Abstract
Understanding the nature of partially folded intermediates transiently populated during protein folding is important for understanding both protein folding and misfolding. These ephemeral species, however, often elude direct experimental characterization. The well-characterized protein ribonuclease H from E. coli populates an on-pathway intermediate identified in both bulk studies and single molecule mechanical unfolding experiments. Here, we set out to trap the transient intermediate of RNase H at equilibrium by selectively destabilizing the region of the protein known to be unfolded in this species. Surprisingly, a single change at Ile 25 (I25A) resulted in the equilibrium population of the intermediate under near-native conditions. The intermediate was undetectable in a series of HSQC's, revealing the dynamic nature of this partially unfolded form on the timescale of NMR detection. This result is in contrast to studies in which the structures of trapped intermediates are solved by NMR, indicating that the they are well-packed and native-like. The dynamic nature of the RNase H intermediate may be important for its role as an on-pathway, productive species that promotes efficient folding.
Partially folded intermediates are known to play an important role in the mechanism of protein folding. For some proteins, intermediates appear to play a productive role, aiding in the formation of the native fold, while for others they constitute an off-pathway species. In addition to their importance in folding mechanisms, partially folded forms may also play crucial biological roles. There are many examples of proteins with unstructured regions whose disordered-ordered folding transitions are important for binding and other regulatory events 1; 2. For example, the active form of the steroidogenic acute regulatory protein appears to be a molten globule 3, a state usually associated with early folding intermediates. Such intermediates have also been implicated in the formation of aggregation-prone species 4. In spite of their clear importance in both folding and function, these partially folded intermediates are usually not amenable to detailed structural and biophysical studies due to their low populations at equilibrium and transient nature.
Here, we set out to trap the intermediate of ribonuclease H from E. coli (RNase H), a small (17.5 kDa), monomeric enzyme known to fold through such a partially folded kinetic intermediate 5, 6; 7 (throughout this manuscript, wild type RNase H refers to the E. coli variant with all three cysteines replaced by alanine). Recent single molecule experiments conclusively demonstrated that this species is on-pathway and obligatory8. Characterization of the intermediate by phi-value analysis and quenched flow hydrogen exchange suggest that the intermediate is composed of native-like topology in the core and a relatively unstructured periphery (Figure 1a) 5, 6. This partially folded ensemble appears to play a robust and important role in the folding trajectory of RNase H, dominating the folding trajectory even in variants that do not transiently populate the state (see accompanying manuscript). Equilibrium native state hydrogen exchange experiments detect a rare high energy partially unfolded form whose structure and energetics mirror this kinetic intermediate, suggesting that the most stable region of the protein is the first to fold 6, 9.
Figure 1.
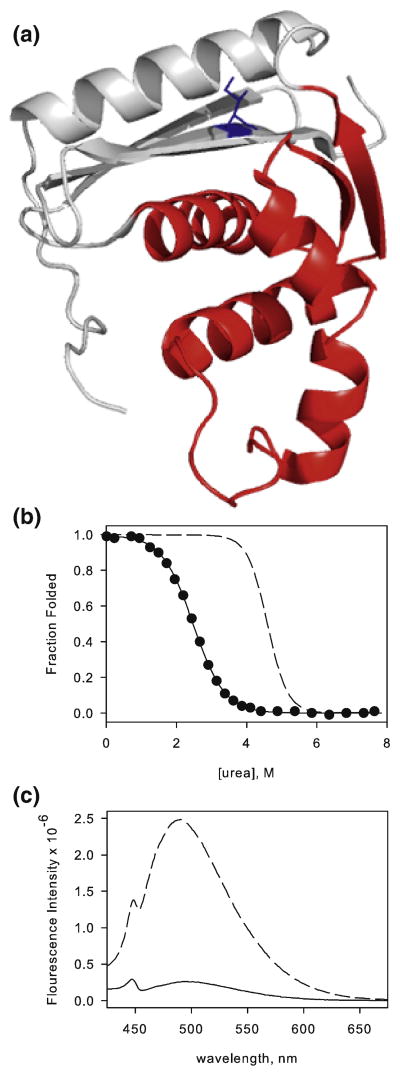
I25A resembles the RNase H intermediate. a) Ribbon representation of RNase H (pdb 1F21) with the core of the protein colored red and the periphery colored grey. Residue I25 is shown as sticks and colored blue. b) Fraction folded of I25A (filled circles) determined from urea melts overlaid with the wild type curve (dashed line). The solid line represents the two-state fit for I25A. c) Fluorescence emission spectra of ANS in the presence of WT (solid line) and I25A (dashed line). The fluorescence emission of ANS in buffer alone has been subtracted from the data.
We have used rational design to selectively destabilize the native state of the protein, enriching the population of the high-energy species. The response of RNase H to mutations highlights the differential distribution of stability in the core and periphery that makes this approach possible. Isoleucine 53, located in the core, was mutated to alanine (I53A), destabilizing the protein by ∼2 kcal/mol (ΔΔGUN ∼ 2kcal mol-1) 10. Native state hydrogen exchange demonstrated that this destabilizing effect was localized to the core, while residues in the periphery were unaffected, resulting in destabilization of the intermediate. A similar study was carried out in the periphery with the stabilizing mutation D10A 11, where, although all secondary elements showed an increase in the free energy of unfolding as measured by native state hydrogen exchange, the difference between unfolding the periphery and the core remained unchanged, suggesting that the two regions are energetically independent. This selective increase in stability causes a decrease in the population of the partially folded form. For both of these variants, the effects in the equilibrium energetics were mirrored in the transient intermediate observed during the folding process. In fact, a severely destabilizing mutation in the core of the protein (I53D) destabilizes the intermediate such that the protein folds in an apparent two-state manner without the obvious accumulation of the intermediate 7.
In principle, then, we should be able to use mutations that destabilize the periphery of the protein without perturbing the stability of the intermediate to selectively destabilize the native state, enhancing the population of the high-energy intermediate such that we can study it more easily. This strategy has been demonstrated by Bai 12; 13; 14; 15; 16; 17 who, based on native state hydrogen exchange data, create mutations to selectively destabilize the regions predicted to be unfolded in the intermediate, thereby selectively destabilizing the native state. Similarly, Radford and coworkers destabilized regions of the immunity protein Im7 based on results from phi-value analysis (monitoring the folding kinetics for engineered site-specific mutations).18
In an alternative approach, Bai and coworkers also recently designed a fragment of T. thermophilus RNase H modeled, in part, on our native state hydrogen exchange data. By removing two strands believed to be disordered in the intermediate, they obtained a well-folded species amenable to high resolution NMR spectroscopy. Their results reveal a fragment that folds into a well-packed native-like species, suggesting that the structure of the partially folded intermediate may be a subset of the native structure. These results are at odds with studies that suggest the intermediate state of RNase H is a heterogeneous ensemble with the hallmarks of a molten globule19, 20. While the fragment approach has the advantage of creating a fragment amenable for high-resolution studies, there are drawbacks. The subjective assumptions about the structure, based on the limited hydrogen exchange data, may omit important structural components of the intermediate. All interpretations are based on the underlying assumption that the fragment is indeed a representative mimic of the high-energy ensemble. Questions therefore remain as to the exact nature of the contacts formed in the high-energy intermediate of the protein, the extent to which tertiary structure has been fixed and the breadth of this species, and which regions contribute to the molten nature of the intermediate.
Here, we set out to trap the intermediate of RNase H by rational design of destabilizing mutants believed to selectively to the native state stability. We make no assumptions as to the structure of the species, but instead use mutagenesis to selectively destabilize the native state to enrich the population of the high-energy species. This approach is completely analogous to Bai's protein engineering approach to uncover hidden intermediates, only in our case, the intermediate is not hidden, but rather a detectable transiently populated species.
To our surprise, the first target for mutation, isoleucine 25, located in Strand II of the periphery and completely buried in a hydrophobic pocket (Figure 1a), was almost sufficient to accomplish our task. This variant, which is part of a series of mutations used to investigate the robustness of the folding trajectory in two-state and three-state versions of the protein (Connell et. al, accompanying manuscript), results in a protein that detectably populates the kinetic intermediate at equilibrium. Although we could not obtain high-resolution structural information, with this mutation we were able to gain valuable insight into the dynamic properties of the intermediate.
Results
I25A populates the intermediate
Equilibrium urea denaturation of I25A was monitored by circular dichroism (CD) at 222 nm and fit with the standard two-state linear extrapolation method (Figure 1b) 21. This analysis results in surprisingly low values for both ΔG and the m-value (the denaturant dependence of ΔGUN). The calculated ΔGUN is 3.4 kcal/mol, indicating an abnormally large destabilization from the wild type protein (WT ΔGUN = 9.7 kcal/mol, see Table 1), and the m-value is 1.3 kcal mol-1 M-1, much lower than the wild type m-value of 2.1 kcal mol-1 M-1 (Table 1). The m-value is believed to correlate with the burial of surface area upon folding 22; therefore, if the overall fold is retained, this parameter should remain unchanged. The m-value we obtained for I25A, while clearly different from that expected based on the native structure, is strikingly similar to that determined for the transient folding intermediate of RNase H 5, suggesting that the native state was unexpectedly destabilized to the extent that the intermediate is now the most stable species.
Table 1.
Stability and m-values for RNase H variants. The values in parentheses represent the standard deviation calculated from at least three individual denaturant melts. The reported ΔGUN for D10A/I25A was obtained by fixing the m-value to 2.1 kcal mol-1 M-1. See accompanying manuscript for details.
| wt6 | I53A5 | D10A5 | I25A | D10A/I25A | ||
|---|---|---|---|---|---|---|
| urea melt | model | |||||
| Δ GUN (kcal mol-1) | 9.7 | 7.6 | 13 | 3.4 (0.3) | 4.5 | 7.86 (0.07) |
| mUN (kcal mol-1 M-1) | 2.1 | 2.1 | 2.1 | 1.3 (0.1) | 2.1* | 2.1 |
| Δ GUI (kcal mol-1) | 3.55 | 1.66 | 4.38 | ND | 3.55* | ND |
| mUI (kcal mol-1 M-1) | 1.24 | 1.24 | 1.45 | ND | 1.24* | ND |
Wild type values assumed for the model
ND: not determined
To further investigate the possibility that the I25A variant populates the intermediate state, 1-anilino-8-napthalene sulphonic acid (ANS) binding was monitored by fluorescence (Figure 1c). The RNase H kinetic intermediate resembles its equilibrium molten globule 6, and one of the trademarks of molten globules is binding of the hydrophobic dye ANS. When ANS binds exposed hydrophobic surface area, its emission maximum blue-shifts and its fluorescence intensity increases 23. Under native conditions, I25A incubated with ANS shows a large increase in fluorescence compared to the wild type enzyme; this increase is similar in magnitude to what is generally observed for molten globules.
I25A RNase H populates the native fold
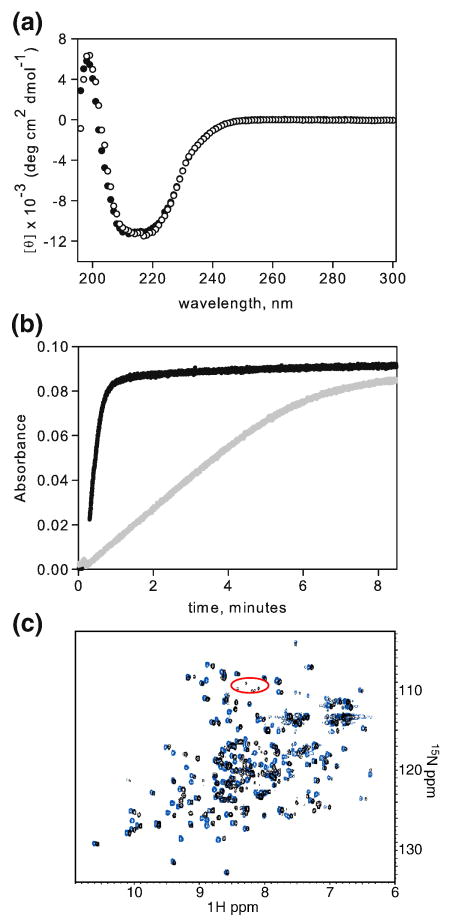
In contrast to the evidence presented above, several studies demonstrate that I25A retains a native-like fold. The CD spectrum of I25A under native conditions is indicative of a folded, α/β protein and resembles that of wild type RNase H (Figure 2a). I25A RNase H is also active, assessed by the loss of the hypochromic effect as the RNA strand is cleaved from a DNA-RNA hybrid (Figure 2b). Preservation of the active enzyme, albeit impaired in comparison to wild type, implies that the protein can access the native fold under these conditions.
Figure 2.
Evidence for an E. coli RNase H fold. a) CD spectrum of I25A (filled circles) overlaid with wild type RNase H (open circles). b) Activity of I25A (grey) compared with that of WT (black). c) 1H-15N HSQC of I25A RNase H (black) overlaid with that of WT (blue) in 20 mM NaOAc, pH 5.5, 50 mM KCl, 10 % D2O.
To obtain higher resolution structural information, a native state 15N-1H HSQC NMR spectrum was obtained (Figure 2c). I25A shows clearly dispersed peaks characteristic of a well-folded protein; in contrast, the spectra of molten globules and unfolded proteins exhibit a collapsed set of peaks in the 1H dimension. Furthermore, the I25A peaks overlay well with those of wild type RNase H, clearly demonstrating that I25A populates the native structure under the conditions of the NMR experiments.
The two-state assumption is not valid for I25A
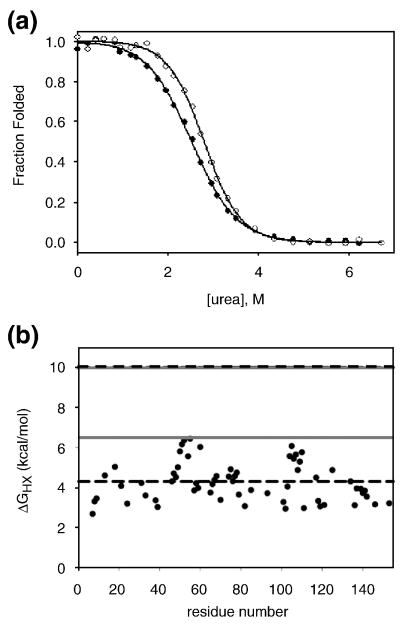
In light of the conflicting behavior of I25A RNase H, intrinsic tryptophan fluorescence was used to further probe equilibrium behavior via urea denaturation. RNase H has six Trp residues. Fluorescence intensity at 340 nm as a function of urea concentration was fit using the two-state approximation allowing a direct comparison between CD and fluorescence. In the case of wild type RNase H, the curves representing the fraction folded as calculated by CD and fluorescence are indistinguishable20. For I25A, however, the fluorescence and CD data are non–coincident (Figure 3a). The fit from the fluorescence results in an m-value of 1.5 kcal mol-1 M-1 and ΔGUN of 4.1 kcal/mol, which, while again significantly lower than wild type values, do not match with those calculated by CD. This non-coincidence of probes suggests that for I25A RNase H the two-state assumption breaks down. In support of this, the emission spectra do not have an isosbestic point, also indicating that more than two species contribute to the signal.
Figure 3.
I25A does not follow two-state behavior at equilibrium. a) Fraction folded of I25A by CD (closed circles) and fluorescence (open circles) determined from urea melts fit to a two-state approximation. Solid lines represent each fit. b) Hydrogen exchange data for I25A. The free energy of exchange of I25A is plotted as a function of residue. The grey solid lines represent the stability of WT and I25A by hydrogen exchange methods, as indicated, and the dashed black lines represent the stability of each protein calculated from the two-state fits to melts acquired in deuterated solvent.
To obtain an estimate of the true global stability of the native conformation without assuming a two-state model, hydrogen exchange was carried out on the I25A variant. By monitoring the rate at which amide hydrogens exchange with solvent deuterium by NMR, we calculated the free energy of exchange for 63 residues (ΔG = -RT ln (kobs/krc)) 24. The ΔGHX values obtained for I25A, shown in Figure 3b, reveal that the regions with the highest protection are the A and D helices, located in the core. These values suggest a global stability of approximately 6 kcal/mol. Since it is known that D2O has an effect on protein stability 25, we repeated the denaturant melt in D2O in order to compare the stability calculated by hydrogen exchange to that obtained by standard urea denaturation (data not shown). The stability of I25A in D2O, assuming a two-state model, is increased by less than 1 kcal/mol, and the m-value remains the same. A large discrepancy, therefore, remains between the stability of I25A calculated assuming a two-state fit and that by hydrogen exchange (Figure 3b). This incongruity is not evident for wild type RNase H. In this case the values for the global stability of the protein determined by native state hydrogen exchange and a two-state fit to the equilibrium denaturation profile are closely matched 9, confirming the validity of the two-state assumption. The inconsistency observed for I25A is additional evidence that the two-state assumption does not hold for this variant.
I25A populates the intermediate under native conditions
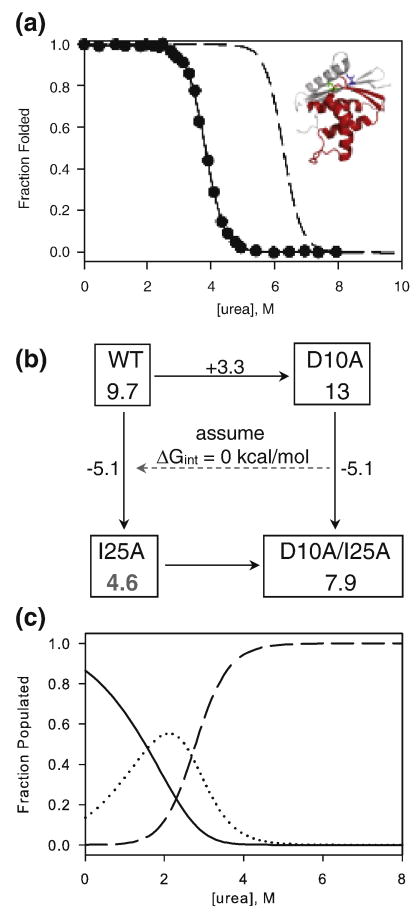
We determined the effect of I25A in the background of D10A, a mutation in the periphery known to stabilize RNase H by 3.3 kcal/mol (ΔGUN = 13.0 kcal/mol) 5. In a more stable background, there is less potential for a destabilizing mutation to affect the apparent two-state equilibrium transition. Equilibrium urea denaturation of D10A/I25A was monitored by CD and fit using the two-state approximation (Figure 4a). The introduction of I25A into D10A destabilizes the protein by 5.1 kcal/mol (ΔGUN = 7.9 kcal/mol) and results in an m-value of 2.1 kcal mol-1 M-1, identical to that of wild type and D10A (Table 1). Assuming additivity provides a theoretical value for the stability of I25A of 4.6 kcal/mol (Scheme 1).
Figure 4.
Determining the effect of I25A. a) Fraction folded of D10A/I25A determined from urea melts (filled circles) overlaid with curves for wild type (dashed line). The solid line represents the two-state fit to the D10A/I25A melt. Inset: Locations of I25 (blue) and D10 (green) shown on the structure of RNase H with the periphery in grey and the core in red. b) A thermodynamic cycle assuming no interaction between residues 10 and 25 was used to calculate the expected stability of I25A. Numbers in each box represent the global stability, ΔGUN (kcal/mol), and the numbers along the arrows indicate the change in stability upon making the indicated mutation. c) A graph of the theoretical populations for the native (solid), intermediate (dotted), and unfolded (dashed) forms of I25A as a function of urea concentration. The model assumes the native form of I25A is 4.6 kcal mol-1 stable with respect to the denatured form and that an m-value of 2.1 kcal mol-1 M-1 is associated with this transition. The intermediate is assumed to be 3.5 kcal mol-1 stable with respect to the denatured form, as it is in the wild type protein, with an m-value of 1.3 kcal mol-1 M-1.
We constructed a model of the populations of native, intermediate, and denatured I25A RNase H as a function of urea using this value for ΔGUN and the wild type values for mUN, ΔGUI and mUI. This model, shown in Figure 4b, reveals that the stability of the native state of I25A is sufficiently close to the stability of the intermediate that a significant population of intermediate exists under native conditions and becomes the dominant species around 2 M urea.
NMR spectra in denaturant reveal two sets of peaks
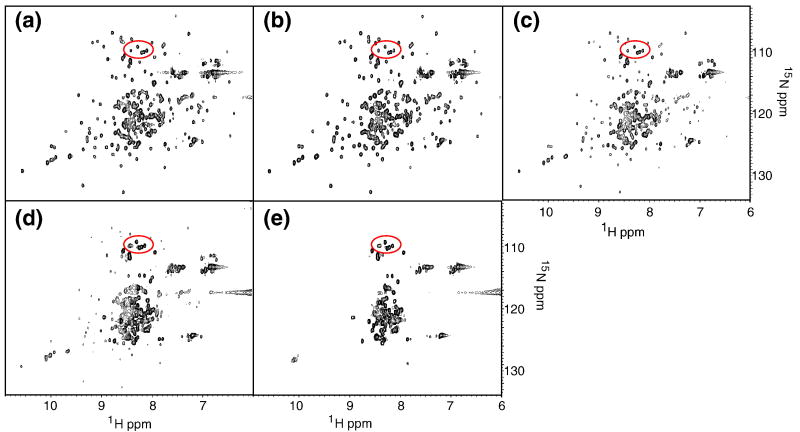
Confident that I25A does in fact populate the intermediate state, we obtained HSQCs at varying urea concentrations in the hopes of obtaining structural information about the intermediate at the residue level (Figure 5a-e). In low urea, the spectrum closely resembles that in no denaturant. Upon closer inspection, especially around the glycine region, however, there is a clear cluster of peaks that is not present in the wild type spectrum under native conditions. These peaks are circled in red in Figure 2c and Figure 5a-3. As urea is added, these peaks intensify and persist to high concentrations of denaturant, indicating that they correspond to residues in the unfolded state.
Figure 5.
Probing the Intermediate by NMR. A series of HSQC's in 20 mM NaOAc, pH 5.5, 50 mM KCl, 10 % D2O and a) 0.5 M, b) 1.0 M, c) 1.5 M, d) 2.0 M, and e) 2.5 M urea.
Peaks in other regions of the spectrum also emerge at increasing urea concentrations. By 1.5 M urea (Figure 5c), there are clearly two sets of peaks. One set corresponds to the dispersed set expected for the native conformation, and these decrease in intensity as denaturant increases. A second set, which builds in intensity with urea concentration, is characterized by a lack of dispersion in the 1H dimension and appears to arise from residues in the unfolded state. This is in stark contrast to what we observe for the wild type protein, where we see one set of peaks at all concentrations of urea.
Discussion
Protein engineering when combined with prior knowledge of the energy landscape of a protein, particularly from hydrogen exchange data, can be a useful tool to manipulate the population of protein conformations. Here, by creating a single destabilizing mutation in the periphery of RNase H (I25A) we show that it can be used to the protein populates the partially folded intermediate known to be important in the folding trajectory of the protein, and thereby allowing us to study this experimentally elusive species at equilibrium.
Modeling the population of each species in I25A
Initial characterization of I25A provided conflicting views of the effect of the mutation. The CD spectrum, activity assay, and 15N-1H HSQC indicate that the overall structure of the protein is not notably perturbed from that of wild type. On the other hand, the significantly lower m-value obtained by the two-state fit of the denaturant melts and the ANS binding assay suggested that I25A resembles the intermediate conformation of RNase H. The non-coincidence of CD and fluorescence and the estimate of global stability obtained by hydrogen exchange indicate that the two-state approximation cannot be applied in this case. Examining the effect of the isoleucine to alanine mutation at position 25 in a more stable background (D10A) allowed us to obtain a better estimate of the effect of this mutation on the native state of RNase H. Surprisingly, this single mutation appears to destabilize the protein by more than 5 kcal/mol. Since the stability of the intermediate of RNase H is approximately 3.5 kcal/mol, a mutation of this magnitude that selectively destabilizes the native state (∼10 kcal/mol) will result in a native state stability close to that of the intermediate and in a significant accumulation of the intermediate at equilibrium. The two-state assumption used to fit the denaturant melt is therefore invalid in this case, resulting in an inaccurate determination of the stability of the protein standard urea-induced denaturation profiles.
Why does this single amino acid change have such a drastic effect on the stability of RNase H? By introducing alanine at position 25, we expected to destabilize the periphery by creating a small cavity in the hydrophobic pocket. Based on the change in hydrophobic surface area, we expect this mutation to destabilize the protein by ∼ 3 kcal/mol 26. At this point, we do not fully understand why the change is so much greater, although such large changes are not without precedent. For example, in T4 lysozyme, small perturbations (leucine to alanine) at buried sites have been known to result in large stability changes when coupled to the formation of a cavity 27. In our case, this effect may also be exacerbated the presence of a nearby salt bridge network. Creating a cavity in the vicinity of these salt bridges may decrease the strength of the interaction, allowing the periphery to unfold more readily.
Our three-state equilibrium model successfully accounts for all of our experimental data. To model our data, the intermediate was assigned a CD signal 80% that of the native state based on kinetic studies on the wild type protein 6. Fitting the simulated melt to the two-state model reproduces the observed data with a calculated free energy of unfolding (3.5 kcal/mol) and the m-value (1.3 kcal mol-1 M-1) of I25A RNase H.
The stability estimate obtained by hydrogen exchange is also consistent with our conclusions. This method should allow us to measure the free energy of hydrogen exchange of individual residues with an upper limit of the stability of I25A in D2O. By looking at the most protected residues in the protein, those in helices A and D, we would estimate the stability of the protein to be ∼ 5 - 6 kcal/mol. Studies of RNase H in the presence of D2O suggest that the solvent increases the stability by just over 1 kcal/mol, which would put our estimate of the stability of I25A at ∼4.5 kcal/mol, consistent with the value derived from assuming additivity with the mutation D10A. We attempted to repeat the urea melts used in this study in D2O to get a more exact number, but urea is not stable for the long equilibration times required for the variants containing D10A. Nonetheless, our expectation of 4.5 kcal/mol is consistent with both the hydrogen exchange and mutant cycle analysis.
The HSQC spectra in varying urea concentrations provide further corroborating evidence that I25A populates the intermediate. The cluster of peaks circled in Figure 2c is not present in the wild type spectrum. They fall in the glycine region, and twelve of the fifteen glycines in RNase H are located in the periphery, a region presumed to be unstructured in the intermediate. These peaks persist in the higher urea concentrations (see peaks circled in red, Figures 2c and 5), where two sets of peaks are evident, one corresponding to folded protein and one characterized by the lack of dispersion typical of unfolded proteins. At these relatively low levels of denaturant, there should be no detectable unfolded protein, suggesting that these peaks arise from the intermediate state. In addition, similar spectra of the wild type protein in varying amounts of urea do not exhibit two sets of peaks. We therefore attribute the glycine peaks and the appearance of the collapsed second set to unfolded regions of the intermediate.
No peaks appear to correspond to structured areas of the intermediate. The dispersed set of peaks appears to arise from the native state. At 2 M urea, where the intermediate should be maximally populated, the dispersed set of peaks overlays well with the native spectrum, and disappear together with the glycine peaks believed to arise from the native periphery. Together, these data suggest that the peaks from the folded core arise from the native structure, not the folded region of the intermediate. Although this set of peaks does exhibit some shift from the native spectrum, these shifts may arise from the addition of urea and are not of the magnitude we would expect if they were to originate from the intermediate. The gradual shift of peaks can also indicate two species in fast exchange, the observed peak being an average of the separate signals from each of the populations rapidly interconverting. The intermediate and native structure are unlikely to be in fast exchange because the rate of the I → N step in H2O is 0.74 s-1, far slower than the NMR fast exchange regime. Instead we attribute the loss of peaks for the structured region of the intermediate to dynamic line broadening.
Insights into the nature of the folding intermediate
The HSQC spectra of molten globules commonly show no peaks that report on the structured regions, with signal building in only as structure is lost 28. Fluctuations throughout the molten globule on the millisecond timescale are cited as the cause of peak broadening. We believe that, likewise, the absence of peaks arising from the intermediate in these experiments indicates that it is a dynamic species. This exchange between conformers suggests that the barriers between them are low and that they lack fixed tertiary interactions; contacts in the core, though native-like, are most likely weak.
This model of the folding intermediate of RNase H as a dynamic species agrees with studies on equilibrium molten globules that are thought to mirror the intermediates formed in the kinetic folding trajectory. The intermediate of apomyoglobin is also thought to be a heterogeneous and dynamic ensemble29. Our results, however, differ from those obtained on other systems where mutations were made to selectively populate the intermediate, such as Im7 and redesigned-apocytochrome b562. Both of these proteins are four-helix proteins with one or more helices unstructured in their intermediates. NMR studies on the models of these intermediates indicate that the folded regions are quite rigid 18; 30 14; 31, although the Im7 intermediate does show increased heterogeneity over the native state. In the cases of both Im7 and redesigned-apocytochrome b562, the authors stress the existence of non-native contacts in the intermediate that need to be broken upon folding to the native state. Instead of utilizing non-native tertiary contacts to stabilize the intermediate, RNase H appears to fluctuate, forming weak native-like interactions that do not persist.
Our results are also at odds with recent NMR studies on a fragment of T. thermophilus RNase H that forms a well-folded native-like subdomain. The fragment was generated by removing regions from the periphery: two internal strands and the final helix of the protein, both of which appear unprotected by hydrogen exchange, while leaving one strand from the periphery (strand 1) which, based on native state hydrogen exchange and mutagenesis studies, also does not appear to be structured in the folding intermediate of E. coli RNase H. The inclusion of this strand may stabilize the fragment, allowing for a well-behaved sample amenable to high resolution NMR. The high-resolution structure reveals only native-like interactions, questioning the nature of the barrier in the progression of folding from the intermediate to the native state and at odds with the observation that mutations in this region affect the transition state stability more than the intermediate 5, 7. The difference between what we find here and the NMR study on this fragement may also be attributed to a difference in the interactions that stabilize the intermediates from these two proteins (E. coli and T. thermophilus) . Another explanation might be that the well-folded fragment is actually a mimic of another identified high-energy intermediate of the T. thermophilus protein where just the last helix appears to be unfolded.
The dynamic nature of the E. coli RNase H intermediate may have important implications for efficient and productive folding in vivo. The intermediate of RNase H is populated prior to the rate-limiting step and its presence appears to aid in folding. Destabilizing the intermediate slows overall folding, suggesting that the interactions that stabilize the native state are present and important in the rate-limiting transition state. If interactions that form as the reaction progresses stabilize the transition state more than the intermediate, the folding reaction will be accelerated. This may be the case for RNase H, for which all evidence points to a productive, on-pathway, obligatory intermediate.
Materials and Methods
Materials
Deuterium oxide, 15N ammonium chloride, deuterated buffers, acids, and bases were purchased from Isotec . All other buffer reagents were purchased from Sigma or Fischer.
Protein Expression and Purification
E. coli BL21 pLysS cells were transformed with the appropriate plasmid and grown at 37°C in LB medium with 200 μg/ml ampicillin. Induction was initiated by the addition of 1 mM IPTG to cells at OD600 ∼0.6. Cells were harvested by centrifugation 3 – 4 hours after induction. I25A expressed in inclusion bodies, and purification was carried out on cell pellets as previously described for other RNase H variants 7. D10A/I25A was expressed solubly and was purified as previously described 20. Purity and molecular weights of all variants were verified by mass spectrometry (data not shown). To express 15N-labeled protein, log-phase cells were transferred to M9 medium containing 15N ammonium chloride as described 32.
All experiments were carried out in 20 mM sodium acetate and 50 mM KCl at pH 5.5 unless otherwise noted. Protein concentrations were determined based on the extinction coefficient, calculated according to the number of Trp and Tyr residues 33.
Equilibrium CD Experiments
Circular dichroism measurements were carried out on an Aviv 62DS spectrometer at 25°C. For denaturant melts, individual samples containing 40 - 50 μg/mL protein at varying urea concentrations were equilibrated overnight. For each sample, CD signal was monitored at 222 nm, and the signal was averaged over a 60 second time period. Data were fit assuming a two-state model and linear dependence of ΔGUN on urea concentration.
ANS Binding
Samples containing 500 μM 1-anilino-8-napthalene sulphonic acid in buffer with and without 2 μM protein were prepared and equilibrated overnight. Fluorescence emission spectra were collected from 425 to 675 nm with an excitation wavelength of 405 nm. The spectrum of ANS in buffer alone was subtracted from those of ANS containing protein.
Activity Assay
Activity of RNase H is monitored by the loss of the hypochromic effect as the RNA strand is cleaved from a DNA-RNA hybrid. The reaction was carried out in 50 mM Tris, 50 mM NaCl, 10 mM MgCl2, and 10 μg/mL rA-dT and was initiated by the addition of enzyme to a final concentration of 5 nM. Absorbance at 260 nm was followed over time on a Cary UV-Vis spectrometer.
Tryptophan Fluorescence Measurements
Urea denaturation was monitored by tryptophan fluorescence using a Fluoromax 3 fluorimeter (JYHoriba) at 25°C. Individual samples of 40 – 50 μg/mL in varying urea concentrations equilibrated overnight. Excitation was at 295 nm, and emission spectra were recorded with both slits at 4 nm. Fluorescence at 340 nm as well as the center of mass were analyzed and fit using a two-state approximation and a linear dependence of ΔGUN on urea concentration.
Hydrogen Exchange
Amide hydrogen exchange was initiated by exchanging protonated 15N I25A RNase H into deuterated buffer (20 mM sodium acetate and 50 mM KCl at pDr 5.6) using a polypropylene spin column (Pierce) packed with Sephadex resin. The sample was immediately transferred to an NMR tube and placed in the instrument; time between initiation of exchange and start of data collection was approximately 25 minutes. 15N-1H HSQC spectra were recorded on a Bruker 600 MHz at 25°C as an average of 16 scans with 1024 points in the direct dimension and 256 complex points in the indirect dimension. HSQC's (∼1 hour each) were collected consecutively for 10 hours and then increasingly spaced out to two weeks. Spectra were processed using Felix 97.0 (Accrelys), and peak height as a function of time was fit to a single exponential decay in SigmaPlot (SSI) to obtain a value for kobs. Wild type peak assignments were used to assign peaks for I25A. The free energy of exchange was calculated as:
where krc is the intrinsic rate of exchange for that residue in a random coil 24.
HSQC's at varying concentrations of urea
Two-dimensional, gradient-enhanced HSQC's were recorded on a Bruker 600 MHz at 25°C. 32 scans were collected with 1024 points in the direct dimension and 128 complex points in the indirect dimension. These data were processed using NMRPipe and viewed in NMRDraw.
Acknowledgments
We thank David E. Wildes for suggestions and development of this project and Rachel Bernstein for thoughtful comments on the manuscript. We also acknowledge David Wemmer for NMR use and Jeff Pelton for guidance in NMR data collection. This work was supported by NIH grant GM50945 to S.M. and an NSF graduate research fellowship to K.B.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 2.Wright PE, Dyson HJ. Intrinsically unstructured proteins: reassessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–31. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 3.Bose HS, Whittal RM, Baldwin MA, Miller WL. The active form of the steroidogenic acute regulatory protein, StAR, appears to be a molten globule. Proc Natl Acad Sci U S A. 1999;96:7250–5. doi: 10.1073/pnas.96.13.7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jahn TR, Parker MJ, Homans SW, Radford SE. Amyloid formation under physiological conditions proceeds via a native-like folding intermediate. Nat Struct Mol Biol. 2006;13:195–201. doi: 10.1038/nsmb1058. [DOI] [PubMed] [Google Scholar]
- 5.Raschke TM, Kho J, Marqusee S. Confirmation of the hierarchical folding of RNase H: a protein engineering study. Nat Struct Biol. 1999;6:825–31. doi: 10.1038/12277. [DOI] [PubMed] [Google Scholar]
- 6.Raschke TM, Marqusee S. The kinetic folding intermediate of ribonuclease H resembles the acid molten globule and partially unfolded molecules detected under native conditions. Nat Struct Biol. 1997;4:298–304. doi: 10.1038/nsb0497-298. [DOI] [PubMed] [Google Scholar]
- 7.Spudich GM, Miller EJ, Marqusee S. Destabilization of the Escherichia coli RNase H kinetic intermediate: switching between a two-state and three-state folding mechanism. J Mol Biol. 2004;335:609–18. doi: 10.1016/j.jmb.2003.10.052. [DOI] [PubMed] [Google Scholar]
- 8.Cecconi C, Shank EA, Bustamante C, Marqusee S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309:2057–60. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- 9.Chamberlain AK, Handel TM, Marqusee S. Detection of rare partially folded molecules in equilibrium with the native conformation of RNaseH. Nat Struct Biol. 1996;3:782–7. doi: 10.1038/nsb0996-782. [DOI] [PubMed] [Google Scholar]
- 10.Spudich G, Lorenz S, Marqusee S. Propagation of a single destabilizing mutation throughout the Escherichia coli ribonuclease HI native state. Protein Sci. 2002;11:522–8. doi: 10.1110/ps.37202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goedken ER, Marqusee S. Native-state energetics of a thermostabilized variant of ribonuclease HI. J Mol Biol. 2001;314:863–71. doi: 10.1006/jmbi.2001.5184. [DOI] [PubMed] [Google Scholar]
- 12.Bai Y. Energy barriers, cooperativity, and hidden intermediates in the folding of small proteins. Biochem Biophys Res Commun. 2006;340:976–83. doi: 10.1016/j.bbrc.2005.12.093. [DOI] [PubMed] [Google Scholar]
- 13.Bai Y, Feng H, Zhou Z. Population and structure determination of hidden folding intermediates by native-state hydrogen exchange-directed protein engineering and nuclear magnetic resonance. Methods Mol Biol. 2007;350:69–81. doi: 10.1385/1-59745-189-4:69. [DOI] [PubMed] [Google Scholar]
- 14.Feng H, Vu ND, Bai Y. Detection and structure determination of an equilibrium unfolding intermediate of Rd-apocytochrome b562: native fold with non-native hydrophobic interactions. J Mol Biol. 2004;343:1477–85. doi: 10.1016/j.jmb.2004.08.099. [DOI] [PubMed] [Google Scholar]
- 15.Feng H, Vu ND, Bai Y. Detection of a hidden folding intermediate of the third domain of PDZ. J Mol Biol. 2005;346:345–53. doi: 10.1016/j.jmb.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 16.Kato H, Feng H, Bai Y. The folding pathway of T4 lysozyme: the high-resolution structure and folding of a hidden intermediate. J Mol Biol. 2007;365:870–80. doi: 10.1016/j.jmb.2006.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato H, Vu ND, Feng H, Zhou Z, Bai Y. The folding pathway of T4 lysozyme: an on-pathway hidden folding intermediate. J Mol Biol. 2007;365:881–91. doi: 10.1016/j.jmb.2006.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spence GR, Capaldi AP, Radford SE. Trapping the on-pathway folding intermediate of Im7 at equilibrium. J Mol Biol. 2004;341:215–26. doi: 10.1016/j.jmb.2004.05.049. [DOI] [PubMed] [Google Scholar]
- 19.Chamberlain AK, Marqusee S. Molten globule unfolding monitored by hydrogen exchange in urea. Biochemistry. 1998;37:1736–42. doi: 10.1021/bi972692i. [DOI] [PubMed] [Google Scholar]
- 20.Dabora JM, Marqusee S. Equilibrium unfolding of Escherichia coli ribonuclease H: characterization of a partially folded state. Protein Sci. 1994;3:1401–8. doi: 10.1002/pro.5560030906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greene RF, Jr, Pace CN. Urea and guanidine hydrochloride denaturation of ribonuclease, lysozyme, alpha-chymotrypsin, and beta-lactoglobulin. J Biol Chem. 1974;249:5388–93. [PubMed] [Google Scholar]
- 22.Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4:2138–48. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Semisotnov GV, Rodionova NA, Razgulyaev OI, Uversky VN, Gripas AF, Gilmanshin RI. Study of the “molten globule” intermediate state in protein folding by a hydrophobic fluorescent probe. Biopolymers. 1991;31:119–28. doi: 10.1002/bip.360310111. [DOI] [PubMed] [Google Scholar]
- 24.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makhatadze GI, Clore GM, Gronenborn AM. Solvent isotope effect and protein stability. Nat Struct Biol. 1995;2:852–5. doi: 10.1038/nsb1095-852. [DOI] [PubMed] [Google Scholar]
- 26.Xu J, Baase WA, Baldwin E, Matthews BW. The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect. Protein Sci. 1998;7:158–77. doi: 10.1002/pro.5560070117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eriksson AE, Baase WA, Zhang XJ, Heinz DW, Blaber M, Baldwin EP, Matthews BW. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science. 1992;255:178–83. doi: 10.1126/science.1553543. [DOI] [PubMed] [Google Scholar]
- 28.Redfield C. Using nuclear magnetic resonance spectroscopy to study molten globule states of proteins. Methods. 2004;34:121–32. doi: 10.1016/j.ymeth.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 29.Nishimura C, Dyson HJ, Wright PE. The apomyoglobin folding pathway revisited: structural heterogeneity in the kinetic burst phase intermediate. J Mol Biol. 2002;322:483–9. doi: 10.1016/s0022-2836(02)00810-0. [DOI] [PubMed] [Google Scholar]
- 30.Gsponer J, Hopearuoho H, Whittaker SB, Spence GR, Moore GR, Paci E, Radford SE, Vendruscolo M. Determination of an ensemble of structures representing the intermediate state of the bacterial immunity protein Im7. Proc Natl Acad Sci U S A. 2006;103:99–104. doi: 10.1073/pnas.0508667102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng H, Zhou Z, Bai Y. A protein folding pathway with multiple folding intermediates at atomic resolution. Proc Natl Acad Sci U S A. 2005;102:5026–31. doi: 10.1073/pnas.0501372102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marley J, Lu M, Bracken C. A method for efficient isotopic labeling of recombinant proteins. J Biomol NMR. 2001;20:71–5. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- 33.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–54. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]