Abstract
Rational vaccines designed to engender T cell responses require intimate knowledge of how epitopes are generated and presented. Recently, we vaccinated 8 Mamu-A*02+ rhesus macaques with every SIV protein except Envelope (Env). Surprisingly, one of the strongest T cell responses engendered was against the Env protein, the Mamu-A*02–restricted epitope, Env788–795RY8. In this paper, we show that translation from an alternate reading frame of both the Rev-encoding DNA plasmid and the rAd5 vector engendered Env788–795RY8-specific CD8+ T cells of greater magnitude than “normal” SIV infection. Our data demonstrate both that the pathway from vaccination to immune response is not well understood and that products of alternate reading frames may be rich and untapped sources of T cell epitopes.
CD8+ T lymphocytes are important for killing both tumor and pathogen-infected cells. Hence, there is immense interest in creating vaccines that elicit these responses. Indeed, because an Ab-based AIDS vaccine is unlikely in the near future, the goal for a vaccine is now to control viral replication and therefore to slow disease progression and prevent transmission. To achieve this, several laboratories are investigating the use of vaccines that solely elicit T cell responses (1, 2). Most of these vaccines employ rDNA and/or viral vectors that encode HIV proteins (or SIV proteins, in the case of rhesus macaque studies). Understanding the pathway from vaccination to T cell response is a critical step toward rational vaccine design.
Despite the importance of T cells in controlling HIV and SIV (3, 4), it is becoming clear that much remains to be learned about these cells and the epitopes they target. For instance, it is now established that the processes of viral gene expression can lead to the translation of viral alternate reading frames (ARFs) and the production of T cell epitopes derived from these “mistranslation” events (5, 6). Similar processes may lead to the translation of portions of functional viral proteins in regions of open reading frame (ORF) overlap and T cell epitopes contained therein. The contribution of these processes to the production of epitopes either during infection or after vaccination is poorly understood.
Recently, we showed that Mamu-A*02+ rhesus macaques vaccinated with a DNA/rAd5 regimen encoding all of the SIV proteins, except Envelope (Env), made strong CD8+ T cell responses against the Mamu-A*02–restricted Env-encoded epitope, Env788–795RY8 (RY8) (7). In this paper, we show that the vaccine-induced RY8-specific response was much stronger than that elicited by “normal” viral infection. Further, we demonstrate that the RY8 epitope was produced by translation of a small portion of the Env protein from both the overlapping Rev-encoding DNA plasmid and the rAd5 viral vector encoding Rev. Together, our data indicate that T cells targeting out-of-frame encoded peptides might be more ubiquitous and important than currently appreciated. These data strongly suggest that there is much to discover about how vaccines are translated to induce immune responses and that rational vaccine design could benefit from a greater understanding of how and when these nontraditional translation events might occur.
Materials and Methods
Detection and analysis of immune responses
We measured cellular immune responses in the vaccinated animals, using IFN-γ ELISPOT as previously described (2, 7), 2 wk after an rAd5 boost in vaccinated animals. Data represent the average of two replicate wells minus the average of all negative (no peptide) wells and are reported as spot-forming cells (SFCs) per million PBMCs, with 100,000 cells added per well. Responses were considered positive if the SFC count was greater than 5 spots (50 SFC per million PBMCs) and greater than twice the background plus 2 SD. These responses were compared with archived data (performed and analyzed in identical fashion) from Mamu-A*02+ animals previously infected with SIVmac239 from other studies. Specifically, these nonvaccinated animals were in the acute (3–10 wk postinfection) or chronic (>6 mo postinfection) phase of SIV infection and expressed a variety of MHC class I (MHC-I) molecules in addition to Mamu-A*02. Comparisons of immune responses were performed using a two-tailed t test.
Plasmid synthesis, mutagenesis, and transfection of B cells
The Rev and Gag plasmids were made as described (7). The Env plasmid was made as described elsewhere (8). Mutation of the Rev plasmid to encode an escape version of RY8 was achieved using the QuikChange XL Kit (Stratagene, La Jolla, CA), according to the manufacturer’s instructions and using the following primers: F_5′-GCAGAACCTTGCTATTGAGAGTATACCACATC-3′ and R_5′-GATCTGGTATACTCTCAATAGCAAGGTTCTGC-3′ (nucleotides introducing mutation are underlined), which were designed using Web-based software, available at www.bioinformatics.org/primerx. MHC-I negative 721.221 cells previously made to stably express either Mamu-A*02 or -B*08 were used as APCs. In each experiment, 2–5 × 106 cells were transfected with 5 μg Rev (wild type or containing RY8 escape mutation), Gag, or Env plasmids using the Nucleofector device (Lonza, Walkersville, MD), with solution C and program G-16. The cells were then cultured in RPMI 1640 with 10% FBS and no antibiotics for 24 h.
Culture and Ad5 infection of monocyte-derived dendritic cells
CD14+ monocytes were isolated from PBMCs using magnetic bead-bound anti-CD14 Abs and LS separation columns (Miltenyi Biotec, Auburn, CA). The cells were cultured according to Ignatius et al. (9), using RPMI supplemented with 1% FBS (R1) and GM-CSF (1000 U/ml) and IL-4 (100 U/ml). After 4 d of culture, 1 × 106 cells were resuspended in 200 ml media and rAd5 viruses were added at ratios of 100:1 (viral particles: cells) and 1000:1 for 90 min. The volumes were then brought up to 1 ml with complete R1 media supplemented with GM-CSF and IL-4 and cultured for 24 or 48 h.
Recognition assays and intracellular cytokine staining
For recognition assays, APCs—either DNA-transfected 721.221 B cells or rAd5-infected monocyte-derived dendritic cells (MDDCs)—were mixed with RY8- or Rev44–51RL8 (RL8)-specific T cells at a ratio of 1:1 (100,000 of each); then intracellular cytokine staining (ICS) for the detection of IFN-γ and TNF-α was performed as previously described (7).
Results
Rhesus macaques vaccinated with all of the SIV sequences, except Env, made robust T cell responses against an Env epitope
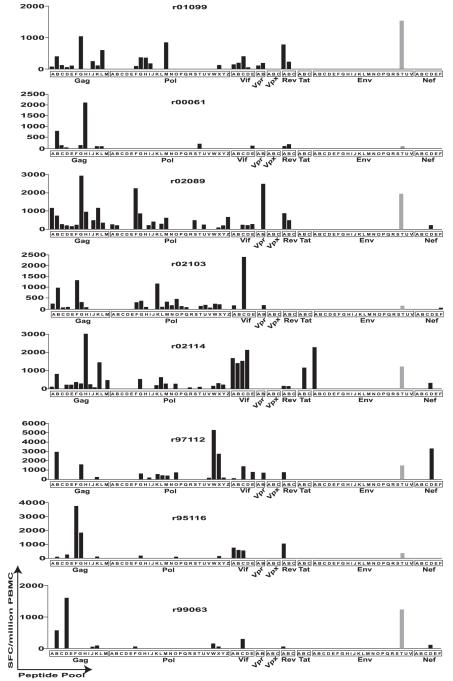
Previously, we vaccinated eight Mamu-A*02+ rhesus macaques with all of the SIV proteins except Env to test the hypothesis that a vaccine that solely elicits cellular immune responses can control AIDS virus replication (7). The vaccine regimen consisted of three injections of DNA (i.m.), with each DNA plasmid synthesized to encode a single viral protein. We then boosted the DNA primed responses with a single injection (i.m.) of rAd5 made to express the same viral proteins except that Vif, Vpr, and Vpx were encoded on the same rAd5 vector. Two weeks after the rAd5 boost, we used IFN-γ ELISPOT with overlapping peptides representing the entire SIV proteome to examine the total cellular immune response elicited by the vaccine regimen. We were surprised to find that all eight animals had mounted responses against a peptide in the Env protein (Fig. 1). This response was mapped to the RY8 epitope, restricted by the MHC-I molecule Mamu-A*02 (10), expressed by all of the animals. In five of the eight vaccinees, the RY8-directed response was among the strongest responses elicited, surpassing 1000 SFCs per million PBMCs (7) (Fig. 1).
FIGURE 1.
Whole-proteome ELISPOT of the eight vaccinated animals. Total cellular immune response was measured using IFN-γ ELISPOT 2 wk after rAd5 administration. Peptides were 15-mers overlapping by 11, spanning the entire known SIVmac239 proteome. ELISPOT responses were considered positive if the mean number of SFCs exceeded background plus 2 SD and was >50 SFCs per million PBMCs. Because total PBMC was assayed, these responses are composed of both CD4 and CD8 responses.
Because the RY8-directed response was so strong, we compared it with that elicited by normal infection with SIVmac239 in Mamu-A*02+ animals. The RY8-specific response surpassed in magnitude the response observed during either acute or chronic SIV infection (Fig. 2). The RY8 response is normally subdominant to the Vif97–104WY8 (WY8) and Nef159–167YY9 (YY9) responses in acute infection and to the YY9 and Gag71–79GY9 (GY9) responses in chronic infection. However, the vaccine induced strong and nearly equivalent responses against all of these epitopes (Fig. 2).
FIGURE 2.
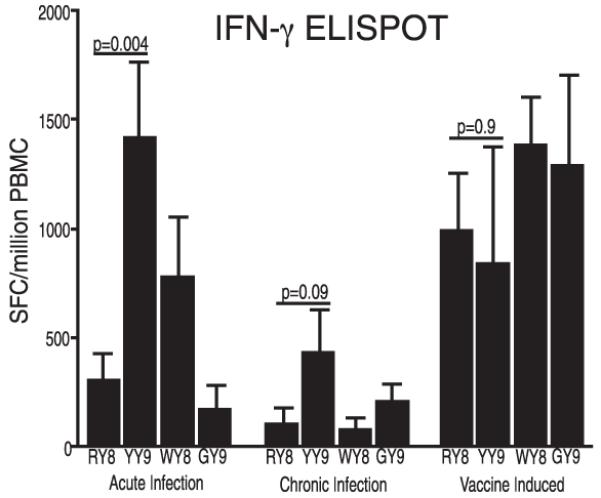
Magnitude of the Env788–795RY8 (RY8)-specific response relative to responses against Vif97–104WY8 (WY8), Nef159–167YY9 (YY9), and Gag71–79GY9 (GY9), as measured by IFN-γ ELISPOT. PBMCs from vaccinated animals were assayed 2 wk after administration of rAd5 vectors. Also shown are data from 11 SIVmac239-infected animals between 3 and 10 wk postinfection (acute phase) and 11 animals in the chronic phase of infection (in this paper defined as >6 mo postinfection). The magnitude of response, shown as SFCs per million PBMCs, is shown on the y-axis. Error bars represent the mean ±SD. ELISPOT responses were considered positive if the mean number of SFC exceeded two times background plus 2 SD and was >50 SFCs per million PBMCs. Statistical analyses (two-tailed t tests) were conducted to determine the relative dominance of the four responses under the three conditions (acute and chronic infection and postvaccination). The p values are shown above each comparison; p values for head-to-head comparisons with RY8 that are not shown are not significant.
The Rev-encoding vectors contain the nucleotide sequence for RY8
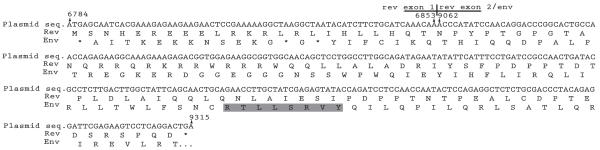
Because all of the animals were seronegative for Abs against the Env protein (published as supplemental data in Ref. 7), we reasoned that the RY8 epitope must be presented from one of the vectors used in the vaccine. Intriguingly, a small portion of the env ORF that overlaps rev exon 2 encodes the RY8 epitope. Hence, the DNA that encodes the RY8 epitope is contained in the rev plasmid (Fig. 3) and might be translated to produce the epitope. We therefore conducted a series of experiments to determine the source of the RY8 epitope.
FIGURE 3.
The sequence of the Rev-encoding plasmid. Shown are the predicted translations in both the rev and the env reading frames. The three stop codons in the env reading frame are present because the first exon of Rev does not overlap with the Env coding region. The shaded box represents the RY8 epitope. Numbering corresponds to the SIVmac239 genome.
The RY8 epitope was presented by translation from an ARF of the Rev plasmid
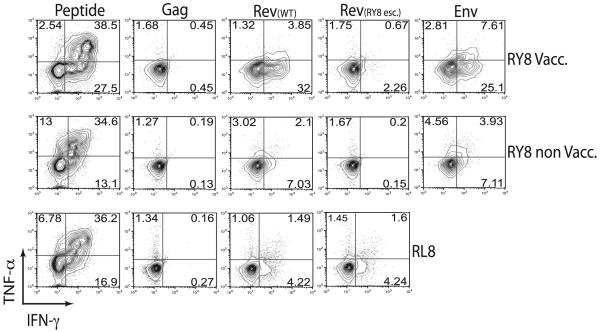
The existence of strong T cell responses against the RY8 epitope in all vaccinated animals, and the fact that the DNA encoding RY8 is contained in the Rev-encoding plasmid, led us to hypothesize that RY8 was translated from an ARF of this plasmid. To test this hypothesis, we first transfected 721.221 cells that stably express Mamu-A*02 with the Rev-encoding plasmid, the Gag-encoding plasmid (as a negative control), or an Env-encoding plasmid. After 24 h, we tested whether these cells could present the RY8 epitope to RY8-specific T cell lines grown both from a vaccinated animal in this study (RY8 Vacc.) and from an SIVmac239-infected Mamu-A*02+ animal infected for another study (RY8 Non-Vacc.) (Fig. 4). As expected, the RY8-specific T cells did not recognize cells transfected with the Gag plasmid. However, they did recognize cells transfected with the Rev and Env plasmids. We also tested whether cells transfected with the Rev plasmid could present the Rev-derived epitope, RL8, restricted by Mamu-B*08 (11), to RL8-specific T cells (Fig. 4) grown from a Mamu-B*08+ animal infected for another study. This epitope was likewise presented, indicating that this single plasmid was translated in both the Rev and the Env reading frames to create immunogenic T cell epitopes.
FIGURE 4.
Recognition of cells transfected with the vaccine plasmid encoding Rev. We transfected 721.221 cells that stably express either Mamu-A*02 or Mamu-B*08 with the plasmid encoding Gag from the vaccine, the Rev plasmid, the Rev plasmid engineered to express an escaped version of RY8, or an Env-encoding plasmid. At 24 h after transfection, we used ICS to see if T cell lines specific for Env788–795RY8 or Rev44–51RL8 could recognize the cells, as measured by TNF-α and IFN-γ production. We tested for recognition by RY8-specific cells grown from an animal vaccinated in this study (RY8 Vacc.), RY8-specific cells grown from an SIV-infected animal from a previous study (RY8 non Vacc.), and RL8-specific cells grown from an SIV-infected animal from a previous study (RL8).
We next transfected cells with a mutant version of the Rev plasmid engineered to encode an escape variant of the RY8 epitope (position 5 S-L) while leaving the Rev ORF untouched (10). The RL8-specific T cell line recognized cells transfected with this plasmid as well as wild type, whereas recognition by RY8-specific T cells was largely abrogated (Fig. 4). It is interesting that recognition was far greater with the RY8-specific cells derived from the vaccinated animal, r02089, than with cells from either of the other animals. However, in our experience, variation is often substantial between cell lines recognizing either infected or transfected cells. Despite this, the recognition depicted in Fig. 3 clearly shows that the Rev plasmid is translated in both the rev and the env reading frames and that T cell epitopes can be derived from both. The above data were collected using a plasmid that was recloned from the original vaccine stock. We repeated the transfection assays using the original vaccine stock and achieved the same results (data not shown). In addition, we attempted to PCR amplify and sequence the plasmid using a series of env-specific primers. Sequence was obtained only when using a primer that aligned within the region of env/rev exon 2 overlap. These controls, along with the data presented above, leave little doubt that the Rev-encoding plasmid was the source of the strong RY8-specific response in vaccinated animals.
The Rev-encoding rAd5 vector produces RY8
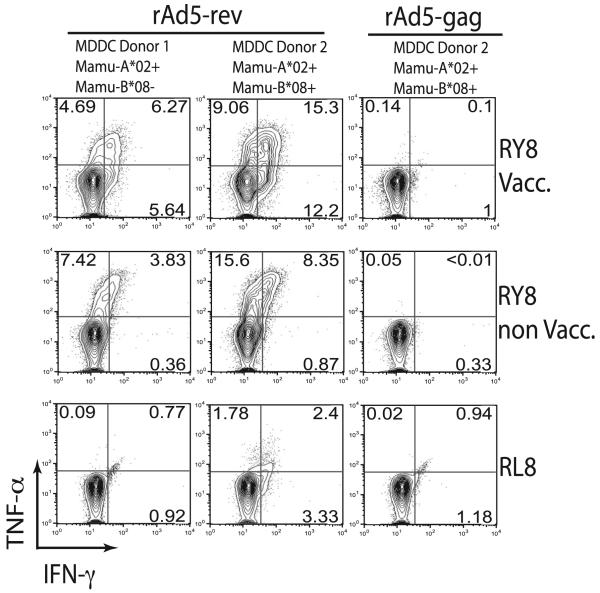
Finally, we tested whether cells infected with the rAd5 vector could present RY8. We first cultured MDDCs from animals that expressed Mamu-A*02, both Mamu-A*02 and Mamu-B*08, or neither. We chose to use dendritic cells because they are excellent APCs and because rAd5 vectors are known to infect them (12). Next, we infected the MDDCs with the rAd5 vectors encoding Rev or Gag and tested whether they could present the RY8 or RL8 epitopes to Ag-specific T cell lines. As with the DNA plasmid data, we found robust recognition of Ad5-infected MDDC after 24 h (data not shown), which became even greater after 48 h (Fig. 5). There was no recognition of MDDC that did not express the correct MHC (data not shown). Together with the DNA data, we have clearly demonstrated that an immunodominant CD8+ T lymphocyte response can be directed against an out-of-frame epitope with no clear mechanism of translation.
FIGURE 5.
Recognition of cells infected with the rAd5 vectors. We infected derived MDDCs with either the Rev- or the Gag-encoding vectors. After 48 h, we used ICS to see if RY8- or RL8-specific T cell lines could recognize their cognate Ag presented on the surface of the MDDCs. As in Fig.3, we tested RY8-specific T cell lines derived from a vaccinated animal in this study (RY8 Vacc.), RY8-specific cells from a nonvaccinated animal from another study (RY8 non Vacc.), and RL8-specific cells from a nonvaccinated animal from another study (RL8). The MDDCs were derived from an animal that was Mamu-A*02+, -B*08−, an animal that expressed both, and an animal that expressed neither (data not shown).
Discussion
Data presented in this study and other recent data from our laboratory (8) paint an emerging picture of the sources of T cell epitopes. The protein source of a given epitope is clearly important, and likely plays a primary role both in the timing of epitope generation (13, 14), with important exceptions (15), and in the ability of that epitope to escape T cell responses (16-18). However, at the very least, caution should be exercised when drawing whole-protein conclusions based on epitope-specific data. In some cases, epitopes can be derived from novel translation events of portions of viral proteins. This model of the sources of T cell epitopes shares fundamental similarities with that of the defective ribosomal product hypothesis, which states that T cell epitopes are primarily derived from defective protein products that do not achieve stable conformation, and are rapidly degraded by the proteasome into T cell epitopes (19). Indeed, the products of these unique translation events may be prime examples of defective ribosomal products.
The important mechanism by which the RY8 epitope is translated remains unknown. We sequenced the Rev-encoding vector and found that three stop codons are present in the env reading frame in the region overlapping rev exon 1, indicating that translation of the env reading frame must occur farther downstream or via an unknown splicing mechanism. In addition, there are no AUG initiation codons in the env reading frame. However, it is possible that translation of the RY8 epitope was due to translation initiation at a CUG codon, as has been described (20-23). Several CUG codons (encoding leucines) are found in the env reading frame upstream of the epitope, including two that contain conserved features of a Kozak consensus, either a position −1 cysteine or a position −3 purine (24). It is also possible that translation occurs by way of a +1 ribosomal frameshift. This phenomenon is difficult to predict but is well documented in yeast (25) and may facilitate translation of a portion of the Env protein containing the RY8 epitope.
An important observation of this study is that the relative dominance of the RY8-directed response was greater than that elicited during either acute or chronic SIVmac239 infection. It is unknown if this is due to its peculiar source of translation or to some other factor associated with this vaccine regimen. Indeed, the GY9-specific response elicited by this vaccine is likewise greater than during normal viral infection. In addition, the vaccinated animals used in this study did not express the MHC-I molecules Mamu-A*01 or -B*17, which restrict immunodominant CD8 T cell responses during SIV infection, whereas some of the SIVmac239-infected animals used for comparison do express one or both of these molecules, which could impact our comparative analysis. It is clear from Fig.1, however, that the vaccinated animals do make very strong T cell responses against epitopes not presented by Mamu-A*02. Hence, the relative dominance of the RY8-specific response, within the Mamu-A*02-restricted repertoire, is indeed greater than that elicited by SIVmac239 infection, and its unique mode of translation and presentation should be investigated further.
Although the total contribution of these unique epitope sources to the AIDS virus-specific cellular immune response is unknown, it is becoming clear that pathogen ARFs are rich sources of epitopes, whether or not the overlapping reading frame encodes a functional protein (or a portion of one). Collectively, our data demonstrate clearly that an epitope contained in an ARF with no obvious mode of translation can elicit a dominant T cell response. Perhaps most interesting, our data also indicate that the unknown mechanism producing RY8 may be a useful way to produce epitopes from a vaccine because the relative dominance of this response was greater than that typically seen during normal viral infection. Further study of this phenomenon is needed. Taken together, these data show beyond doubt that there is still much to understand about the processes of translation, epitope generation, and how to elicit T cell responses with vaccination.
Acknowledgments
We thank A.T. Bean and M.B. Buechler for help with cell culture and T.C. Friedrich for helpful discussions.
This work was supported by National Institutes of Health Grants R01 AI052056, R01 AI049120, and R24 RR015371 (to D.I.W.) and Grant P51 RR000167 from the National Center for Research Resources, a component of the National Institutes of Health (to the Wisconsin National Primate Research Center, University of Wisconsin–Madison). This research was conducted in part at a facility constructed with support from Research Facilities Improvement Program Grants RR15459 01 and RR020141-01.
Abbreviations used in this paper
- ARF
alternate reading frame
- Env
Envelope
- ICS
intracellular cytokine staining
- MDDC
monocyte-derived dendritic cell
- MHC-I
MHC class I
- ORF
open reading frame
- RY8
Env788–795RY8
- SFC
spot-forming cell
Footnotes
This publication’s contents are solely the responsibility of the authors and do not necessarily represent the official views of National Center for Research Resources or National Institutes of Health.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Liu J, O’Brien KL, Lynch DM, Simmons NL, La Porte A, Riggs AM, Abbink P, Coffey RT, Grandpre LE, Seaman MS, et al. Immune control of an SIV challenge by a T-cell-based vaccine in rhesus monkeys. Nature. 2009;457:87–91. doi: 10.1038/nature07469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson NA, Reed J, Napoe GS, Piaskowski S, Szymanski A, Furlott J, Gonzalez EJ, Yant LJ, Maness NJ, May GE, et al. Vaccine-induced cellular immune responses reduce plasma viral concentrations after repeated low-dose challenge with pathogenic simian immunodeficiency virus SIVmac239. J. Virol. 2006;80:5875–5885. doi: 10.1128/JVI.00171-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedrich TC, Valentine LE, Yant LJ, Rakasz EG, Piaskowski SM, Furlott JR, Weisgrau KL, Burwitz B, May GE, León EJ, et al. Subdominant CD8+ T-cell responses are involved in durable control of AIDS virus replication. J. Virol. 2007;81:3465–3476. doi: 10.1128/JVI.02392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardinaud S, Moris A, Février M, Rohrlich PS, Weiss L, Langlade-Demoyen P, Lemonnier FA, Schwartz O, Habel A. Identification of cryptic MHC I-restricted epitopes encoded by HIV-1 alternative reading frames. J. Exp. Med. 2004;199:1053–1063. doi: 10.1084/jem.20031869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maness NJ, Valentine LE, May GE, Reed J, Piaskowski SM, Soma T, Furlott J, Rakasz EG, Friedrich TC, Price DA, et al. AIDS virus specific CD8+ T lymphocytes against an immunodominant cryptic epitope select for viral escape. J. Exp. Med. 2007;204:2505–2512. doi: 10.1084/jem.20071261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson NA, Keele BF, Reed JS, Piaskowski SM, MacNair CE, Bett AJ, Liang X, Wang F, Thoryk E, Heidecker GJ, et al. Vaccine-induced cellular responses control simian immunodeficiency virus replication after heterologous challenge. J. Virol. 2009;83:6508–6521. doi: 10.1128/JVI.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maness NJ, Sacha JB, Piaskowski SM, Weisgrau KL, Rakasz EG, May GE, Buechler MB, Walsh AD, Wilson NA, Watkins DI. Novel translation products from simian immunodeficiency virus SIVmac239 Env-encoding mRNA contain both Rev and cryptic T-cell epitopes. J. Virol. 2009;83:10280–10285. doi: 10.1128/JVI.00138-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ignatius R, Marovich M, Mehlhop E, Villamide L, Mahnke K, Cox WI, Isdell F, Frankel SS, Mascola JR, Steinman RM, Pope M. Canarypox virus-induced maturation of dendritic cells is mediated by apoptotic cell death and tumor necrosis factor α secretion. J. Virol. 2000;74:11329–11338. doi: 10.1128/jvi.74.23.11329-11338.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loffredo JT, Sidney J, Wojewoda C, Dodds E, Reynolds MR, Napoé G, Mothé BR, O’Connor DH, Wilson NA, Watkins DI, Sette A. Identification of seventeen new simian immunodeficiency virus-derived CD8+ T cell epitopes restricted by the high frequency molecule, Mamu-A*02, and potential escape from CTL recognition. J. Immunol. 2004;173:5064–5076. doi: 10.4049/jimmunol.173.8.5064. [DOI] [PubMed] [Google Scholar]
- 11.Loffredo JT, Friedrich TC, León EJ, Stephany JJ, Rodrigues DS, Spencer SP, Bean AT, Beal DR, Burwitz BJ, Rudersdorf RA, et al. CD8+ T cells from SIV elite controller macaques recognize Mamu-B*08-bound epitopes and select for widespread viral variation. PLoS One. 2007;2:e1152. doi: 10.1371/journal.pone.0001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loré K, Adams WC, Havenga MJ, Precopio ML, Holterman L, Goudsmit J, Koup RA. Myeloid and plasmacytoid dendritic cells are susceptible to recombinant adenovirus vectors and stimulate polyfunctional memory T cell responses. J. Immunol. 2007;179:1721–1729. doi: 10.4049/jimmunol.179.3.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sacha JB, Chung C, Rakasz EG, Spencer SP, Jonas AK, Bean AT, Lee W, Burwitz BJ, Stephany JJ, Loffredo JT, et al. Gag-specific CD8+ T lymphocytes recognize infected cells before AIDS-virus integration and viral protein expression. J. Immunol. 2007;178:2746–2754. doi: 10.4049/jimmunol.178.5.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sacha JB, Chung C, Reed J, Jonas AK, Bean AT, Spencer SP, Lee W, Vojnov L, Rudersdorf R, Friedrich TC, et al. Pol-specific CD8+ T cells recognize simian immunodeficiency virus-infected cells prior to Nef-mediated major histocompatibility complex class I downregulation. J. Virol. 2007;81:11703–11712. doi: 10.1128/JVI.00926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sacha JB, Reynolds MR, Buechler MB, Chung C, Jonas AK, Wallace LT, Weiler AM, Lee W, Piaskowski SM, Soma T, et al. Differential antigen presentation kinetics of CD8+ T-cell epitopes derived from the same viral protein. J. Virol. 2008;82:9293–9298. doi: 10.1128/JVI.00749-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedrich TC, Dodds EJ, Yant LJ, Vojnov L, Rudersdorf R, Cullen C, Evans DT, Desrosiers RC, Mothé BR, Sidney J, et al. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat. Med. 2004;10:275–281. doi: 10.1038/nm998. [DOI] [PubMed] [Google Scholar]
- 17.Friedrich TC, Frye CA, Yant LJ, O’Connor DH, Kriewaldt NA, Benson M, Vojnov L, Dodds EJ, Cullen C, Rudersdorf R, et al. Extraepitopic compensatory substitutions partially restore fitness to simian immunodeficiency virus variants that escape from an immunodominant cytotoxic-T-lymphocyte response. J. Virol. 2004;78:2581–2585. doi: 10.1128/JVI.78.5.2581-2585.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang YE, Li B, Carlson JM, Streeck H, Gladden AD, Goodman R, Schneidewind A, Power KA, Toth I, Frahm N, Alter G, Brander C, Carrington M, Walker BD, Altfeld M, Heckerman D, Allen TM. Protective HLA class I alleles that restrict acute-phase CD8+ T-cell responses are associated with viral escape mutations located in highly conserved regions of human immunodeficiency virus type 1. J. Virol. 2009;83:1845–1855. doi: 10.1128/JVI.01061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yewdell JW, Antón LC, Bennink JR. Defective ribosomal products (DRiPs): a major source of antigenic peptides for MHC class I molecules? J. Immunol. 1996;157:1823–1826. [PubMed] [Google Scholar]
- 20.Malarkannan S, Horng T, Shih PP, Schwab S, Shastri N. Presentation of out-of-frame peptide/MHC class I complexes by a novel translation initiation mechanism. Immunity. 1999;10:681–690. doi: 10.1016/s1074-7613(00)80067-9. [DOI] [PubMed] [Google Scholar]
- 21.Schwab SR, Li KC, Kang C, Shastri N. Constitutive display of cryptic translation products by MHC class I molecules. Science. 2003;301:1367–1371. doi: 10.1126/science.1085650. [DOI] [PubMed] [Google Scholar]
- 22.Schwab SR, Shugart JA, Horng T, Malarkannan S, Shastri N. Unanticipated antigens: translation initiation at CUG with leucine. PLoS Biol. 2004;2:e366. doi: 10.1371/journal.pbio.0020366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Starck SR, Ow Y, Jiang V, Tokuyama M, Rivera M, Qi X, Roberts RW, Shastri N. A distinct translation initiation mechanism generates cryptic peptides for immune surveillance. PLoS One. 2008;3:e3460. doi: 10.1371/journal.pone.0003460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pande S, Vimaladithan A, Zhao H, Farabaugh PJ. Pulling the ribosome out of frame by +1 at a programmed frameshift site by cognate binding of aminoacyl-tRNA. Mol. Cell. Biol. 1995;15:298–304. doi: 10.1128/mcb.15.1.298. [DOI] [PMC free article] [PubMed] [Google Scholar]