Abstract
The human superorganism is a conglomerate of mammalian and microbial cells, with the latter estimated to outnumber the former by ten to one and the microbial genetic repertoire (microbiome) to be approximately 100-times greater than that of the human host. Given the ability of the immune response to rapidly counter infectious agents, it is striking that such a large density of microbes can exist in a state of synergy within the human host. This is particularly true of the distal gastrointestinal (GI) tract, which houses up to 1000 distinct bacterial species and an estimated excess of 1 × 1014 microorganisms. An ever-increasing body of evidence implicates the GI microbiota in defining states of health and disease. Here, we review the literature in adult and pediatric GI microbiome studies, the emerging links between microbial community structure, function, infection and disease, and the approaches to manipulate this crucial ecosystem to improve host health.
Keywords: gastrointestinal tract, inflammation, microneme, probiotics
Role of microbial community composition in defining host health
The application of culture-independent tools has dramatically improved our ability to interrogate the vast diversity of unculturable or fastidious microbial species present in disparate environments and has led to significant advances in our understanding of ecosystem functioning [1–17]. Many of the tools developed for environmental microbial studies have recently been applied to human samples, providing a more comprehensive view of our microbial inhabitants in a number of discreet host niches, including the respiratory, gastrointestinal (GI) and urogenital tracts [7,18,19–24]. Recent studies have demonstrated that bacterial community composition is dramatically altered in diseases such as obesity and periodontal disease, with healthy subjects typically exhibiting distinct, diverse and temporally stable bacterial consortia at these sites when compared with patients displaying disease symptoms [6,25,26,27–29]. A number of studies have also demonstrated that bacterial community structure plays a key role in defining its functionality; compared with lean individuals, obese subjects exhibit a dramatic tenfold shift in the ratio of Firmicutes to Bacteroidetes (from 3:1 to 35:1), two of the major phyla present in the human GI tract [7]. This altered community structure is associated with a shift in function, resulting in increased energy harvest from ingested food; unexpended excess energy is deposited as adipose tissue [17]. Diet is a complex confounding factor in such studies [30,31] in that it can dramatically impact the composition of the gut microbial community [31]. A high-fat diet has been associated with an increase in Firmicutes and Proteobacteria and a concomitant decrease in Bacteroidetes in both wild-type mice and in isogenic resistin-like molecule-β animals that are resistant to high-fat-induced obesity, indicating diet to be a key determinant of gut microbiome composition [31]. In humans, a high-fat diet resulted in a similar phylogenetic shift in the GI microbiome associated with obesity [6]; this restructuring is largely due to dietary selective pressure, which promotes organisms optimally poised to metabolize and import readily available carbon sources, particularly simpler sugars, such as glucose, fructose and sucrose [32]. In support of this, a separate study of C57BL/6J mice fed a high-fat/high-sugar Western diet exhibited GI microbiome domination by the class Mollicutes (within the Firmicutes phylum), which was associated with increased body fat and upregulation of metabolic pathways involved in the import and fermentation of simple sugars and host glycans [32]. Interestingly, a high-fat diet has also been shown to reduce the abundance of Bifidobacteria [33], which are traditionally thought of as beneficial species in the gut microbiome.
The shifts in microbiome elicited by diet and other factors (described later) are key to host health, particularly because the structure of the GI microbial population has been associated with protection against pathogens. Recently, Dong and colleagues demonstrated that aseptic mosquitoes (Anopheles gambiae), the natural vector for Plasmodium falciparum (the causative agent of malaria), were susceptible to this parasite once their natural gut microbiome was disrupted through antibiotic treatment [34]. This effect was ameliorated by feeding or injecting the insects with live bacterial species, which was associated with a reduction in the number of oocysts produced by P. falciparum [34]. The authors further established that this protective effect was elicited indirectly through microbiome-dependent manipulation of the insect's immunity, involving upregulation of several anti-Plasmodium factors and the antimicrobial peptides cecropins 1 (Cec1) and 3 (Cec3), Defensin 1 (Def1), as well as other basal immune response factors such as lysozyme c-1 [34]. Other murine studies have also examined gut microbiome destructuring (mediated by antibiotic administration) and infection by GI pathogens, and have also demonstrated a key role for native bacterial species in controlling the behavior and physiology of infectious agents [35,36]. Following administration of antibiotics that dramatically alter the composition of the gut microbiome, Salmonella enterica serovar Typhimurium or Clostridium difficile-infected mice exhibit a supershedder phenotype (shed >108 CFU/g of pathogenic cells in stool), resulting in rapid transmission of the infectious agent [35,36]. These studies demonstrate the complexity of the host–microbiome interaction, and the key role played by the microbial community in modulating the host immune response and controlling the behavior and outgrowth of pathogenic species.
The importance of microbial immunomodulation is further exemplified by a recent study demonstrating that mice mono-colonized with a murine gut commensal anaerobe, segmented filamentous bacterium (SFB), exhibited induction of CD4+ T-helper cells producing a Th17 cytokine (IL-22 and IL-17) profile [37]. Expression analysis of SFB-colonized mice demonstrated an upregulation of serum amyloid A, which specifically induces a dendritic cell-mediated Th17 cell-inducing environment in the gut. In addition, SFB colonization also reduced the severity of Citrobacter rodentium infection; mice colonized with SFB did not exhibit penetration of the colonic wall by C. rodentium and had significantly less colonic inflammation when compared with noncolonized animals [37]. These findings are of particular interest given recent reports that aberrant Th17 populations are associated with a number of chronic diseases such as inflammatory bowel disease (IBD) [38], lupus, multiple sclerosis, psoriasis, and rheumatoid arthritis (reviewed in [39]), disorders that are believed to be linked to GI dysbiosis.
Prenatal microbial exposures & early immune development
Recent studies have suggested that predisposition to disease may, at least in part, be determined in utero. Maternal exposure to environmental stimuli, particularly microbes during pregnancy, appears to play an important role in postnatal immune functioning and, in particular, the subsequent development of allergic disease [40–42]. Schaub et al. demonstrated that mothers exposed to farms and farm animals during pregnancy were less likely to have children who developed allergies and asthma (Figure 1) [43]. These prenatal exposures impacted immune responses and were associated with increased number and function of cord blood T-regulatory (Treg) cells, which are linked to lower Th2 cytokine secretion (increased Th2 cytokine secretion is a characteristic of an allergic response). The authors speculated that maternal prenatal exposure to farm-associated microbes could provide a form of natural immunotherapy, potentially shaping the child's immune development during the gestational period. The work of Ege and colleagues [40] provides further support for this hypothesis, demonstrating that children whose mothers had been exposed to stables during their prenatal period, exhibited higher expression levels of the innate immune components, Toll-like receptor (TLR2, TLR4 and CD14), which specifically recognize and facilitate response to both Gram-positive and Gram-negative bacteria. Moreover, the authors demonstrated that this response was dose dependent; for every extra farm-associated species the mother encountered, expression levels increased by 1.16-fold [40].
Figure 1. Factors that influence the infant gut microbiome and early immune development.
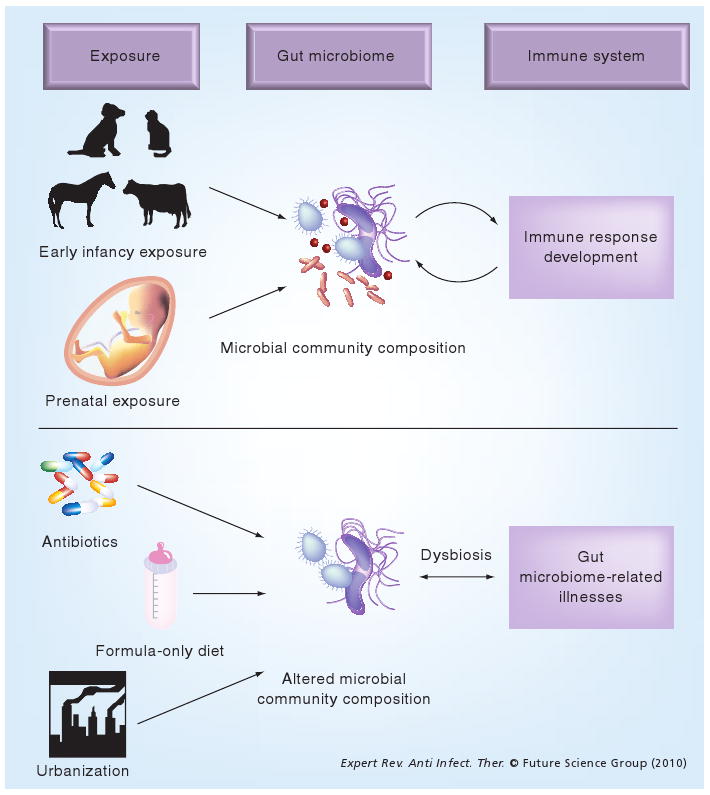
Exposure to farm animals and pets, vaginal birth and breast milk, all of which have a potential microbial link, are putatively associated with a beneficial effect on the developing gut microbiome and host immune response. Factors such as urbanization (lack of microbial exposures), formula-only diet and antibiotic administration during the neonatal stage are associated with the development of subsequent chronic diseases such as asthma and atopy, putatively through the development of aberrant gut microbiomes.
The fact that these exposures mediate their effect via microbes is supported by the finding that bacterial species such as Acinetobacter lwofii and Lactococcus lactis, isolated from farming communities, have specifically been shown to reduce allergic responses in murine models [44]. Both A. lwofii and L. lactis isolated from cow sheds demonstrated the ability to polarize T-cell maturation towards a Th1 response by stimulating dendritic cell IL-12 production, resulting in abrogation of allergic inflammation and improved airway responsiveness [44]. Thus, there is evidence for the potential of these farm-associated microbial species to manipulate the immune response; however, whether they directly or indirectly (via maternal immune manipulation) impact the immune response of the developing fetus remains to be determined.
Protective prenatal exposures do not appear to be restricted to farm animals; maternal prenatal exposure to household pets (cats and dogs) has also been shown to protect against allergic disease development [42,45–47] and it is hypothesized that this protection is, as with farm animal exposures, mediated via microbes. Pet exposure is associated with lower cord blood IgE levels [47], which is particularly pertinent given the crucial role that IgE plays in fetal immune system functioning [48] and that elevated cord blood IgE levels have previously been associated with subsequent development of allergic disorders [49–51]. Wegienka and colleagues also demonstrated that exposure of nonallergic pregnant women to pets is associated with increased Treg cell numbers [42], suggesting that the protective effect of pet exposure against allergic disease development may be through the induction of Treg cell populations, which are known to play a central role in immune homeostasis [43]. The concept that microbial exposures are key to defining the developing immune response in utero is also supported by a study demonstrating that maternal prenatal exposure to antibiotics (which dramatically impact the human microbiome; see later) resulted in a dose-dependent increased risk for childhood asthma [52].
Again, whether such prenatal exposures directly (via exposure of the developing fetus to microbial products) or indirectly impact fetal immune response development is unclear; however, a recent study suggests that microbial exposure can occur in utero and can impact postnatal infant health. Amniotic fluid, which is typically sterile in healthy pregnant women [53], has recently been shown to be a source of direct microbial exposure for the developing fetus. Leptotrichia spp. and other related bacterial species were detected using culture-independent molecular approaches in the amniotic fluid of women in preterm labor with a strong dose-dependent relationship between bacterial abundance in the amniotic fluid and gestational age at delivery [54]. A separate study also detected bacterial species in the amniotic fluid of preterm pregnancies and further demonstrated that women in this study whose amniotic fluid was PCR-positive for bacteria exhibited elevated levels of IL-6, histological chorioamnionitis and funisitis, which were strongly associated with the development of neonatal sepsis [53]. The authors did not comment on the causative agent of sepsis in these neonates, making it difficult to determine whether prenatal exposures were directly or indirectly responsible for the subsequent infection. Nonetheless, the study provides evidence for direct in utero microbial exposure and the possibility that introduction of the developing fetus to microbial products may play a role in shaping postnatal immune development.
Postnatal GI microbial colonization & immune response development
Prenatal maternal exposures clearly influence early infant immune responses and therefore presumably also regulate postnatal microbial colonization, an area of research that has recently become the focus of intense study. Exposures that shape the gut microbial community composition have come under particular scrutiny (Figure 1). The diverse ecosystem of the human gut microbiome encodes genes for essential functions that the human host is incapable of performing, such as vitamin production and metabolism of indigestible dietary polysaccharides [55–57]. Thus, the host immune system must strike a balance between providing a favorable environment for this vital community while protecting against invasion or outgrowth of pathogenic species. Insights into how the immune system initially develops the ability to discriminate between harmful and beneficial microbial species is becoming more apparent and appears to be based, at least in part, on both prenatal maternal exposures (discussed earlier) and postnatal infant GI colonization events [56,58,59]. GI mucosal defense and homeostasis are typically mediated by two distinct mechanisms, immune exclusion – mediated by secretory antibodies at the mucosal surface – and immunosuppression to prevent inappropriate responses to ‘friendly’ antigens by recognizing both pathogenic and commensal bacteria via TLRs [60–64]. The ability to discriminate between microbial ‘friend’ and ‘foe’ is primarily dependent on postnatal immune development, which is increasingly associated with appropriate microbial colonization of the GI tract [59,65,66]. Studies of germ-free mice have demonstrated deficiencies in immune development in the absence of GI microbial colonization [67–69]. The importance of early microbial exposure is emphasized in a mouse study in which neonatal mice were exposed to lipopolysaccharides (LPS) or ovalbumin [70]. Mice exposed to LPS developed T-cell populations expressing CD25+ and IL-10 upon antigen challenge and exhibited reduced airway hypersensitivity in comparison to those exposed exclusively to ovalbumin (who demonstrated a strong Th1-dominated response [70]). Several other animal models and comparisons of human gut communities have reinforced the key role that appropriate microbial colonization plays in gut-associated lymphoid tissue (GALT) development [71], specific aspects of immune system development [14,72,73] and the integrity of the mucosal barrier [69,72,74]. Culture-independent studies have provided greater insight into the temporal fluctuations in bacterial community diversity as it develops during an infant's first year of life, including dramatic decreases in diversity upon antimicrobial administration [11]. However, by approximately 12 months of age, this gut community begins to resemble that of an adult-like microbiome, dominated by the bacterial phyla Firmicutes and Bacteroides and possessing members of the Proteobacteria, Actinobacteria and Verrucomicrobia, amongst others [11]. Thus, it has been suggested that this initial period of colonization, which coincides with immune response development, represents a crucial window during which aberrations in colonization patterns may impact appropriate immune maturation [75].
Much of the evidence for the link between early microbial colonization events in the GI tract and subsequent development of immune disorders come from studies of asthma and allergy, which are regarded as a failure in the development of a balanced immune response [42,75–78]. The hygiene hypothesis, originally based on the observation that children with older siblings exhibit a reduced incidence of allergic disease, was postulated to be due to exposure to viral infections [79]. However, a more recent evolution of this hypothesis is that a lack of exposure to microbes in early infancy due to improved living conditions leads to development of a skewed immune response [80]. Recent studies provide strong evidence for a link between early GI colonization events and the subsequent development of allergic disease [75,81]. A microbiological examination of almost 1000 stool samples from 1-month-old infants demonstrated that a high abundance of Escherichia coli was associated with the subsequent development of eczema, while infants colonized with large numbers of Clostridium difficile were associated with a higher risk for eczema, recurrent wheeze, allergic sensitization and atopic dermatitis [75]. These data suggest that microbiome deviations, particularly outgrowth of specific species even at this early stage of life, are associated with subsequent immune disease development.
Key factors that impact gut microbiome composition in the early stages of infancy and which have been associated with subsequent childhood asthma and allergy development include Caesarian delivery (rather than vaginal birth), formula-based diet (in lieu of breast milk), hospitalization, gestational age (preterm) and antibiotic administration [41]. These findings are supported by several other investigations. An independent epidemiological study associated antibiotic use in the first year of life with increased risk of atopy, the risk being greater in subgroups of children who were breast-fed for 4 months or longer or had two or fewer pets in the home [82]. The authors concluded that antibiotic use in infants could change the gut microbiota, which may negatively impact immune system development and increase the risk of atopy in specific groups of children [82]. A meta-analysis of studies on the mode of birth and allergic diseases demonstrated a 20% increase in the development of asthma and allergy in children delivered by Caesarian section compared with those delivered vaginally [81]. Vaginal delivery results in exposure to the maternal vaginal microbiome, typically composed of commensal organisms commonly found in the lower GI tract [83]. These infants typically possess higher abundances of certain Bifidobacterium and Bacteroides species, which have been associated with health-promoting effects, including downregulation of inflammatory responses [77,84,85]. In contrast, infants delivered by Caesarian section exhibit a delayed and deviant GI bacterial community, dominated by Staphylococcus spp., Streptococcus spp. [86,87] and C. difficile [41] (associated with an increased risk of allergic disease development [78]). Indeed Penders and colleagues recently demonstrated that full-term infants born vaginally at home, who were exclusively breastfed, exhibited the most ‘beneficial’ gut microbiota, characterized by high numbers of Bifidobacteria and reduced abundances of E. coli and C. difficile [41].
As in adulthood, mode of nutrition also plays a key role in shaping the developing GI microbiome over the first year of life. In addition to the nutritional support breast milk provides, it also facilitates transfer of bioactive agents, for example, maternal secretory IgA, which provides passive immunoprotection [88,89]. In addition, IgA has been shown to sequester commensal species in the neonatal intestine and promote biofilm formation [88,89], an aspect that has been argued to facilitate colonization by protective native gut bacteria (immune inclusion) and prevent colonization by pathogenic species (immune exclusion) [90,91]. In mouse studies, a reciprocal relationship between the concentration of maternal IgA and bacterial colonization has been previously demonstrated, suggesting that breast milk IgA may delay the development of a diverse gut microbiome [92], providing a window for the development of protective biofilms of commensal organisms along the GI mucosa. Other components of breast milk modulate the developing mucosal immune response, while the presence of indigestible oligosaccharides promotes the growth of specific bacterial families such as the Bifidobacteriaceae and may act as decoy ligands for pathogens, preventing their mucosal attachment [93]. Thus the components of breast milk serve to both directly and indirectly enhance mucosal barrier function and shape immune development [93]. In addition to protection against allergic disease development, breastfeeding has also been associated with defense against neonatal diarrhea [94], necrotizing colitis [95], obesity (meta-analysis is provided in [96]) and Type II diabetes [97]. The fact that an association between early nutritional status (which clearly impacts the developing microbiome) and subsequent development of chronic adult diseases exists, argues strongly for the role of GI microbes in the development and modulation of these disorders. It also demonstrates that predisposition to these diseases may originate from initial GI colonization events and immune system programming during the early years of life, suggesting a role for early GI microbial colonization events in determining subsequent inflammatory disease outcomes.
Infants who are exclusively formula-fed exhibit altered GI microbiota with higher levels of E. coli and C. difficile, two species that have previously been linked to the development of eczema in infants [75]. Although it has been reported that there is no significant difference in the numbers of Bifidobacteria detected in stools of exclusively formula or breast milk-fed infants [41,98], the functional gene expression and type of Bifidobacteria present in infant stools has been shown to differ according to feeding mode [99]. Klaassens and colleagues used a Bifidobacterium mixed-species expression microarray to examine the differences between infants who were exclusively breast- or formula-fed. They demonstrated that diet explained 44% of the expression profile variation in Bifidobacteria in formula- or breast-fed infants and that each treatment group exhibited specific expression profiles [99]. For example, glycobiome-associated enzymes were more highly expressed in breast-fed infants who consume more diverse and complex breast milk oligosaccharides, providing these infants with the increased potential to metabolize these sugars [99]. These results suggest that not only is the presence and abundance of a specific microbial species or family important, but that microbiome expression profiles are strongly influenced by external influences, such as mode of feeding.
These and other studies have indicated that Bifidobacteria are beneficial to human health, which is currently a contentious issue. Nonetheless, several Bifidobacterium species including Bifidobacterium bifidum, Bifidobacterium breve and Bifidobacterium infantis have demonstrable anti-inflammatory properties that protect the epithelial cells from toxins [77,100]. These three Bifidobacteria species have been shown to induce the anti-inflammatory cytokine IL-10 [100]. In addition, soluble factors from B. breve inhibit LPS-induced TNF-α secretion by immune cells [101] and reduce chemokine, other proinflammatory molecule release and epithelial chloride secretion putatively by targeting serine/threonine kinase activity [102]. Sequence analysis of the commensal bacterial species Bifidobacterium longum, which is highly abundant in the breast-fed infant gut, revealed the blueprint of an organism specialized to competitively utilize the indigestible sugars in breast milk [103]. Possessing a high abundance of Bifidobacteria also appears to be beneficial in old age. In a recent study, Bifidobacterium species were isolated from Chinese centenarians from Bama, a village known for having a substantially higher than normal proportion of residents over 100 years old who exhibit a low occurrence of age-related inflammatory diseases. Compared with control animals, mice that received daily supplementation with a Bifidobacterium adolescentis strain isolated from the centenarians exhibited altered intestinal morphology, including a significant increase in villus height and crypt depth, features believed to enhance digestive efficacy. In addition, duodenal secretory IgA was significantly increased in animals receiving a high dose of this strain (2 × 1010 CFUs daily) compared with controls [104]. In a separate study, Sjögren and colleagues examined the relationships between mucosal and systemic immune responses and a number of species that have been implicated in protection against, or development of, allergic diseases [105]. They demonstrated that Bifidobacteria diversity was associated with accelerated maturation of the mucosal secretory IgA system and that increased abundance of Bacteroides fragilis at the early stages of GI colonization reduced LPS responsiveness [105]. Although the mechanism by which these species provide protection is not fully understood, what is clear is that their loss, reduction or altered gene expression in the GI tract during the critical early stages of immune maturation leads to the subsequent development of disease [73,75,106,107]. Based on these and other observations, it is becoming increasingly clear that early events in GI colonization, particularly those events that may alter this process, play a crucial role in the development and maintenance of the host immune system and predisposition to subsequent development of disease.
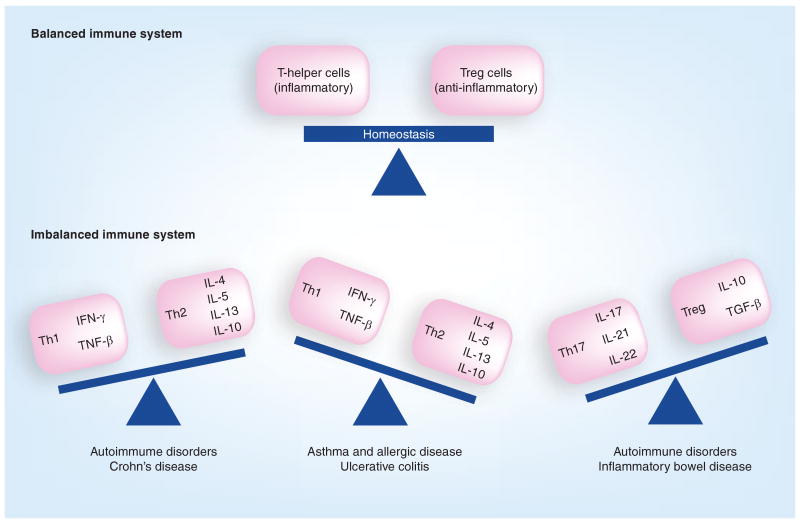
Diseases & disorders due to aberrations in the human gut microbiome
As discussed earlier, development of the GI microbiota over the first year of life appears to be intimately linked to subsequent disease susceptibility. Aberrations in the adult gut microbiota have also been associated with a number of diseases and disorders including allergic disease development [73,75], colon cancer [14], and even progression and severity of HIV (Table 1) [108]. Disruption of the gut microbiome, termed dysbiosis [18], is frequently accompanied by overgrowth of pathogenic bacteria or fungi, in conjunction with significant loss of microbial diversity or key functional groups [9,14,109–123] and an inflammatory response by the host [9,39,115,121,123–125], which contributes to disease development [108]. Dysbiosis has been associated with an imbalance between populations of inflammation-mediating T-helper cells (Th1, Th2 and Th17) and anti-inflammatory Treg cells. Prolonged overproduction of Th1- and Th17-associated cytokines has been linked with IBD (overproduction of Th1 for Crohn's disease [CD] [126] and Th17 for both CD and ulcerative colitis [UC] [38,127]) and autoimmune disorders such as lupus, multiple sclerosis, psoriasis and rheumatoid arthritis, while a Th2 skew is linked with asthma, allergic disorders and UC (Figure 2) [39,128,129]. Such chronic inflammatory responses set up a ‘vicious circle’, disrupting the GI microbiota [115], eliminating subsets of beneficial bacteria and permitting opportunistic colonizers, typically pathogens, to compete in this niche [115] and maintain a persistent inflammatory state. Members of the Enterobacteriaceae, which includes Salmonella enterica serovar Typhimurium, a bacterium linked to gastroenteritis [109,115,121], appear to use this strategy. In an attempt to eliminate the pathogen, the host response disrupts the native gut microbiota, providing the pathogen with the opportunity to proliferate [109], emphasizing the key role microbiome homeostasis plays in protection against pathogen overgrowth.
Table 1. Diseases and disorders associated with human gut microbiome aberrations.
| Disease/disorder | Potential role of the microbiome | Recent findings (2004–present) |
|---|---|---|
| Atopy and asthma |
|
|
| Candida infection |
|
|
| Celiac disease |
|
|
| CC |
|
|
| Type I diabetes |
|
|
| Type II diabetes |
|
|
| HIV |
|
|
| IBD |
Crohn's disease (IBDC)
|
|
| IBS |
|
|
| Gastroenteritis |
|
|
| NEC |
|
|
| Obesity |
|
|
| Rheumatoid arthritis |
|
ADP: Asymptomatic diabetes prone; CC: Colon cancer; DGGE: Density gradient gel electrophoresis; DP: Diabetes-prone; DR: Diabetes-resistant; GI: Gastrointestinal; IBD: Inflammatory bowel disease; IBDC: Irritable bowel disease–Crohn's disease; IBDU: Irritable bowel disease–ulcerative colitis; IBS: Irritable bowel syndrome; NEC: Necrotizing enterocolitis.
Figure 2. Consequences of immune system imbalances.
Immune system homeostasis involves a regulated balance of inflammatory and anti-inflammatory cells; however, skewed responses putatively through lack of microbial exposure or outgrowth of pathogenic species may contribute to imbalances associated with the development of chronic inflammatory disorders.
Many of the diseases and disorders associated with adult gut microbiome dysbiosis exhibit an overall reduction of bacterial diversity (which may, in part, be attributed to disease-related diarrhea) and appears to be a characteristic shared by the majority of chronic inflammatory diseases (Table 1). To date, only colon cancer patients exhibit a significantly higher microbial diversity compared with that of healthy individuals; however, these assemblages exhibit dramatic temporal instability [14], an emerging characteristic of chronic inflammatory diseases [25,26]. Scanlan et al. emphasized that although the microbial diversity was increased in colon cancer patients, it was unclear which species proliferated, and suggested that the community may be composed of a multitude of organisms capable of toxin production that outcompete the native beneficial microbes [14].
It is beyond the scope of this review article to present an in depth discussion of all of the diseases and disorders associated with gut microbiome dysbiosis. Instead we have presented a summary in Table 1 and focus here on one, IBD, which encompasses many of the issues associated with chronic inflammatory diseases. Characterized by chronic or recurrent inflammation of the GI mucosa [130], the GI tract of the IBD patient appears unable to reprogram towards a noninflammatory state even after depletion of the primary pathogen [131]. This suggests that low levels of certain microbial species maintain a proinflammatory state, the microbiome is irreversibly altered and incapable of appropriate reassembly, or a combination of these factors contribute to this chronic inflammatory state. Interestingly, a recent study has proposed a role for the appendix as a ‘safe-house’ for commensal bacterial species, which exist in protected biofilms that may detach and serve to repopulate the GI tract following perturbation of the natural microbiome [132]. Although it has previously been suggested that there are no discernable long-term effects of appendectomies in westernized countries [132], appendectomy has been associated with progression and severity of IBD [133]. The impact of appendectomy (or indeed appendix colonization by inappropriate microbial species) on the ability to ‘re-seed’ the GI microbiome, facilitating community reassembly following perturbation, particularly in patients with inflammatory GI disorders, is an intriguing concept that has not been comprehensively studied.
Crohn's disease and UC, the two main disorders associated with IBD, typically require care for the length of the patient's life. Patients with CD suffer from patchy inflammation that may occur anywhere along the GI tract and is characterized by large transmural ulcerations [134]. UC patients typically possess more superficial ulcerations restricted to the colonic mucosa and inflammation that characteristically extends continuously from the rectum throughout the large intestine [134]. Like other chronic inflammatory disorders, there are several risk genes (IL23R, IL12B, HLA, NKX2–3 and MST1) associated with increased susceptibility to both CD and UC [135]. Specific additional genetic components exclusive to CD such as NOD2/CARD15, FcGRIIIA, ATG16L1 and IRGM [136–141] have been identified, however, risk genes exclusive to UC remain poorly defined [138,142–145]. Several environmental factors have been shown to impact progression and severity of the disease, including smoking, diet, drugs, stress, appendectomy, geography and social status [133]. While aberrations in the gut microbiota have been associated with IBD [131], its etiology and whether the associated dysbiosis is a cause or result, remains to be determined [146,147]. Nonetheless, patients with IBD typically exhibit lower GI bacterial diversity compared with healthy individuals [5,9,14], an increase in E. coli [123], and lower counts of specific Firmicutes [9,112] and Bacteroides species [111,118,148]. However it should be noted that some species of Bacteroides have been shown to increase in relative abundance in CD patients [5], suggesting that specific functions associated with particular species or strains may contribute to the niche-specific pathophysiology of UC and CD.
At a functional level, the butyrate-producers, which belong to the Clostridium leptum group of the Firmicutes, are less abundant in the GI microbiome of IBD patients. Butyrate is an essential regulator of gene expression, inflammation, differentiation and apoptosis, and is a major energy source for the mucosa-associated microbial community [149–151]. Faecalibacterium prausnitzii, a butyrate-producing bacterium with anti-inflammatory properties [122,123,147], is significantly reduced in abundance in the ileum of CD patients [111,116,123,152] in parallel with increased numbers of E. coli [123]. This emphasizes the key role a number of distinct bacterial species play in maintaining inflammatory homeostasis and that loss of key functional organisms leads to pathogen overgrowth associated with chronic inflammatory illness.
To further determine the basis for host health and disease, several groups have turned to high-resolution functional profiling of microbial communities. Metabolomic studies are becoming more prevalent in dysbiosis-related diseases; several metabolic pathways implicated in CD have now been identified [153]. In addition to better understanding the pathogenesis of this disease, these pathways are also useful for differentiating between patients and healthy subjects. A total of 2155 discriminating phenotypes for ileal-specific CD, 3113 for colon-specific CD and 2650 that were characteristic of healthy individuals were identified in this study [153]. An independent study using metabolic profiling demonstrated lower levels of butyrate, methylamine and trimethylamine in CD patients in comparison to UC patients [10]. In addition, elevated levels of amino acids in fecal samples were detected in IBD patients, which implicated malabsorption (presumably a consequence of mucosal inflammation), as an additional component of the disease [10]. In a separate functional profiling study, in vitro high-resolution proton magnetic resonance spectroscopy was used to measure levels of amino acids in the colonic mucosa of patients with active or inactive IBD and demonstrated that decreased amino acid levels were associated with reduced carbohydrate and protein metabolism [154]. Probably due to a shift in GI microbiota composition, this aspect of active IBD may result in lower energy and altered mucosal integrity. In comparison, samples from inactive IBD patients exhibited comparable amino acid concentrations with healthy controls [154]. These studies serve to highlight the complexity of IBD and the key role of microbial communities, functional gene expression and host responses in inflammatory disease exacerbation and remission.
Manipulation of the GI microbiome: implications for host health
Antibiotic administration
While antibiotics have revolutionized our ability to combat infectious diseases, their widespread use has led to a dramatic rise in the prevalence of antibiotic-resistant microbes, and recent concerns have been raised regarding the potential for adverse effects on host microbiota. In the process of eliminating the pathogenic agent, antibiotic administration dramatically impacts the native microbial community, leading to an unintentional state of dysbiosis [18]. The impact of these therapeutics appears to be more pronounced in infants 1 year of age or less, likely due to their influence on microbial colonization of the gut during the initial stages of immune response development (discussed earlier). A study of clamoxyl administration to mice demonstrated altered microbial colonization of the gut with the near depletion of Lactobacillus species coinciding with a reduction in the total aerobic and anaerobic bacteria, notably Enterobacteriaceae spp. and Enterococcus spp. [15]. In addition, administration of the antibiotic downregulated expression of genes involved in antigen presentation and the innate immune host defense [15], presumably because of changes in the microbial colonization pattern of these animals. Palmer and colleagues demonstrated dramatic decreases in GI bacterial community diversity upon the administration of antibiotics to infants in the first year of life [11], which has specifically been associated (in a separate study) with lower numbers of Bifidobacteria and Bacteroides [41]. These two bacterial genera appear to be critical for immune development, and their loss has been associated with the development of allergic disease, an aspect attributed to the antibiotic's effect on the gut microbiota [155].
Another aspect of gut microbiome manipulation by antibiotics is antibiotic-associated diarrhea (AAD), which affects 11% of children in the USA, is more frequent in those who are less than 2 years of age and appears to be dependent on the type of antibiotic received [156]. Surawicz reported that the administration of broad-spectrum antibiotics that target anaerobic bacteria is associated with a higher risk of AAD development [157]. Interestingly, a recent study examining factors predictive of development of Clostridium difficile-associated diarrhea (CDAD), including gut microbiome profiles, found that the resident microbiota and not the class of antibiotic administered provided the highest predictive power for CDAD development [4]. Despite the dramatic impact antibiotic administration has on the gut microbiota, the community in healthy adults appears to be relatively resilient; once administration of antibiotics has ceased, the gut microbiome largely returns to a pretherapy consortium after approximately 4 weeks [19]. However, subtle persistent changes were evident – a number of bacterial species had not returned to preantibiotic levels 6 months post-treatment [19]. The impact of the loss of these species on host health and the potential for long-term issues are currently unknown [19]. In a murine study, following treatment with the antibiotic enrofloxacin, changes in three key functions were associated with depletion of the bacterial gut community: loss of acetate due to reduced microbial metabolism of sugars and polysaccharides; decreased trimethylamine-N-oxide due to deficient microbial catabolism of choline (also associated with CD [10]); and an increase in creatine due to a lack of microbial enzyme degradation [13]. In addition, a loss of amino acids produced by microbial proteases, the reduction of metabolites from lactate-utilizing microbes and an increase in urea from the depletion of microbial ureases were also detected [13]. Therefore, although the microbial community may appear to return to a community that largely resembles the pretreatment consortium, the possibility remains that the impact of the loss of even a small number of specific microbial species may be more profound at the functional level. Determining the impact of such subtle alterations necessitates the use of high-resolution microbial community profiling tools, such as phylogenetic microarrays, high-throughput sequencing and metabolomic or metaproteomic approaches to determine the microbial community structure, function and host response.
Microbial/nutritional manipulation of the GI microbiome
Owing to the increased prevalence of diseases and disorders associated with gut microbiota imbalances (Table 1) and the fact that traditional treatments such as antibiotic administration appear to have the potential for long-term disruption, microbial manipulation of the host microbiome to treat chronic diseases has become the focus of recent renewed interest. Manipulation may be elicited through pro-, pre- or synbiotics. First suggested by Metchnikoff in 1907 [158], probiotic therapy represents alteration of the gut microbiota by supplementation with live microorganisms that function to inhibit pathogen adherence to the mucosa [159,160], improve the intestinal epithelial and mucosal barrier function [161], produce bacteriocins [162,163], increase IgA production [164] and downregulate proinflammatory cytokine secretion [165–167]. Prebiotics, originally defined as nondigestible food ingredients that improve host health by stimulating the growth or activity of colonic bacteria [168], have now been reclassified to include components that are resistant to gastric acidity, hydrolysis by host enzymes and absorption by the upper gastrointestine, are fermented by the gut microbiota, and stimulate growth of microbial species beneficial to the host's health [169]. Finally, synbiotics are supplements composed of a combination of both probiotics and prebiotics [168]. An exhaustive review of the use of probiotics, prebiotics and synbiotics is beyond the scope of this review; instead, we refer the reader to other excellent recent reviews on this topic [170,171] and focus on their use in a limited number of cases to highlight efficacy and concerns associated with these therapies.
Recent studies have demonstrated a direct link between the development of IBD and GI bacteria [172,173], and probiotic therapies have become a focus for various GI dysbiosis-based diseases. Successful use of probiotics for treatment of IBD appears to be dependent on the bacterial strain used, the stage of disease progression and the type of pathology used for analysis (Figure 3) [174]. Probiotics appear to be more successful for treatment of UC than CD, however, probiotic intervention studies of CD patients have typically been small and few have been randomized double-blind controlled trials [175,176], making it difficult to definitively characterize probiotic efficacy in this patient population. However, there may be a physiological basis for enhanced probiotic efficacy in UC patients. For example, defensins are typically downregulated in UC patients, a feature that can be ameliorated by probiotic species such as E. coli Nissle 1917 [177], but cannot be corrected in defensin-defective CD patients. In addition, the ability of probiotic species to colonize specific niches along the GI tract may be variable due to physiologically distinct features such as mucus coverage and the density of antimicrobial peptide producing Peyer's patches at specific sites, potentially impacting their efficacy in UC and CD patient populations.
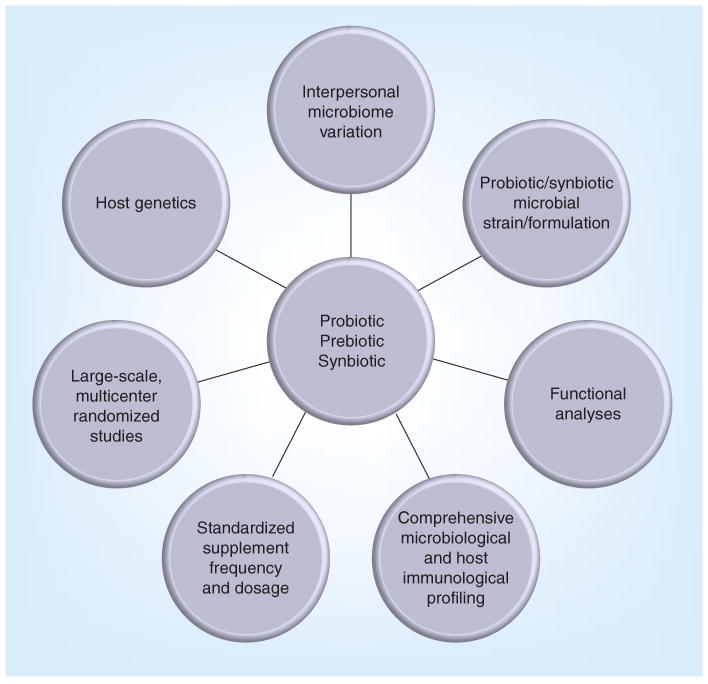
Figure 3. Factors critical for successful probiotic, prebiotic and synbiotic trials.
Consideration of interpersonal genetic and microbiome variations, standardized trials with defined outcomes and characterized strains coupled with high-resolution microbial phylogeny, microbiome functional profiles and host responses, are necessary to fully understand the potential impact of microbiome manipulation through probiotic, prebiotic or symbiotic supplementation.
A clinical trial testing the efficacy of probiotic preparation VSL#3 (a mix of eight probiotic bacterial species) on patients with mild to moderately active UC, demonstrated the probiotic treatment induced remission in significantly more patients than those treated with mesalazine (5-aminosalicylic acid; standard of care [178]). Furthermore, 53% of patients who exhibited persistent symptoms despite receiving mesalazine or corticosteroids for their UC exhibited remission of symptoms following VSL#3 administration [179]. Bibiloni and colleagues also reported that the bacterial species in the VSL#3 cocktail could be detected specifically at the site of inflammation by molecular methods [179], suggesting a direct role for the probiotic species in mucosal colonization and modulation of the local immune response. A separate study reported that patients receiving VSL#3 had a significant reduction in UC symptom severity following 6 weeks of daily supplementation and that more patients were in remission after 12 weeks compared with the placebo group [180]. A pilot study of UC patients receiving a Saccharomyces boulardii-based probiotic demonstrated that 68% of the patients receiving the fungal supplement reported remission of symptoms [181]. This probiotic also appears to be promising for individuals with CD, where it has been shown to induce remission in significantly more patients compared with those treated with mesalazine [182]. Probiotics also appear to be promising for preventing and diminishing severity of necrotizing enterocolitis (NEC) in neonates [183–189]. However, these trials have been criticized because the timing, dosage and type of organisms were not optimal [184,186,187] and there were insufficient data to examine the long- and short-term effects [184,186,189].
Insights into how probiotic supplementation may impact the gut microbiome has come from a recent study of infants in the Trial of Infant Probiotic Supplementation (TIPS) study, in which infants at high risk for asthma development are fed a probiotic species (Lactobacillus casei Rhamnosus GG [LGG]) or placebo [190]. The bacterial community composition of stool samples from a subset of infants in this trial were examined using a high-resolution microarray, the 16S rRNA PhyloChip (Affymetrix, CA, USA) [191], which can detect approximately 8500 bacterial taxa (defined as strains or species that share ≥97% 16S rRNA sequence homology) in a single assay. Although the investigators remained blinded to the identity of the samples, clear differences in the abundance of LGG were evident and analysis of the data demonstrated that a high abundance of the probiotic species was associated with a distinct bacterial community in which many other known beneficial species, including members of the Lactobacillaceae and Bifidobacteriaceae, were also present in high abundance [190]. These communities were composed of species that were phylogenetically related, suggesting a high level of functional redundancy, a characteristic previously associated with stable microbial assemblages resistant to pathogen outgrowth [2,16]. By contrast, communities with low levels of LGG tended to possess a more variable community composition and high abundances of species previously associated with allergic disease development. Hence, it appears that the beneficial effects of probiotic supplementation are not simply due to a high abundance of the probiotic species itself, but rather to a restructuring of the gut microbiome toward a commensal rich, functionally redundant consortium. Although not examined in this study, it is likely (based on previous studies [13,16]) that this large shift in community composition underlies altered functional gene expression and impacts host immune response, a factor which may impact asthma development, one of the primary outcomes being measured in this trial.
Federal regulation has recently been mandated for probiotic supplements because some preparations do not comprehensively list the species present or contain additional species that are potentially pathogenic [186], an aspect that has led to supplement variation in trials [184]. Although it must be stressed that adverse events involving probiotic supplements are rare, these precautions are being implemented because their use has been linked to higher mortality rates for ICU patients with pancreatitis [192] and Lactobacillus sepsis in immunocompromised patients [193,194]. Negative effects have not been found in NEC probiotic trials, but this is probably because the power of these trials is too small to detect side effects [189].
Prebiotics also represent a promising approach for the management of inflammatory diseases. They have the advantage of promoting subsets of existing native GI bacterial community members (e.g., Bifidobacteria) capable of degrading them and can increase production of important anti-inflammatory compounds, such as butyrate, without the conventional caveats of probiotic competitiveness or colonization efficiency. For example, oligofructose (enzymatic product of hydrolyzed inulin) was found to increase butyrate production in both dextran sodium sulfate (DSS) mice and humans [195,196], as well as limiting murine DSS damage and accelerating the healing process [195]. Although the mechanisms remain unclear, prebiotic germinated barley foodstuff has also been shown to significantly decrease IFN-γ expression [197]. IFN-γ is known to induce colitis and the proinflammatory cytokine IL-6, and increase TGF-β production (excess TGF-β has specifically been associated with colitis pathology [197]). Thus, while there are clear functional effects of feeding prebiotic compounds, their mechanism of action, which probably involves structural and functional shifts in the GI microbiota due to the selective pressure of supplementation with complex carbohydrate nutritional sources, remains to be fully elucidated.
Use of synbiotic supplementation is still in its infancy for GI dysbiosis, but some promising signs are starting to emerge in colon cancer treatment. Synbiotic therapy enhanced apoptosis in carcinogen-damaged cells and to genotoxic carcinogens in animal models [198,199] and reduced the number of tumor and lesion occurrences [199–201] by moderating Peyer's patch immune functions [200]. In humans, Rafter et al. reported that there were significant differences in the levels of Bifidobacterium species in fecal samples from synbiotic-treated colon cancer patients, but they did not find any effect on apoptosis, colonic inflammation or tumor cell invasion [202]. Synbiotic approaches for IBD include a supplement and a fungal probiotic therapy. A pilot study using a combination of B. longum and fructo-oligosaccharide/inulin found that those subjects receiving the synbiotic had significantly reduced inflammation compared with the placebo-treated group [203]. Synbiotics also appear to be promising in the treatment of CD; a supplement of B. breve, B. longum and L. casei, and the prebiotic psyllium was examined in ten CD patients who were not responding to standard therapies [204]. Although patients were permitted to choose when and how much of the synbiotic they self-administered, six of the patients exhibited remission of symptoms with substantially reduced CD activity [204].
Aside from genetic and environmental factors, given the broad interpersonal variability in the human host GI microbiome and the capacity for a wide variety of potential perturbations to this ecosystem, it is not surprising that a pre-, pro- or synbiotic therapy that demonstrates efficacy in one patient population may not exhibit the same effect in another. Although initial results are promising, randomized, large-scale studies with well-characterized patient populations who are supplemented with controlled dosages of specific well-characterized supplements, and whose samples are examined using high-resolution microbiome composition and functional analyses, are necessary to fully appreciate the therapeutic effects of GI microbiota manipulation (Figure 3). In the current era where patient-tailored care is being touted as the future of medicine, understanding the complex interactions between microbes and immune response that underpin host health is fundamental to truly providing such a level of care.
The overall indication is that gut microbial communities play a key role in the development of inflammatory disease and that these communities are amenable to manipulation to prevent or abrogate disease development. As the GI microbiome and specific pathways responsible for improved host health continue to be identified with newly developed high-resolution culture-independent tools, our understanding of the delicate balance between the human host and its microbial inhabitants continues to unfold. With these insights, novel therapies may be developed to restore or promote beneficial microbial communities to help combat a myriad of chronic diseases.
Expert commentary
From the influence of prenatal and early postnatal microbial exposures on the developing immune response to gut microbiome aberrations in adulthood associated with chronic inflammatory diseases, recent studies of the human microbiome have revealed the complexity of our dynamic relationship with microbes. Relatively recent changes in lifestyle, diet and the use of antimicrobials are just some of the factors implicated in increased prevalence of a range of inflammatory disorders that have a demonstrated basis in altered GI microbial community structure and function.
Five-year view
Given the cumulative data and, in particular, the recent microbiome studies, the maintenance of a diverse, functionally redundant microbial community encoding a core set of functional genes appears key to human host health. Further mouse and human studies of microbial community composition in parallel with immunophenotyping and long-term outcome measures, in well-defined cohorts, are necessary to fully understand the role of the human microbiome in defining states of health and disease. Given the microbiome initiatives underway globally and the increasingly sophisticated tools to interrogate samples, a more comprehensive understanding of the impact of therapeutics and other microbiome-manipulating agents and their long-term effect on host health will be better defined. In parallel, the development of culture-independent diagnostics based on these findings and using these new profiling approaches will vastly improve diagnosis and permit surveillance of at-risk populations.
Key issues
The gastrointestinal microbiome is associated with host health status.
Structure and composition of the microbiome defines functional gene expression of the community, pathogen abundance and physiology, and the host response.
Prenatal and early postnatal microbial exposures impact immune response development and define predisposition to the development of inflammatory diseases.
Specific microbes have demonstrated roles in immune response modulation.
Manipulation of the microbiome through pro-, pre- or synbiotic supplementation may prove an alternative approach for improving host health status.
Acknowledgments
Susan V Lynch is supported by NIH awards AI075410, HL098964, T004732 and DK083990, and by the Rainin Foundation. Michael D Cabana is supported by NIH awards A112997, AT004732 and HL080074. NA Slusher is supported by a MBRS RISE award 5R25GM-059298.
Footnotes
For reprint orders, please contact reprints@expert-reviews.com
Financial & competing interests disclosure: The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Contributor Information
Kei E Fujimura, Colitis and Crohn's Disease Center, Gastroenterology Division, Department of Medicine, University of California, San Francisco, San Francisco, CA 94143, USA.
Nicole A Slusher, Colitis and Crohn's Disease Center, Gastroenterology Division, Department of Medicine, University of California, San Francisco, San Francisco, CA 94143, USA.
Michael D Cabana, Division of General Pediatrics, Department of Pediatrics, University of California, San Francisco, 505 Parnassus Avenue, San Francisco, CA 94143, USA.
Susan V Lynch, Email: susan.lynch@ucsf.edu, Colitis and Crohn's Disease Center, Gastroenterology Division, Department of Medicine, University of California, San Francisco, 513 Parnassus Ave., Box 0538, San Francisco, CA 94143, USA, Tel.: +1 415 476 6784.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA. 2007;104(3):979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host–bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 3.Blaut M, Clavel T. Metabolic diversity of the intestinal microbiota: implications for health and disease. J Nutr. 2007;137(3):751S–755S. doi: 10.1093/jn/137.3.751S. [DOI] [PubMed] [Google Scholar]
- 4.De La Cochetière M, Durand T, Lalande V, Petit J, Potel G, Beaugerie L. Effect of antibiotic therapy on human fecal microbiota and the relation to the development of Clostridium difficile. Microb Ecol. 2008;56(3):395–402. doi: 10.1007/s00248-007-9356-5. [DOI] [PubMed] [Google Scholar]
- 5.Dicksved J, Halfvarson J, Rosenquist M, et al. Molecular analysis of the gut microbiota of identical twins with Crohn's disease. ISME J. 2008;2(7):716–727. doi: 10.1038/ismej.2008.37. [DOI] [PubMed] [Google Scholar]; • Genetics, environmental exposure during development of the gut microbial composition and shifts in the gut microbiota are interwoven factors that govern the occurrence of Crohn's disease.
- 6.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102(31):11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 8.Li M, Wang B, Zhang M, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci USA. 2008;105(6):2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55(2):205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marchesi JR, Holmes E, Khan F, et al. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J Proteome Res. 2007;6(2):546–551. doi: 10.1021/pr060470d. [DOI] [PubMed] [Google Scholar]
- 11.Palmer C, Bik EM, Digiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5(7):e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Comprehensively demonstrated temporal development of the infant gut microbiota and convergence toward an adult-like microbial community by the end of the first year of life. Twins were more similar temporally, suggesting that environmental exposure is critical in gut microbiota development.
- 12.Roesch LFW, Lorca GL, Casella G, et al. Culture-independent identification of gut bacteria correlated with the onset of diabetes in a rat model. ISME J. 2009;3(5):536–548. doi: 10.1038/ismej.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romick-Rosendale LE, Goodpaster AM, Hanwright PJ, et al. NMR-based metabonomics analysis of mouse urine and fecal extracts following oral treatment with the broad-spectrum antibiotic enrofloxacin (Baytril) Magn Reson Chem. 2009;47(S1):S36–S46. doi: 10.1002/mrc.2511. [DOI] [PubMed] [Google Scholar]
- 14.Scanlan PD, Shanahan F, Clune Y, et al. Culture-independent analysis of the gut microbiota in colorectal cancer and polyposis. Environ Microbiol. 2008;10(3):789–798. doi: 10.1111/j.1462-2920.2007.01503.x. [DOI] [PubMed] [Google Scholar]
- 15.Schumann A, Nutten S, Donnicola D, et al. Neonatal antibiotic treatment alters gastrointestinal tract developmental gene expression and intestinal barrier transcriptome. Physiol Genomics. 2005;23(2):235–245. doi: 10.1152/physiolgenomics.00057.2005. [DOI] [PubMed] [Google Scholar]
- 16.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Gut microbial community composition is highly variable and possibly unique to individuals, but a core set of functional genes are fundamental to human health. Deviation from this functional core was associated with diseases such as obesity.
- 17.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 18.Collins S, Denou E, Verdu E, Bercik P. The putative role of the intestinal microbiota in the irritable bowel syndrome. Dig Liver Dis. 2009;41(12):850–853. doi: 10.1016/j.dld.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 19.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6(11):e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Culture-independent analysis of how antibiotic administration reduced intestinal microbial diversity, which also demonstrated a rapid return to the pretreatment microbiota following cessation of treatment, underscoring the resiliency of the adult gut microbiota to perturbations.
- 20.Harris JK, De Groote MA, Sagel SD, et al. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc Natl Acad Sci USA. 2007;104(51):20529–20533. doi: 10.1073/pnas.0709804104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyman RW, Fukushima M, Diamond L, Kumm J, Giudice LC, Davis RW. Microbes on the human vaginal epithelium. Proc Natl Acad Sci USA. 2005;102(22):7952–7957. doi: 10.1073/pnas.0503236102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sibley CD, Parkins MD, Rabin HR, Duan K, Norgaard JC, Surette MG. A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proc Natl Acad Sci USA. 2008;105(39):15070–15075. doi: 10.1073/pnas.0804326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yi S, Leiqiong C, Jianqian T, Chen X. Preliminary characterization of vaginal microbiota in healthy Chinese women using cultivation-independent methods. J Obstet Gynaecol Res. 2009;35(3):525–532. doi: 10.1111/j.1447-0756.2008.00971.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhou X, Brown CJ, Abdo Z, et al. Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J. 2007;1(2):121–133. doi: 10.1038/ismej.2007.12. [DOI] [PubMed] [Google Scholar]
- 25.Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J Clin Microbiol. 2006;44(10):3665–3673. doi: 10.1128/JCM.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakamoto M, Huang Y, Ohnishi M, Umeda M, Ishikawa I, Benno Y. Changes in oral microbial profiles after periodontal treatment as determined by molecular analysis of 16S rRNA genes. J Med Microbiol. 2004;53(Pt 6):563–571. doi: 10.1099/jmm.0.45576-0. [DOI] [PubMed] [Google Scholar]
- 27.Jenzsch A, Eick S, Rassoul F, Purschwitz R, Jentsch H. Nutritional intervention in patients with periodontal disease: clinical, immunological and microbiological variables during 12 months. Br J Nutr. 2009;101(6):879–885. doi: 10.1017/S0007114508047776. [DOI] [PubMed] [Google Scholar]
- 28.Branch-Mays GL, Dawson DR, Gunsolley JC, et al. The effects of a calorie-reduced diet on periodontal inflammation and disease in a non-human primate model. J Periodontol. 2008;79(7):1184–1191. doi: 10.1902/jop.2008.070629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reynolds MA, Dawson DR, Novak KF, et al. Effects of caloric restriction on inflammatory periodontal disease. Nutrition. 2009;25(1):88–97. doi: 10.1016/j.nut.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cani PD, Amar J, Iglesias MA, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56(7):1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 31.Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137(5):1716–1724. e1–2. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3(4):213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nadal I, Santacruz A, Marcos A, et al. Shifts in clostridia, bacteroides and immunoglobulin-coating fecal bacteria associated with weight loss in obese adolescents. Int J Obes. 2008;33(7):758–767. doi: 10.1038/ijo.2008.260. [DOI] [PubMed] [Google Scholar]
- 34.Dong Y, Manfredini F, Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009;5(5):e1000423. doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawley TD, Bouley DM, Hoy YE, Gerke C, Relman DA, Monack DM. Host transmission of Salmonella enterica serovar typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect Immun. 2008;76(1):403–416. doi: 10.1128/IAI.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawley TD, Clare S, Walker AW, et al. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect Immun. 2009;77(9):3661–3669. doi: 10.1128/IAI.00558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobayashi T, Okamoto S, Hisamatsu T, et al. IL23 differentially regulates the Th1/Th17 balance in ulcerative colitis and Crohn's disease. Gut. 2008;57(12):1682–1689. doi: 10.1136/gut.2007.135053. [DOI] [PubMed] [Google Scholar]
- 39.Chow J, Mazmanian SK. Getting the bugs out of the immune system: do bacterial microbiota “fix” intestinal T cell responses? Cell Host Microbe. 2009;5(1):8–12. doi: 10.1016/j.chom.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 40.Ege MJ, Bieli C, Frei R, et al. Prenatal farm exposure is related to the expression of receptors of the innate immunity and to atopic sensitization in school-age children. J Allergy Clin Immunol. 2006;117(4):817–823. doi: 10.1016/j.jaci.2005.12.1307. [DOI] [PubMed] [Google Scholar]
- 41.Penders J, Thijs C, Vink C, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118(2):511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 42.Wegienka G, Havstad S, Zoratti EM, et al. Regulatory T cells in prenatal blood samples: variability with pet exposure and sensitization. J Reprod Immunol. 2009;81(1):74–81. doi: 10.1016/j.jri.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaub B, Liu J, Höppler S, et al. Maternal farm exposure modulates neonatal immune mechanisms through regulatory T cells. J Allergy Clin Immunol. 2009;123(4):774–782. doi: 10.1016/j.jaci.2009.01.056. [DOI] [PubMed] [Google Scholar]
- 44.Debarry J, Garn H, Hanuszkiewicz A, et al. Acinetobacter lwoffii and Lactococcus lactis strains isolated from farm cowsheds possess strong allergy-protective properties. J Allergy Clin Immunol. 2007;119(6):1514–1521. doi: 10.1016/j.jaci.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 45.de Meer G, Janssen NAH, Brunekreef B. Early childhood environment related to microbial exposure and the occurrence of atopic disease at school age. Allergy. 2005;60(5):619–625. doi: 10.1111/j.1398-9995.2005.00746.x. [DOI] [PubMed] [Google Scholar]
- 46.Duramad P, Harley K, Lipsett M, et al. Early environmental exposures and intracellular Th1/Th2 cytokine profiles in 24-month-old children living in an agricultural area. Environ Health Perspect. 2006;114(12):1916–1922. doi: 10.1289/ehp.9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aichbhaumik N, Zoratti EM, Strickler R, et al. Prenatal exposure to household pets influences fetal immunoglobulin E production. Clin Exp Allergy. 2008;38(11):1787–1794. doi: 10.1111/j.1365-2222.2008.03079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prescott SL, Macaubas C, Holt BJ, et al. Transplacental priming of the human immune system to environmental allergens: universal skewing of initial T cell responses toward the Th2 cytokine profile. J Immunol. 1998;160(10):4730–4737. [PubMed] [Google Scholar]
- 49.Bergmann RL, Edenharter G, Bergmann KE, et al. Predictability of early atopy by cord blood-IgE and parental history. Clin Exp Allergy. 1997;27:752–760. [PubMed] [Google Scholar]
- 50.Illi S, von Mutius E, Lau S, et al. The pattern of atopic sensitization is associated with the development of asthma in childhood. J Allergy Clin Immunol. 2001;108(5):709–714. doi: 10.1067/mai.2001.118786. [DOI] [PubMed] [Google Scholar]
- 51.Tariq SM, Arshad SH, Matthews SM, Hakim EA. Elevated cord serum IgE increases the risk of aeroallergen sensitization without increasing respiratory allergic symptoms in early childhood. Clin Exp Allergy. 1999;29(8):1042–1048. doi: 10.1046/j.1365-2222.1999.00594.x. [DOI] [PubMed] [Google Scholar]
- 52.McKeever TM, Lewis SA, Smith C, Hubbard R. The importance of prenatal exposures on the development of allergic disease: a birth cohort study using the West Midlands general practice database. Am J Respir Crit Care Med. 2002;166(6):827–832. doi: 10.1164/rccm.200202-158OC. [DOI] [PubMed] [Google Scholar]
- 53.Han YW, Shen T, Chung P, Buhimschi IA, Buhimschi CS. Uncultivated bacteria as etiologic agents of intra-amniotic inflammation leading to preterm birth. J Clin Microbiol. 2009;47(1):38–47. doi: 10.1128/JCM.01206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DiGiulio DB, Romero R, Amogan HP, et al. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One. 2008;3(8):e3056. doi: 10.1371/journal.pone.0003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Conly JM, Stein K, Worobetz L, Rutledge-Harding S. The contribution of vitamin K2 (menaquinones) produced by the intestinal microflora to human nutritional requirements for vitamin K. Am J Gastroenterol. 1994;89(6):915–923. [PubMed] [Google Scholar]
- 56.Hill MJ. Intestinal flora and endogenous vitamin synthesis. Eur J Cancer Prev. 1997;6(2):S43–S45. doi: 10.1097/00008469-199703001-00009. [DOI] [PubMed] [Google Scholar]
- 57.Xu J, Bjursell MK, Himrod J, et al. A genomic view of the human–Bacteroides thetaiotaomicron symbiosis. Science. 2003;299(5615):2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 58.Michelsen KS, Arditi M. Toll-like receptors and innate immunity in gut homeostasis and pathology. Curr Opin Hematol. 2007;14(1):48–54. doi: 10.1097/00062752-200701000-00010. [DOI] [PubMed] [Google Scholar]
- 59.Rakoff-Nahoum S, Medzhitov R. Innate immune recognition of the indigenous microbial flora. Muc Immunol. 2008;1(Suppl. 1):S10–S14. doi: 10.1038/mi.2008.49. [DOI] [PubMed] [Google Scholar]
- 60.Kett K, Baklien K, Bakken A, Kral JG, Fausa O, Brandtzaeg P. Intestinal B-cell isotype response in relation to local bacterial load: evidence for immunoglobulin A subclass adaptation. Gastroenterology. 1995;109(3):819–825. doi: 10.1016/0016-5085(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 61.Apperloo-Renkema HZ, Jagt TG, Tonk RH, van der Waaij D. Healthy individuals possess circulating antibodies against their indigenous faecal microflora as well as against allogenous faecal microflora: an immunomorphometrical study. Epidemiol Infect. 1993;111(2):273–285. doi: 10.1017/s0950268800056983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kimura K, McCartney AL, McConnell MA, Tannock GW. Analysis of fecal populations of bifidobacteria and lactobacilli and investigation of the immunological responses of their human hosts to the predominant strains. Appl Environ Microbiol. 1997;63(9):3394–3398. doi: 10.1128/aem.63.9.3394-3398.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kawai T, Akira S. Pathogen recognition with Toll-like receptors. Curr Opin Immunol. 2005;17(4):338–344. doi: 10.1016/j.coi.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 64.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell. 2004;118(2):229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 65.Butler JE, Weber P, Sinkora M, et al. Antibody repertoire development in fetal and neonatal piglets VIII Colonization is required for newborn piglets to make serum antibodies to T-dependent and type 2 T-independent antigens. J Immunol. 2002;169(12):6822–6830. doi: 10.4049/jimmunol.169.12.6822. [DOI] [PubMed] [Google Scholar]
- 66.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313(5790):1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hazebrouck S, Przybylski-Nicaise L, Ah-Leung S, et al. Allergic sensitization to bovine β-lactoglobulin: comparison between germ-free and conventional BALB/c mice. Int Arch Allergy Immunol. 2009;148(1):65–72. doi: 10.1159/000151507. [DOI] [PubMed] [Google Scholar]
- 68.Hrncir T, Stepankova R, Kozakova H, Hudcovic T, Tlaskalova-Hogenova H. Gut microbiota and lipopolysaccharide content of the diet influence development of regulatory T cells: studies in germ-free mice. BMC Immunol. 2008;9:65. doi: 10.1186/1471-2172-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288(5474):2222–2226. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- 70.Wang Y, McCusker C. Neonatal exposure with LPS and/or allergen prevents experimental allergic airways disease: development of tolerance using environmental antigens. J Allergy Clin Immunol. 2006;118(1):143–151. doi: 10.1016/j.jaci.2006.03.020. [DOI] [PubMed] [Google Scholar]; • Demonstrated that neonatal exposure to appropriate allergens may reduce the probability of subsequent allergy development during childhood.
- 71.Cebra JJ. Influences of microbiota on intestinal immune system development. Am J Clin Nutr. 1999;69(5):1046S–1051S. doi: 10.1093/ajcn/69.5.1046s. [DOI] [PubMed] [Google Scholar]
- 72.Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host–microbial relationships in the intestine. Science. 2001;291(5505):881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 73.Penders J, Stobberingh EE, van den Brandt PA, Thijs C. The role of the intestinal microbiota in the development of atopic disorders. Allergy. 2007;62(11):1223–1236. doi: 10.1111/j.1398-9995.2007.01462.x. [DOI] [PubMed] [Google Scholar]
- 74.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003;4(3):269–273. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 75.Penders J, Thijs C, van den Brandt PA, et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut. 2007;56(5):661–667. doi: 10.1136/gut.2006.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Infant gut microbial colonization events at 1 month are associated with the subsequent development of atopic diseases in childhood.
- 76.Alenius H, Pakarinen J, Saris O, et al. Contrasting immunological effects of two disparate dusts – preliminary observations. Int Arch Allergy Immunol. 2009;149(1):81–90. doi: 10.1159/000176310. [DOI] [PubMed] [Google Scholar]
- 77.Macdonald TT, Monteleone G. Immunity inflammation, and allergy in the gut. Science. 2005;307(5717):1920–1925. doi: 10.1126/science.1106442. [DOI] [PubMed] [Google Scholar]
- 78.Schaub B, Liu J, Schleich I, Hoppler S, Sattler C, von Mutius E. Impairment of T helper and T regulatory cell responses at birth. Allergy. 2008;63(11):1438–1447. doi: 10.1111/j.1398-9995.2008.01685.x. [DOI] [PubMed] [Google Scholar]
- 79.Strachan DP. Hay fever, hygiene, and household size. Br Med J. 1989;299(6710):1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yoo J, Tcheurekdjian H, Lynch SV, Cabana M, Boushey HA. Microbial manipulation of immune function for asthma prevention: inferences from clinical trials. Proc Am Thorac Soc. 2007;4(3):277–282. doi: 10.1513/pats.200702-033AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thavagnanam S, Fleming J, Bromley A, Shields MD, Cardwell CR. A meta-analysis of the association between Caesarean section and childhood asthma. Clin Exp Allergy. 2008;38(4):629–633. doi: 10.1111/j.1365-2222.2007.02780.x. [DOI] [PubMed] [Google Scholar]
- 82.Johnson CC, Ownby DR, Alford SH, et al. Antibiotic exposure in early infancy and risk for childhood atopy. J Allergy Clin Immunol. 2005;115(6):1218–1224. doi: 10.1016/j.jaci.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 83.Tannock GW, Fuller R, Smith SL, Hall MA. Plasmid profiling of members of the family Enterobacteriaceae, Lactobacilli, and Bifidobacteria to study the transmission of bacteria from mother to infant. J Clin Microbiol. 1990;28(6):1225–1228. doi: 10.1128/jcm.28.6.1225-1228.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kelly D, Campbell JI, King TP, et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-γ and RelA. Nat Immunol. 2004;5(1):104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 85.Corr SC, Hill C, Gahan CG. Chapter 1 understanding the mechanisms by which probiotics inhibit gastrointestinal pathogens. Adv Food Nutr Res. 2009;56:1–15. doi: 10.1016/S1043-4526(08)00601-3. [DOI] [PubMed] [Google Scholar]
- 86.Adlerberth I, Hanson LA, Wold AE. Ontogeny of the intestinal flora. In: Sanderson I, Walker W, editors. Development of the Gastrointestinal Tract. BC Decker; Hamilton, ON, Canada: 1999. pp. 279–292. [Google Scholar]
- 87.Wagner CL, Taylor SN, Johnson D. Host factors in amniotic fluid and breast milk that contribute to gut maturation. Clin Rev Allergy Immunol. 2008;34(2):191–204. doi: 10.1007/s12016-007-8032-3. [DOI] [PubMed] [Google Scholar]
- 88.Kramer DR, Cebra JJ. Role of maternal antibody in the induction of virus specific and bystander IgA responses in Peyer's patches of suckling mice. Int Immunol. 1995;7(6):911–918. doi: 10.1093/intimm/7.6.911. [DOI] [PubMed] [Google Scholar]
- 89.Kramer DR, Cebra JJ. Early appearance of ‘natural’ mucosal IgA responses and germinal centers in suckling mice developing in the absence of maternal antibodies. J Immunol. 1995;154(5):2051–2062. [PubMed] [Google Scholar]
- 90.Bollinger RR, Everett ML, Palestrant D, Love SD, Lin SS, Parker W. Human secretory immunoglobulin A may contribute to biofilm formation in the gut. Immunology. 2003;109(4):580–587. doi: 10.1046/j.1365-2567.2003.01700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bollinger RR, Everett ML, Wahl SD, Lee YH, Orndorff PE, Parker W. Secretory IgA and mucin-mediated biofilm formation by environmental strains of Escherichia coli: role of type 1 pili. Mol Immunol. 2006;43(4):378–387. doi: 10.1016/j.molimm.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 92.Inoue R, Otsuka M, Ushida K. Development of intestinal microbiota in mice and its possible interaction with the evolution of luminal IgA in the intestine. Exp Anim. 2005;54(5):437–445. doi: 10.1538/expanim.54.437. [DOI] [PubMed] [Google Scholar]
- 93.Rautava S, Walker WA. Academy of Breastfeeding Medicine founder's lecture 2008: breastfeeding – an extrauterine link between mother and child. Breastfeed Med. 2009;4(1):3–10. doi: 10.1089/bfm.2009.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ruiz-Palacios GM, Calva JJ, Pickering LK, et al. Protection of breast-fed infants against Campylobacter diarrhea by antibodies in human milk. J Pediatr. 1990;116(5):707–713. doi: 10.1016/s0022-3476(05)82652-6. [DOI] [PubMed] [Google Scholar]
- 95.McGuire W, Anthony MY. Donor human milk versus formula for preventing necrotising enterocolitis in preterm infants: systematic review. Arch Dis Child Fetal Neonatal Ed. 2003;88(1):F11–F14. doi: 10.1136/fn.88.1.F11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Owen CG, Martin RM, Whincup PH, Smith GD, Cook DG. Effect of infant feeding on the risk of obesity across the life course: a quantitative review of published evidence. Pediatrics. 2005;115(5):1367–1377. doi: 10.1542/peds.2004-1176. [DOI] [PubMed] [Google Scholar]
- 97.Owen CG, Martin RM, Whincup PH, Smith GD, Cook DG. Does breastfeeding influence risk of type 2 diabetes in later life? A quantitative analysis of published evidence. Am J Clin Nutr. 2006;84(5):1043–1054. doi: 10.1093/ajcn/84.5.1043. [DOI] [PubMed] [Google Scholar]
- 98.Songjinda P, Nakayama J, Tateyama A, et al. Differences in developing intestinal microbiota between allergic and nonallergic infants: a pilot study in Japan. Biosci Biotech Bioch. 2007;71(9):2338–2342. doi: 10.1271/bbb.70154. [DOI] [PubMed] [Google Scholar]
- 99.Klaassens ES, Boesten RJ, Haarman M, et al. Mixed-species genomic microarray analysis of fecal samples reveals differential transcriptional responses of bifidobacteria in breast- and formula-fed infants. Appl Environ Microbiol. 2009;75(9):2668–2676. doi: 10.1128/AEM.02492-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.He F, Morita H, Ouwehand AC, et al. Stimulation of the secretion of proinflammatory cytokines by Bifidobacterium strains. Microbiol Immunol. 2002;46(11):781–785. doi: 10.1111/j.1348-0421.2002.tb02765.x. [DOI] [PubMed] [Google Scholar]
- 101.Ménard S, Candalh C, Bambou JC, Terpend K, Cerf-Bensussan N, Heyman M. Lactic acid bacteria secrete metabolites retaining anti-inflammatory properties after intestinal transport. Gut. 2004;53(6):821–828. doi: 10.1136/gut.2003.026252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Heuvelin E, Lebreton C, Grangette C, Pot B, Cerf-Bensussan N, Heyman M. Mechanisms involved in alleviation of intestinal inflammation by Bifidobacterium breve soluble factors. PLoS One. 2009;4(4):e5184. doi: 10.1371/journal.pone.0005184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sela DA, Chapman J, Adeuya A, et al. The genome sequence of Bifidobacterium longum subsp infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci USA. 2008;105(48):18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang HY, Liu SL, Ibrahim SA, et al. Oral administration of live Bifidobacterium substrains isolated from healthy centenarians enhanced immune function in BALB/c mice. Nutr Res. 2009;29(4):281–289. doi: 10.1016/j.nutres.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 105.Sjögren YM, Jenmalm MC, Böttcher MF, Björkstén B, Sverremark-Ekström E. Altered early infant gut microbiota in children developing allergy up to 5 years of age. Clin Exp Allergy. 2009;39(4):518–526. doi: 10.1111/j.1365-2222.2008.03156.x. [DOI] [PubMed] [Google Scholar]
- 106.Remes ST, Koskela HO, Iivanainen K, Pekkanen J. Allergen-specific sensitization in asthma and allergic diseases in children: the study on farmers' and non-farmers' children. Clin Exp Allergy. 2005;35(2):160–166. doi: 10.1111/j.1365-2222.2005.02172.x. [DOI] [PubMed] [Google Scholar]
- 107.von Ehrenstein OS, von Mutius E, Illi S, Baumann L, Böhm O, von Kries R. Reduced risk of hay fever and asthma among children of farmers. Clin Exp Allergy. 2000;30(2):187–193. doi: 10.1046/j.1365-2222.2000.00801.x. [DOI] [PubMed] [Google Scholar]
- 108.Gori A, Tincati C, Rizzardini G, et al. Early impairment of gut function and gut flora supporting a role for alteration of gastrointestinal mucosa in human immunodeficiency virus pathogenesis. J Clin Microbiol. 2008;46(2):757–758. doi: 10.1128/JCM.01729-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Barman M, Unold D, Shifley K, et al. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun. 2008;76(3):907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Baumgart M, Dogan B, Rishniw M, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J. 2007;1(5):403–418. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- 111.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104(34):13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ. Differences between tissue-associated intestinal microfloras of patients with Crohn's disease and ulcerative colitis. J Clin Microbiol. 2006;44(11):4136–4141. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kassinen A, Krogius-Kurikka L, Mäkivuokko H, et al. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology. 2007;133(1):24–33. doi: 10.1053/j.gastro.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 114.Koh AY, Köhler JR, Coggshall KT, Van Rooijen N, Pier GB. Mucosal damage and neutropenia are required for Candida albicans dissemination. PLoS Pathog. 2008;4(2):e35. doi: 10.1371/journal.ppat.0040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lupp C, Robertson ML, Wickham ME, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2(2):119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 116.Martinez-Medina M, Aldeguer X, Lopez-Siles M, et al. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E coli (AIEC) in Crohn's disease. Inflamm Bowel Dis. 2009;15(6):872–882. doi: 10.1002/ibd.20860. [DOI] [PubMed] [Google Scholar]
- 117.Nadal I, Donant E, Ribes-Koninckx C, Calabuig M, Sanz Y. Imbalance in the composition of the duodenal microbiota of children with coeliac disease. J Med Microbiol. 2007;56(12):1669–1674. doi: 10.1099/jmm.0.47410-0. [DOI] [PubMed] [Google Scholar]
- 118.Ott SJ, Musfeldt M, Ullmann U, Hampe J, Schreiber S. Quantification of intestinal bacterial populations by real-time PCR with a universal primer set and minor groove binder probes: a global approach to the enteric flora. J Clin Microbiol. 2004;42(6):2566–2572. doi: 10.1128/JCM.42.6.2566-2572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sokol H, Lepage P, Seksik P, Dore J, Marteau P. Temperature gradient gel electrophoresis of fecal 16S rRNA reveals active Escherichia coli in the microbiota of patients with ulcerative colitis. J Clin Microbiol. 2006;44(9):3172–3177. doi: 10.1128/JCM.02600-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105(43):16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stecher B, Robbiani R, Walker AW, et al. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5(10):e244. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vaahtovuo J, Munukka E, Korkeamäki M, Luukkainen R, Toivanen P. Fecal microbiota in early rheumatoid arthritis. J Rheumatol. 2008;35(8):1500–1505. [PubMed] [Google Scholar]
- 123.Willing B, Halfvarson J, Dicksved J, et al. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn's disease. Inflamm Bowel Dis. 2009;15(5):653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- 124.Claud EC, Walker WA. Bacterial colonization, probiotics, and necrotizing enterocolitis. J Clin Gastroenterol. 2008;42(Suppl. 2):S46–S52. doi: 10.1097/MCG.0b013e31815a57a8. [DOI] [PubMed] [Google Scholar]
- 125.Heilbronn LK, Campbell LV. Adipose tissue macrophages, low grade inflammation and insulin resistance in human obesity. Curr Pharm Des. 2008;14(12):1225–1230. doi: 10.2174/138161208784246153. [DOI] [PubMed] [Google Scholar]
- 126.Fuss IJ, Neurath M, Boirivant M, et al. Disparate CD4+ lamina propria (LP) lymphokine secretion profiles in inflammatory bowel disease: Crohn's disease LP cells manifest increased secretion of IFN-γ, whereas ulcerative colitis LP cells manifest increased secretion of IL-5. J Immunol. 1996;157(3):1261–1270. [PubMed] [Google Scholar]
- 127.Fujino S, Andoh A, Bamba S, et al. Increased expression of interleukin 17 in inflammatory bowel disease. Gut. 2003;52(1):65–70. doi: 10.1136/gut.52.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fuss IJ, Heller F, Boirivant M, et al. Nonclassical CD1d-restricted NK T cells that produce IL-13 characterize an atypical Th2 response in ulcerative colitis. J Clin Invest. 2004;113(10):1490–1497. doi: 10.1172/JCI19836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Heller F, Florian P, Bojarski C, et al. Interleukin-13 is the key effector Th2 cytokine in ulcerative colitis that affects epithelial tight junctions, apoptosis, and cell restitution. Gastroenterology. 2005;129(2):550–564. doi: 10.1016/j.gastro.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 130.Papadakis KA, Targan SR. Role of cytokines in the pathogenesis of inflammatory bowel disease. Annu Rev Med. 2000;51(1):289–298. doi: 10.1146/annurev.med.51.1.289. [DOI] [PubMed] [Google Scholar]
- 131.Hanauer SB. Inflammatory bowel disease: epidemiology, pathogenesis, and therapeutic opportunities. Inflamm Bowel Dis. 2006;12(5):S3–S9. doi: 10.1097/01.mib.0000195385.19268.68. [DOI] [PubMed] [Google Scholar]
- 132.Bollinger RR, Barbas AS, Bush EL, Lin SS, Parker W. Biofilms in the large bowel suggest an apparent function of the human vermiform appendix. J Theor Biol. 2007;249(4):826–831. doi: 10.1016/j.jtbi.2007.08.032. [DOI] [PubMed] [Google Scholar]
- 133.Danese S, Sans M, Fiocchi C. Inflammatory bowel disease: the role of environmental factors. Autoimmun Rev. 2004;3(5):394–400. doi: 10.1016/j.autrev.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 134.Baumgart DC, Sandborn WJ. Inflammatory bowel disease: clinical aspects and established and evolving therapies. Lancet. 2007;369(9573):1641–1657. doi: 10.1016/S0140-6736(07)60751-X. [DOI] [PubMed] [Google Scholar]
- 135.Fisher SA, Tremelling M, Anderson CA, et al. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn's disease. Nat Genet. 2008;40(6):710–712. doi: 10.1038/ng.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411(6837):599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 137.Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411(6837):603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 138.Latiano A, Palmieri O, Valvano MR, et al. Evaluating the role of the genetic variations of PTPN22, NFKB1, and FcGRIIIA genes in inflammatory bowel disease: a meta-analysis. Inflamm Bowel Dis. 2007;13(10):1212–1219. doi: 10.1002/ibd.20185. [DOI] [PubMed] [Google Scholar]
- 139.Hampe J, Franke A, Rosenstiel P, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39(2):207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 140.Rioux JD, Xavier RJ, Taylor KD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39(5):596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Glas J, Török H-P, Tonenchi L, et al. Role of the NFKB1 -94ins/delATTG promoter polymorphism in IBD and potential interactions with polymorphisms in the CARD15/NOD2, IKBL, and IL-1RN genes. Inflamm Bowel Dis. 2006;12(7):606–611. doi: 10.1097/01.ibd.0000225346.23765.6b. [DOI] [PubMed] [Google Scholar]
- 143.Borm MEA, Bodegraven AA, Mulder CJJ, Kraal G, Bouma G. A NFKB1 promoter polymorphism is involved in susceptibility to ulcerative colitis. Int J Immunogenet. 2005;32(6):401–405. doi: 10.1111/j.1744-313X.2005.00546.x. [DOI] [PubMed] [Google Scholar]
- 144.Mirza MM, Fisher SA, Onnie C, et al. No association of the NFKB1 promoter polymorphism with ulcerative colitis in a British case control cohort. Gut. 2005;54(8):1205–1206. doi: 10.1136/gut.2005.070029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Karban AS, Okazaki T, Panhuysen CIM, et al. Functional annotation of a novel NFKB1 promoter polymorphism that increases risk for ulcerative colitis. Hum Mol Genet. 2004;13(1):35–45. doi: 10.1093/hmg/ddh008. [DOI] [PubMed] [Google Scholar]
- 146.Macfarlane GT, Blackett KL, Nakayama T, Steed H, Macfarlane S. The gut microbiota in inflammatory bowel disease. Curr Pharm Des. 2009;15(13):1528–1536. doi: 10.2174/138161209788168146. [DOI] [PubMed] [Google Scholar]
- 147.Tamboli CP, Neut C, Desreumaux P, Colombel JF. Dysbiosis in inflammatory bowel disease. Gut. 2004;53(1):1–4. doi: 10.1136/gut.53.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Conte MP, Schippa S, Zamboni I, et al. Gut-associated bacterial microbiota in paediatric patients with inflammatory bowel disease. Gut. 2006;55(12):1760–1767. doi: 10.1136/gut.2005.078824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27(2):104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 150.Pajak B, Orzechowski A, Gajkowska B. Molecular basis of sodium butyrate-dependent proapoptotic activity in cancer cells. Adv Med Sci. 2007;52:83–88. [PubMed] [Google Scholar]
- 151.Scheppach W, Weiler F. The butyrate story: old wine in new bottles? Curr Opin Clin Nutr Metab Care. 2004;7(5):563–567. doi: 10.1097/00075197-200409000-00009. [DOI] [PubMed] [Google Scholar]
- 152.Sokol H, Seksik P, Furet JP, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15(8):1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 153.Jansson J, Willing B, Lucio M, et al. Metabolomics reveals metabolic biomarkers of Crohn's disease. PLoS One. 2009;4(7):e6386. doi: 10.1371/journal.pone.0006386. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• First metabolomic study to demonstrate differences in the types and concentrations of specific metabolites characteristic of Crohn's disease.
- 154.Balasubramanian K, Kumar S, Singh RR, et al. Metabolism of the colonic mucosa in patients with inflammatory bowel diseases: an in vitro proton magnetic resonance spectroscopy study. Magn Reson Imaging. 2009;27(1):79–86. doi: 10.1016/j.mri.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 155.Alm B, Erdes L, Mollborg P, et al. Neonatal antibiotic treatment is a risk factor for early wheezing. Pediatrics. 2008;121(4):697–702. doi: 10.1542/peds.2007-1232. [DOI] [PubMed] [Google Scholar]
- 156.Alam S, Mushtaq M. Antibiotic associated diarrhea in children. Indian Pediatr. 2009;46(6):491–496. [PubMed] [Google Scholar]
- 157.Surawicz CM. Antibiotic-associated diarrhea and pseudomembranous colitis: are they less common with poorly absorbed antimicrobials? Chemotherapy. 2005;51(Suppl. 1):81–89. doi: 10.1159/000081993. [DOI] [PubMed] [Google Scholar]
- 158.Caramia G. Metchnikoff and the centenary of probiotics: an update of their use in gastroenteric pathology during the age of development. Minerva Pediatr. 2008;60(6):1417–1435. [PubMed] [Google Scholar]
- 159.Resta-Lenert S, Barrett KE. Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC) Gut. 2003;52(7):988–997. doi: 10.1136/gut.52.7.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Servin AL, Coconnier MH. Adhesion of probiotic strains to the intestinal mucosa and interaction with pathogens. Best Pract Res Clin Gastroenterol. 2003;17(5):741–754. doi: 10.1016/s1521-6918(03)00052-0. [DOI] [PubMed] [Google Scholar]
- 161.Resta-Lenert SC, Barrett KE. Modulation of intestinal barrier properties by probiotics: role in reversing colitis. Ann NY Acad Sci. 2009;1165:175–182. doi: 10.1111/j.1749-6632.2009.04042.x. [DOI] [PubMed] [Google Scholar]
- 162.Dembele T, Obdrzalek V, Votava M. Inhibition of bacterial pathogens by lactobacilli. Zentralbl Bakteriol. 1998;288(3):395–401. doi: 10.1016/s0934-8840(98)80013-3. [DOI] [PubMed] [Google Scholar]
- 163.Ocana VS, Elena Nader-Macias M. Production of antimicrobial substances by lactic acid bacteria II: screening bacteriocin-producing strains with probiotic purposes and characterization of a Lactobacillus bacteriocin. Methods Mol Biol. 2004;268:347–353. doi: 10.1385/1-59259-766-1:347. [DOI] [PubMed] [Google Scholar]
- 164.Kaila M, Isolauri E, Soppi E, Virtanen E, Laine S, Arvilommi H. Enhancement of the circulating antibody secreting cell response in human diarrhea by a human Lactobacillus strain. Pediatr Res. 1992;32(2):141–144. doi: 10.1203/00006450-199208000-00002. [DOI] [PubMed] [Google Scholar]
- 165.Di Caro S, Tao H, Grillo A, et al. Effects of Lactobacillus GG on genes expression pattern in small bowel mucosa. Dig Liver Dis. 2005;37(5):320–329. doi: 10.1016/j.dld.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 166.Dotan I, Rachmilewitz D. Probiotics in inflammatory bowel disease: possible mechanisms of action. Curr Opin Gastroenterol. 2005;21(4):426–430. [PubMed] [Google Scholar]
- 167.Yan F, Polk DB. Probiotic bacterium prevents cytokine-induced apoptosis in intestinal epithelial cells. J Biol Chem. 2002;277(52):50959–50965. doi: 10.1074/jbc.M207050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gibson GR, Roberfroid MB. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr. 1995;125(6):1401–1412. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- 169.Gibson GR, Probert HM, Loo JV, Rastall RA, Roberfroid MB. Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr Res Rev. 2004;17(02):259–275. doi: 10.1079/NRR200479. [DOI] [PubMed] [Google Scholar]
- 170.Saulnier DMA, Spinler JK, Gibson GR, Versalovic J. Mechanisms of probiosis and prebiosis: considerations for enhanced functional foods. Curr Opin Biotechnol. 2009;20(2):135–141. doi: 10.1016/j.copbio.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Preidis GA, Versalovic J. Targeting the human microbiome with antibiotics, probiotics, and prebiotics: gastroenterology enters the metagenomics era. Gastroenterology. 2009;136(6):2015–2031. doi: 10.1053/j.gastro.2009.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sellon RK, Tonkonogy S, Schultz M, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66(11):5224–5231. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Taurog JD, Richardson JA, Croft JT, et al. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J Exp Med. 1994;180(6):2359–2364. doi: 10.1084/jem.180.6.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mileti E, Matteoli G, Iliev ID, Rescigno M. Comparison of the immunomodulatory properties of three probiotic strains of lactobacilli using complex culture systems: prediction for in vivo efficacy. PLoS One. 2009;4(9):e7056. doi: 10.1371/journal.pone.0007056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Fedorak RN. Probiotics in the management of inflammatory bowel diseases? Am J Gastroenterol. 2007;102(s1):S22–S28. [Google Scholar]
- 176.Isaacs K, Herfarth H. Role of probiotic therapy in IBD. Inflamm Bowel Dis. 2008;14(11):1597–1605. doi: 10.1002/ibd.20465. [DOI] [PubMed] [Google Scholar]
- 177.Fellermann K, Wehkamp J, Herrlinger KR, Stange EF. Crohn's disease: a defensin deficiency syndrome? Eur J Gastroenterol Hepatol. 2003;15:627–634. doi: 10.1097/00042737-200306000-00008. [DOI] [PubMed] [Google Scholar]
- 178.Tursi A, Brandimarte G, Giorgetti GM, Forti G, Modeo ME, Gigliobianco A. Low-dose balsalazide plus a high-potency probiotic preparation is more effective than balsalazide alone or mesalazine in the treatment of acute mild-to-moderate ulcerative colitis. Med Sci Monit. 2004;10(11):PI126–PI131. [PubMed] [Google Scholar]
- 179.Bibiloni R, Fedorak RN, Tannock GW, et al. VSL#3 probiotic-mixture induces remission in patients with active ulcerative colitis. Am J Gastroenterol. 2005;100(7):1539–1546. doi: 10.1111/j.1572-0241.2005.41794.x. [DOI] [PubMed] [Google Scholar]
- 180.Sood A, Midha V, Makharia GK, et al. The probiotic preparation, VSL#3 induces remission in patients with mild-to-moderately active ulcerative colitis. Clin Gastroenterol H. 2009;7(11):1202–1209. doi: 10.1016/j.cgh.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 181.Guslandi M, Giollo P, Testoni PA. A pilot trial of Saccharomyces boulardii in ulcerative colitis. Eur J Gastroenterol Hepatol. 2003;15(6):697–698. doi: 10.1097/00042737-200306000-00017. [DOI] [PubMed] [Google Scholar]
- 182.Guslandi M, Mezzi G, Sorghi M, Testoni PA. Saccharomyces boulardii in maintenance treatment of Crohn's disease. Dig Dis Sci. 2000;45(7):1462–1464. doi: 10.1023/a:1005588911207. [DOI] [PubMed] [Google Scholar]
- 183.AlFaleh K, Anabrees J, Bassler D. Probiotics reduce the risk of necrotizing enterocolitis in preterm infants: a meta-analysis. Neonatology. 2009;97(2):93–99. doi: 10.1159/000235684. [DOI] [PubMed] [Google Scholar]
- 184.Barclay AR, Stenson B, Simpson JH, Weaver LT, Wilson DC. Probiotics for necrotizing enterocolitis: a systematic review. J Pediatr Gastroenterol Nutr. 2007;45(5):569–576. doi: 10.1097/MPG.0b013e3181344694. [DOI] [PubMed] [Google Scholar]
- 185.Bin-Nun A, Bromiker R, Wilschanski M, et al. Oral probiotics prevent necrotizing enterocolitis in very low birth weight neonates. J Pediatr. 2005;147(2):192–196. doi: 10.1016/j.jpeds.2005.03.054. [DOI] [PubMed] [Google Scholar]
- 186.Caplan MS. Probiotic and prebiotic supplementation for the prevention of neonatal necrotizing enterocolitis. J Perinatol. 2009;29(S2):S2–S6. doi: 10.1038/jp.2009.21. [DOI] [PubMed] [Google Scholar]
- 187.Deshpande G, Rao S, Patole S. Probiotics for prevention of necrotising enterocolitis in preterm neonates with very low birthweight: a systematic review of randomised controlled trials. Lancet. 2007;369(9573):1614–1620. doi: 10.1016/S0140-6736(07)60748-X. [DOI] [PubMed] [Google Scholar]
- 188.Lin HC, Su BH, Chen AC, et al. Oral probiotics reduce the incidence and severity of necrotizing enterocolitis in very low birth weight infants. Pediatrics. 2005;115(1):1–4. doi: 10.1542/peds.2004-1463. [DOI] [PubMed] [Google Scholar]
- 189.Martin CR, Walker WA. Probiotics: role in pathophysiology and prevention in necrotizing enterocolitis. Semin Perinatol. 2008;32(2):127–137. doi: 10.1053/j.semperi.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 190.Cox MJ, Huang YJ, Fujimura KE, et al. Lactobacillus casei abundance is associated with profound shifts in the infant gut microbiome. PLoS One. 2010;5(1):e8745. doi: 10.1371/journal.pone.0008745. [DOI] [PMC free article] [PubMed] [Google Scholar]; • First study to demonstrate global shifts, including promotion of other phylogenetically related probiotic species in the gut microbiome upon supplementation with a probiotic species (Lactobacillus casei Rhamnosus GG).This study suggests that the protective effects of L. casei GG against development of allergic disease may be from the formation of stable, even and functionally redundant communities composed of commensal species.
- 191.Brodie EL, DeSantis TZ, Joyner DC, et al. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl Environ Microbiol. 2006;72(9):6288–6298. doi: 10.1128/AEM.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Besselink MG, van Santvoort HC, Buskens E, et al. Probiotic prophylaxis in predicted severe acute pancreatitis: a randomised, double-blind, placebo-controlled trial. Lancet. 2008;371(9613):651–659. doi: 10.1016/S0140-6736(08)60207-X. [DOI] [PubMed] [Google Scholar]
- 193.Kunz AN, Noel JM, Fairchok MP. Two cases of Lactobacillus bacteremia during probiotic treatment of short gut syndrome. J Pediatr Gastroenterol Nutr. 2004;38(4):457–458. doi: 10.1097/00005176-200404000-00017. [DOI] [PubMed] [Google Scholar]
- 194.Land MH, Rouster-Stevens K, Woods CR, Cannon ML, Cnota J, Shetty AK. Lactobacillus sepsis associated with probiotic therapy. Pediatrics. 2005;115(1):178–181. doi: 10.1542/peds.2004-2137. [DOI] [PubMed] [Google Scholar]
- 195.Winkler J, Butler R, Symonds E. Fructo-oligosaccharide reduces inflammation in a dextran sodium sulphate mouse model of colitis. Digest Dis Sci. 2007;52(1):52–58. doi: 10.1007/s10620-006-9224-z. [DOI] [PubMed] [Google Scholar]
- 196.Lewis S, Brazier J, Beard D, Nazem N, Proctor D. Effects of metronidazole and oligofructose on faecal concentrations of sulphate-reducing bacteria and their activity in human volunteers. Scand J Gastroenterol. 2005;40(11):1296–1303. doi: 10.1080/00365520510023585. [DOI] [PubMed] [Google Scholar]
- 197.Kanauchi O, Oshima T, Andoh A, Shioya M, Mitsuyama K. Germinated barley foodstuff ameliorates inflammation in mice with colitis through modulation of mucosal immune system. Scand J Gastroenterol. 2008;43(11):1346–1352. doi: 10.1080/00365520802245411. [DOI] [PubMed] [Google Scholar]
- 198.Le Leu RK, Hu Y, Brown IL, Woodman RJ, Young GP. Synbiotic intervention of Bifidobacterium lactis and resistant starch protects against colorectal cancer development in rats. Carcinogenesis. 2010;31(2):246–251. doi: 10.1093/carcin/bgp197. [DOI] [PubMed] [Google Scholar]
- 199.Pool-Zobel BL. Inulin-type fructans and reduction in colon cancer risk: review of experimental and human data. Br J Nutr. 2005;93(Suppl. S1):S73–S90. doi: 10.1079/bjn20041349. [DOI] [PubMed] [Google Scholar]
- 200.Roller M, Pietro Femia A, Caderni G, Rechkemmer G, Watzl B. Intestinal immunity of rats with colon cancer is modulated by oligofructose-enriched inulin combined with Lactobacillus rhamnosus and Bifidobacterium lactis. Br J Nutr. 2004;92(6):931–938. doi: 10.1079/bjn20041289. [DOI] [PubMed] [Google Scholar]
- 201.Rowland IR, Rumney CJ, Coutts JT, Lievense LC. Effect of Bifidobacterium longum and inulin on gut bacterial metabolism and carcinogen-induced aberrant crypt foci in rats. Carcinogenesis. 1998;19(2):281–285. doi: 10.1093/carcin/19.2.281. [DOI] [PubMed] [Google Scholar]
- 202.Rafter J, Bennett M, Caderni G, et al. Dietary synbiotics reduce cancer risk factors in polypectomized and colon cancer patients. Am J Clin Nutr. 2007;85(2):488–496. doi: 10.1093/ajcn/85.2.488. [DOI] [PubMed] [Google Scholar]
- 203.Furrie E, Macfarlane S, Kennedy A, et al. Synbiotic therapy (Bifidobacterium longum/Synergy 1) initiates resolution of inflammation in patients with active ulcerative colitis: a randomised controlled pilot trial. Gut. 2005;54(2):242–249. doi: 10.1136/gut.2004.044834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Fujimori S, Tatsuguchi A, Gudis K, et al. High dose probiotic and prebiotic cotherapy for remission induction of active Crohn's disease. J Gastroenterol Hepatol. 2007;22(8):1199–1204. doi: 10.1111/j.1440-1746.2006.04535.x. [DOI] [PubMed] [Google Scholar]
- 205.Alfvén T, Braun-Fahrländer C, Brunekreef B, et al. Allergic diseases and atopic sensitization in children related to farming and anthroposophic lifestyle – the PARSIFAL study. Allergy. 2006;61(4):414–421. doi: 10.1111/j.1398-9995.2005.00939.x. [DOI] [PubMed] [Google Scholar]
- 206.Bråbäck L, Hjern A, Rasmussen F. Trends in asthma, allergic rhinitis and eczema among Swedish conscripts from farming and non-farming environments. A nationwide study over three decades. Clin Exp Allergy. 2004;34(1):38–43. doi: 10.1111/j.1365-2222.2004.01841.x. [DOI] [PubMed] [Google Scholar]
- 207.Brussee JE, Smit HA, van Strien RT, et al. Allergen exposure in infancy and the development of sensitization, wheeze, and asthma at 4 years. J Allergy Clin Immunol. 2005;115(5):946–952. doi: 10.1016/j.jaci.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 208.Korppi M, Hyvarinen M, Kotaniemi-Syrjanen A, Piippo-Savolainen E, Reijonen T. Early exposure and sensitization to cat and dog: different effects on asthma risk after wheezing in infancy. Pediatr Allergy Immunol. 2008;19(8):696–701. doi: 10.1111/j.1399-3038.2008.00758.x. [DOI] [PubMed] [Google Scholar]
- 209.Perzanowski MS, Chew GL, Divjan A, et al. Cat ownership is a risk factor for the development of anti-cat IgE but not current wheeze at age 5 years in an inner-city cohort. J Allergy Clin Immunol. 2008;121(4):1047–1052. doi: 10.1016/j.jaci.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Radon K, Schulze A, Nowak D. Inverse association between farm animal contact and respiratory allergies in adulthood: protection, underreporting or selection? Allergy. 2006;61(4):443–446. doi: 10.1111/j.1398-9995.2006.00995.x. [DOI] [PubMed] [Google Scholar]
- 211.Roponen M, Hyvärinen A, Hirvonen M-R, Keski-Nisula L, Pekkanen J. Change in IFN-γ-producing capacity in early life and exposure to environmental microbes. J Allergy Clin Immunol. 2005;116(5):1048–1052. doi: 10.1016/j.jaci.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 212.Schafer T, Stieger B, Polzius R, Krauspe A. Associations between cat keeping, allergen exposure, allergic sensitization and atopic diseases: results from the Children of Lubeck Allergy and Environment Study (KLAUS) Pediatr Allergy Immunol. 2009;20(4):353–357. doi: 10.1111/j.1399-3038.2008.00806.x. [DOI] [PubMed] [Google Scholar]
- 213.Takkouche B, Gonzalez-Barcala FJ, Etminan M, Fitzgerald M. Exposure to furry pets and the risk of asthma and allergic rhinitis: a meta-analysis. Allergy. 2008;63(7):857–864. doi: 10.1111/j.1398-9995.2008.01732.x. [DOI] [PubMed] [Google Scholar]
- 214.Maitra A, Sherriff A, Strachan D, Henderson J. Mode of delivery is not associated with asthma or atopy in childhood. Clin Exp Allergy. 2004;34(9):1349–1355. doi: 10.1111/j.1365-2222.2004.02048.x. [DOI] [PubMed] [Google Scholar]
- 215.Mikami K, Takahashi H, Kimura M, et al. Influence of maternal bifidobacteria on the establishment of bifidobacteria colonizing the gut in infants. Pediatr Res. 2009;65(6):669–674. doi: 10.1203/PDR.0b013e31819ed7a8. [DOI] [PubMed] [Google Scholar]
- 216.Turroni F, Marchesi JR, Foroni E, et al. Microbiomic analysis of the bifidobacterial population in the human distal gut. ISME J. 2009;3(6):745–751. doi: 10.1038/ismej.2009.19. [DOI] [PubMed] [Google Scholar]
- 217.Campo P, Kalra HK, Levin L, et al. Influence of dog ownership and high endotoxin on wheezing and atopy during infancy. J Allergy Clin Immunol. 2006;118(6):1271–1278. doi: 10.1016/j.jaci.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Pohlabeln H, Jacobs S, Bohmann J. Exposure to pets and the risk of allergic symptoms during the first 2 years of life. J Investig Allergol Clin Immunol. 2007;17(5):302–308. [PubMed] [Google Scholar]
- 219.Adlerberth I, Lindberg E, Aberg N, et al. Reduced enterobacterial and increased staphylococcal colonization of the infantile bowel: an effect of hygienic lifestyle? Pediatr Res. 2006;59(1):96–101. doi: 10.1203/01.pdr.0000191137.12774.b2. [DOI] [PubMed] [Google Scholar]
- 220.Bertini I, Calabrò A, De Carli V, et al. The metabonomic signature of celiac disease. J Proteome Res. 2008;8(1):170–177. doi: 10.1021/pr800548z. [DOI] [PubMed] [Google Scholar]
- 221.Wen L, Ley RE, Volchkov PY, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455(7216):1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Dumas ME, Barton RH, Toye A, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci USA. 2006;103(33):12511–12516. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Brugman S, Klatter FA, Visser JT, et al. Antibiotic treatment partially protects against type 1 diabetes in the Bio-Breeding diabetes-prone rat Is the gut flora involved in the development of type 1 diabetes? Diabetologia. 2006;49(9):2105–2108. doi: 10.1007/s00125-006-0334-0. [DOI] [PubMed] [Google Scholar]
- 224.Bibiloni R, Membrez M, Chou CJ. Gut microbiota, obesity and diabetes. Ann Nestle (English ed) 2009;67(1):39–47. [Google Scholar]
- 225.Cani P, Neyrinck A, Fava F, et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. 2007;50(11):2374–2383. doi: 10.1007/s00125-007-0791-0. [DOI] [PubMed] [Google Scholar]
- 226.Cani PD, Bibiloni R, Knauf C, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57(6):1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 227.Cani PD, Delzenne NM. Gut microflora as a target for energy and metabolic homeostasis. Curr Opin Clin Nutr Metab Care. 2007;10(6):729–734. doi: 10.1097/MCO.0b013e3282efdebb. [DOI] [PubMed] [Google Scholar]
- 228.Bäckhed F. Changes in intestinal microflora in obesity: cause or consequence? J Pediatr Gastroenterol Nutr. 2009;48:S56–S57. doi: 10.1097/MPG.0b013e3181a11851. [DOI] [PubMed] [Google Scholar]
- 229.Membrez M, Blancher F, Jaquet M, et al. Gut microbiota modulation with norfloxacin and ampicillin enhances glucose tolerance in mice. FASEB J. 2008;22(7):2416–2426. doi: 10.1096/fj.07-102723. [DOI] [PubMed] [Google Scholar]
- 230.Monteleone G, Monteleone I, Fina D, et al. Interleukin-21 enhances T-helper cell type I signaling and interferon-γ production in Crohn's disease. Gastroenterology. 2005;128(3):687–694. doi: 10.1053/j.gastro.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 231.Bibiloni R, Mangold M, Madsen KL, Fedorak RN, Tannock GW. The bacteriology of biopsies differs between newly diagnosed, untreated, Crohn's disease and ulcerative colitis patients. J Med Microbiol. 2006;55(8):1141–1149. doi: 10.1099/jmm.0.46498-0. [DOI] [PubMed] [Google Scholar]
- 232.Macfarlane S, Furrie E, Kennedy A, Cummings JH, Macfarlane GT. Mucosal bacteria in ulcerative colitis. Br J Nutr. 2005;93(Suppl. S1):S67–S72. doi: 10.1079/bjn20041347. [DOI] [PubMed] [Google Scholar]
- 233.Sokol H, Seksik P, Rigottier-Gois L, et al. Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm Bowel Dis. 2006;12(2):106–111. doi: 10.1097/01.MIB.0000200323.38139.c6. [DOI] [PubMed] [Google Scholar]
- 234.Zhang M, Liu B, Zhang Y, Wei H, Lei Y, Zhao L. Structural shifts of mucosa-associated lactobacilli and Clostridium leptum subgroup in patients with ulcerative colitis. J Clin Microbiol. 2007;45(2):496–500. doi: 10.1128/JCM.01720-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Lin PW, Nasr TR, Stoll BJ. Necrotizing enterocolitis: recent scientific advances in pathophysiology and prevention. Semin Perinatol. 2008;32(2):70–82. doi: 10.1053/j.semperi.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 236.Henry MCW, Moss RL. Necrotizing enterocolitis. Annu Rev Med. 2009;60(1):111–124. doi: 10.1146/annurev.med.60.050207.092824. [DOI] [PubMed] [Google Scholar]
- 237.Wang Y, Hoenig JD, Malin KJ, et al. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 2009;3(8):944–954. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Zhang H, DiBaise JK, Zuccolo A, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA. 2009;106(7):2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]