Abstract
PHR (Pam-Highwire-RPM-1) proteins are conserved, large multi-domain E3 ubiquitin ligases with modular architecture. PHR proteins presynaptically control synaptic growth and axon guidance and postsynaptically regulate endocytosis of glutamate receptors. Dysfunction of neuronal ubiquitin-mediated proteasomal degradation is implicated in various neurodegenerative diseases. PHR proteins are characterized by the presence of two PHR domains near the N-terminus, which are essential for proper localization and function. Structures of both the first and second PHR domains of Mus musculus (mouse) Phr1 (MYC binding protein 2, Mycbp2) have been determined, revealing a novel β sandwich fold composed of 11 anti-parallel β-strands. Conserved loops decorate the apical side of the first PHR domain (MmPHR1) yielding a distinct conserved surface feature. The surface of the second PHR domain (MmPHR2), in contrast, lacks significant conservation. Importantly, the structure of MmPHR1 provides insights into a loss of function mutation, Gly1092→Glu, observed in the C. elegans ortholog RPM-1.
Keywords: PHR domain, Phr1, RPM-1, Mycbp2, synaptic development and axon guidance
Targeting proteins for proteasome mediated degradation through poly-ubiquitination (ubiquitin proteasome system, UPS) represents an important mechanism controlling various aspects of cellular life.1 Dysfunction of the neuronal UPS has been implicated in neurodegenerative diseases.2 PHR proteins are one of several E3 ubiquitin ligases that function within the nervous system. In recent years, numerous publications have illustrated the critical roles PHR proteins play in proper formation and function of synapses at neuron muscular junctions and in axon guidance during embryonic development.3-5 PHR (protein associated with Myc (PAM) from human; Highwire (HIW) from Drosophila; Regulator of Presynaptic Morphology 1 (RPM-1) of C. elegans) proteins are huge, multi-domain E3 ubiquitin ligases.6-9 Phr1 and Esrom are well characterized, conserved PHR orthologs from mouse and zebrafish, respectively.10,11 Impairment of RPM-1 activity in C. elegans leads to decreased number of synapses and improper organization of presynaptic terminals. RPM-1 also postsynaptically regulates endocytosis of the AMPA-type glutamate receptor.12 hiw mutants of Drosophila, in contrast to rpm-1 mutants, show increased numbers of synaptic boutons, albeit smaller in size, and more extensive synaptic branches. In vertebrates, Phr1 and Esrom play critical roles in both nerve and synapse development and in axon growth and guidance.13-15 Phr1 associates with the microtubule cytoskeleton and regulates microtubule stability, which is necessary for appropriate neurite outgrowth.13 More recently, differential purification of neurite and soma using transwells containing 3.0μm porous filters combined with quantitative proteome analysis revealed that Phr1 is 12 fold enriched within the neurite,16 underscoring its functional importance in neurite outgrowth and axon guidance. Interestingly, Phr1 is also localized in the pseudopodium, but not the cell body of the migrating Cos-7 cell.17 Pseudopodial targeting of Phr1 and its interaction with the microtubule cytoskeleton suggest that Phr1 plays a role in regulating pseudopodium formation, which is necessary for proper cell migration.
PHR proteins regulate different MAP kinase signaling pathways in various organisms.13,18,19 RPM-1 and HIW have been shown to function in an SCF-like (Skp1, Cullin, and FBox) E3 ubiquitin ligase complex. RPM-1 interacts with FSN-1 in the SKR-1:CUL-1:FSN-1 complex20 and negatively regulates the PMK-3 pathway (a C. elegans ortholog of p38 kinase) by targeting the upstream dual leucine-zipper bearing kinase-1 (DLK-1), a MAPKKK, for degradation.18 Similarly, HIW interacts with the F-Box protein DFsn to target Wallenda for degradation, the closest Drosophila ortholog of DLK-1,21 although the downstream substrate of Wallenda is JNK kinase.19 Human PAM plays a role in regulating mTOR (mammalian target of rapamycin) signaling through its interaction with tuberin, a component of Tuberous Sclerosis Complex.22 Other targets of PHR proteins include ALK,20 a C. elegans ortholog of mammalian receptor tyrosine kinase, Myc,6 Medea,23 and cyclic AMP metabolism.24 RPM-1 also positively regulates the Rab GTPase GLO-1 via a ubiquitin ligase independent mechanism.25
PHR proteins show similarity both in their modular architecture and individual domain sequences. They contain a RHD (regulator of chromosome condensation 1, RCC1-homology) domain and two PHR repeats within the N-terminal portion, a Myc-binding region, an HHD (histone-binding protein homology domain) in the middle portion, and a RING finger domain and two C2H2-type Zinc binding motifs within the C-terminal portion.6 All PHR proteins known to date are characterized by the presence of two PHR domains. In addition to the PHR family proteins, BTBD1 and BTBD2 proteins contain “PHR-like domains”.26 Mechanistic insights into the role of PHR domains have been obtained only recently. Abrams et al. (2008)27 showed that C. elegans RPM-1 mutants lacking PHR domains accumulate in cell bodies, and that the PHR domains are both necessary and sufficient for proper localization of RPM-1 in synapses, which is essential for function.
Structures of the first PHR (MmPHR1, residues 1229-1390) and the second PHR (MmPHR2, residues 1723-1883) domains were determined at 1.3Å and 2.25Å resolution, respectively. These structures provide the first atomic views of PHR domains, which share a novel β sandwich architecture. Mapping of sequence conservation revealed unique surface features that could be important for the function of PHR proteins. Moreover, structural insights into the Gly1092→Glu loss of function mutation of RPM-1, the C. elegans homologue of MmPhr1, have been obtained.
Structures of MmPHR1 and MmPHR2
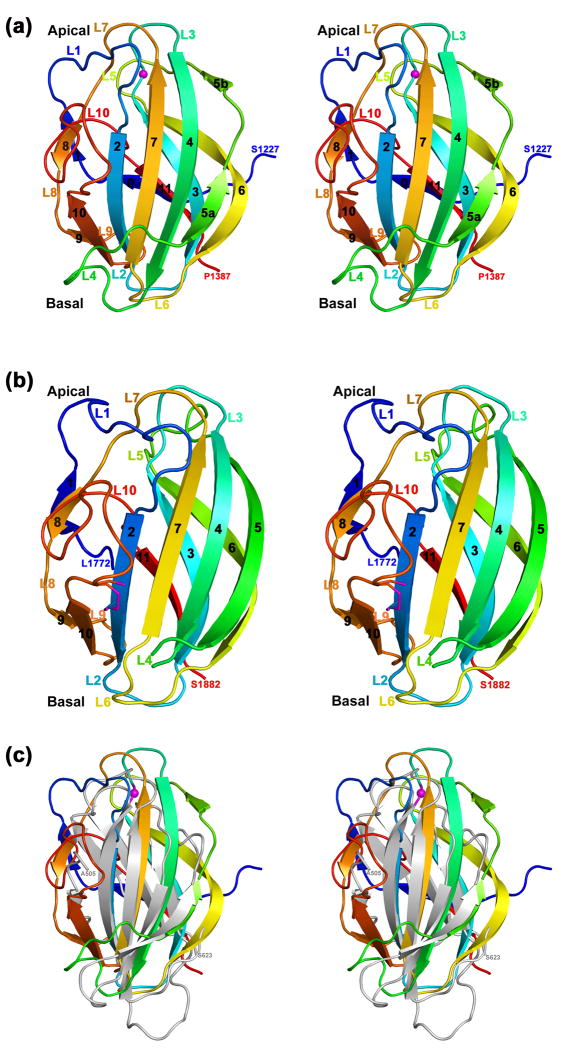
The structure of MmPHR1 (PDB Code 3GBW) reveals a β-sandwich fold composed of 11 anti-parallel strands (Fig. 1a). The N-terminal β0 strand is derived from residues falling outside the canonical PHR domain as defined by PFam (http://pfam.sanger.ac.uk/). The overall structure of MmPHR1 is made up of three anti-parallel β-sheets: (i) an anterior β-sheet made up of six strands (β9-β10-β2-β7-β4-β5a from left to right; Fig. 1a) with β5b making additional interactions with β4; (ii) a posterior β-sheet made up of three strands (β11-β3-β6); and (iii) a short two stranded β-sheet formed by β1 and β8. The β strands are connected by short and long loops both on the apical and basal surfaces of the domain. Loops L1, L3, L5, L7, and L10 form the apical surface, and loops L2, L4, L6, and L9 form the basal surface of MmPHR1 (Fig. 1a).
Fig. 1. Overall PHR domain structure.
(a) Stereoview of the overall fold of MmPHR1 with secondary structural elements colored in blue to red from N- to C-terminus. The Cα atom of Gly1271 of the mouse Phr-1, the equivalent residue of the loss of function RPM-1 mutant Gly1092→Glu, is shown as a magenta sphere. (b) Stereoview of the overall fold of MmPHR2 with secondary structural elements colored as above. A short 310-helix preceding the β2 strand is depicted as a loop for clarity. The partially formed disulfide link between Cys1745, from strand β2, and Cys1860, from loop L10, is shown as a stick figure. (c) Superposition of MmPHR1 (PDB Code 3GBW) and HA3 subcomponent of C. botulinum 16S progenitor toxin (HA3, PDB Code 2ZOE, residues 505-623). Cartoons of MmPHR1 and HA3 are colored as rainbow and in gray, respectively.
Structure of MmPHR1 was determined via single wavelength anomalous dispersion. Se atoms were located with SHELXD35 and phases were calculated using SHELXE as implemented in HKL2MAP.36 The 1.3Å resolution experimental electron density map was of excellent quality. Initial model building was carried out with ARP/wARP37, followed by manual adjustment with COOT.30 The atomic model of MmPHR1 was refined using REFMAC5,38 as implemented in CCP4,34 using anisotropic modeling of individual atomic displacement parameters (B-values). The electron density corresponding to the polypeptide backbone of MmPHR1 was everywhere continuous from Glu1229 to Pro1387. Our poly-alanine model of MmPHR1 domain was used as a search model to determine the structure of MmPHR2 domain via molecular replacement. Automated rotational and translational searching with PHASER39 permitted placement of two molecules of MmPHR2 in the asymmetric unit (Table 1). The initial atomic model built with ARP/wARP37 was subjected to several rounds of manual rebuilding in COOT30 and refined to convergence with REFMAC5.38 The final refined atomic models of MmPHR1 and MmPHR2 have excellent stereochemistry (Table 1). Illustrations were prepared using PyMol (http://pymol.sourceforge.net). Sequence alignments were carried out using ClustalW240 and color coded with ESPript.41 Sequence conservation was mapped on solvent-accessible molecular surfaces using ConSurf.42
The overall fold of MmPHR2 (PDB Code 3HWJ) is similar to that of MmPHR1 [(Fig. 1b); root-mean square deviation=1.3Å for 144 Cα atomic pairs with sequence identity=28%]. The three anti-parallel β-sheets are arranged in identical fashion and loops L1, L3, L5, L7, and L10 form the apical surface, while loops L2, L4, L6, and L9 form the basal surface. Notable differences in the structural features of MmPHR1 versus MmPHR2 are as follows: (i) The β5 strand of MmPHR2, which is part of the anterior β-sheet is continuous in contrast to that of MmPHR1 (Fig. 1a, b); (ii) loops L1 and L4 are longer in MmPHR2; (iii) the central 10-12 residues of L4 of MmPHR2 are disordered; and (iv) MmPHR2 contains a disulfide bond between Cys1745 and Cys1860. These two Cys residues are conserved in 10 out of the 14 PHR2 sequences analyzed (see below). The disulfide bridge observed in the X-ray structure of MmPHR2 is only partially formed, presumably because of the presence of 5mM dithiothreitol in the protein storage buffer. The biological significance of this disulfide bridge for PHR protein function is not known.
PHR domain adopts a novel fold
A DALI28 search was performed to identify possible structural homologues of MmPHR1. No clear cut structural homologues were detected (i.e., those with Z-scores>10). Our DALI search identified 46 non-PHR polypeptide chains with Z-scores between 6.0 and 7.3. The r.m.s. deviations for these superpositions fall between 2.8Å and 4.1Å over 94 to 119 Cα atomic pairs with sequence identities of 5-13%. One of the closest possible structural homologues of MmPHR1 was Clostridium botulinum Type C 16S progenitor toxin nonhemagglutinin HA3 subcomponent (HA3, PDB Code 2ZOE, residues 505-623)29 with a Z-score of 7.1 and r.m.s. deviation of 2.8Å for 94 Cα atomic pairs with sequence identity=9%. Visual inspection of the superposition of MmPHR1 and HA3 reveals both similarities and differences in the overall folds of these two polypeptide chains (Fig. 1c). Strands β2-β7-β4 -β5a (front β-sheet) and β11-β3 (back β-sheet) of MmPHR1 do have counterparts in HA3. However, strands corresponding to β6, β9, and β10 are absent in HA3. In addition, HA3 lacks the two stranded sheet formed by β1-β8 of MmPHR1 (Fig. 1c). The C-terminal domain of CocE/NonD family hydrolase from Staphylococcus aureus (SACOL2612, PDB Code 3IB3) was also identified as a possible structural homologue of PHR1 with a Z-score of 7.3 and r.m.s. deviation 3.5Å for 177 Cα atomic pairs with sequence identity=13%. Again, while some features of the front and rear β-sheets are similar between MmPHR1 and SACOL2612, there are profound differences (data not shown). Given the low sequence identities in these alignments and many structural differences, we believe the observed structural similarities simply reflect β-strand packing constraints. We conclude, therefore, that the overall structure of the PHR domain revealed by our work on MmPHR1 and MmPHR2 represents a previously undescribed protein fold. Both crystal forms reveal the two PHR domains to be monomers, which is consistent with the results of size exclusion chromatography (data not shown).
Sequence conservation among PHR1 and PHR2 domains
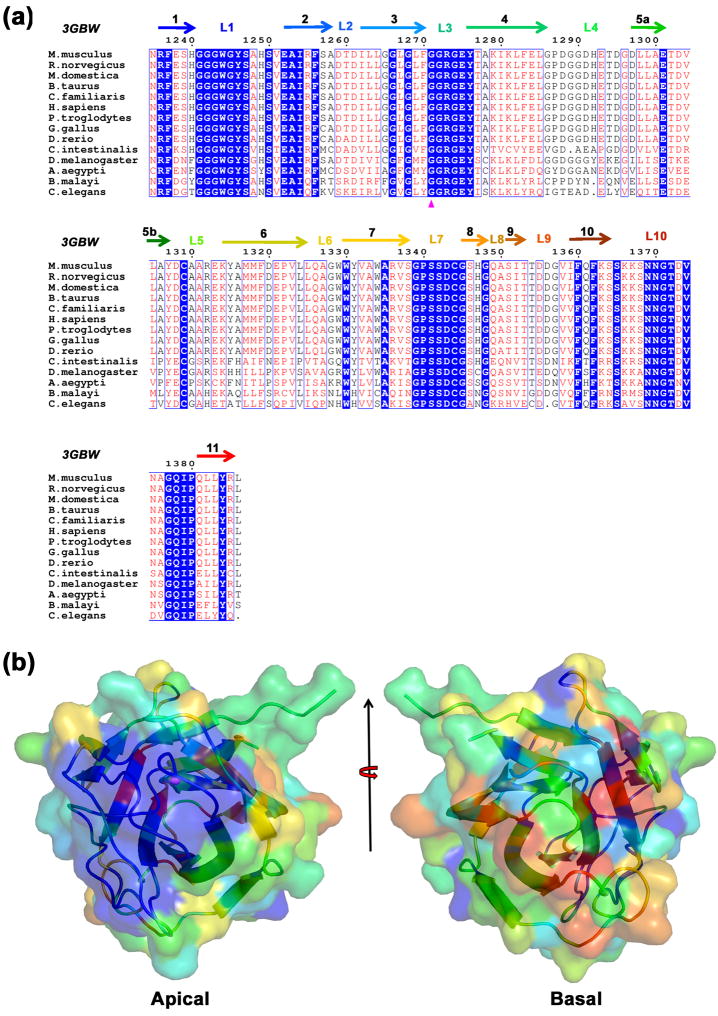
Sequences of the first and second PHR domains from PHR proteins of 14 organisms were compared to analyze sequence conservation within this new protein fold family. A striking feature of PHR1 sequences of chordata, insect, and nematoda origins is absolute conservation of L1, L3 and L7 loop residues (Fig. 2a). In addition, most (10 out of 16) of the residues of L10 loop are also conserved. This finding reflects the presence of a conserved feature on the surface of MmPHR1 (Fig. 2b). Loops showing absolute sequence conservation decorate the apical surface of PHR1 (Figs. 1a and 2b). Sequence conservation is relatively low in the L2, L4, L5, L6, and L9 loops. Hence, the basal surface of MmPHR1, in contrast to the apical surface, is devoid of such surface conservation. The distinct conservation of the apical surface across phylogeny suggests that L1, L3, L5, L7, and L10 loop residues could play important roles in PHR1 domain function.
Fig. 2. Sequence and surface conservation of PHR1 domains.
(a) Sequence alignment of the first PHR (PHR1) domains of PHR proteins from fourteen organisms. Secondary structural elements are color coded as in Fig. 1a and Gly1271 (corresponding to the location of the loss of function mutation of the C. elegans ortholog) is indicated by a magenta triangle. (b) Sequence conservation of PHR1 mapped on the surface of the MmPHR1 structure. Absolutely conserved to least conserved residues are color coded from blue to red. The apical surface is shown by 90° rotation of Fig. 1a about the horizontal axis and the basal surface is shown by subsequent 180° rotation of the apical surface about the vertical axis.
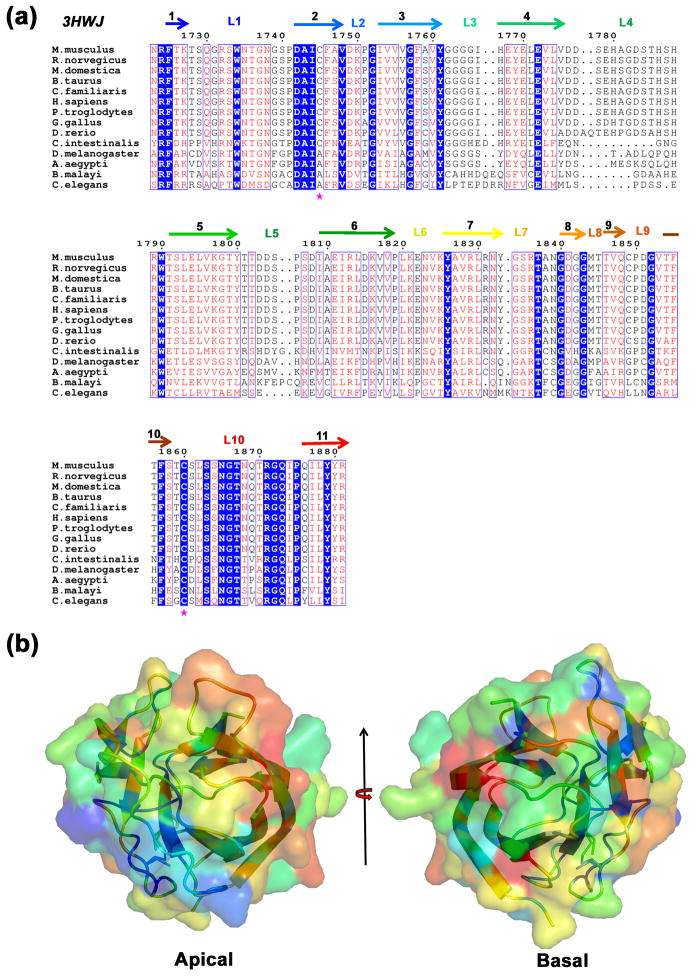
Overall, PHR2 domains show lower sequence conservation when compared to PHR1 sequences (Fig. 3a). Sequence conservation is low even among the loops decorating the apical surface, except for L10 (9 out of 17 residues from L10 are absolutely conserved among both PHR1 and PHR2 sequences). The low degree of overall sequence conservation is due to differences in PHR2 sequences of chordata, insect, and nematoda origin. Therefore, when underlying sequence conservation is plotted on the surface of MmPHR2 no distinct features are apparent (Fig. 3b). Within the phylum of chordate, however, the apical loops of PHR2 domains are highly conserved (Fig. 3a).
Fig. 3. Sequence and surface conservation of PHR2 domains.
(a) Sequence alignment of the second PHR (PHR2) domains of PHR proteins from fourteen organisms. Secondary structural elements are color coded as in Fig. 1b. Cys1745 and Cys1860 are labeled in magenta asterisks. (b) Sequence conservation of PHR2 mapped on the surface of the MmPHR2 structure, using the same color coding and views as in Fig. 2.
These analyses give rise to the following possibilities with regard to the function of PHR domains: (i) loops decorating the apical surface are crucial for the function of PHR domains; and (ii) the second PHR (PHR2) domain of insect and nematoda may have diverged from the first PHR (PHR1) domain to support different functional requirements. In addition, loop L10 residues and β-strand 11 show a high degree of conservation across both the PHR1 and PHR2 domains (Figs. 2a and 3a), which suggests that “N-G-T-X4-G-Q-I/L-P-X-φ2-Y” present in loop L10 could represent a defining motif for the PHR domains (where φ is a hydrophobic residue). Remarkably, the “N-G-T-X4-G-Q-I/L-P-X-φ2-Y/F” motif is also found in BTBD proteins (Fig. S1, supplementary material), which contain N-terminal POZ and BACK domains followed by a single “PHR-like” domain at the C-terminus. Extreme C-terminal residues of BTBD1 and BTBD2 proteins interact with the core subdomain II of human topoisomerase.26 PHR domains are likely play a structural role because they are not known to posses any catalytic activity per se and are thought to mediate interactions between PHR/BTBD proteins and their substrates/binding partners. With the structures of MmPHR1 and MmPHR2 in hand, systematic mutational studies of this motif and/or deletion of loop L10 and other conserved surface features can be contemplated.
Structure of the PHR domain explains the loss of function mutation in RPM-1
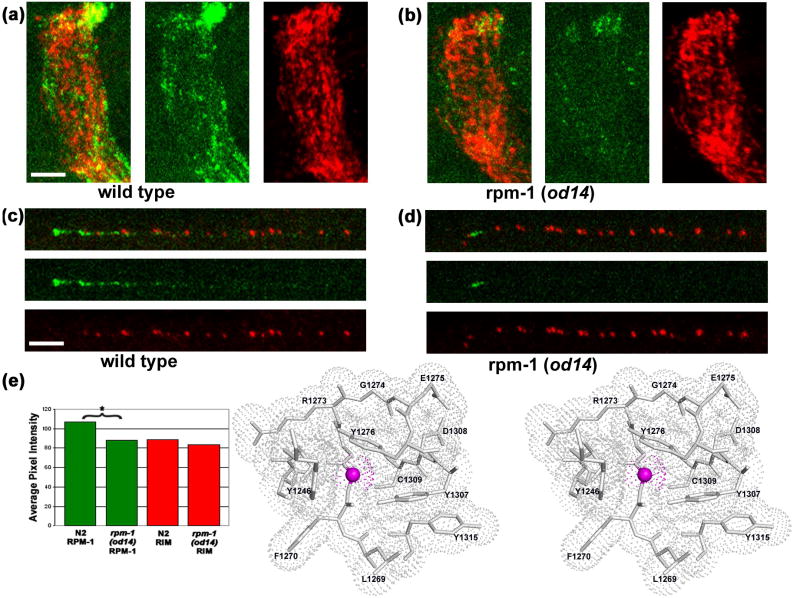
In C. elegans, RPM-1 regulates trafficking of the AMPA-type glutamate receptor GLR-1 between synapses and endosomes.12 RPM-1 regulates GLR-1 trafficking by repressing the PMK-3/p38 MAP kinase cascade via ubiquitinylation/proteasomal degradation of DLK-1. Distribution of GLR-1 at the synapses is also controlled by LIN-10, C. elegans homolog of Mint in mammals. LIN-10 deficient mutants exhibit accumulation of GLR-1 in large non-synaptic sites throughout neurites, with decreased levels of normal punctate synaptic GLR-1 distribution. od14, an allele of C. elegans rpm-1, enhances the lin-10 phenotype. The od14 allele corresponds to the Gly1092→Glu mutant of C. elegans RPM-1.12 Despite this mutation within the first PHR domain of C. elegans RPM-1, the product of the od14 allele of rpm-1 localizes normally to synaptic regions and the tip of the head motor neurons, albeit with decreased intensity (Fig. 4a-e). Therefore, the Gly1092→Glu mutation affects RPM-1 function as it obtains to GLR-1 accumulation (i.e., the lin-10 phenotype) without affecting RPM-1 localization, which depends on the presence of both PHR domains.27
Fig. 4. Structural insights into the loss of function mutation.
(a, c) Wild type (N2) worms stained with anti-RPM-1 (green) and anti-RIM (red), a presynaptic active zone protein,43 show strong and punctate staining with each antibody. (a) Wild type nerve ring. (c) Wild type SAB neuron. (b, d) rpm-1(od14) animals show localization similar to that of wild type with a significant reduction in average RPM-1 signal intensity. (b) rpm-1(od14) nerve ring. (d) rpm-1(od14) SAB neuron. (e) Quantification27 of nerve ring staining intensity for RPM-1 (green) and RIM (red) in N2 and rpm-1(od14), * indicates a statistically significant difference (p<0.001). Scale bar 5μm. (f) Gly1271 of MmPHR1 is equivalent to residue Gly1092 of RPM-1, the C. elegans ortholog of mouse Phr-1. Surrounding residues are shown as atomic stick figures, with overlying van der Waals dot surface. The surface above Cα of Gly1271 is colored magenta.
Whole-mount staining was performed using a Finney-Ruvkun based protocol,44 with a borate buffer (pH 9.3) reduction step. Rat anti-RPM-1 antibodies were used at 1:300 (reference 27), Rabbit anti-RIM antibodies were used at 1:1000.43 Anti-rat Alexa 594 antibody was used with RPM-1 and anti-rabbit Cy5 was used with RIM. Both secondary antibodies were used at 1:1000. Images were acquired on a Zeiss LSM 5 Pascal confocal microscope as Z-stacks. Detector gain was tuned to minimize pixel saturation and maximize detection range. All images were acquired with exactly the same channel and detector settings for each genotype. Projections were made of the z-stack images using Zeiss Pascal software, exported as tif files and processed in parallel using Adobe Photoshop. For quantitative analyses the same size region (as shown) was used for each nerve ring intensity measurement. Average pixel intensity measurements were performed on each region for each channel using Metamorph (Molecular Devices Corporation, Chicago IL) n=9 for each genotype. Measurements were compared using a Welch Two Sample T-test performed in “R” (see The Foundation for Statistical Computing, http://www.R-project.org).
Examination of the structural environment of MmPHR1 Gly1271, the equivalent residue in murine Phr-1, provides insights into this C. elegans RPM-1 loss of function of mutation. Gly1271 is located at the N-terminus of loop L3. All residues comprising L3 (1271GGRGERY1276) are absolutely conserved across available PHR1 sequences (Fig. 2a). L3 is one of several loops of MmPHR1 that decorate the conserved apical surface (Fig. 2b). Gly1271 is situated in a pocket lined by residues derived from loops L1, L3, and L5. This pocket is lined by both main chain and sidechain atoms of Tyr1246, Phe1270, Gly1272, Arg1273, Gly1274, Glu1275, Tyr1276, Tyr1307, Asp1308, and Cys1309. The Cα atom of Gly1271 fits tightly into this pocket (Fig. 4f). Not surprisingly, COOT30 modeling of the Gly1271→Glu mutation resulted in severe steric clashes with residues lining the pocket for all allowed rotamers of the Glu sidechain. For example, the CG, CD, and OE2 atoms of Glu1271 clash with the sidechains of Tyr1307 for the most probable rotamer. Similarly, modeling the second most probable rotamer of Glu1271 gave rise to a clash of CG with the sidechain of Tyr1276.
Given the tight fit of the Gly residue into this conserved pocket, the Glu mutant is expected to disturb the topography of the conserved apical surface of RPM-1 and we believe this disruption affects protein-protein interactions that are necessary for proper function of RPM-1 in the nematode nervous system. The Gly1271 backbone torsion angles fell within the allowed region for non-glycine residues. It is unlikely that the Gly1092Glu mutation unfolds the PHR1 domain of RPM-1, because antibody staining of the Gly1092Glu mutant of RPM-1 showed essentially normal protein localization (Fig. 4), which depends on the presence of both PHR domains.27 In summary, the structures of the PHR domains from murine Phr1, described herein, explain a loss of function mutation in the C. elegans ortholog of murine Phr1 and set the stage for future research on PHR proteins focused on conserved surface residues/features.
The DNA template required production of MmPHR1 encoding residues 1213-1402 was designed for codon-optimal expression in E. coli and synthesized (DNA 2.0 Inc., CA, USA). The synthetic DNA template for MmPHR2 encoding residues 1716-1898 was also obtained via total gene synthesis. Sequence numbering is based on Genbank NP_997098 (geneID: 105689). The desired constructs (residues 1213-1390, 1213-1402, 1229-1390, and 1229-1402 of MmPHR1 and 1716-1883, 1716-1898, 1723-1883, and 1723-1898 of MmPHR2) were PCR amplified using appropriate primers and subsequently TOPO® (Invitrogen, USA) cloned into pSGX3, a derivative of pET26 b(+), giving rise to recombinant proteins with non-cleavable C-terminal hexa histidine tags. Plasmids were transformed into BL21(DE3)-Condon+RIL (Invitrogen, USA) cells for overexpression. Expression of Se-Met proteins was carried out in 1L of HY media, containing 50μg/ml of kanamycin and 35μg/ml of chloramphenicol, at 22°C with IPTG induction. Cells were harvested after 21 hours by centrifugation at 4°C.
For purification, the E. coli cell pellet was resuspended in 30mL of cold buffer, containing 20mM Tris HCl pH 8.0, 500mM NaCl, 25mM Imidazole, and 0.1% (w/v) Tween20, and cells were lysed with a sonication robot. Debris was removed by centrifugation at 4°C. The decanted supernatant was applied to a 5mL HisTrapHP column (GE Health Care, USA), charged with nickel and pre-equilibrated with 20mM Tris HCl pH 8.0, 500mM NaCl, 10% (v/v) glycerol, and 25mM Imidazole. The sample was washed with 5 column volumes (CV) of 20mM Tris HCl pH 8.0, 500mM NaCl, 10% (v/v) glycerol, and 40mM imidazole, and subsequently eluted with 2 CV of the same buffer with an imidazole concentration of 250mM. Eluted protein was passed over a 120ml Superdex 75 column, equilibrated with 10mM HEPES pH 7.5, 150mM NaCl, 10% (v/v) glycerol, and 5mM DTT. SDS-PAGE analysis showed greater than 95% purity and protein fractions corresponding to the symmetric portion of the size exclusion chromatography profile were pooled for concentration using AMICON spin filters. Concentrated protein aliquots were frozen in liquid nitrogen and stored at -80°C.
Protein samples were subjected to initial crystallization screening with the Classics, Classics II, and PEG kits (Qiagen, USA) using a Phoenix Liquid Handling System (Art Robins, USA) with equal volumes (0.3μL) of protein and reservoir solution in 96-well sitting drop format at 21°C. MmPHR1 crystals (SL+residues 1229-1390+EGHHHHHH, protein concentration=12.2 mg/ml) grew in 100mM Sodium Acetate pH 4.6, 25% PEG8000 (Qiagen PEG Condition 40) and were cyro protected with addition of 20% (v/v) ethylene glycol and flash frozen by immersion in liquid nitrogen. For MmPHR2 (SL+residues 1723-1883+EGHHHHHH, protein concentration=9.5mg/ml) initial crystals grew in 2.1M D,L-malic acid pH 7.0 (Qiagen Classics II Condition 23) containing 2100mM D,L-malic acid (pH 7.0). Diffraction quality crystals of MmPHR2 were obtained via sitting drop vapor diffusion by mixing 2μL of protein solution with 1μL of 2M D,L-malic acid (pH 7.0). Diffraction data were recorded using LRL-CAT 31-ID beamline at the APS and processed with MOSFLM31 and SCALA32.
Supplementary Material
Sequence alignment of the first and second PHR domains of PHR proteins from 14 organisms with the human BTBD1, BTBD2, BTBD3 and BTBD6 proteins. Absolutely conserved residues are shaded in blue.
Table 1.
Crystallographic statistics.
| Data Collection | MmPHR1 | MmPHR2 |
|---|---|---|
| PDB code: | 3GBW | 3HWJ |
| Space group: | P212121 | C2221 |
| Unit-cell dimensions (Å): | a=41.7, b=57.8, c=64.6 | a=93.9, b=99.9, c=83.1 |
| Matthew’s coefficient (Å3/Da): | 2.22 | 2.57 |
| Solvent content (%): | 45 | 52 |
| Resolution (Å): | 41.70-1.32 (1.39-1.32)* | 42.80-2.25 (2.37-2.25)* |
| Number of unique reflections: | 37295 (5369) | 18914 (2732) |
| Completeness (%): | 99.7 (100.0) | 100.0 (100.0) |
| Rsym (%): | 5.2 (19.5) | 13.0 (64.3) |
| Multiplicity: | 11.6 (10.8) | 7.3 (7.3) |
| < I/σ(I) >: | 23.2 (16.4) | 8.7 (3.7) |
| Refinement | ||
| Resolution (Å): | 18.45-1.32 | 31.94-2.25 |
| Number of unique reflections: | 37210 | 18890 |
| Number of reflection for Rfree: | 1864 | 972 |
| Rcryst (%): | 16.2 | 19.8 |
| Rfree (%): | 18.9 | 25.6 |
| RMS deviations from ideal values | ||
| Bond angles (Å): | 0.009 | 0.017 |
| Bond angles (°) | 1.32 | 1.72 |
| Average B-factors (Å2) | ||
| Protein | 12.7 | 42.8 |
| Water molecules | 25.8 | 44.9 |
| Ligands | None | 60.5 |
| Ramachandran Plot32: | ||
| MolProbity33 Residues in | ||
| Favored region (%): | 97.7 | 97.1 |
| Allowed region (%): | 100.0 | 99.7 |
Values in parenthesis correspond to the highest-resolution shell
Acknowledgments
This work was carried out at SGX Pharmaceuticals, Inc. and Eli Lilly and Company. Funding for this work was provided by NIH Grants U54 GM074945 (PI: S.K. Burley) and NS035546 (PI: Y.Jin). Use of the Advanced Photon Source (APS) was supported by the U.S. Department of Energy, Office of Basic Energy Sciences. Use of the LRL-CAT beam line facilities at Sector 31 of the APS was provided by Eli Lilly and company, which operates the facility.
Footnotes
Accession numbers Coordinates and structure factors of MmPHR1 were deposited to the Protein Data Bank (PDB, http://www.rcsb.org) on February 20, 2009 with accession code 3GBW and those of MmPHR2 were deposited on June 17, 2009 with accession code 3HWJ. The New York SGX Research Center for Structural Genomics (NYSGXRC) target identifier for mouse Phr-1 in TargetDB (http://targetdb.pdb.org) is “NYSGXRC-13170b”. Clone sequences and further experimental information are available in PepcDB (http://pepcdb.pdb.org/). BTBD1 corresponds to “NYSGXRC-10601a”, BTBD2→“NYSGXRC-10602a”, BTBD3→“NYSGXRC-10603a”, and BTBD6→“NYSGXRC-10606a”.
Authors declare no financial interest related to the work described in this communication.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Jung T, Catalgol B, Grune T. The proteasomal system. Mol Aspects of Med. 2009;30:191–296. doi: 10.1016/j.mam.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Hass KF, Broadie K. Roles of ubiquitination at the synapse. Biochim Biophy Acta. 2008;1779:495–506. doi: 10.1016/j.bbagrm.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fulga TA, Van Vactor D. Synapses and growth cones on two sides of a Highwire. Neuron. 2008;57:339–344. doi: 10.1016/j.neuron.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 5.Segref A, Hoppe T. Think locally: control of ubiquitin-dependent protein degradation in neurons. EMBO Reports. 2009;10:44–50. doi: 10.1038/embor.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo O, Xie J, Dang CV, Liu ET, Bishop JM. Identification of a large Myc-binding protein that contains RCC1-like repeats. Proc Natl Acad Sci USA. 1998;95:9172–9177. doi: 10.1073/pnas.95.16.9172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wan HI, DiAntonia A, Fetter RD, Bergstrom K, Strauss R, Goodman CS. Highwire regulates synaptic growth in Drosophila. Neuron. 2000;26:313–329. doi: 10.1016/s0896-6273(00)81166-6. [DOI] [PubMed] [Google Scholar]
- 8.Zhen M, Huang X, Bamber B, Jin Y. Regulation of presynaptic terminal organization by C. elegans RPM-1, a putative guanine nucleotide exchanger with a RING-H2 finger domain. Neuron. 2000;26:331–343. doi: 10.1016/s0896-6273(00)81167-8. [DOI] [PubMed] [Google Scholar]
- 9.Schaefer AM, Hadwiger GD, Nonet ML. rpm-1, A conserved neuronal gene that regulates targeting and synaptogenesis in. C elegans Neuron. 2000;26:345–356. doi: 10.1016/s0896-6273(00)81168-x. [DOI] [PubMed] [Google Scholar]
- 10.Burgess RW, Peterson KA, Johnson MJ, Roix JJ, Welsh IC, O’Brien TP. Evidence for a conserved function in synapse formation reveals Phr1 as candidate gene for respiratory failure in newborn mice. Mol Cell Biol. 2004;24:1096–1105. doi: 10.1128/MCB.24.3.1096-1105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Souza J, Hendricks M, Le Guyader S, Subbjuraju S, Grunewald B, Scholich K, Jesuthasan S. Formation of the retinotectal projection requires Esrom, an ortholog of PAM (protein associated with Myc) Development. 2004;132:247–256. doi: 10.1242/dev.01578. [DOI] [PubMed] [Google Scholar]
- 12.Park EC, Glodowski DR, Rongo C. The ubiquitin ligase RPM-1 and the p38 MAPK PMK-3 regulate AMPA receptor trafficking. PLoS One. 2009;4:e4284. doi: 10.1371/journal.pone.0004284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewcock JW, Genoud N, Lettieri K, Pfaff SL. The ubiquitin ligase Phr1 regulates axon outgrowth through modulation of microtubule dynamics. Neuron. 2007;56:604–620. doi: 10.1016/j.neuron.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 14.Hendricks M, Jesuthasan S. PHR regulates growth cone pausing at intermediate targets through microtubule disassembly. J Neurosci. 2009;29:6593–6598. doi: 10.1523/JNEUROSCI.1115-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bloom AJ, Miller BR, Sanes JR, DiAntonio A. The requirement for Phr1 in CNS axon tract formation reveals the corticostriatal boundary as a choice point for cortical axons. Genes & Develop. 2007;21:2593–2606. doi: 10.1101/gad.1592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pertz OC, Wang Y, Yang F, Wang W, Gay LJ, Gristenko MA, Clauss TR, Anderson DJ, Liu T, Auberry KJ, Camp DG, II, Smith RD, Klemke RL. Spatial mapping of the neurite and soma proteomes reveals a functional Cdc42/Rac regulatory network. Proc Natl Acad Sci USA. 2008;105:1931–1936. doi: 10.1073/pnas.0706545105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Ding S-J, Wang W, Jacobs J, Qian W-J, Morre RJ, Yang F, Camp DG, II, Smith RD, Klemke RL. Profiling signaling polarity in chemotactic cells. Proc Natl Acad Sci USA. 2007;104:8328–8333. doi: 10.1073/pnas.0701103104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakata K, Abrams B, Grill B, Goncharov A, Huang X, Chishom AD, Jin Y. Regulation of a DLK-1 and p38 MAP kinase pathway by the ubiquitin ligase RPM-1 is required for presynaptic development. Cell. 2005;120:407–420. doi: 10.1016/j.cell.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 19.Collins CA, Wairkar YP, Johnson SL, DiAntonio A. Highwire restrains synaptic growth by attenuating a MAP kinase signal. Neuron. 2006;51:57–69. doi: 10.1016/j.neuron.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 20.Liao EH, Hung W, Abrams B, Zhen M. An SCF-like ubiquitin ligase complex that controls presynaptic differentiation. Nature. 2004;430:345–350. doi: 10.1038/nature02647. [DOI] [PubMed] [Google Scholar]
- 21.Wu C, Daniels RW, DiAntonio A. DFsn collaborates with Highwire to down-regulate the Wallenda/DLK kinase and restrain synaptic terminal growth. Neural Develop. 2007;2:16. doi: 10.1186/1749-8104-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han S, Witt RM, Santos TM, Polizzano C, Sabatini BL, Ramesh V. Pam (Protein associated with Myc) functions as an E3 ubiquitin ligase and regulates TSC/mTOR signaling. Cellular Signaling. 2008;20:1084–1091. doi: 10.1016/j.cellsig.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCabe BD, Hom S, Aberle H, Fetter RD, Marques G, Haerry TE, Wan H, O’Connor MB, Goodman CS, Haghighi AP. Highwire regulates presynaptic BMP signaling essential for synaptic growth. Neuron. 2004;41:891–905. doi: 10.1016/s0896-6273(04)00073-x. [DOI] [PubMed] [Google Scholar]
- 24.Pierre SC, Hausler J, Birod K, Geisslinger G, Scholich K. PAM mediates sustained inhibition of cAMP signaling by sphingosine-1-phosphate. EMBO J. 2004;23:3031–3040. doi: 10.1038/sj.emboj.7600321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grill B, Bienvenut WV, Brown HM, Ackley BD, Quadroni M, Jin Y. C. elegans RPM-1 regulates axon termination and synaptogenesis through the Rab GEF GLO-4 and the Rab GTPase GLO-1. Neuron. 2007;55:587–601. doi: 10.1016/j.neuron.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 26.Xu L, Yang L, Hashimoto K, Anderson M, Kohlhagen G, Pommier Y, D’Arpa P. Characterization of BTBD1 and BTBD2, two similar BTB-domain-containing Kelch-like proteins that interact with Topoisomerase I. BMC Genomics. 2003;3:1. doi: 10.1186/1471-2164-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abrams B, Grill B, Huang X, Jin Y. Cellular and molecular determinants targeting the Caenorhabditis elegans PHR protein RPM-1 to perisynaptic regions. Develop Dynamics. 2008;237:630–639. doi: 10.1002/dvdy.21446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura T, Kotani M, Tonozuka T, Ide A, Oguma K, Nishikawa A. Crystal structure of the HA3 subcomponent of Clostridium botulinum type C progenitor toxin. J Mol Biol. 2009;385:1193–1206. doi: 10.1016/j.jmb.2008.11.039. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Ramakrishnan C, Ramachandran GN. Stereochemical criteria for polypeptide and protein chain conformations. II. Allowed conformations for a pair of peptide units. Biophys J. 1965;5:909–933. doi: 10.1016/S0006-3495(65)86759-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, III, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucl Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leslie AGW, Brick P, Wonacott AJ. An improved program package for the measurement of oscillation photographs. CCP4 News. 1986;18:33–39. [Google Scholar]
- 34.Collaborative Computing Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallog Sect D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 35.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallog Sect D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 36.Pape T, Schneider TR. HKL2MAP: a graphical user interface for phasing with SHELX program. J Appl Crystallog. 2004;37:843–844. [Google Scholar]
- 37.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 38.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the Maximum-Likelihood Method. Acta Crystallogr Sect D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 39.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. PHASER crystallographic software. J Appl Crystallog. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. ClustalW and ClustalX version2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. http://www.ebi.ac.uk/Tools/clustalw2/index.html. [DOI] [PubMed]
- 41.Gouet P, Robert X, Courcelle E. ESPript/ENDscirpt: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucl Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi. [DOI] [PMC free article] [PubMed]
- 42.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucl Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. http://consurf.tau.ac.il. [DOI] [PMC free article] [PubMed]
- 43.Koushika SP, Richmond JE, Hadwiger G, Weimer RM, Jorgensen EM, Nonet ML. A post-docking role for active zone protein Rim. Nat Neurosci. 2001;4:997–1005. doi: 10.1038/nn732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Finney M, Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C elegans. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignment of the first and second PHR domains of PHR proteins from 14 organisms with the human BTBD1, BTBD2, BTBD3 and BTBD6 proteins. Absolutely conserved residues are shaded in blue.