using high resolution genomic arrays may be complementary to other genomic deletion screens and resequencing efforts in identifying new tumor suppressor genes.
Introduction
Classical tumor suppressor gene discovery involved loss-of-heterozygosity (LOH) screens, followed by detailed mapping of tumor-dependent allelic losses (1), while subsequent efforts used genome-wide PCR-based methods to detect rare homozygous deletions (2, 3), array-based hybridization technologies (4, 5) and most recently whole genome exon resequencing (6, 7). However, the complexity of these efforts has limited the number of tumors that can be screened by each method, a critical consideration since the recurrence of a gene-targeting event across different cancers is key to distinguishing driver from passenger mutations.
Here, we performed a high resolution array-based gene copy number analysis of 684 cancer cell lines, with the goal of identifying rare homozygous deletions across a broad panel of diverse cancer types. While initial arrays based on BACs or cDNAs contained relatively sparse genomic probes, the advent of oligonucleotide arrays with every increasing probe density has paved the way for simultaneously interrogating large numbers of tumors at increased resolution for discrete regions of genomic loss (8, 9). However, genomic instability in epithelial cancers has complicated attempts at finding recurrent deletions using copy number screens, leading to algorithms that readily reveal large, frequent amplifications and deletions, possibly at the expense of focal or less frequent variation in allelic dosage (10, 11). In contrast, we reasoned that the use of a very large cancer cell line panel would enable a focused screen for small intragenic deletions, and that recurrence of such focal deletions across different cancers would serve as evidence of functional significance. In addition, the primary screen provides the key target cells in which to test reconstitution of the deleted gene.
Materials and Methods
Human cancer cell lines
Sources for human cancer cell lines are listed in Supplementary Table 1. To minimize the chance of cross contamination or mis-identification, genomic DNA for array hybridization was prepared from each cell line within 6 months of receipt and as soon as possible after initial culture (always within one month). Methods of cell line verification performed by the individual repositories include STR DNA-typing, cytogenetic analysis and immunophenotyping. Additional details of verification steps are described in Supplementary Information online.
Primary tumors
Frozen and paraffin-embedded tumor samples were obtained from the MGH and University of Chicago Departments of Pathology (SCCHN) and the MGH Brain Tumor Bank (GBM). The presence of tumor within each tumor tissue was confirmed by a pathologist (P.M.S. and M.W.L. for SCCHN, D.N.L. for GBM) by Hematoxylin-and-Eosin staining of representative tissue sections. In cases where tumor cells comprised <70% of the tissue, micro dissection was employed to isolate a homogeneous tumor cell population.
Array hybridization
Sample processing for genomic DNA, complexity reduction, amplification, purification, labeling and hybridization to GeneChip Human Mapping 250K Sty Arrays (http://www.affymetrix.com Part # 901188) were performed as described in the Affymetrix GeneChip Human Mapping 500k assay manual (P/N 701930 Rev. 2). Genomic DNA from six EBV-transformed B-lymphocyte cell lines were included as normal controls for copy number calculations (Coriell Institute for Medical Research, Camden, NJ). Array hybridization, data acquisition and analysis for Agilent Human 105K oligonucleotide arrays were based on published methods (12). Additional methods describing preparation of nucleic acid and data acquisition and initial processing is available in Supplementary Information online.
Copy number analysis and identification of candidate tumor suppressor loci
The method for identifying homozygous deletions is outlined in Supplementary Fig. 1b and described in detail in Supplementary Information online. Briefly, after calculation of copy number for each SNP probe using dCHIP (http://www.hsph.harvard.edu/~cli/complab/dchip/), a copy number threshold equal to < 0.3 was applied to select candidate deleted regions (this threshold was found empirically to correctly capture common tumor suppressor genes known to be inactivated by deletion (e.g. CKDN2A/B, PTEN) (Supplementary Table 2c and Supplementary Fig. 2). Next, the boundaries of each deletion were adjusted such that deleted probes occurring within 0.1Mb of each other in the same sample were initially considered as a single deleted region. Then, deletions that did not disrupt exons of protein coding genes (using the March 2004 hg17 human reference sequence) and were not recurrent were eliminated. This left 706 recurrent deletions constituting 211 unique genes (from 2 to 145 deletions per gene) (Supplementary Table 2). To select genes for further analysis, we focused on genes disrupted by deletion in multiple samples (see Supplementary Data for calculation of the number of recurrences likely to be significant), as well as small deletions containing genes not clearly implicated in tumorigenesis and without extensive copy number polymorphisms (according to the Database of Genomic Variants, http://projects.tcag.ca/variation/). Among these, PARD3 was selected for further analysis.
PCR-based exon re-sequencing and RT-PCR
See Supplementary Information online.
Fluorescence In Situ Hybridization (FISH)
All BAC probes were obtained from BAC/PAC Resources (Children’s Hospital, Oakland, CA). PARD3 gene copy number was determined by two-color FISH on 5 μm thick sections following published protocols (13). Images were acquired using an Olympus BX61 fluorescent microscope equipped with a CCD camera, and analysis was performed with Genus software (Applied Imaging, San Jose, CA). At least 100 nuclei were scored for each sample. Non-tumor tissue samples were used as controls (Cybrdi Inc., Frederick, Maryland).
DNA and shRNA constructs and lentivirus expression
The cDNA for PARD3 was generously provided by Ian Macara, University of Virginia, Charlotsville, VA. See Supplementary Information online for additional details of cDNA and shRNA vector construction and expression.
Immunoblotting and Immunofluorescence
See Supplementary Information online.
Growth Assays
Cells were plated at 50,000–100,000 per well in triplicate in 6-well plates. At each time point, cells were detached with trypsin, stained with 1:10 dilution of 0.1% trypan blue and counted using a Cellometer Auto T4 (Nexcelom Bioscience LLC, Lawrence, MA). Media was refreshed on the remaining wells every 3 days.
Results
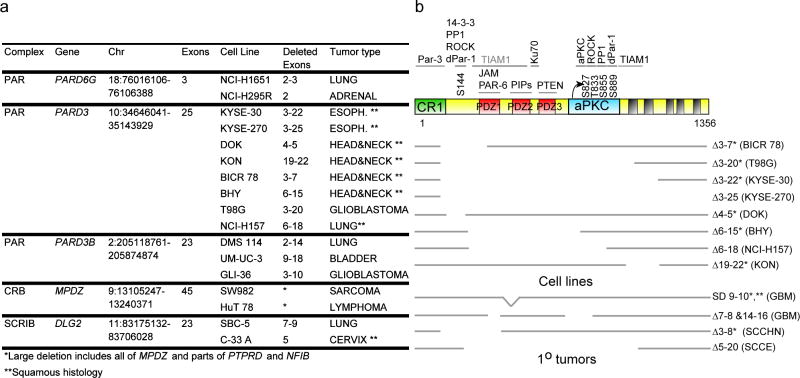
Analysis of 684 specimens with 250,000 SNP probes identified 11,328 regions with a reduction in copy number consistent with a homozygous deletion (<0.3) (Supplementary Fig. 1, Supplementary Table 1 online). Filtering for overlap with exons of protein coding genes and involvement of the candidate gene by at least one other deletion in an unrelated cancer cell line produced 211 genes identified as being inactivated by focal, recurrent homozygous deletions. These included 9 bona fide tumor suppressor genes (thereby validating the approach), 6 putative tumor suppressor genes located within highly polymorphic regions of the genome and 196 novel candidate genes (Supplementary Table 2 and Supplementary Fig. 2 online). Among these are five genes (PARD6G, PARD3, PARD3B, MPDZ, and DLG2) that encode components of the PAR, SCRB and CRB protein complexes implicated in apical-basal cell polarity (Fig. 1a) (14). The most commonly targeted polarity gene, PARD3 (chromosomal locus 10p11.21), was selected for detailed analysis.
Fig. 1.
Deletions in polarity complex genes in human tumor samples. (a) Human tumor cell lines with homozygous deletions in polarity complex genes. (b) Mapping of deleted regions to the domain structure of PARD3. *samples with aberrant transcripts encoding frameshift mutations. **splice site point mutation leads to aberrant splicing.
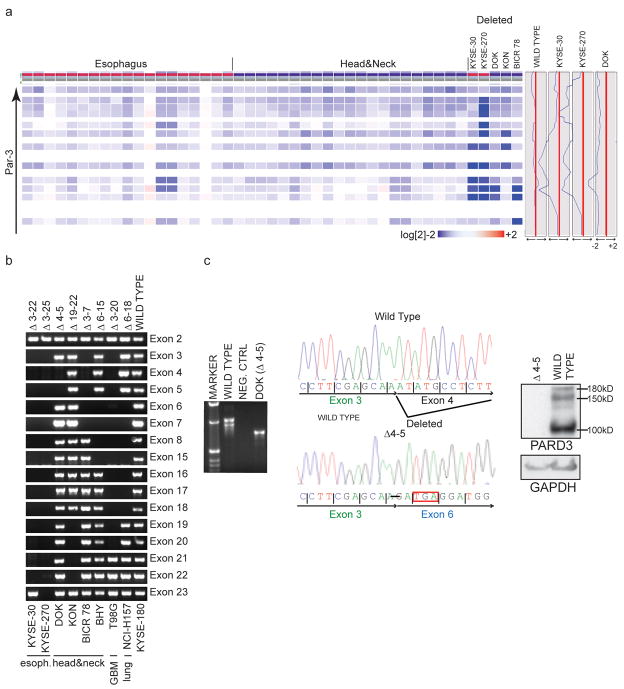
Microdeletions in PARD3 were evident in 8 samples, removing 2 to 23 of the 25 coding exons, without affecting neighboring genes (Figs 1b and 2a). Remarkably, PARD3 deletions were limited to two very distinct types of cancer: squamous carcinomas (2/20 esophagus (SCCE), 4/35 head and neck (SCCHN), 1/14 lung) as well as glioblastoma (GBM, 1/16). All deletions were confirmed by PCR amplification of genomic DNA (Fig. 2b). RT-PCR, nucleotide sequencing and immunoblot analysis showed absence of wild-type PARD3 mRNA, fusion of the deletion-flanking exons, with frameshift and loss of detectable protein expression (Figs. 2c, Supplementary Fig. 3 online).
Fig. 2.
Intragenic PARD3 deletions in human cancer cell lines. (a) Heat map (left) and graphical display (right) demonstrating intragenic, homozygous PARD3 deletions in esophagus and head and neck squamous carcinoma cell lines. Deviations to the left of the red line denoting normal copy number (to ~ log[2] −2) represent deleted probes. (b) PCR of genomic DNA confirms the location of the missing exons. (c) RT-PCR of SCCHN cell line DOK (genomic deletion exons 4–5) reveals a truncated transcript from fusion of exons 3 and 6 (left), leading to a frameshift, stop codon (middle) and loss of PARD3 expression (right).
To extend the cell line analysis to primary tumor specimens, we analyzed 21 SCCHN, 29 SCCE and 43 GBM. Somatic, intragenic PARD3 mutations were observed in four tumors: two homozygous deletions (GBM and SCCE), one heterozygous deletion (SCCHN) and one splice site mutation (GBM) (Fig. 3, Supplementary Fig. 4 online). Analysis of the SCCHN with a heterozygous deletion revealed a truncated PARD3 mRNA leading to a frameshift and stop codon (Fig. 3a). Analysis of the GBM with a splice site mutation (this tumor had loss of the other chromosome 10) demonstrated aberrant mRNA splicing, removing regions encoding PDZ domains 1 and 2 that are critical for effector binding (Figs. 1b, 3c). In addition to these definitive mutations, previously unreported missense mutations were identified in 4 cases (Supplementary Fig. 5a online). The combined mutation analysis of cell lines and primary tumors suggests that definitive PARD3 alterations occur in up to 9% of SCCHN, 6% of SCCE and 5% of GBMs (Supplementary Fig. 5b online).
Fig. 3.
PARD3 mutations in human primary tumors. (a) FISH identifies a heterozygous intragenic deletion within PARD3 in a primary SCCHN (left). Only one of two red signals for BAC RP11-198D23 is present in tumor nuclei (T, upper right panel, 44% nuclei) compared to matched normal tissue bordering the tumor (N, upper left--two signals in 94.6% nuclei). FISH with PARD3 BACs surrounding the deleted region demonstrates two signals for each probe (lower panels). RT-PCR (upper right) detects a truncated transcript, the result of out-of-frame fusion of exons 3 and 9 with loss of the intervening exons (lower right). (b) Homozygous deletion of exons 7–8 and 14–16 in a primary glioblastoma. A and B represent two different primer sets for exons 7 and 15. (c) A somatic G-A mutation present in GBM tumor DNA (asterix) but not DNA from matched normal blood (left sequence traces) leads to a truncated transcript (agarose gel image), the likely result of aberrant splicing that joins sequences within exons 9 and 10, with loss of the intervening sequence (right traces).
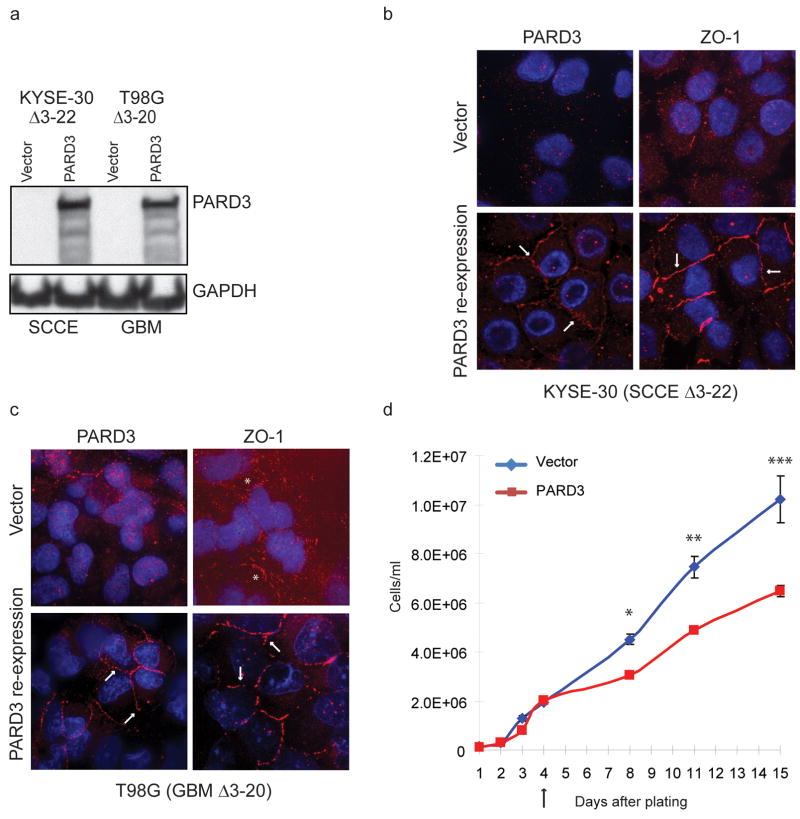
The identification of PARD3 deletions in cancer cell lines made it possible to test the consequences of re-expression of the wild type cDNA (Fig. 4a). Both squamous cancer and GBM cells with PARD3 deletions demonstrated reduced localization of ZO-1 to regions of cell-cell contact (Fig. 4b,c). Reconstitution of PARD3 expression resulted in re-localization of ZO-1 to appropriate intercellular junctions (Fig. 4b,c). The consequence of PARD3 re-expression on cell proliferation appeared to be specific to intercellular contact, in that PARD3-defective or reconstituted cells proliferated at comparable rates when seeded at low density, but a clear enhancement of PARD3-null cell proliferation emerged once cell density allowed for cell-cell contact (Fig. 4d). shRNA-mediated knockdown of PARD3 in other cancer cells with a wild type endogenous gene resulted in reduced localization of ZO-1 to cell-cell contacts and enhanced proliferation (Supplementary Fig. 6 online). Re-expression of PARD3 decreased migration in BHY cells (Δ6-15) but not KYSE-30 cells (Δ3-22) (Supplementary Fig. 7 online). There was no consistent effect of PARD3 re-expression on markers of squamous differentiation (Supplementary Fig. 8 online).
Fig. 4.
Effect of gain-of-function of PARD3. (a) Restoration of PARD3 expression in squamous carcinoma and glioblastoma cell lines with endogenous deletions. (b,c) Re-expression of PARD3 relocalizes ZO-1 to cell-cell contacts. Arrows indicate location of PARD3 or ZO-1. Asterixes indicate discontinuous ZO-1 staining at some cell borders in control cells. (d) Restoring PARD3 expression in KYSE-30 SCCE cells retards cell growth. The arrow indicates when the cells first appeared to make cell-cell contacts. Cells were plated at 1e5 cells/well in 6-well plates in triplicate. Every 3–4 days, cells were trypsinized and counted. Mean and standard error of the mean (n=3) for one of two representative experiments is shown. *P=0.004. **P=0.005. ***P=0.018 (alpha = 0.05, two-sided, unpaired student’s t-test).
Discussion
PARD3 is thought to be a master regulator of apical-basal cell polarity, a process that has been indirectly implicated in tumorigenesis (15, 16). Recently, SCRIB (also known as Scribble) was proposed as a tumor suppressor by virtue of its mislocalization in human breast cancers (17). However, inactivating mutations in SCRIB have not been identified, and it is possible that mutational inactivation of PARD3 (and potentially other polarity complex family members) provides a genetic mechanism that explains dysregulation of cell polarity in a subset of human cancers. While this manuscript was in preparation, Zen et al reported finding PARD3 deletions in the SCCE cell lines also described here (e.g. KYSE-30 and −270) (18). As shown in our two studies, loss of PARD3 in SCCE cells leads to abnormal cellular contacts, consistent with disruption of the PAR polarity complex. The apparent tissue specificity of PARD3 deletions uncovered in our study, affecting both diverse squamous epithelial cancers and GBMs, highlights a somewhat unexpected parallel role for the PAR complex in these very distinct cell types. The loss of tight junctions may permit contact-inhibited cells to loosen from neighboring cells and proliferate. Notably, genomic losses affecting chromosome 10p have been demonstrated in primary GBMs, suggesting the presence of a novel tumor suppressor(s) (in addition to PTEN on 10q) (19).
The strategy that we employed to identify targeted homozygous deletions is complementary to that of ongoing cancer genome sequencing efforts. Cell line analysis offers the advantage of a homogeneous tumor cell population free of contaminating stromal cells that could mask deletions. However, to rule out potential cell culture-related genetic events, it is critical to demonstrate the presence of inactivating mutations in primary tumor samples. As we have shown for PARD3 and potentially other tumor suppressors, deletion of one or a few exons can have dramatic consequences for gene expression. It is generally assumed that tumor suppressor genes are more frequently targeted by point mutations than by deletion events, although for PARD3, as well as other well established tumor suppressor genes like CDKN2A/B, chromosome deletions appear to be predominant. Consistent with this observation, PARD3 mutations were not identified in a recent genome re-sequencing analysis of 22 human GBMs (20). Moreover, the small homozygous PARD3 deletions also went undetected in other gene copy number screens using oligonucleotide arrays, perhaps due to primary tumor heterogeneity or algorithms that filter the deleted segments by size and frequency (8). As the density of genomic probes increases, optimizing these approaches to allow detection of focal homozygous deletions, and using a very large panel of cancer cell lines to increase the chance of detecting recurrent lesions across diverse cancer types, thus offers a potentially important tool for new cancer gene discovery.
Supplementary Material
Acknowledgments
Funding was provided by the Howard Hughes Medical Institute (D.A.H., M.N.R.) and National Institutes of Health Grants RO1 CA207186 (D.A.H.) and RO1 CA57683 (D.N.L.). S.M.R. is supported by the MGH Tosteson Postdoctoral Fellowship and a T32 Institutional Ruth L. Kirstein National Research Service Award. M.N.R. is supported by National Institutes of Health Grant K08 DK080175 and the Burroughs Wellcome Fund.
Footnotes
AUTHOR CONTRIBUTIONS
S.M.R, G.M and M.N.R. directed the study, performed functional work and interpreted the data. D.C.W. performed PCR, RT-PCR and sequencing analysis. P.G. performed copy number analysis. M.N. performed FISH analysis. G.S., M.J.U. and B.W.B. performed sequencing analysis. P.M.S, D.N.L. and M.W.L. contributed samples and performed pathologic analysis. R.M.P. and A.T. performed functional work. X.J.M and M.E. performed oligonucleotide array hybridization. D.C.S performed laser capture microdissection. J.W.R., E.E.W.C. and D.N.L. contributed samples and comments on the manuscript. S.M.R, G.M., M.N.R, D.N.L., J.S, and D.A.H conceived the study. S.M.R. and D.A.H. wrote the manuscript.
Note: Please refer to Supplementary Information for additional methods and data.
References
- 1.Dryja TP, Rapaport JM, Joyce JM, Petersen RA. Molecular detection of deletions involving band q14 of chromosome 13 in retinoblastomas. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:7391–4. doi: 10.1073/pnas.83.19.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lisitsyn NA, Segre JA, Kusumi K, et al. Direct isolation of polymorphic markers linked to a trait by genetically directed representational difference analysis. Nature genetics. 1994;6:57–63. doi: 10.1038/ng0194-57. [DOI] [PubMed] [Google Scholar]
- 3.Schutte M, da Costa LT, Hahn SA, et al. Identification by representational difference analysis of a homozygous deletion in pancreatic carcinoma that lies within the BRCA2 region. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:5950–4. doi: 10.1073/pnas.92.13.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pinkel D, Segraves R, Sudar D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nature genetics. 1998;20:207–11. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 5.Pollack JR, Perou CM, Alizadeh AA, et al. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nature genetics. 1999;23:41–6. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 6.Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sjoblom T, Jones S, Wood LD, et al. Science. Vol. 314. New York, NY: 2006. The consensus coding sequences of human breast and colorectal cancers; pp. 268–74. [DOI] [PubMed] [Google Scholar]
- 8.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao X, Li C, Paez JG, et al. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer research. 2004;64:3060–71. doi: 10.1158/0008-5472.can-03-3308. [DOI] [PubMed] [Google Scholar]
- 10.Hupe P, Stransky N, Thiery JP, Radvanyi F, Barillot E. Bioinformatics. Vol. 20. Oxford, England: 2004. Analysis of array CGH data: from signal ratio to gain and loss of DNA regions; pp. 3413–22. [DOI] [PubMed] [Google Scholar]
- 11.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Biostatistics. Vol. 5. Oxford, England: 2004. Circular binary segmentation for the analysis of array-based DNA copy number data; pp. 557–72. [DOI] [PubMed] [Google Scholar]
- 12.Gabeau-Lacet D, Engler D, Gupta S, et al. Genomic profiling of atypical meningiomas associates gain of 1q with poor clinical outcome. Journal of Neuropathology Experimental Neurology. 2009;68:1155–65. doi: 10.1097/NEN.0b013e3181ba3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohapatra G, Betensky RA, Miller ER, et al. Glioma test array for use with formalin-fixed, paraffin-embedded tissue: array comparative genomic hybridization correlates with loss of heterozygosity and fluorescence in situ hybridization. J Mol Diagn. 2006;8:268–76. doi: 10.2353/jmoldx.2006.050109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macara IG, Mili S. Polarity and differential inheritance--universal attributes of life? Cell. 2008;135:801–12. doi: 10.1016/j.cell.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bilder D, Li M, Perrimon N. Science. Vol. 289. New York, NY: 2000. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors; pp. 113–6. [DOI] [PubMed] [Google Scholar]
- 16.Pagliarini RA, Xu T. Science. Vol. 302. New York, NY: 2003. A genetic screen in Drosophila for metastatic behavior; pp. 1227–31. [DOI] [PubMed] [Google Scholar]
- 17.Zhan L, Rosenberg A, Bergami KC, et al. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell. 2008;135:865–78. doi: 10.1016/j.cell.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zen K, Yasui K, Gen Y, et al. Defective expression of polarity protein PAR-3 gene (PARD3) in esophageal squamous cell carcinoma. Oncogene. 2009;28:2910–8. doi: 10.1038/onc.2009.148. [DOI] [PubMed] [Google Scholar]
- 19.Ichimura K, Schmidt EE, Miyakawa A, Goike HM, Collins VP. Distinct patterns of deletion on 10p and 10q suggest involvement of multiple tumor suppressor genes in the development of astrocytic gliomas of different malignancy grades. Genes, chromosomes & cancer. 1998;22:9–15. doi: 10.1002/(sici)1098-2264(199805)22:1<9::aid-gcc2>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 20.Parsons DW, Jones S, Zhang X, et al. Science. Vol. 321. New York, NY: 2008. An integrated genomic analysis of human glioblastoma multiforme; pp. 1807–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.