Abstract
The killer cell immunoglobulin-like receptors (KIR) interact with major histocompatibility complex (MHC) class I ligands to regulate the functions of natural killer cells and T cells. Like human leukocyte antigens class I, human KIR are highly variable and correlated with infection, autoimmunity, pregnancy syndromes, and transplantation outcome. Limiting the scope of KIR analysis is the low resolution, sensitivity, and speed of the established methods of KIR typing. In this study, we describe a first-generation single nucleotide polymorphism (SNP)-based method for typing the 17 human KIR genes and pseudogenes that uses analysis by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. It is a high-throughput method that requires minute amounts of genomic DNA for discrimination of KIR genes with some allelic resolution. A study of 233 individuals shows that the results obtained by the SNP-based KIR/MALDI-TOF method are consistent with those obtained with the established sequence-specific oligonucleotide probe or sequence-specific polymerase chain reaction methods. The added sensitivity of the KIR/MALDI-TOF method allowed putative novel alleles of the KIR2DL1, KIR3DL1, KIR2DS5, and KIR2DL5 genes to be identified. Sequencing the KIR2DL5 variant proved it was a newly discovered allele, one that appears associated with Hispanic and Native American populations. This KIR/ MALDI-TOF method of KIR typing should facilitate population and disease-association studies that improve knowledge of the immunological functions of KIR–MHC class I interactions.
Keywords: KIR, HLA, MALDI-TOF, Genotyping, SNP
Introduction
Killer immunoglobulin-like receptors (KIR) are a family of inhibitory and activating major histocompatibility complex (MHC) class I receptors expressed on natural killer (NK) cells and subpopulations of T cells (Bottino et al. 2005; Moretta 2002). The inhibitory KIR, in partnership with CD94:NKG2A, ensure that NK cells are tolerant of healthy autologous cells and responsive to cells with compromised MHC class I expression (Carrington and Norman 2003), as occurs frequently in virus-infected and tumor cells. Although their ligands and functions are less clearly defined, the activating KIR are hypothesized to contribute to the activation of NK cells in response to infection and malignancy.
The KIR gene family is part of the leukocyte receptor complex, located on human chromosome 19q13.4. The KIR complex comprises a tandem array of highly homologous genes, which exhibits haplotypic variation in gene content as well as polymorphism of the individual KIR genes. As a consequence of these variations, unrelated individuals usually differ in KIR genotype. Specific KIR, either alone or in combination with human leukocyte antigens (HLA) class I, have been correlated with a variety of diseases and syndromes. These include the outcome of bone marrow transplantation (Cook et al. 2004; Gagne et al. 2002; Parham and McQueen 2003; Ruggeri et al. 2002), the progress and severity of virally transmitted disease (Christiansen et al. 2003; Khakoo et al. 2004; Martin et al. 2002; Trachtenberg et al. 2004), the probability of preeclampsia during pregnancy (Hiby et al. 2004), and susceptibility to autoimmune diseases such as psoriatic arthritis, scleroderma, sarcoidosis, and type 1 diabetes (Mizuki et al. 2000; Momot et al. 2004; Nelson et al. 2004; van der Slik et al. 2003). Collectively, these studies make a convincing case that interactions between variable HLA class I ligands and variable KIR have considerable impact on human health. Consequently, better understanding of the underlying mechanisms could influence future clinical practice.
Limiting interpretation of the published studies on clinical associations and population comparisons of KIR are the small cohorts analyzed and the low resolution of the KIR genotyping, which has mainly involved determination of the presence and absence of KIR genes. The key problems are that the established methods of KIR genotyping are labor intensive and require more DNA (300 ng–5 μg) than is compatible with many of the samples that are available and informative to study. What are needed are new methods that combine speed, economical consumption of DNA, and a resolution that distinguishes all KIR alleles. To this end, we have explored the use of matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS), as a technique that can accurately and rapidly detect single nucleotide polymorphisms (SNPs), the dominant feature of KIR variation. In this study, we describe the development of a MALDI-TOF MS-based method for highly accurate high-throughput KIR genotyping, its comparison to the established KIR typing methods, and its application to epidemiological studies and the discovery of novel KIR alleles.
Materials and methods
Assay design and development
The SEQUENOM™ MALDI-TOF MS-based homogenous mass extend (hME) platform was used to develop the high-throughput SNP-based KIR/MALDI genotyping assay. The instrumentation and related assay design and genotyping software comprise the MassARRAY™ MALDI-TOF system (San Diego, CA).
SNPs that differentiate the individual KIR genes were identified by inspection in silico of the 17 locus KIR sequence alignment in the Immuno Polymorphism Database (IPD; http://www.ebi.ac.uk/ipd/kir). A region of ∼400 bp surrounding the SNP of interest was targeted for capture after evaluation for homology in the flanking regions and base composition of the extension primer site. The targeted sequence was uploaded to the SEQUENOM™ MassArray Assay Design software, which assisted in the design of primers, typically 17–21 bp in length, to capture a ∼150–250 bp region surrounding the SNP (Table 1). The “capture primers” were designed with a 5′ 10-mer tag to keep them out of the 5–10 kDa window of detection used for this application; without the 10-mer tag, the capture primers would have similar mass to the extended and unextended hME products and confound the analysis. The 10-mer tag also helped to balance the primers so that they functioned well in multiplex reactions, which were initially identified using the SEQUENOM™ Design software, and then optimized empirically. Because of the high degree of homology between the KIR loci, some assays could not be multiplexed.
Table 1.
Primer sequences and plex level and composition for the KIR/MALDI genotyping assay
| Well number | SNP assay name | 5′ Capture primer (5′ 10-mer tag not shown) | 3′ Capture primer (5′ 10 mer tag not shown) | Extend primer |
|---|---|---|---|---|
| W1 4 plex | 2DS3.D1.G | CTGTGATCACGATGTCCAG | AAGGCCAACTTCTCCATCGG | CACTCCCCCTATCAGTT |
| 2DL2.D2.S | GAGCTCCTATGACATGTACC | GCCTGGAATGTTCCGTTGACCTTG | CCCTGCAGAGAACCTAC | |
| 3DL3.D1.G | GATGACTAAGGACCCCTTGC | TCATGGGACCCATGGAATAG | AATAGTTGACCTGGGAACCC | |
| 3DL3.D2.G | AGAATGTGACCTTGTCCTGC | CAGTGAGCCTAAGTTCACCG | GGATAGATGGTAAATGTCAAA | |
| W2 3 plex | 2DL4.DO.G | TGCCGACCACTCAGTGGG | CCCTGAGCTCTACAACAGAA | TGGAACAGTTTCCTCAT |
| 3DL2.TC.G | GATGAACAAGACCCTCAGGAGGTG | TACACGCTGGTATCTGTT | GCCTCTGAGAAGGGCGA | |
| 3DS1.3DL1.D1.S | CAAGGCCAATTTCTCCATCG | GGGAGCTGACAACTGATAGG | CTGTAGGTCCCTGCAAGGGCA | |
| W3 4 plex | 2DL5.D2.G | TGACAGAAACAAGCAGTGGG | GACTTTCCTCTGGGCCCTG | CCACGGAGGGACCTACA |
| 2DL5.TC.G | CTTGGGCCTCTGAGAAGGG | CAAGACCCTCAGGAGGTGAC | CACTGCGTTTTCACACAGA | |
| 2DS4.D1.G | AGAGACAGTCATCCTGCAATG | ATGGAGAAGTTGGCCTTGGA | GAAGTGCTCAAACATGACATC | |
| 3DL2.D1.G | GGAGCTGACAACTGATAGGG | CCAAGGCCAACTTCTCCATC | CTTGCAGGAACCTACAGATG | |
| W4 3 plex | 2DL1.D2.G.no004 | GACTTTGACCACTCGTAT | CAGGGCCCAAGGTCAACG | ATGCTTCGGCTCTTTCC |
| 2DL4.TC.G | ATCTGTTGAGGGTCTCTTGC | AGGTGACATACGCACAGTTG | CACAGTTGGATCACTGC | |
| 2DL2.004.TC.G | GGCCGAGGAGTACCTACCT | GTAATGGACCAAGAGTCTGC | GAAACAGAACAGCGAATA | |
| W5 2 plex | 2DS2.D1.G | AGAAGTTGGCCTTGGAGACC | CCTGCAATGTTGGTCAGATG | GCACAGAGAGGGGAAGT |
| 3DL1.TC.S | ATGGGCAGGAGACAACTTTG | CACTGCGTTTTCACACAGAG | GAGGCCCAAGACACCCCC | |
| W6 2 plex | 2DS5.D2.G | AGGCCCATGAACGTAGGCTCC | AAGAGCCGAAGCATCTGTAG | CTCCGTGGGTGGCAGGG |
| 2DL1.no005.2DL2. 004.TC.S | GTAATGGACCAAGAGTCTGC | CGGGCCGAGGAGTACCTACCT | CGCTATTCGCTGTTCTGTT | |
| W7 uniplex | 2DL3.2DL2.D1.S | GAGTCCACAGAAAACCTTCCCTCC | AGTGTCCTTAAACTTCCCTTCTC | CTTCTGATTTCACCAGG |
| W8 2 plex | 2DS3.D2.S | AGGTCAACGGAACATTCCAGGCCG | AAGAGCCGAAGCATCTGTAG | CATCTGTAGGTTCCTCC |
| 3DS1.TC.G.INT | AACTGCTATGATTAGCTTC | GATGAAGGAGAAAGAAGAGGAGGA | GAATGTGCAGGTGTCTG | |
| W9 5 plex | 3DP1.D2.G | GAGCTGCAGGACAAGGTCAC | TGGGAAACCTTCTCTCTCAGCC | CTCTCTCAGCCCAGCCG |
| 3DS1.DO.S | TCATGCTATACAAAGAAGAC | TGTGTAGTTCCCTGCATGTG | AGGGCTCATGTTGAAGC | |
| 2DL5sub5 | GATCTTGGCTTAGCATTTGG | CTGCGTTTTCACACAGAC | CTTCTCAGAGGCCCAAG | |
| 2DL5sub4 | CCACGGAGGGACCTACAC | GTGACAGAAACAAGCAGTGG | GTGAGTCATGGAGAGAGC | |
| 2DL5sub1 | AGGACAAGCCCTTGCTGTCT | CAAGACGAGAGCGACACA | GTCCTCCTCGAGGCACCACAG | |
| W10 2 plex | 2DS5.D1.G | ACACTTTGCGCCTCATTGGAG | GTGAGTAACAGAACCGTAG | GACCGATGGAGAAGTTG |
| 2DP1.D0.G | GGGTTTAACAACTTCAGTCTGT | TGTGCTGGGGTCACAGGGCC | ATTCTGTTGTAGAGCTCAG | |
| W11 2plex | 2DS2.D2.G | GTCTATATGAGAAACCTTC | GGACAAGGTCACGCTCTCTC | CACGCTCTCTCCTGCCA |
| 2DL1.2DS1.D1.S.tri | AAGGCCAACTTCTCCATCA | GTGAGTAACAGAACCGTAGC | GGTCCCTGCCAGGTCTTGC | |
| W12 2 plex | 2DL5sub3 | GACATGAGTCCTCTGACCTG | CCCTGAGCTCTACAACAA | GCAACCCCCTGGTGATC |
| 2DL5sub2 | GACATGAGTCCTCTGACCTG | CCCTGAGCTCTACAACAA | CGCTCCCCCATTGAGTGGTC | |
| W13 2 plex | 2DS4del.sub | TTGACCACTCGTAGGGAGC | CGGTTCAGGCAGGAGAGAAT | CCTTGTCCTGCAGCTCC |
| 2DL5sub6 | TAAGGTGGCGCCTCCTTCTC | CAAGACGAGAGCGACACA | AGCAAGGGCTTGTCCTG | |
| W14 2plex | 2DS4.D2.S | GAGCTCTGTGACGGAAACAA | GCATCAACGGAACATTCCAGGCC | TCGGCTCTTTCCGTGAC |
| 2DL3.TC.S.INT | CTGCTTCGTGAGACTTACTT | GTAACCCCAGACACCTGCATG | TCTCCTTCATCGCTGGTGCT | |
| W15 uniplex | 2DL2.001.2.3.D1.G | GGAGCTGACAACTGATAGGG | CCTGCAATGTTGGTCAGATG | CATGATGGGGTCTCCAA |
| W16 uniplex | 2DS1.D2.G | ACTTGACTTTGACCACTCGT | CCTATGACATGTACCATCTA | CAACGGAACATTCCAGGCC |
The assays are named according to the following convention: “Targeted Locus.Targeted Domain.Specificity (G or S).comment.” If more than one locus is targeted, the convention is “Targeted Locus. 2nd Targeted Locus.Targeted Domain.Specificity (G or S)”. A “universal” 10-mer tag (ACGTTGGATG) is added to the 5′ end of each of the capture primers to keep the capture primers out of the “mass window” where the extension primers are measured.
W Well, e.g., W1 well no. 1; INT intronic; G generic, i.e., primers are designed to capture more than the targeted locus; S specific, i.e., primers were designed to exclude most of the KIR genes; D0 domain 0; D1 domain 1; D2 domain 2; Trans/Cyt transmembrane/cytoplasmic domain; sub subtyping primer; tri triallelic choice
Extension primers that query the chosen SNP were designed to terminate at the base immediately adjacent to the SNP (Table 1). Because of regions of high homology in the KIR genes, the designer software required extensive manipulation to obtain the primer designs necessary to create the specificity needed for a working KIR assay. Both capture primers and extension primers were chosen with an eye towards simultaneous identification of a particular locus (locus-specific assay) and a highly conserved region (positive control assay).
Sample populations and DNA preparation
Samples for validation of the method consisted of previously KIR-genotyped International Histocompatibility Working Group (IHWG) samples (Cook et al. 2003; Gomez-Lozano and Vilches 2002) and other controls from Stanford University (Uhrberg et al. 1997; Valiante et al. 1997 and unpublished). The KIR/MALDI method was further validated using two population-based sample groups, which were analyzed by both the KIR/MALDI and at least one other KIR genotyping method. These were 120 samples (59 donor/recipient pairs and two donors) from the National Marrow Donor Program (NMDP) repository and 98 samples from the Chicago Multicenter AIDS Cohort Study (MACS). All samples were genotyped in-house or previously genotyped by other laboratories using sequence-specific oligonucleotide probe (SSOP; Crum et al. 2000; Trachtenberg, unpublished) and/or sequence-specific polymerase chain reaction (PCR; SSP) methodologies (Gomez-Lozano and Vilches 2002; Uhrberg et al. 1997). Data from these previous analyses were used in the validation of the KIR/MALDI method. Whole blood or lymphocyte cell pellet samples were manually extracted using Qiagen’s 96-well block extraction method (QIAamp 96 DNA Blood Kit) or robotically with Genovision’s GenoM-6 magnetic bead-based extraction method. They were then quantitated using Picogreen (Invitrogen, Carlsbad, CA) fluorescence methodology and normalized to a final concentration of 2 ng/ μL. Approximately 10.0% of the samples yielded concentrations below 2 ng/μL and were, therefore, typed using lower DNA concentrations.
KIR genotyping assay
Primary PCR for capture of the region surrounding the SNP was performed on an Applied Biosystems GeneAmp 9700 in a 384-well format in a volume of 5 μL with the following profile: denaturation at 95°C for 15 min, followed by 45 cycles of denaturation at 95°C for 20 s, annealing at 56°C for 30 s, and extension at 72°C for 1 min, and a final extension at 72°C for 3 min. Final concentrations of each component were as follows: 1.25× Qiagen HotStar 10× PCR Buffer, 3.5 mM MgCl2, 500 μM deoxyribonucleotide triphosphate (dNTPs; each), 100 nM PCR primers each, 0.15 U/reaction of Qiagen HotStar Taq, and 2 ng genomic DNA.
To inactivate the primary PCR by dephosphorylating any unicorporated dNTPs, a shrimp alkaline phosphatase (SAP) cocktail consisting of nanopure water, 10× hME buffer (SEQUENOM™), and 0.3 U SAP/reaction was distributed to the 384-well reaction plates using a Beckman Multimek 9600. Plates were then cycled on an Applied Biosystems GeneAmp PCR System 9700 with the following profile: dNTP terminal phosphate cleavage at 37°C for 20 min, followed by heat inactivation of the SAP enzyme at 85°C for 5 min, and a 4°C final hold.
To set up the primer extension reactions, 2 μL of hME cocktail was added directly to the PCR reactions using the Beckman Multimek 9600, bringing the final concentration of each reaction to ∼1 μM each hME primer, 50 μM each d/ddNTP termination mixes, and 1.25 U Thermosequenase (Amersham). Extension reactions were then cycled on an Applied Biosystems GeneAmp PCR System 9700 according to the following profile: denaturation for 2 min at 94°C, followed by 99 cycles of denaturation at 94°C for 5 s, annealing at 52°C for 5 s, and extension at 72°C for 5 s, and a final hold at 4°C. After primer extension, 16 μL of nanopure water was distributed to each reaction using the Beckman Multimek 9600, followed by manual distribution of 6 mg of SpectroClean ion-exchange resin (SEQUE NOM™). Plates were sealed with MJ Research (Hercules,CA) Microseal B adhesive sealers (#MSB-1001), rotated for approximately 15 min, and centrifuged at 3,000 rpm for 5 min. The reactions along with a three-point calibrant were spotted onto SpectroChip 384 chips with a Samsung MassArray NanoDispenser at a dispense speed of 65 mm/s.
The Assay Editor module within SEQUENOM™’s Mass-ARRAY Typer software was used to enter sample information and to map the chip in regards to name of assay, termination mix used, multiplex level, and genotyping rules for each hME.
MALDI-TOF MS analysis
Samples were analyzed on SEQUENOM™’s MassArray Compact MALDI-TOF MS, which processed each 384-well chip in approximately half an hour. Data was automatically retrieved by the SEQUENOM™ SpectroAC-QUIRE software and reviewed using the TrafficLights module of the MassARRAY Typer software.
Detailed information on peak heights for each assay and a probability value for each call, based on signal-to-noise ratios and peak probability statistics, were reviewed for individual samples as necessary. In this system, probability was automatically calculated and parsed into three levels of stringency, resulting in conservative, moderate, and aggressive calls. Conservative calls resulted in the lowest rate of uncalled genotypes, while aggressive calls resulted in a higher error rate (<1%). Low probability calls were excluded. Spectral data were reviewed for individual samples (Fig. 1) when necessary. The Cluster Plot software module was used to visualize atypical heterozygous and homozygous SNP states (Fig. 1).
Fig. 1.
Spectral data (left) and cluster plots (right) illustrating single nucleotide and double nucleotide calls with the hME assay “3DS1.3DL1.D1.S,” which discriminates between KIR3DS1 and KIR3DL1. a and c show spectral data and cluster plots for the single nucleotide call T (3DL1) and G (3DS1), respectively. b shows data for the double nucleotide call T/G (KIR3DL1 and KIR3DS1 both present). Arrows indicate specific data points illustrated in spectral graphs. For all panels, the lowest mass peak (6447.2 Da) represents the unextended primer. The peak produced when the polymerase pauses leading to incorporation of an unexpected dNTP (pausing peak) is also shown—Cluster plots (right) illustrate the intensity of the high mass product (T) peak vs intensity of the low mass product (G) peak for a given sample. The separation between clusters and the tightness within a cluster are indicative of the accuracy and specificity of the assay
Genotype analysis
Because the KIR genotype of any one sample is based on results from many different SNP assays, a second level of automated calling was incorporated for high-throughput genotyping. Our genotyping program “KIR Genotype Caller” was developed to capture the output data from the SpectroAnalyzer module of the SEQUENOM™ software suite to generate a genotype from the individual SNP assays based on a hierarchical SNP-pattern table (Table 2) and simultaneously tag anomalous calls and problematic samples for user inspection. For each run, the KIR Genotype Caller created the following:
Table 2.
Expected SNP for extension primers for 17 KIR genes
| D0
|
D1
|
D2
|
Trans/cytoplasmic
|
subtyping
|
||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2DL4. | 3DS1. | 2DP1. | 2DL1. | 2DL2. | 2DL3. | 2DS2. | 2DS3. | 2DS4. | 2DS5. | 3DL2. | 3DL3. | 3DS1. | 2DL1. | 2DL2. | 2DL5. | 2DS1. | 2DS2. | 2DS3. | 2DS4. | 2DS5. | 3DL3. | 3DP1. | 2DL1. | 2DL2. | 2DL3. | 2DL4. | 2DL5. | 3DL1. | 3DL2. | 3DS1. | 2DL5 | 2DL5 | 2DL5 | 2DL5 | 2DL5 | 2DL5 | 2DS4 | |
| DO.G | DO.S | D0.G | 2DS1. | 001.2. | 2DL2. | D1.G | D1.G | D1.G | D1.G | D1.G | D1.G | 3DL1. | D2.G. | D2.S | D2.G | D2.G | D2.G | D2.S | D2.S | D2.G | D2.G | D2.G | no005. | OO4. | TC.S. | TC.G | TC.G | TC.S | TC.G | TC.G. | sub1 | sub2 | sub3 | sub4 | sub5 | sub6 | del. | |
| D1. | 3.D1. | D1.S | D1.S | no004 | 2DL2. | TC.G | INT | INT | sub | |||||||||||||||||||||||||||||
| S.tri | G | 004. | ||||||||||||||||||||||||||||||||||||
| TC.S | ||||||||||||||||||||||||||||||||||||||
| 2DL1*001 | C | C | A | G | C | G | A | G | G | A | ||||||||||||||||||||||||||||
| 2DL1*002 | C | C | A | G | C | G | A | G | G | A | ||||||||||||||||||||||||||||
| 2DL1*003 | C | C | A | G | C | G | A | G | G | A | ||||||||||||||||||||||||||||
| 2DL1*004 | C | C | G | G | T | C | G | A | G | G | A | |||||||||||||||||||||||||||
| 2DL1*005 | C | C | A | G | C | G | G | G | G | A | ||||||||||||||||||||||||||||
| 2DL2*001 | A | G | T | G | T | C | C | T | G | G | C | G | A | |||||||||||||||||||||||||
| 2DL2*002 | A | G | T | G | T | C | C | T | G | G | C | G | A | |||||||||||||||||||||||||
| 2DL2*003 | A | G | T | G | T | C | C | T | G | G | C | G | A | |||||||||||||||||||||||||
| 2DL2*004 | G | C | T | G | T | C | C | T | A | T | C | G | A | |||||||||||||||||||||||||
| 2DL3*001 | G | C | T | G | C | C | C | C | T | G | G | G | A | C | ||||||||||||||||||||||||
| 2DL3*002 | G | C | T | G | C | C | C | C | T | G | G | G | A | C | ||||||||||||||||||||||||
| 2DL3*003 | G | C | T | G | C | C | C | C | T | G | G | G | A | C | ||||||||||||||||||||||||
| 2DL3*004 | G | C | T | G | C | C | C | C | T | G | G | G | A | C | ||||||||||||||||||||||||
| 2DL3*005 | G | C | T | G | C | C | C | C | T | G | G | G | A | C | ||||||||||||||||||||||||
| 2DL3*006 | G | C | T | G | C | C | C | C | T | G | G | G | A | C | ||||||||||||||||||||||||
| 2DL4*001 | T | A | ||||||||||||||||||||||||||||||||||||
| 2DL4*002 | T | A | ||||||||||||||||||||||||||||||||||||
| 2DL4*003 | T | A | ||||||||||||||||||||||||||||||||||||
| 2DL4*004 | T | A | ||||||||||||||||||||||||||||||||||||
| 2DL4*005 | T | A | ||||||||||||||||||||||||||||||||||||
| 2DL4*006 | T | A | ||||||||||||||||||||||||||||||||||||
| 2DL4*007 | T | A | ||||||||||||||||||||||||||||||||||||
| 2DL5A*001 | C | C | G | A | G | G | A | T | ||||||||||||||||||||||||||||||
| 2DL5B*002 | C | C | G | G | G | A | A | A | ||||||||||||||||||||||||||||||
| 2DL5B*003 | C | C | G | A | A | G | A | T | ||||||||||||||||||||||||||||||
| 2DL5B*004 | C | C | A | A | G | G | A | T | ||||||||||||||||||||||||||||||
| 2DS1*001 | G | G | C | C | T | C | C | |||||||||||||||||||||||||||||||
| 2DS1*002 | A | G | C | C | T | C | C | |||||||||||||||||||||||||||||||
| 2DS1*003 | A | G | C | C | T | C | C | |||||||||||||||||||||||||||||||
| 2DS1*004 | A | G | C | C | T | C | C | |||||||||||||||||||||||||||||||
| 2DS2*001 | G | A | C | C | T | C | C | |||||||||||||||||||||||||||||||
| 2DS2*002 | G | A | C | C | T | C | C | |||||||||||||||||||||||||||||||
| 2DS2*003 | G | A | C | C | T | C | C | |||||||||||||||||||||||||||||||
| 2DS2*004 | G | A | C | C | T | C | C | |||||||||||||||||||||||||||||||
| 2DS2*005 | G | A | C | C | T | C | C | |||||||||||||||||||||||||||||||
| 2DS3*001 | T | C | C | C | A | C | ||||||||||||||||||||||||||||||||
| 2DS4*001 | G | C | G | G | C | C | ||||||||||||||||||||||||||||||||
| 2DS4*002 | G | C | G | G | C | C | ||||||||||||||||||||||||||||||||
| 2DS4*003 | G | C | G | G | C | A | ||||||||||||||||||||||||||||||||
| 2DS5*001 | G | C | A | C | ||||||||||||||||||||||||||||||||||
| 2DS5*002 | G | C | A | C | ||||||||||||||||||||||||||||||||||
| 2DS5*003 | G | C | A | C | ||||||||||||||||||||||||||||||||||
| 3DL1*001 | A | T | T | C | ||||||||||||||||||||||||||||||||||
| 3DL1*002 | A | T | T | C | ||||||||||||||||||||||||||||||||||
| 3DL1*003 | A | T | T | C | ||||||||||||||||||||||||||||||||||
| 3DL1*004 | A | T | T | C | ||||||||||||||||||||||||||||||||||
| 3DL1*005 | A | T | T | C | ||||||||||||||||||||||||||||||||||
| 3DL1*006 | A | T | T | C | ||||||||||||||||||||||||||||||||||
| 3DL1*007 | A | T | T | C | ||||||||||||||||||||||||||||||||||
| 3DL1*008 | A | T | T | C | ||||||||||||||||||||||||||||||||||
| 3DL1*009 | G | T | T | C | ||||||||||||||||||||||||||||||||||
| 3DS1*010 | G | G | C | T | ||||||||||||||||||||||||||||||||||
| 3DS1*011 | G | G | C | T | ||||||||||||||||||||||||||||||||||
| 3DS1*012 | G | G | C | T | ||||||||||||||||||||||||||||||||||
| 3DS1*013 | G | G | C | T | ||||||||||||||||||||||||||||||||||
| 3DS1*014 | G | G | C | T | ||||||||||||||||||||||||||||||||||
| 3DL2*001 | G | G | A | T | G | C | G | G | ||||||||||||||||||||||||||||||
| 3DL2*002 | G | G | A | T | G | C | G | G | ||||||||||||||||||||||||||||||
| 3DL2*003 | G | G | A | T | G | C | G | G | ||||||||||||||||||||||||||||||
| 3DL2*004 | G | G | A | T | G | C | G | G | ||||||||||||||||||||||||||||||
| 3DL2*005 | G | G | A | T | G | C | G | G | ||||||||||||||||||||||||||||||
| 3DL2*006 | G | G | A | T | G | C | G | G | ||||||||||||||||||||||||||||||
| 3DL2*007 | G | G | A | T | G | C | G | G | ||||||||||||||||||||||||||||||
| 3DL2*008 | G | G | A | T | G | C | G | G | ||||||||||||||||||||||||||||||
| 3DL2*009 | G | G | A | T | G | C | G | G | ||||||||||||||||||||||||||||||
| 3DL2*010 | G | G | A | T | G | C | G | G | ||||||||||||||||||||||||||||||
| 3DL2*011 | G | G | A | T | G | C | G | G | ||||||||||||||||||||||||||||||
| 3DL2*012 | G | G | A | T | G | C | G | G | ||||||||||||||||||||||||||||||
| 3DL3*001 | C | C | G | G | G | C | ||||||||||||||||||||||||||||||||
| 3DL3*002 | C | C | G | G | G | C | ||||||||||||||||||||||||||||||||
| 3DL3*003 | C | C | G | G | G | C | ||||||||||||||||||||||||||||||||
| 3DL3*004 | C | C | G | G | G | C | ||||||||||||||||||||||||||||||||
| 3DP1*001 | C | A | C | |||||||||||||||||||||||||||||||||||
| 3DP1*002 | C | A | C | |||||||||||||||||||||||||||||||||||
| 3DP1*003 | C | A | C | |||||||||||||||||||||||||||||||||||
| 2DP1*001 | A | G | ||||||||||||||||||||||||||||||||||||
| 2DP1*002 | A | G | ||||||||||||||||||||||||||||||||||||
Variant are shown in bold, consensus in italics
Conflict log—which checked for agreement between replicates of the same assay applied to the same sample in a given data set (duplicate check)
A partial match log—which checked for agreement between assays that typed for the same gene in different domains and generated a potential recombinants list
A control check—which checked the SNP profiles of previously characterized samples against an index of expected results for these samples
Tables 3 and 4 present the hME hit pattern used to distinguish between KIR3DL1 and KIR3DS1 as an example of how KIR Genotype Caller used the hit patterns from the targeted domains to determine genotype.
Table 3.
hMEs used to distinguish KIR3DL1 from KIR3DS1
| Domain allele | D0 | D1 | D2 | Trans/cyt | No. SNPs |
|---|---|---|---|---|---|
| 3DL1 | 3DS1.3DL1. D1.S | 3DL1.TC.S | 2 | ||
| 3DS1 | 3DS1. D0.S | 3DS1.3DL1. D1.S | 3DS1.TC. G.INT | 3 |
This table presents the hME pattern used to distinguish between KIR3DL1 and KIR3DS1 as an example of how the SNP patterns are used in our genotyping program “KIR Genotype Caller”.
Table 4.
SNP patterns used to distinguish KIR3DL1 from KIR3DS1
| Domain | D0 | D1 | TC | TC |
|---|---|---|---|---|
| 3DS1. | 3DS1.3DL1. | 3DL1. | 3DS1.TC.G. | |
| D0.S | D1.S | TC.S | INT | |
| 3DL1 | ||||
| 3DL1*001 | T | T | ||
| 3DL1*002 | T | T | ||
| 3DL1*004 | T | T | ||
| 3DL1*005 | T | T | ||
| 3DL1*006 | T | T | ||
| 3DL1*007 | T | T | ||
| 3DL1*008 | T | T | ||
| 3DL1*009 | G | T | T | |
| 3DS1 | ||||
| 3DS1*010 | G | G | T | |
| 3DS1*011 | G | G | T | |
| 3DS1*012 | G | G | T | |
| 3DS1*013 | G | G | T | |
| 3DS1*014 | G | G | T | |
This table presents the SNP pattern used to distinguish between KIR3DL1 and KIR3DS1 as an example of how the SNP patterns are used in our genotyping program “KIR Genotype Caller”.
Locus-specific genotyping using SSOP
Samples were amplified at KIR domains D0, D1, and D2 and a transmembrane–cytoplasmic region using ∼100 ng of genomic DNA per amplification and assayed in an SSOP format with 39 biotinylated probes designed to identify 14 KIR genes and some alleles (Trachtenberg, unpublished; Crum et al. 2000). The specific constellation of 39 SSO probes utilized did not distinguish KIR2DL5 subtypes A and B. Amplified PCR products were denatured and vacuum blotted onto replicate 96-sample nylon membranes. Replicate membranes were hybridized to SSO probes, washed under stringent conditions to remove unbound probe, and developed using nonradioactive detection methods. KIR probe hybridization patterns were individually decoded using a computer program developed in-house.
Cloning and sequencing of a novel KIR2DL5 allele
Genomic DNA was extracted from the 10th International Histocompatibility Workshop (IHW) OLGA B cell line using the Qiagen Genomic-tip 20/G kit. Primers LFcon63 and LRg1769 were used to amplify the entire coding region of KIR2DL5 (Vilches et al. 2000). OLGA is presumed homozygous consanguineous for KIR2DL1, KIR2DL3, KIR3DL1, and KIR3DL2 loci (Garcia et al. 2004), but KIR2DL5A and KIR2DL5B have also been reported for this sample (Gomez-Lozano and Vilches 2002), and in this study, OLGA typed as KIR3DL1 and KIR3DS1 positive. A total of 100 ng of DNA was amplified in 20-μL reactions containing 0.4 U Phusion DNA Polymerase (New England BioLabs, Beverly, MA), 1× GC buffer, and 10 pmol of each primer. The following conditions were used for long-range PCR: initial denaturation at 98°C for 45 s, 35 cycles of 98°C for 10 s, 67°C for 30 s, and 72°C for 5 min, followed by a final extension at 72°C for 8 min and a 4°C hold. The PCR products were electrophoresed in a 0.8% agarose gel, and the DNA was visualized by crystal violet staining. The 9.3-kb fragment was purified using a SNAP column (Invitrogen, Carlsbad, CA). 3′-A overhangs were added to the purified fragment using 2.5 U Taq DNA polymerase (Roche) and 2′-deoxyadenosine 5′-triphosphate (dATP) and incubation at 72°C for 20 min. The fragment was subsequently inserted into the pCR-XL-TOPO cloning vector (Invitrogen, Carlsbad, CA). Plasmids were isolated from individual colonies using the Qiaprep Spin miniprep kit and sent to the UC Berkeley DNA Sequencing Facility (Berkeley, CA) for complete sequencing of exons 1 through 9. Three clones were sequenced in forward and reverse directions, including two clones with complete genomic sequence of all introns and exons and one clone sequenced at exons only. Primers were chosen based on genomic sequence for KIR2DL5 (http://genome.ucsc.edu).
Results
A method for high-throughput KIR genotyping using MALDI-TOF mass spectrometry
Study of the functional and clinical consequences of KIR variation is currently limited by the sensitivity, resolution, and speed of the assays used for KIR genotyping. To address these limitations, we developed a high-throughput SNP-based MS method for KIR genotyping that is based on the SEQUENOM™ MALDI-TOF MS platform and hME chemistry (outlined in Fig. 2).
Fig. 2.
A high-throughput SNP-based MS method for KIR genotyping that is based on the SEQUENOM™ MALDI-TOF MS platform and homogenous MassExtend (hME) chemistry
A set of 38 SNPs that distinguish the 17 KIR genes and pseudogenes was identified by inspection in silico of the aligned KIR sequences from the IPD (http://www.ebi.ac.uk/ipd/kir). The two pseudogenes (KIR2DP1 and KIR3DP1) were included in the analysis to help identify recombinants and to assist with haplotype analysis. Primer pairs were developed (Table 1) to ‘capture’ these SNPs in ∼100–250-bp PCR products amplified from genomic DNA samples. After SAP treatment, the amplification products were subjected to a primer–extension reaction (called an hME) using a primer (Table 1) that terminates adjacent to the targeted SNP, which is extended through the SNP by incorporating deoxy and/or dideoxy nucleotides. The particular termination mixes used are dictated by the targeted SNP and its surrounding sequence. The rationale for a particular mixture being that by choosing an appropriate mixture of deoxy and dideoxy nucleotides, one can determine not only which extension products will be produced but which product will be extended by one, two, or three nucleotides. The MALDI-TOF MS is sensitive enough to distinguish between these nucleotide extensions. Because recombination within these tandemly arrayed homologous genes is a source of novel alleles (Rajalingam et al. 2004; Shilling et al. 1998; Witt et al. 1999), the assay was designed to enable identification of recombinants or novel alleles. To accomplish this, the hME reactions for each functional KIR gene were designed to be redundant: SNPs from a minimum of two different domain-coding exons were targeted. The primer–extension products from the hME reactions were analyzed by MALDI-TOF MS, which measures the mass of the extension products and uses the mass differences to determine which nucleotides were incorporated to identify gene presence and absence and to detect novel variants.
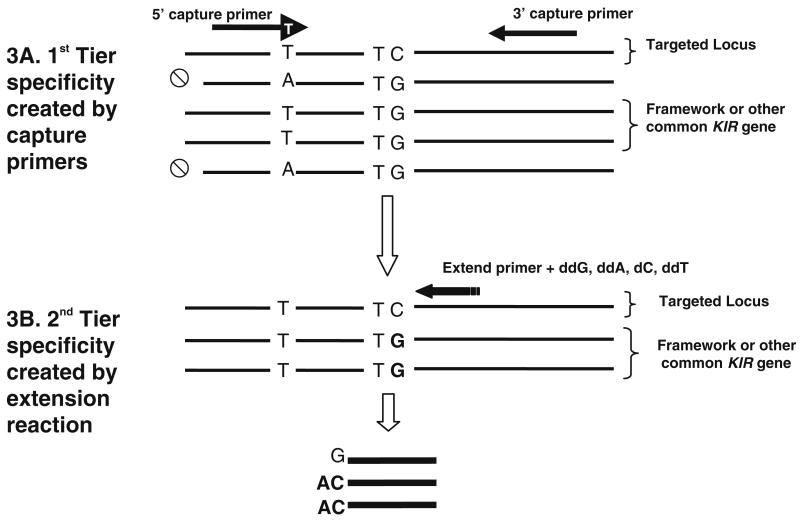
This method for KIR genotyping provides two levels of specificity. The capture reaction limits the subsequent analysis to the inclusion of a single queried KIR gene and the conserved region in a framework or common gene, which provides a positive control for the PCR reaction (Fig. 3a). In the hME reaction, the choice of SNP, and the design of extension primers and dideoxy/deoxy nucleotide reaction mixes provide the second level of specificity and the potential for distinguishing allelic variants as well as the presence or absence of the targeted gene (Fig. 3b). The SNP-based KIR/MALDI-TOF assay uses 38 capture primer sets and 38 SNP extension primers multiplexed into 16 reactions to distinguish all 15 functional KIR genes and two pseudogenes, as well as certain KIR2DL1, KIR2DS4, and KIR2DL5 variants. For the MALDI-TOF MS analysis, the hMEs are distributed on 384-array microchips that are analyzed in less than 30 min. The data generated is automatically deposited into an Oracle database, which can be accessed to visualize the spectra and assess the SNP calls. Data from the Oracle database is then sent to our KIR Genotype Caller program for automated KIR genotyping and data archiving. Using the KIR/MALDI system, ∼384 KIR genotypes per technologist can easily be determined in 1 week.
Fig. 3.
Two tiers of specificity are built into the KIR/MALDI assay. a The first tier of specificity is created by the “capture” primers used in the initial PCR. In this example, allelic forms containing an “A” fail to amplify (⊘) while those containing “T” are captured (and amplified) by the primers during the PCR. One of the KIR genes that is present in all haplotypes (“framework” gene) or another common KIR gene will also be captured with the same set of primers, thereby serving as an internal control for the PCR. b The design and directionality of the extension primer, along with the chosen deoxy (d)/dideoxy (dd) mixture provide the second tier of specificity. In this example, ddG, ddA, dC, and ddT are used in the extension reaction. When ddG is incorporated during the primer extension reaction using the capture product from the targeted locus (the uppermost product shown in both a and b) and framework gene, the reaction is terminated by the incorporation of the dideoxynucleotide. When dC is incorporated during primer extension using the capture product from the targeted locus and framework gene, the extension reaction proceeds until a dideoxy nucleotide is incorporated. The choice of the extension primer and deoxy/dideoxy mixes allows the creation of extension products that differ by length as well as composition. The extension products are measured by the MALDI-TOF MS, which can easily differentiate between products that differ in length by one or more bases
Concordance between KIR/MALDI-TOF gene-content typing and established methods
In developing the SNP-based KIR/MALDI-TOF KIR genotyping assay, we analyzed a panel of 15 individuals who encompass most of the published variability at the KIR locus, as shown by analysis with the established SSOP and/or SSP methods. The set of 38 hME assays proved necessary and sufficient to define the presence or absence of the 15 expressed KIR genes and the two KIR pseudogenes. At this level of resolution, there was complete agreement between the results obtained with KIR /MALDI-TOF genotyping and those obtained by SSOP and SSP (Table 5).
Table 5.
Concordance between KIR/MALDI genotyping and established methods: results for 15 samples at 17 KIR loci
| 2DL1 | 2DL2 | 2DL3 | 2DL4 | 2DL5 generic | 2DL5 A&B | 3DL1 | 3DL2 | 3DL3 | 2DS1 | 2DS2 | 2DS3 | 2DS4 generic | 2DS4 *del | 2DS5 | 3DS1 | 2DP1 | 3DP1 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of probes/locus | 3 | 4 | 2 | 2 | 2 | 5 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 3 | 1 | 1 | |
| BM16 | Exp | + | + | + | + | + | + | + | + | + | |||||||||
| Obs | + | + | + | + | + | + | + | + | + | + | |||||||||
| PITOUT | Exp | + | + | + | + | + | + | + | + | + | + | + | |||||||
| Obs | + | + | + | + | + | + | + | + | + | + | + | + | |||||||
| BM92 | Exp | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| Obs | + | + | + | + | + | A+B | + | + | + | + | + | + | + | + | + | + | + | ||
| OLGA | Exp | + | + | + | + | A+B | + | + | + | + | + | + | + | + | + | ||||
| Obs | + | + | + | + | NEW | + | + | + | + | + | + | + | + | + | + | ||||
| RML | Exp | + | + | + | + | + | A | + | + | + | + | + | + | + | + | + | + | ||
| Obs | + | + | + | + | + | A | + | + | + | + | + | + | + | + | + | + | |||
| HS | Exp | + | + | + | + | + | + | + | + | + | + | ||||||||
| Obs | + | + | + | + | + | + | + | + | + | + | |||||||||
| NV | Exp | + | + | + | + | A+B | + | + | + | + | + | + | + | + | + | + | + | ||
| Obs | + | + | + | + | A+B | + | + | + | + | + | + | + | + | + | + | + | |||
| WC | Exp | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||
| Obs | + | + | + | + | + | B | + | + | + | + | + | + | + | + | + | + | |||
| WT47 | Exp | + | + | + | + | A+B | + | + | + | + | + | + | + | + | + | ||||
| Obs | + | + | + | + | A+B | + | + | + | + | + | + | + | + | + | |||||
| RR | Exp | + | + | + | + | + | B | + | + | + | + | + | + | + | + | + | |||
| Obs | + | + | + | + | + | B | + | + | + | + | + | + | + | + | + | ||||
| YW | Exp | + | + | + | + | + | + | + | + | + | + | ||||||||
| Obs | + | + | + | + | + | + | + | + | + | + | |||||||||
| T7527 | Exp | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||
| Obs | + | + | + | + | + | B | + | + | + | + | + | + | + | + | + | + | |||
| HOR | Exp | + | + | + | + | A | + | + | + | + | + | + | + | ||||||
| Obs | + | + | + | + | A | + | + | + | + | + | + | + | |||||||
| DU145 | Exp | + | + | + | A | + | + | + | + | + | + | + | |||||||
| Obs | + | + | + | A | + | + | + | + | + | + | + | ||||||||
| FC | Exp | + | + | + | + | + | + | + | + | + | + | + | + | ||||||
| Obs | + | + | + | + | + | + | + | + | + | + | + | + | |||||||
Abbreviations : Exp = expected; Obs = observed
A “+” in the expected row signifies that the locus is expected to be present; blank signifies that locus is not expected to be present. Filled gray indicates that the sample had not been previously typed at allelic level resolution. “NEW” is a putative novel allele found for “OLGA” at KIR 2D L 5 A/B .
Expected KIR genotypes for BM16, PITOUT, BM92, OLGA, RML, T7527 and HOR are as shown in Cook et al. 2003. Expected KIR genotypes for RML, WT47, OLGA, HOR, NV, and DU145 are as shown in Gomez-Lazano and Vilches 2002 and 2004. Expected KIR genotypes for HS, WC, RR, YW and FC are unpublished results from PP.
A generic assay for the presence of KIR 2DL 5 is combined with a higher resolution assay capable of resolving KIR 2DL 5A and KIR 2DL 5B . A generic assay for KIR 2DS 4 is combined with an assay for detection of the deletion variants KIR 2DS 4*003 /*004/*006.
A panel of 98 individuals from the Chicago MACS population that we had previously genotyped for KIR using the SSOP and/or SSP methods was analyzed using the KIR/ MALDI-TOF method. Again, there was complete concordance in the determination of the KIR gene content. This analysis shows that in terms of its precision and resolution, the KIR/MALDI-TOF is the equal of the established SSOP and SSP methods. It combines the advantage of allowing high-throughput analysis with no loss in either accuracy or sensitivity.
Discovery of novel KIR alleles by KIR/MALDI-TOF typing
Certain KIR allele differences have profound functional consequences. For example, common KIR2DS4 alleles are inactivated by a deletion of 18 nucleotides. Our KIR/ MALDI-TOF typing system was designed to distinguish the 2DS4 deletion variants from the full-length forms and gave results concordant with those obtained by the SSOP and SSP methods (Table 5).
A more complicated situation is presented by KIR2DL5, for which the gene can variably be found in either the centromeric or telomeric parts of KIR haplotypes or in both parts. Although these genes have been designated as KIR2DL5B and KIR2DL5A according to their centromeric or telomeric position, respectively, their sequences show no A-specific or B-specific character and form a single lineage of seven ‘alleles.’ Consequently, to distinguish the two KIR2DL5A subtypes from the five KIR2DL5B subtypes, it was necessary to perform allele-specific typing. We designed hME subtyping assays to distinguish the KIR2-DL5A and KIR2DL5B subtypes, based on six SNPs, previously used in SSP typing to discriminate KIR2DL5 variants (Gomez-Lozano and Vilches 2002). In the 15-member panel, seven samples were previously subtyped for KIR2DL5A and KIR2DL5B, and these included individuals who had only KIR2DL5A or only KIR2DL5B. For six of the donors, there was concordance between KIR/MALDI-TOF and SSP typing (Table 5). The seventh donor, OLGA, had previously been characterized as having both KIR2DL5A and KIR2DL5B (Gomez-Lozano et al. 2002). Only one nucleotide was detected at each of the five SNP positions analyzed at 2DL5 for OLGA using the KIR/MALDI system. Furthermore, this combination of SNPs did not correspond to any of the known KIR2DL5A or KIR2DL5B alleles. Moreover, WT47 (IHWG sample) had also been typed as KIR2DL5A and KIR2DL5B (Gomez-Lozano et al. 2002) and did show the expected SNP patterns for the previously reported type. These data indicated that OLGA had a novel variant of KIR2DL5.
To test this hypothesis, the KIR2DL5 gene from OLGA was isolated and characterized. Long-range PCR amplification of genomic DNA was used to obtain a fragment spanning the 5′ untranslated region (UTR) through to the 3′’UTR. This 9.3-kb fragment, including exons 1 through 9, was cloned and sequenced (GenBank accession #EF102434). The sequence analysis showed that OLGA had a KIR2DL5 allele with a combination of nucleotide substitutions in exons 3 and 5 not seen in previously characterized KIR2DL5 subtypes (Table 6). The OLGA KIR2DL5 novel allele was also observed in a Hispanic individual from the National Marrow Donor Program study and in one sample from the MACS cohort.
Table 6.
Nucleotides that distinguish KIR2DL5*OLGA from other KIR2DL5 alleles
| Exon 3
|
Exon 5
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Nucleotide | 139 | 173 | 364 | 385 | 410 | 581 | 647 | |
| Sub6 | Sub1 | Sub2 | Sub3 | Sub4 | ||||
| 2DL5A | 001 | T | G | A | A | G | A | G |
| 2DL5B | 002 | A | G | G | G | G | G | A |
| 003 | T | G | A | A | A | A | G | |
| 004 | T | A | A | A | G | A | G | |
| 005 | A | G | G | G | G | G | A | |
| 006 | T | G | A | A | G | A | G | |
| 007 | T | G | A | A | A | A | G | |
| OLGA | T | G | A | G | G | A | G | |
Nucleotides positions shown in bold were queried by KIR/MALDI method, using indicated KIR2DL5 subtyping primers. Numbering system follows Vilches et al. (2000).
Additional novel KIR alleles were identified from analysis of 120 individuals from the sample repository of the NMDP. At the level of the KIR gene content, KIR/ MALDI-TOF typing gave results that were in full agreement with the KIR types we had previously obtained using the SSOP method. The increased resolution of the KIR/ MALDI-TOF analysis identified four novel combinations of SNPs that correspond to putative novel alleles at the KIR2DL1, KIR3DL1, KIR2DS5, and KIR2DL5 genes (Tables 7 and 8). The average putative novel allele frequency for the MACS and NMDP populations and IHW samples was 4% (9/233) using the KIR/MALDI method.
Table 7.
Expected/observed MALDI-TOF MS SNP patterns for putative novel alleles of KIR2DL1, KIR3DL1, and KIR2DS5: unexpected SNP patterns observed through locus-specific resolution two-SNP/two-domain strategy
| Domain | D0 | D1 | D2 | Trans/cyt |
|---|---|---|---|---|
| 2DL1 expected SNP pattern | + | + | + | |
| Putative novel 2DL1 allele | + | missing | missing | |
| 3DL1 expected SNP pattern | + | + | ||
| Putative novel 3DL1 allele | + | missing | ||
| 2DS5 expected SNP pattern | + | + | ||
| Putative novel 2DS5 allele | + | missing |
indicates expected hME SNP pattern, and missing indicates aberrant hME SNP pattern.
Table 8.
Expected SNP pattern for known KIR2DL5 alleles with intermediate resolution subtyping hME’s
| Probe | 2DL5sub1 | 2DL5sub2 | 2DL5sub3 | 2DL5sub4 | 2DL5sub5 | 2DL5sub6 |
|---|---|---|---|---|---|---|
| Expected SNP pattern for all known KIR2DL5 alleles with six subtyping assays | ||||||
| Nt position | 173 | 385 | 410 | 947 | 1325 | 139 |
| 2DL5A*001 | G | A | G | G | A | T |
| 2DL5B*002 | G | G | G | A | A | A |
| 2DL5B*003 | G | A | A | G | A | T |
| 2DL5B*004 | A | A | G | G | A | T |
| 2DL5A*005 | G | G | G | A | A | A |
| 2DL5B*006 | G | A | G | G | A | T |
| 2DL5B*007 | G | A | A | G | A | T |
| Novel SNP pattern found in OLGA that identified a putative new allele | ||||||
| 2DL5*novel | G | G | G | G | A | T |
Discussion
To date, most KIR genotypes have been obtained using sequence-specific priming of PCR, the first method developed for KIR genotyping (Dupont et al. 1997; Selvakumar et al. 1997a, b; Shilling et al. 2002; Shilling et al. 2003; Uhrberg et al. 1997). In this assay, genomic DNA is amplified using a battery of primer pairs in separate reactions and the products visualized by gel electrophoresis. The requirement for many different and independent PCRs makes this method both machine and labor intensive. It also requires relatively large quantities (∼3–5 μg) of good quality DNA. For high-throughput KIR genotyping, the SSOP blot method has offered some advantage over SSP because it is amenable to analysis of hundreds of samples concurrently and requires less DNA (<1 μg), an important factor for the study of rare archival samples.
In this study, we describe a first generation assay for high-throughput SNP-based KIR genotyping using primer extension PCR and MALDI-TOF MS. This method detects the presence of 17 KIR genes and pseudogenes and has some capacity for discriminating KIR alleles and for discovering new KIR alleles. The method uses 38 capture primer sets and 38 SNP extension primers multiplexed into 16 reactions that are carried out under identical conditions in a 384-well format. In this configuration, 24 samples can be run per 384-well SpectroChip. The method also has the flexibility to run each of the 16 multiplexed reactions on separate chips, allowing a total of 384 samples to be genotyped per 16 chips. The latter format easily allows the addition of new assays and is ideal for higher-throughput studies. The MALDI-TOF analyzes two 384-well chips (48 samples) in ∼1 h.
Comparative analysis of 233 samples shows excellent agreement between the results obtained by SSP, SSOP, and MALDI-TOF. However, KIR typing by the SNP-based MALDI-TOF method has major advantages over the SSP and SSOP methods. Mass spectrometric analysis uses at least an order of magnitude less DNA (<40 ng), with much higher throughput and greater sensitivity and accuracy than these other methods. The inclusion of a primer extension step after PCR also increases precision and accommodates DNA samples of poorer quality. The primer extension’s advantage is that the primer that queries a SNP anneals not to the polymorphic position but to the adjacent conserved sequence. Consequently, an extension product is made regardless of the nucleotide at the SNP. It is thus possible to determine all the nucleotides present at a given SNP position from one primer extension assay. As a result, the SNP-based MALDI-TOF approach can give greater precision with fewer assays than SSP and SSOP. This property is illustrated by the new KIR2DL1, KIR3DL1, KIR2DS5, and KIR2DL5 alleles identified during MALD-TOF analysis of the 233 samples.
Mounting evidence suggests that interactions between KIR and their HLA ligands influence bone marrow transplant outcome (Gagne et al. 2002; Parham 2003; Parham and McQueen 2003; Ruggeri et al. 2002), viral disease outcome (Christiansen et al. 2003; Khakoo et al. 2004; Martin et al. 2002; Trachtenberg et al. 2003), and the etiology of several autoimmune diseases (Mizuki et al. 2000; Momot et al. 2004; Nelson et al. 2004; van der Slik et al. 2003). Because both KIR and HLA are so highly polymorphic, large cohorts will be necessary to have power sufficient to define many of these interactions. Such studies demand a methodology for KIR typing that is efficient in the use of genomic DNA and combines high resolution with high-throughput. The KIR/MALDI-TOF method described here has all these attributes. This rapid and sensitive method is currently being refined for allelic level resolution of all the KIR genes.
Acknowledgments
We wish to express our gratitude to Drs. Derek Middleton and Carlos Vilches and their laboratories for their generosity and gracious sharing of expertise. Finally, we are grateful for the technical and administrative contributions from J Agraz, K Saeteurn, N Bose, E Sanseau, K Guttierrez, J-C Cossec, and M Vinson. This work was supported by grants R21 AI 65254-01A1, P01 CA 111412, and UO1 AI067068-01 from the National Institutes of Health and the My Brother Joey Foundation to Elizabeth Trachtenberg. The MALDI-TOF MassARRAY system was acquired through NIH-NCRR #1S10RR 16703-01. The investigation was conducted at the Children’s Hospital and Research Center Oakland in facilities constructed with support from Research Facilities Improvement Program Grant Number CO6RR-16226 from the National Center for Research Resources, National Institutes of Health.
References
- Bottino C, Castriconi R, Moretta L, Moretta A. Cellular ligands of activating NK receptors. Trends Immunol. 2005;26:221–226. doi: 10.1016/j.it.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Carrington M, Norman PJ. NCBI Bookshelf. 2003. The KIR gene cluster. [Google Scholar]
- Christiansen FT, Gaudieri S, De Santis D, Moore CB, Witt CS, James IR. NK Receptor genes are predictors of HIV progression. Hum Immunol. 2003;64(Suppl 1):S12. [Google Scholar]
- Cook MA, Norman PJ, Curran MD, Maxwell LD, Briggs DC, Middleton D, Vaughan RW. A multi-laboratory characterization of the KIR genotypes of 10th International Histocompatibility Workshop cell lines. Hum Immunol. 2003;64:567–571. doi: 10.1016/s0198-8859(03)00042-9. [DOI] [PubMed] [Google Scholar]
- Cook MA, Milligan DW, Fegan CD, Darbyshire PJ, Mahendra P, Craddock CF, Moss PA, Briggs DC. The impact of donor KIR and patient HLA-C genotypes on outcome following HLA-identical sibling hematopoietic stem cell transplantation for myeloid leukemia. Blood. 2004;103:1521–1526. doi: 10.1182/blood-2003-02-0438. [DOI] [PubMed] [Google Scholar]
- Crum KA, Logue SE, Curran MD, Middleton D. Development of a PCR-SSOP approach capable of defining the natural killer cell inhibitory receptor (KIR) gene sequence repertoires. Tissue Antigens. 2000;56:313–326. doi: 10.1034/j.1399-0039.2000.560403.x. [DOI] [PubMed] [Google Scholar]
- Dupont B, Selvakumar A, Steffens U. The killer cell inhibitory receptor genomic region on human chromosome 19q13.4. Tissue Antigens. 1997;49:557–563. doi: 10.1111/j.1399-0039.1997.tb02802.x. [DOI] [PubMed] [Google Scholar]
- Gagne K, Brizard G, Gueglio B, Milpied N, Herry P, Bonneville F, Cheneau ML, Schleinitz N, Cesbron A, Follea G, Harrousseau JL, Bignon JD. Relevance of KIR gene polymorphisms in bone marrow transplantation outcome. Hum Immunol. 2002;63:271–280. doi: 10.1016/s0198-8859(02)00373-7. [DOI] [PubMed] [Google Scholar]
- Garcia CA, Robinson J, Shilling HG, Guethlein LA, Parham P, Madrigal JA, Marsh SG. KIR gene characterisation of HLA homozygous cell lines, HLA 2004. 13th IHWS NK/KIR Joint Report.2004. [Google Scholar]
- Gomez-Lozano N, Vilches C. Genotyping of human killer-cell immunoglobulin-like receptor genes by polymerase chain reaction with sequence-specific primers: an update. Tissue Antigens. 2002;59:184–193. doi: 10.1034/j.1399-0039.2002.590302.x. [DOI] [PubMed] [Google Scholar]
- Gomez-Lazano N, Vilches C. Erratum. Tissue Antigens. 2004;64:629. [Google Scholar]
- Gomez-Lozano N, Gardiner CM, Parham P, Vilches C. Some human KIR haplotypes contain two KIR2DL5 genes: KIR2-DL5A and KIR2DL5B. Immunogenetics. 2002;54:314–319. doi: 10.1007/s00251-002-0476-2. [DOI] [PubMed] [Google Scholar]
- Hiby SE, Walker JJ, O’Shaughnessy KM, Redman CW, Carrington M, Trowsdale J, Moffett A. Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J Exp Med. 2004;200:957–965. doi: 10.1084/jem.20041214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, Cheng J, Goedert JJ, Vlahov D, Hilgartner M, Cox S, Little AM, Alexander GJ, Cramp ME, O’Brien SJ, Rosenberg WM, Thomas DL, Carrington M. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, Wilson M, O’Brien SJ, Carrington M. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- Mizuki M, Eklund A, Grunewald J. Altered expression of natural killer cell inhibitory receptors (KIRs) on T cells in bronchoalveolar lavage fluid and peripheral blood of sarcoidosis patients. Sarcoidosis Vasc Diffuse Lung Dis. 2000;17:54–59. [PubMed] [Google Scholar]
- Momot T, Koch S, Hunzelmann N, Krieg T, Ulbricht K, Schmidt RE, Witte T. Association of killer cell immunoglobulin-like receptors with scleroderma. Arthritis Rheum. 2004;50:1561–1565. doi: 10.1002/art.20216. [DOI] [PubMed] [Google Scholar]
- Moretta A. Natural killer cells and dendritic cells: rendezvous in abused tissues. Nat Rev Immunol. 2002;2:957–964. doi: 10.1038/nri956. [DOI] [PubMed] [Google Scholar]
- Nelson GW, Martin MP, Gladman D, Wade J, Trowsdale J, Carrington M. Cutting edge: heterozygote advantage in autoimmune disease: hierarchy of protection/susceptibility conferred by HLA and killer Ig-like receptor combinations in psoriatic arthritis. J Immunol. 2004;173:4273–4276. doi: 10.4049/jimmunol.173.7.4273. [DOI] [PubMed] [Google Scholar]
- Parham P. Killer Cell Immunoglobulin like receptors: diverse species specific components of innate immunity. Immunology Suppl. 2003;110:21. [Google Scholar]
- Parham P, McQueen KL. Alloreactive killer cells: hindrance and help for haematopoietic transplants. Nat Rev Immunol. 2003;3:108–122. doi: 10.1038/nri999. [DOI] [PubMed] [Google Scholar]
- Rajalingam R, Parham P, Abi-Rached L. Domain shuffling has been the main mechanism forming new hominoid killer cell Ig-like receptors. J Immunol. 2004;172:356–369. doi: 10.4049/jimmunol.172.1.356. [DOI] [PubMed] [Google Scholar]
- Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, Posati S, Rogaia D, Frassoni F, Aversa F, Martelli MF, Velardi A. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295:2097–2100. doi: 10.1126/science.1068440. [DOI] [PubMed] [Google Scholar]
- Selvakumar A, Steffens U, Dupont B. Polymorphism and domain variability of human killer cell inhibitory receptors. Immunol Rev. 1997a;155:183–196. doi: 10.1111/j.1600-065x.1997.tb00951.x. [DOI] [PubMed] [Google Scholar]
- Selvakumar A, Steffens U, Palanisamy N, Chaganti RS, Dupont B. Genomic organization and allelic polymorphism of the human killer cell inhibitory receptor gene KIR103. Tissue Antigens. 1997b;49:564–573. doi: 10.1111/j.1399-0039.1997.tb02803.x. [DOI] [PubMed] [Google Scholar]
- Shilling HG, Lienert-Weidenbach K, Valiante NM, Uhrberg M, Parham P. Evidence for recombination as a mechanism for KIR diversification. Immunogenetics. 1998;48:413–416. doi: 10.1007/s002510050453. [DOI] [PubMed] [Google Scholar]
- Shilling HG, Guethlein LA, Cheng NW, Gardiner CM, Rodriguez R, Tyan D, Parham P. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol. 2002;168:2307–2315. doi: 10.4049/jimmunol.168.5.2307. [DOI] [PubMed] [Google Scholar]
- Shilling HG, McQueen KL, Cheng NW, Shizuru JA, Negrin RS, Parham P. Reconstitution of NK cell receptor repertoire following HLA-matched hematopoietic cell transplantation. Blood. 2003;101:3730–3740. doi: 10.1182/blood-2002-08-2568. [DOI] [PubMed] [Google Scholar]
- Trachtenberg E, Korber B, Sollars C, Kepler TB, Hraber PT, Hayes E, Funkhouser R, Fugate M, Theiler J, Hsu YS, Kunstman K, Wu S, Phair J, Erlich H, Wolinsky S. Advantage of rare HLA supertype in HIV disease progression. Nat Med. 2003;9:928–935. doi: 10.1038/nm893. [DOI] [PubMed] [Google Scholar]
- Trachtenberg EA, Sollars C, Korber B, Erlich EH, Wolinsky S, Kepler T. Natural killer cell immunogloblin-like receptors (KIR) are associated with protective effect in HIV-1 transmission and disease progression in the Chicago Multicenter AIDS Cohort Study (MACS). 8th Annual Meeting for the Society of Natural Immunity and the 20th International Natural Killer Cell Workshop; Noordwijkerhout, The Netherlands. 2004. [Google Scholar]
- Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- Valiante NM, Lienert K, Shilling HG, Smits BJ, Parham P. Killer cell receptors: keeping pace with MHC class I evolution. Immunol Rev. 1997;155:155–164. doi: 10.1111/j.1600-065x.1997.tb00948.x. [DOI] [PubMed] [Google Scholar]
- van der Slik AR, Koeleman BP, Verduijn W, Bruining GJ, Roep BO, Giphart MJ. KIR in type 1 diabetes: disparate distribution of activating and inhibitory natural killer cell receptors in patients versus HLA-matched control subjects. Diabetes. 2003;52:2639–2642. doi: 10.2337/diabetes.52.10.2639. [DOI] [PubMed] [Google Scholar]
- Vilches C, Rajalingam R, Uhrberg M, Gardiner CM, Young NT, Parham P. KIR2DL5, a novel killer-cell receptor with a D0–D2 configuration of Ig-like domains. J Immunol. 2000;164:5797–5804. doi: 10.4049/jimmunol.164.11.5797. [DOI] [PubMed] [Google Scholar]
- Witt CS, Dewing C, Sayer DC, Uhrberg M, Parham P, Christiansen FT. Population frequencies and putative haplotypes of the killer cell immunoglobulin-like receptor sequences and evidence for recombination. Transplantation. 1999;68:1784–1789. doi: 10.1097/00007890-199912150-00024. [DOI] [PubMed] [Google Scholar]