Abstract
O-linked beta-N-acetylglucosamine (O-GlcNAc) modification of nuclear and cytoplasmic proteins is important for many cellular processes, and the number of proteins that contain this modification is steadily increasing. This modification is dynamic and reversible, and in some cases competes for phosphorylation of the same residues. O-GlcNAc modification of proteins is regulated by cell cycle, nutrient metabolism, and other extracellular signals. Compared to protein phosphorylation, which is mediated by a large number of kinases, O-GlcNAc modification is catalyzed only by one enzyme called O-linked N-acetylglucosaminyl transferase or OGT. Removal of O-GlcNAc from proteins is catalyzed by the enzyme beta-N-acetylglucosaminidase (O-GlcNAcase or OGA). Altered O-linked GlcNAc modification levels contribute to the establishment of many diseases, such as cancer, diabetes, cardiovascular disease, and neurodegeneration. Many transcription factors have been shown to be modified by O-linked GlcNAc modification, which can influence their transcriptional activity, DNA binding, localization, stability, and interaction with other co-factors. This review focuses on modulation of transcription factor function by O-linked GlcNAc modification.
Keywords: O-GlcNAc, transcription factors, OGT, OGA, hexosamine, transcription
1. Introduction
O-linked GlcNAc modification is an important post-translational modification that modulates the function of many nuclear and cytoplasmic proteins. Proteins are modified at serine or threonine residues by attachment of a single N-acetylglucosamine (GlcNAc) molecule catalyzed by the enzyme O-linked N-acetylglucosaminyl transferase OGT [1-4]. OGT is encoded by a single gene on the X-chromosome and its function is critical for mouse development, since OGT knock out mice are embryonically lethal [5, 6]. In general, OGT is ubiquitously expressed with high transcript levels in macrophages, pancreas, and the nervous system [7]. Although, there is only one OGT enzyme, the OGT gene encodes for several splice variants, which differ in the length of the N-terminal tetratricopeptide (TPR) repeats and are targeted to the cytosol, nucleus, and mitochondria [8, 9]. The specificity of OGT may be regulated by posttranslational modifications and by its association with different targeting subunits.
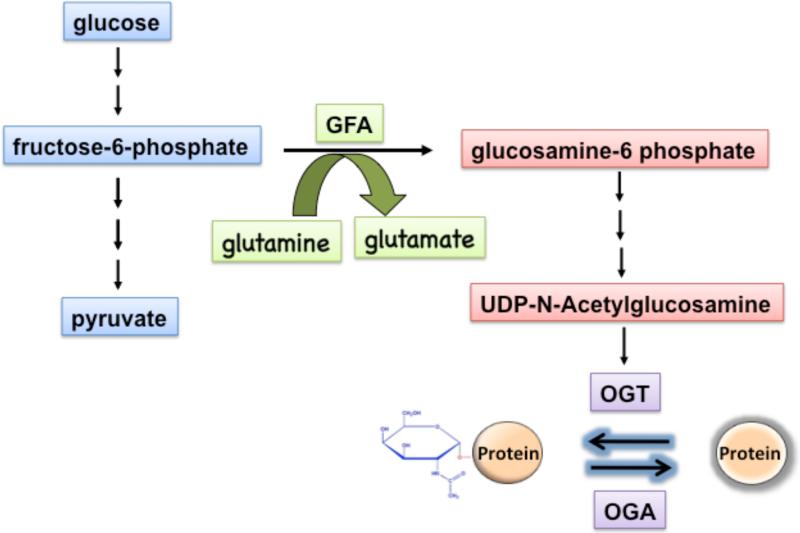
The substrate UDP-GlcNAc for OGT is synthesized by the hexosamine biosynthetic pathway (HBP), which uses the glycolytic metabolite fructose-6-phosphate and glutamine (Fig. 1). Only a small fraction of glucose (2-5%) enters the HBP as fructose 6-phosphate [10, 11]. The HBP together with O-linked GlcNAc modification of proteins has been suggested to function as a nutrient sensor for the cell [12, 13]. Consistent with this idea, exposure to high glucose leads to increased flux via the HBP and results in elevated levels of O-GlcNAc modified proteins. However, recent data suggest that O-GlcNAc modification of a number of proteins, including glycogen synthase is stimulated during nutrient deprivation via upregulation of OGT expression [14-16]. This suggests that different classes of proteins are modified during glucose excess versus glucose deprivation.
Fig. 1. O-GlcNAc modification is linked to glycolysis via the hexosamine biosynthetic pathway (HBP).
Only a small fraction of the glucose (2-5%) enters the HBP, which starts with the conversion of the glycolytic metabolite fructose-6-phosphate and glutamine to glucosamine-6-phosphate and glutamate by the rate-limiting enzyme GFA. The end product of HBP is UDP-N-acetylglucosamine, which is used by OGT as substrate to modify proteins by O-GlcNAc linkages. This modification is reversible, and proteins are deglycosylated by the O-GlcNAcase (OGA).
O-linked GlcNAc modification of nuclear and cytoplasmic proteins is dynamic and reversible. It has been suggested that O-linked GlcNAcylation and phosphorylation play a reciprocal role in regulation of protein function by competing for modification of the same serine or threonine residues. This reciprocal relationship has been demonstrated for several proteins, including RNA Pol II, estrogen receptor beta, and c-myc [17, 18]. However, there are many examples where O-GlcNAc modification has been shown to be promixal or distant from important phosphorylation sites within the same protein [19]. Nevertheless, there is an extensive crosstalk between O-GlcNAc modification and phosphorylation in regulation of protein function.
Altered O-linked GlcNAc modification has been linked to various human diseases, including cardiovascular disease [20, 21], neurodegenerative disorders [22-24], diabetes mellitus [18, 25-27], and cancer [28]. A single nucleotide polymorphism in OGlcNAcase (OGA; MGEA5) has been associated with increased susceptibility to type 2 diabetes in Mexican Americans [29]. Interestingly, the human OGT gene is localized on the chromosome X q13.1 region that has been linked to X-linked Dystonia Parkinsonism [30, 31].
Many nuclear proteins that are modified by O-linked GlcNAc include transcription factors, such as Pdx-1 [32, 33], Sp1 [34-36], c-myc [28, 37], NF-κB [38-40], NFAT [39], p53 [41, 42] STAT5A [43], FoxO-1 [44, 45], and co-activators CRTC2 [46] and PGC-1α [47]. In fact, over 25% of the O-GlcNAc modified proteins are involved in transcriptional regulation. O-GlcNAc modification of transcription factors is important in regulation of gene expression in various tissues [48]. Many transcription factors are modified by O-linked GlcNAcylation in response to physiological stimuli, cell cycle stage, and developmental stage, and this modification can modulate their function in different ways [49-51]. O-linked GlcNAc moieties on transcription factors may be recognized by various components of the transcriptional machinery, serve as a nuclear localization signal, antagonize the action of protein kinases by masking the potential serine and threonine sites for phosphorylation, modulate the DNA binding activity or the half-life, and increase the stability of transcription factors in the cell.
There are already several excellent reviews focusing on detection of O-GlcNAc modification on proteins [52-54] and on the regulation of signal transduction pathways by O-GlcNAc cycling [7, 55-58]. Thus, this review focuses on O-linked GlcNAc modification of transcriptional regulators and the role of O-linked GlcNAcylation in modulation of transcription factor function. We will discuss the function of O-linked GlcNAc modification in regulating the stability, localization, protein-protein interaction, and DNA binding ability of transcription factors. Although a large number of transcription factors have been demonstrated to be O-GlcNAc modified, this review will focus only on a selected number of transcriptional regulators, where the role of O-GlcNAc modification has been studied in detail.
2. Regulation of protein-protein interaction by O-GlcNAc modification
2.1. NF-κB
The transcription factor NF-κB (nuclear factor-kappaB) serves as a critical regulator of cytokine production, lymphocyte activation, and proliferation [59, 60]. NF-κB is present as a dimer consisting of p65 (RelA) and p50 subunits in most cell types. This dimer is localized to the cytoplasm and binds the inhibitor IκB (Fig. 2). Treatment with TNFα or other activating agents stimulate IκB kinase (IKK), which phosphorylates IκB and thereby induces its degradation. This leads to dissociation and translocation of NF-κB into the nucleus and activation of target genes [59, 61].
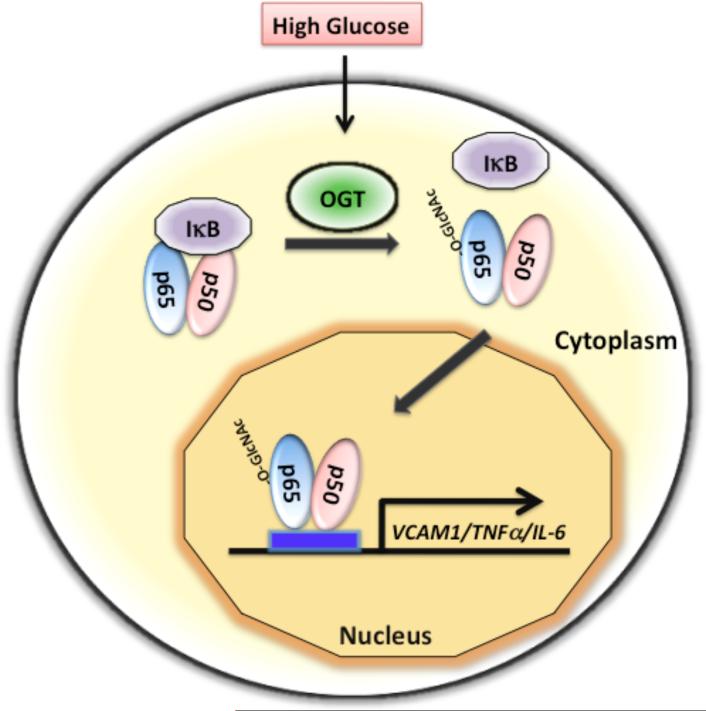
Fig. 2. High glucose mediated O-GlcNAc modification of NF-κB disrupts its interaction with the inhibitor IκB and causes nuclear translocation of NF-κB.
The transcription factor NF-κB, consisting of two subunits (p50 and p65), is normally sequestered in the cytoplasm by its interaction with the inhibitor IκB. O-linked GlcNAc modification of NF-κB p65 subunit in response to high glucose disrupts its interaction with IκB, leading to nuclear accumulation and activation of NF-κB target genes, such as VCAM-1, TNFα, and IL-6.
Activation of NF-κB requires posttranslational modifications, including phosphorylation and acetylation. O-linked GlcNAc modification of NF-κB regulates its nuclear localization by disrupting its interaction with the inhibitor IκB [39, 40]. In T lymphocytes, NF-κB p65 subunit has been shown to be O-GlcNAc modified, which causes its translocation into the nucleus [38, 39]. Furthermore, a recent report demonstrates that modification of NF-κB by O-GlcNAc decreases its binding to IκBα in vascular smooth muscle cells (VSMCs) and increases its transcriptional activity in response to hyperglycemia [40]. The modification sites within NF-κB have been identified as Thr-322 and Thr-352. O-GlcNAc modification of NF-κB at Thr-352 in response to high glucose has been shown to inhibit the interaction of NF-κB with IκB, causing the nuclear translocation of NF-κB and activation of its target genes [40].
NF-κB has also been shown to be O-GlcNAc modified in mesangial cells and accumulates in the nucleus by treatment with high glucose or glucosamine [38]. This leads to activation of NF-κB-dependent genes, such as VCAM-1, TNF-α, and IL-6 (Fig. 2). Overexpression of GFA (glutamine:fructose-6-phosphate amidotransferase) also leads to NF-κB dependent gene expression, indicating a positive role for O-GlcNAc in regulation of NF-κB function in mesangial cells [38]. It is likely that O-GlcNAc modification of NF-κB in mesangial cells disrupts the interaction of NF-κB with IκB in the cytoplasm and causes it nuclear accumulation as observed in VSMCs. Thus, the primary function of O-GlcNAc modification of NF-κB is to disrupt its interaction with IκB, which results in increased nuclear accumulation of NF-κB and stimulation of NF-κB-dependent gene expression (Fig. 2). However, it is possible that O-GlcNAc modification of NF-κB not only disrupts the interaction with IκB, but may also be directly involved in nuclear translocation of NF-κB.
2.2. Stat5a
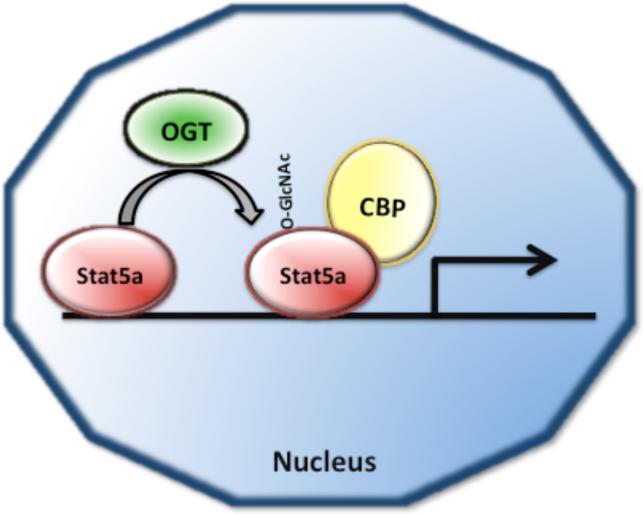
Signal transducer and activator of transcription (STAT) proteins mediate cellular responses to cytokines, hormones, and growth factors [62]. Mammalian cells contain seven isoforms of Stat proteins (Stat1, Stat2, Stat3, Stat4 Stat5a, Stat5b and Stat6), which are involved in regulation of development, cell differentiation, and inflammation [62-64]. STAT proteins are activated by tyrosine phosphorylation catalyzed by Janus Kinase (Jak) and Src kinases [62]. Tyrosine-phosphorylated Stat proteins form dimers and translocate to the nucleus to induce target gene transcription [62]. In addition to tyrosine phosphorylation at Tyr-694, Stat5a is also O-GlcNAc modified at Thr-92 in HC11 mammary epithelial cells [65]. Mutation of Thr-92 to an alanine results in the loss of Stat5a glycosylation and loss of activation of a reporter gene. The mutant Stat5a-Thr-92Ala is able to bind DNA and translocate into the nucleus, but is unable to interact with the transcriptional co-activator CBP (CREB binding protein) (Fig. 3). The loss of interaction of Stat5a with CBP results in lack of Stat5-mediated gene transcription [65]. This demonstrates that O-GlcNAc modification mediates the interaction of Stat5a with the co-activator CBP to induce Stat5-dependent gene transcription (Fig. 3). The exact mechanism(s) by which O-GlcNAc modification enhances Stat5a interaction with CBP remains to be established.
Fig. 3. O-GlcNAc modification of Stat5a enhances its association with the co-activator CREB binding protein CBP.
Posttranslational modification of Stat5a with O-GlcNAc has been proposed to enhance transactivation of Stat5a dependent gene expression by promoting the association of Stat5a with the transcriptional co-activator CBP.
2.3. CREB
CREB (cAMP response element-binding) protein is a transcription factor that binds to DNA sequences known as cAMP response elements (CREs), and thereby regulates the transcription of target genes. CREB is involved in regulation of metabolism [66], the formation of long-term memory in the brain [67], and proliferation of cancer cells [68]. CREB knockout mice die shortly after birth, suggesting a critical role for CREB in promoting cell survival [69]. CREB has been shown to be modified by O-GlcNAc in the rat brain [70]. Using mass spectrometry, two residues within the region of amino acids 256-261 of CREB were found to be O-GlcNAc modified. Further studies suggested that O-GlcNAc modification inhibits CREB binding to TAFII130 and thereby interferes with CREB-mediated transcription [70].
2.4. YY1
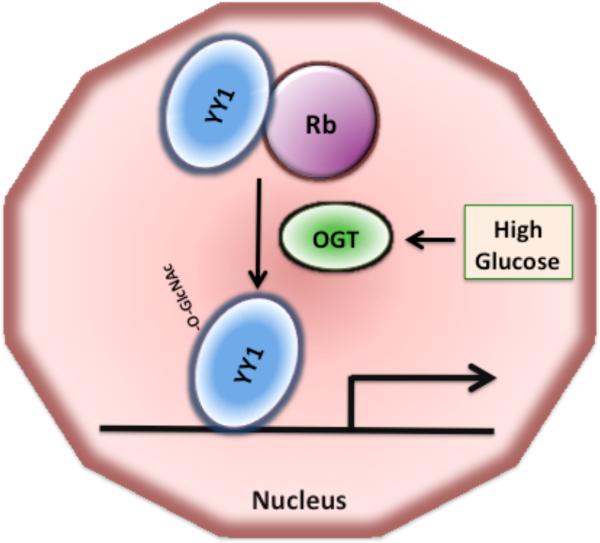
YY1 (Yin Yang 1) is a zinc finger transcription factor that regulates the transcription of many genes, involved in replication, cell growth, and differentiation [71-73]. YY1 functions as an activator as well as a repressor and has an essential role in development [73]. Nuclear YY1 has been shown to be O-GlcNAc modified at several residues in a cell-cycle independent manner [74] (Fig. 4). Exposure to glucose increases the O-GlcNAc modification of YY1. During the G1 phase YY1 is associated with the hypophosphorylated retinoblastoma protein Rb, which inhibits YY1 binding to DNA (Fig. 4). O-GlcNAc modification of YY1 disrupts its association with Rb, thereby favoring the binding of YY1 to target gene promoters [74]. O-GlcNAc modification of YY1 has no effect on its ability to bind DNA and to initiate transcription. In the presence of high glucose, the O-GlcNAc modified YY1 is mostly free of Rb, indicating that O-GlcNAc modification disrupts the YY1-Rb complex formation [74] (Fig. 4). In the absence of glucose, YY1 is not modified by O-GlcNAcylation, which enhances the YY1-Rb complex formation. These data suggest that O-linked GlcNAc modification may affect cell cycle by inhibiting the interaction of YY1 with Rb and that YY1-regulated transcription may be regulated by glucose metabolism. Nevertheless, the role of glucose in induction of O-GlcNAc modification of YY1 and how O-GlcNAc modification disrupts the YY1-Rb complex remain to be determined.
Fig. 4. The interaction of YY1 with Rb is disrupted by O-GlcNAc modification of YY1.
Association of YY1 with the retinoblastoma protein Rb inhibits YY1 binding to DNA. O-GlcNAc modification of YY1 increases upon exposure to high glucose and disrupts YY1 interaction with Rb, increasing the ability of YY1 to bind to DNA and to activate transcription.
2.5. PGC-1α
PGC-1α (peroxisome proliferator-activated receptor gamma, co-activator 1 alpha) is a transcriptional co-activator that regulates the expression of genes involved in energy metabolism by interacting with various transcription factors [75]. PGC-1α is phosphorylated by various kinases [76, 77], methylated by the protein arginine methyltransferase 1 (PRMT1) [78], and acetylated [79]. A recent study shows that PGC-1α is also O-GlcNAc modified at Ser-333, which is within a putative inhibitory domain of PGC-1α [47]. PGC-1α has been demonstrated to complex with OGT and to target OGT to the FoxO-1 transcription factor [47], which results in increased glycosylation and thereby enhancing the tansactivation capability of FoxO-1 [44]. This indicates that one of the mechanisms by which the co-activator PGC-1α activates FoxO-dependent gene expression is via targeting OGT to FoxO transcription factors. Although Ser-333 of PGC-1α was determined to be O-GlcNAc modified by mass spectrometry, no mutational analysis of this site was carried out to confirm the importance of this site in the recruitment and targeting of OGT to FoxO-1. Further studies are required to determine whether PGC-1α targets OGT to other transcription factors in order to enhance their transcriptional capability. Interestingly, PGC-1α is not only the co-activator for FoxO-1, but its expression is also under the control of FoxO-1 [80]. Therefore, treatment with glucosamine or PUGNAc also increases PGC-1α expression via FoxO-1 activation [81]. This indicates that O-linked GlcNAc modification of PGC-1α results in activation of PGC-1α expression via FoxO-1.
3. O-GlcNAc modification affects protein stability of transcription factors
3.1. p53
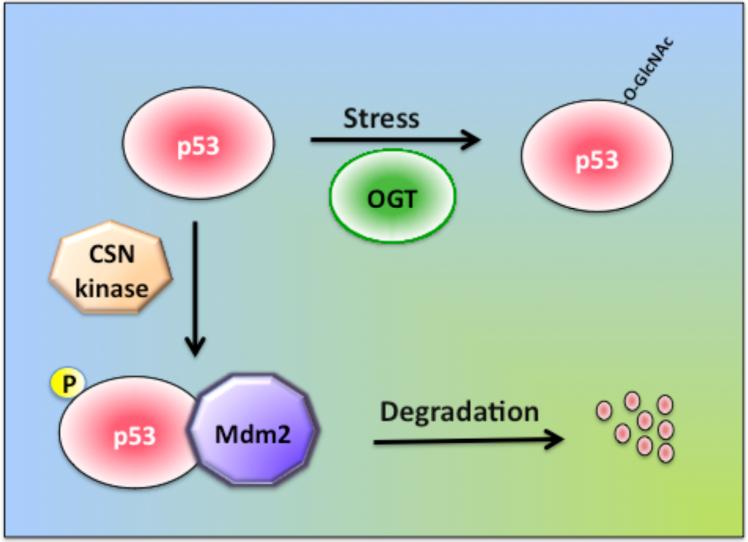
The tumor suppressor protein p53 is a regulator of cell cycle, and mutations in p53 cause cancer [82]. Normally, p53 levels are low due to continuous degradation by ubiquitin-dependent proteolysis. In unstressed cells, p53 interacts with Mdm2, which acts as an ubiquitin ligase, leading to degradation of p53 by the proteasome [83, 84] (Fig. 5). p53 becomes activated in response to various types of stress, including DNA damage, which leads to stabilization and accumulation of p53 in stressed cells. Stabilization of p53 is mediated via phosphorylation by the ATM and Rad3-related kinase at its N-terminal domain, which disrupts Mdm2 binding and prevents p53 degradation [85]. On the other hand, phosphorylation of p53 by COP9 signalosome CSN-associated kinases at Thr-155 promotes p53 degradation by the ubiquitin–proteasome pathway [86]. Phosphorylation-independent mechanisms, including acetylation [87], sumoylation [88, 89], and O-GlcNAc modification [41, 42] have been shown also to stabilize p53 (Fig. 5).
Fig. 5. The stability of the tumor suppressor protein p53 is regulated by O-GlcNAc modification.
In unstressed cells, p53 is phosphorylated by CSN (COP9 signalosome)-associated kinases, and phosphorylated p53 interacts with the ubiquitin ligase Mdm2, which promotes the proteasomal degradation of p53. In stressed cells, O-GlcNAc modification of p53 prevents its degradation by the proteasome.
The mechanism by which O-GlcNAc modification enhances p53 stability has been recently established by the identification of the O-GlcNAc residue within p53 using mass spectrometry. Activation of p53 by stress involves the O-GlcNAc modification of Ser-149, and modification of this site interferes with phosphorylation at Thr-155 by the CSN-associated kinases. Lowering phosphorylation at Thr-155 weakens the interaction of p53 with Mdm2 and decreases p53 ubiquitination/proteolysis, resulting in higher stability of the p53 protein [41] (Fig. 5). However, mutating Ser-149 to Ala does not significantly decrease the O-GlcNAc modification of p53 [41], suggesting that in addition to Ser-149, there are other residues within p53 that become O-GlcNAc modified. One caveat with this study is that streptozotocin (STZ) was used to increase O-GlcNAc modification of p53, and studies on O-GlcNAc modification in the brain suggest that STZ treatment can inhibit the proteasome by increasing O-GlcNAc levels [90]. Intraventricular administration of STZ leads to accumulation of O-GlcNAc modified proteins and of p53 in the brain [90]. Thus, it needs to be further clarified whether the stabilization of p53 by increases in O-GlcNAc modification is a direct or an indirect effect due to inhibition of the proteasome.
3.2. Estrogen receptor alpha (ER-α)
The estrogen receptor alpha (ER-α) is a ligand-inducible transcription factor and a central component of estrogen regulation [91, 92]. Ligand or estrogen binding leads to conformational changes in the ER, which enables ER to bind to the ER-response elements on target genes and to initiate transcription. Activated ER is subject to hyperphosphorylation by several kinases [93, 94]. ER contains several PEST domains and is rapidly degraded by the proteasome after ligand-induced activation [95]. A subpopulation of ER-α protein (about 10%) isolated from various sources has been shown to be O-GlcNAc modified. The major site of O-GlcNAc modification on mouse ER-α (mER-α) expressed in insect cells is Thr-575, located within the PEST domain near the carboxyl terminus [96]. However, mutation of Thr-575 does not completely abolish O-GlcNAc modification of ER-α[96]. Utilizing this mutant ER-α receptor, two additional O-GlcNAc modification sites (Ser-10 and Thr-50) on mER-α in insect cells were identified [97]. All of the identified O-GlcNAc modification sites are located near or within the PEST domains of mER-α, which have been implicated in mediating the rapid degradation of mER-α protein through the proteasome pathway [95, 98]. Thus, it is proposed that O-GlcNAcylation of the mER-α PEST regions may prevent mER-α degradation by the proteasome. However, it remains to be demonstrated that mutating the O-GlcNAc modified sites (Ser-10 and Thr-50) indeed leads to enhanced degradation of the ER-α protein.
3.3. Estrogen Receptor β (ER-β
The estrogen receptor ER-β is highly homologous to ER-α within the DNA and ligand binding domains, but differs from ER-α by having a distinct N-terminal domain called activation function 1 (AF1) region [99-101]. ER-β has also a very different tissue distribution and ligand responsiveness compared to ER-α [101, 102]. The fact that ER-β is highly expressed in many tissues, including brain, cardiovascular system, thymus, bone, kidney, and lung suggests an important role for estrogen in various tissues. Mouse ER-β is modified by O-GlcNAcylation at Ser-16, which is also phosphorylated [103, 104]. Ser-16 is within the transactivation domain, in a region with a high PEST score.
Further studies revealed that mutations in Ser-16 abolish O-linked GlcNAcylation of ER-β completely, suggesting the idea that Ser-16 is the major site for O-GlcNAc modification [104]. Moreover, mutation of Ser-16 to glutamic acid, which mimics constitutive phosphorylation, resulted in accelerated degradation of ER-β. On the other hand, mutation of Ser-16 to alanine caused a slow-down of ER-β degradation [104]. Mutations in Ser-16 also affected the ability of ER-β to activate transcription from a Luciferase reporter driven by an ER-responsive element, while the DNA binding activity and nuclear localization of ER-β was unaffected [104]. Recent data suggest that O-GlcNAc modification and phosphorylation at Ser-16 induce different conformational changes, which may explain the different effects that post-translational modifications have on ER-β function [105, 106]. In summary, O-GlcNAc modification of Ser-16 increases ER-β stability, but reduces its transactivation capability, while phosphorylation of the same residue has been shown to increase ER-β degradation and transcriptional activity [105].
4. Nucleo-cytoplasmic shuttling of transcription factors mediated by O-GlcNAc modification
4.1. NeuroD1
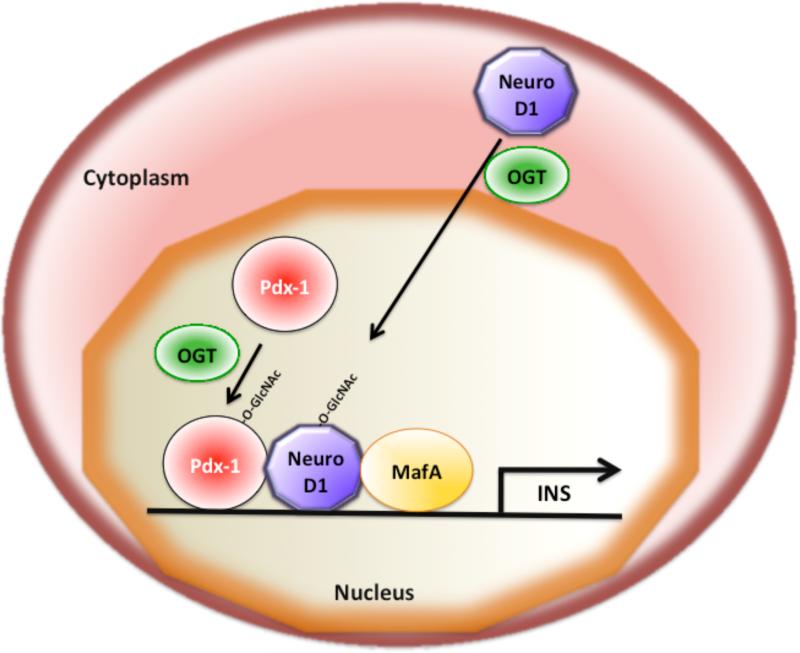
NeuroD1 (neurogenic differentation 1) or Beta-2 is a transcription factor that is expressed in neurons and in pancreatic beta cells and mediates neuronal development and insulin gene transcription, respectively [107, 108]. Increasing levels of glucose stimulates insulin gene transcription by the synergistic action of the transcription factors NeuroD1, MafA, and Pdx-1 [109, 110] (Fig. 6). Changes in glucose levels have been shown to regulate the nucleo-cytoplasmic shuttling of NeuroD1 [111, 112]. While NeuroD1 is mainly localized in the cytoplasm under low or normal glucose conditions, exposure to high glucose causes NeuroD1 to translocate into the nucleus and thereby to activate insulin gene transcription in pancreatic beta cells (Fig 6). The nuclear translocation of NeuroD1 on high glucose is mediated by O-linked GlcNAc modification of NeuroD1 itself [111]. Inhibition of O-GlcNAc modification by treatment with OGT siRNA abolishes the nuclear translocation of NeuroD1 in pancreatic beta cells. Increased O-GlcNAc modification by treatment with PUGNAc (an inhibitor of OGA, Fig. 1) mediates the nuclear translocation of NeuroD1 in the absence of high glucose [111]. Taken together, these findings indicate that increases in glucose levels mediate the O-GlcNAc modification of NeuroD1, which then translocates from the cytoplasm into the nucleus and participates in insulin gene expression (Fig. 6). Further work is required to map the O-GlcNAc modified residues within NeuroD1 and to confirm their role in regulation of subcellular localization of NeuroD1.
Fig. 6. The function of the insulin gene transcription factors Pdx-1, NeuroD1, and MafA is regulated by O-GlcNAc modification.
Activation of insulin gene transcription in pancreatic beta cells requires the synergistic interaction of the transcription factors Pdx-1, NeuroD1, and MafA. O-GlcNAc modification of Pdx-1 enhances the DNA binding capability of Pdx-1. NeuroD1 is normally localized to the cytoplasm, but translocates into the nucleus in response to high glucose mediated by O-GlcNAc modification. MafA expression is induced by high glucose and requires O-GlcNAc modification of an unknown transcriptional regulator.
4.2. TORC2/ CRTC2
The transcriptional co-activator TORC2 or CRTC2 (transducer of regulated cyclic adenosine monophosphate response element-binding protein) is involved in regulation of gluconeogenesis in liver [66]. In fed animals, CRTC2 is sequestered in the cytoplasm, while during fasting CRTC2, translocates into the nucleus, where it interacts with the transcription factor CREB to mediate the expression of gluconeogenic genes. The nuclear translocation of CRTC2 requires the dephosphorylation of Ser-171 [113]. CRTC2 levels are also regulated by ubiquitin-dependent degradation, which occurs in the presence of insulin [113].
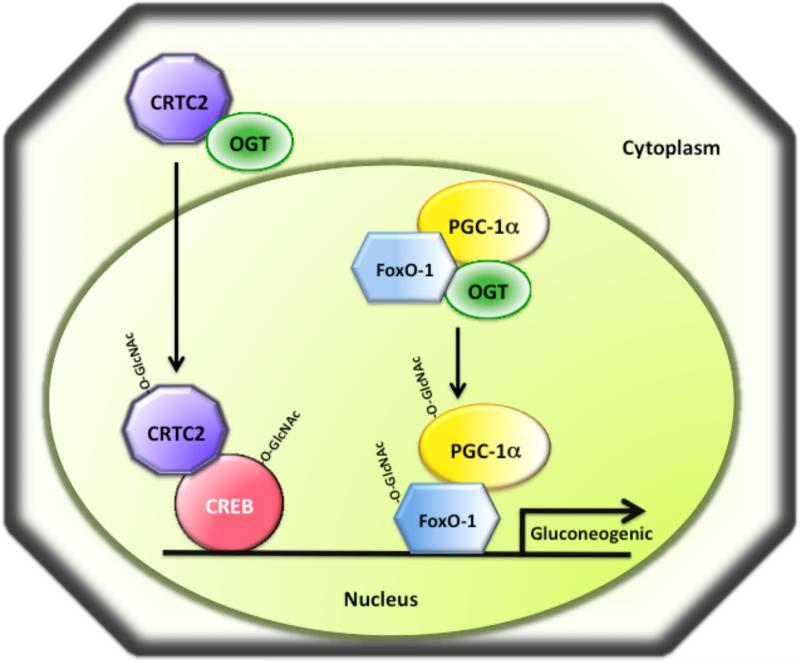
CRTC2 has been recently shown to be O-GlcNAc modified in hepatocytes and this modification regulates the localization of CRTC2 [46]. Upon exposure to chronic hyperglycemia, CRTC2 becomes O-GlcNAc modified in hepatocytes and translocates into the nucleus, where it associates with CREB to upregulate the expression of glucose-6-phosphatase and other gluconeogenic genes [46] (Fig. 7). Mapping of the O-GlcNAc modified sites by mass spectrometry identified six modified residues on CRTC2, of which Ser-70 and Ser-171 have been shown to be the most important ones. Interestingly, both of these sites are phosphorylated by AMP-activated kinases, which causes CRTC2 association with 14-3-3 proteins leading to its sequestration in the cytoplasm [114]. Thus, O-GlcNAcylation of CRTC2 at Ser-70 and Ser-171 by OGT in liver blocks CRTC2 phosphorylation at these two sites, causing the shuttling of CRTC2 into the nucleus [114, 115]. The same study found that the levels of O-GlcNAc modified CRTC2 are increased in obese diabetic db/db mice, which is likely to be responsible for the paradoxically increased gluconeogenic gene expression in these diabetic mice [46].
Fig. 7. O-GlcNAc modification regulates gluconeogenic gene expression in liver during diabetic conditions.
Chronic hyperglycemia or diabetes causes induction of gluconeogenic gene expression by O-GlcNAc modification of the transcription factors CREB and FoxO-1 and their co-activators CRTC2 and PGC-1α, respectively. O-GlcNAc modification of CRTC2 under diabetic conditions causes CRTC2 translocation into the nucleus where it interacts with CREB to activate gluconeogenic gene expression. The co-activator PGC-1α has been demonstrated to be O-GlcNAc modified and to recruit OGT to FoxO-1, promoting O-GlcNAc modification of FoxO-1 and activation of gluconeogenic genes.
4.3. NFATc1
NFATc1 (nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1) is predominantly expressed in T-cells and induces the transcription of various cytokines during immune response [116]. NFATc1 interacts directly with OGT and becomes O-GlcNAc modified in human T-cells [39]. Shortly after lymphocyte stimulation, O-GlcNAc modification of NFATc1 increases and declines within 15 minutes, indicating that O-GlcNAc modification of NFATc1 is transient [38, 39]. Nuclear fractionation experiments in T cells indicate that the levels of O-GlcNAc modified NFATc1 increase in the nucleus following T-cell activation. Consistent with a role for OGT in regulating NFATc1 nuclear translocation, silencing of OGT expression leads to impaired T-cell activation [39]. Previous data have demonstrated that dephosphorylation of NFATc1 by calcineurin upon activation of T-cells mediates the translocation of NFATc1 into the nucleus [117]. Although it is not known how O-GlcNAc modification promotes NFATc1 nuclear translocation, it is proposed that OGlcNAc modification may prevent the phosphorylation of NFATc1 or alternatively, change the conformation of NFATc1 and thereby unmask the nuclear localization signal.
4.4. Elf-1
The Elf-1 (E74-like factor 1) transcription factor belongs to the Ets (E twenty-six specific) family of proteins and regulates gene expression in hematopoietic cells [118]. The expected molecular mass for Elf-1 is 68 kDa, but it displays a molecular mass of 80kD and 98kD. The discrepancy in Elf-1 molecular mass is due to posttranslational modification of Elf-1 by phosphorylation mediated by protein kinase C and by O-GlcNAc modification [119]. Phosphorylation by PKC mediates the conversion of the 80kD to the 98kD form, while both the 80kD and 98kD forms are O-GlcNAc modified. The 80-kDa form of Elf-1 resides mainly in the cytoplasm. Modification by phosphorylation and O-GlcNAc converts the 80kD to the 98kDa form, which translocates into the nucleus of T cells. Once in the nucleus the 98kDa form binds to the TCR ζ-chain promoter and activates its transcription in Jurkat and T cells [120]. Interestingly it has been found that the 98kDa form of Elf-1 is more sensitive to proteasomal degradation than 80kDa form [119]. Collectively, O-GlcNAc modification and phosphorylation regulate the sub-cellular localization, DNA binding, and degradation of Elf-1.
5. O-GlcNAc modification of transcriptional activity
5.1. c-Myc
c-Myc (v-myc myelocytomatosis viral oncogene homolog), a helix-loop-helix leucine zipper transcription factor, contributes to the development of various malignancies [121-123]. In a normal resting cell, c-Myc levels are very low. However upon mitogenic stimulation, c-myc expression levels are highly increased, and c-myc heterodimerizes with Max to regulate transcription of proliferation and cell differentiation genes [124]. c-Myc is phosphorylated by casein kinase II [125] and MAP kinase [126] and can also be O-GlcNAc modified [37]. The O-GlcNAc modification of c-Myc has been identified both in mammalian (rabbit reticulocyte lysate and Chinese hamster ovary (CHO) cell line) and in insect cell systems. O-GlcNAc modified sites within c-myc were originally found to be located close to the transcriptional activation domain [37]. A later study identified Thr-58, an in vivo phosphorylation site in the transactivation domain, as the major site of O-GlcNAc modification [127]. This suggests a mutually exclusive modification of c-myc by either phosphorylation or O-GlcNAc modification. The transactivation domain of c-myc associates with the tumor suppressor retinoblastoma protein Rb and the Rb-related protein p107 in vitro [128]. Thus, the presence of O-GlcNAc modification on c-myc may result in altered interaction with Rb and Rb-related protein p107, thereby interfering with transactivation by c-myc. Interestingly, Thr-58 is located within a mutational hot spot in lymphomas, suggesting that this region is associated with increased tumorigenicity.
5.2. FoxO1
FoxO-1 (forkhead box other 1, FKHR) transcription factor is important for regulation of apoptosis, cell cycle, metabolism, and oxidative stress [129, 130]. FoxO-1 is modified by phosphorylation, acetylation, and ubiquitination. Phosphorylation of FoxO-1 at Thr-24, Ser-256 and, Ser-319 by Akt, which is mediated by insulin, leads to nuclear exclusion [131] and subsequent ubiquitination and degradation of FoxO-1 [132]. FoxO-1 has also been recently shown to be O-GlcNAc modified in response to increased glucose levels, and this modification is implicated in regulation of FoxO-1 transcriptional activity [44, 45] (Fig. 7). Phosphorylation and O-GlcNAc modification of FoxO-1 occurs at different sites and are not functionally reciprocal. This suggests that O-GlcNAc modification is not involved in regulation of nuclear localization of FoxO-1 [45]. A detailed analysis of the O-GlcNAc modified residues within FoxO-1 resulted in identification of four residues, and only the O-GlcNAc modification of Thr-317 proved to be important for transcriptional activation by FoxO-1 [44] (Fig. 7). However, in this study Thr-317 was mutated to alanine, thus it cannot be completely excluded that phosphorylation at Thr-317 may also be important for FoxO-1 activation. Interestingly, a new study suggests that the co-activator PGC-1α binds to OGT and targets it to the FoxO transcription factors, leading to increased O-GlcNAc modification and enhanced transcriptional activity [47] (Fig. 7).
6. Regulation of DNA binding activity by O-GlcNAc modification
6.1. Pdx-1
Pdx-1 (pancreatic and duodenal homeobox 1), a homeodomain transcription factor, is mainly expressed in pancreatic β cells and regulates insulin gene transcription in response to glucose [109]. Elevated concentrations of glucose lead to O-GlcNAc modification of Pdx-1 in pancreatic beta cells [32, 33]. O-GlcNAc modification regulates Pdx-1 binding to the insulin promoter and thereby influences insulin secretion in the mouse insulinoma cell line MIN6 [32]. Therefore, O-GlcNAc modification of Pdx-1 appears to be important for Pdx-1 DNA binding to the insulin promoter and for activation of insulin gene expression (Fig. 6). However, the exact mechanism(s) by which O-GlcNAc modification enhances Pdx-1 DNA binding activity and the Pdx-1 residues that are O-GlcNAc modified remain to be identified. Increased O-GlcNAc modification of Pdx-1 has also been observed in diabetic Goto-Kakizaki (GK) rats and was associated with decreased insulin secretion from pancreatic beta cells [33].
6.2. C/EBPβ
C/EBPβ (CCAAT enhancer-binding protein) belongs to the family of basic region leucine zipper (bZIP) transcription factors and is important for adipocyte differentiation, and its expression is increased during adipogenesis [133-136]. C/EBPβ has also been shown to be expressed in various other tissues, including hepatocytes [137] and epithelial cells [138]. C/EBPβ is activated by phosphorylation of Thr-179, Ser-184, and Thr-188 by MAPK and GSK3β [139]. Phosphorylation at Thr-188 by MAPK or CDK2 is a perquisite for phosphorylation of Thr-179 and Ser-184 by GSK3β, which causes a conformational change important for facilitating C/EBPβ DNA binding [139-141].
C/EBPβ is also modified with O-GlcNAc at Ser-180 and Ser-181, which inhibits the phosphorylation of the neighboring Thr-179, Ser-184, and Thr-188 [142]. Since phosphorylation is essential for C/EBPβ binding to target gene promoters, O-GlcNAc modification of Ser-180 and Ser-181 results in a delay of adipogenesis [142]. Ser-180 and Ser-181 to alanine mutations increases the transcriptional activity of C/EBPβ via enhanced phosphorylation at Thr-179, Ser-184, and Thr-188 [142]. In summary, O-GlcNAc modification of the residues Ser-180 and Ser-181 within C/EBPβ interferes with the expression of C/EBPβ target genes by preventing its phosphorylation and thereby inhibiting its DNA binding activity. Thus, O-GlcNAc modification and phosphorylation regulate the DNA binding activity of C/EBPβ in a reciprocal and competitive manner.
7. Induction of transcription factor expression by O-linked GlcNAc modification
7.1. MafA
The MafA (v-maf musculoaponeurotic fibrosarcoma oncogene homolog A) transcription factor is important for glucose regulation of insulin gene expression, and MafA knock-out mice display reduced insulin gene transcription and age-dependent diabetes [109]. MafA expression is induced by high glucose and requires increased flux through the hexosamine biosynthetic pathway (HBP) and an O-linked GlcNAc modification event [143]. Knockdown of OGT using siRNA oligonucleotides abolishes the induction of MafA expression by high glucose in pancreatic beta cells. Treatment of pancreatic beta cells with PUGNAc, an inhibitor of O-GlcNAcase, induces MafA expression even in the absence of high glucose. These data suggest that high glucose induction of MafA expression may be mediated by O-GlcNAc modification of an unknown transcriptional regulator(s) that is required for activation of MafA expression [143].
7.2. Id2
Id2 (inhibitor of differentiation 2) belongs to the family of helix-loop-helix (HLH) transcriptional regulators and modulates gene expression indirectly by antagonizing basic helix-loop-helix transcription factors (E proteins) [144-146]. It is involved in regulation of differentiation and apoptosis. Id2 levels are regulated by glucose in J774.2 macrophages via the HBP pathway [147, 148] (Fig. 1). Consistent with the idea that induction of Id2 levels by high glucose requires HBP, treatment with glucosamine, which increases the flux via HBP, mimics high glucose induction of Id2. Furthermore, inhibition of GFA, the rate-limiting enzyme of HBP, blocks the increase in Id2 in response to glucose, while overexpression of GFA by adenoviral gene transfer increases Id2 levels [147]. Thus, high glucose increases the levels of the transcriptional repressor Id2, via increased flux through HBP pathway, and this is likely to be dependent on O-GlcNAc modification .
7.3. USF
High glucose concentrations have been shown to cause an accumulation of the upstream stimulatory factors (USF) in the nucleus of mesangial cells, leading to upregulation of TGF–β1 (transforming growth factor-β1) expression via enhanced binding of USF proteins to the TGF-β1 promoter [149, 150]. TGF-β1 induces the production of extracellular matrix proteins, leading to the thickening of glomerular basement membranes [151]. Upregulation of TGF-β1 expression by hyperglycemia in renal cells has been linked to diabetic nephropathy in humans [152].
Increasing the flux via the HBP by addition of glutamine or by overexpression of GFA results in increased expression of USF-2 [149]. Treatment of mesangial cells with streptozotocin (STZ), an analog of GlcNAc, which inhibits the O-GlcNAcase and thereby increases the level of O-GlcNAc modified proteins, results also in elevated USF-2 mRNA levels. The USF proteins themselves are not O-GlcNAc modified indicating that increases in O-GlcNAc modification stimulates the expression of USF proteins. Thus, increased O-GlcNAc modification of proteins under conditions of chronic hyperglycemia may contribute to diabetic nephropathy by causing increased expression of USF proteins, which then stimulate TGF-β1 expression leading to expansion of glomerular mesangium. The exact mechanisms by which O-GlcNAc modification increases USF gene expression remain to be elucidated.
8. Modulation of transcription factor function by multiple mechanisms
8.1. Sp1
The Sp1 (specificity protein 1) transcription factor is an ubiquitously expressed C2H2-type zinc finger protein that regulates the transcription of many genes [153]. Sp1 can function as an activator as well as a repressor of gene expression, and its activity is regulated by posttranslational modifications, such as phosphorylation, acetylation, and O-GlcNAc modification [35, 36, 49]. These modifications regulate the intracellular localization as well as Sp1 interaction with other transcriptional regulators.
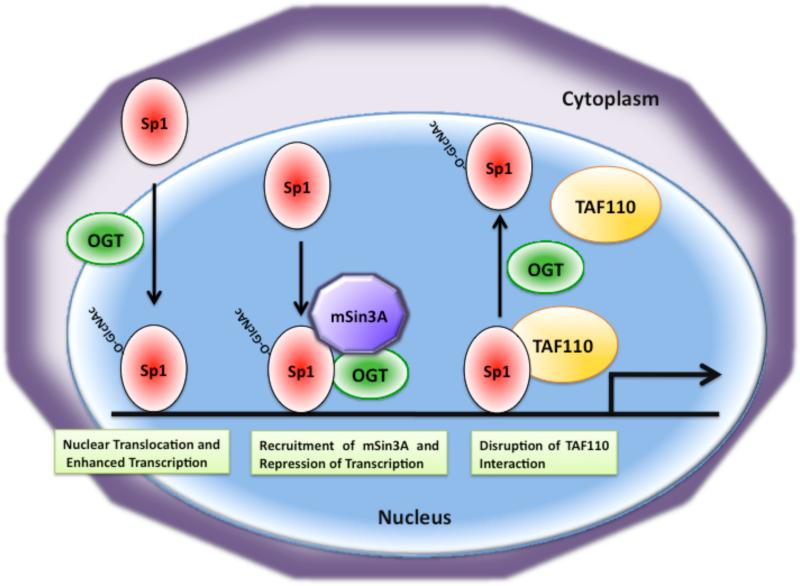
Studies on O-GlcNAc modification of Sp1 in H411E liver cells indicate that insulin induces sequential O-GlcyNAcylation and phosphorylation of Sp1, which causes its nuclear accumulation and thereby increases Sp1-dependent gene expression [154] (Fig. 8). O-GlcNAc modification of Sp1 in rat lymphoma cells has also been shown to target Sp1 to the nucleus [155]. However, in a different study, O-GlcNAc modification of Sp1 has been shown to have a negative role by interfering with Sp1 transcriptional capability in the pancreatic HIT-T15 beta-cell line [156]. Furthermore, it has been shown that OGT recruits the transcriptional co-repressor mSin3A to inhibit activation of transcription by Sp1 [157] (Fig. 8). Moreover, modification of Sp1 by O-GlcNAc has been implicated in regulation of Sp1 stability. It has been demonstrated that treatment of cells with glucose or glucosamine results in hyperglycosylation of Sp1, which in turn blocks Sp1 degradation suggesting the idea that O-GlcNAc modification increases Sp1 protein stability [34].
Fig. 8. Modulation of Sp1 function by O-GlcNAc modification involves multiple mechanisms.
Modification of Sp1 with O-GlcNAc has been shown to cause its nuclear localization thereby leading to enhanced transcription. O-GlcNAc modification can also lead to transcriptional repression by Sp1 by recruitment of the co-repressor mSin3A via OGT. There is also evidence that O-GlcNAc modification of Sp1 disrupts its interaction with the TATA binding protein-associated factor TAF110.
Using the glutamine-rich transactivation domain of Sp1, it has been demonstrated that the role of O-GlcNAc modification is to inhibit the interaction of Sp1 with the TATA-binding-protein-associated factor TAF110 [158] (Fig. 8). Further evidence that O-GlcNAc modification of Sp1 inhibits its interaction with various transcription factors comes from recent studies, where Sp1 interaction with Elf-1, Oct1, and NF-Y is blocked by O-GlcNAc modification [159-161].
In summary, O-GlcNAcylation of Sp1 has been shown to regulate Sp1 nuclear localization, transactivation capability, protein stability, and its interaction with other transcriptional regulators. However, it is not clear if regulation of Sp1 function by multiple mechanisms requires O-GlcNAc modification of distinct sites within Sp1. It is possible that O-GlcNAc modification regulates Sp1 function differently in a tissue-specific manner.
9. Summary and future directions
Although O-GlcNAc modification of proteins has been discovered over two decades ago, our understanding of the exact role of this modification in regulating cell function still remains incomplete. The number of proteins that are modified with O-GlcNAc has been steadily increasing, but it is still puzzling how OGT or OGA can modify so many different proteins in a regulated manner. Some of this may be explained by the fact that OGT has been shown to have splice variants that are localized to the nucleus, cytoplasm, and mitochondria, however detailed studies on the function of the OGT splice variants are lacking. It is also not known if the expression, stability, or activity of OGT and OGA is regulated in a tissue-specific manner by different pathways. Recent findings suggest that OGT may interact with different targeting subunits to achieve specificity.
O-GlcNAc modification of many transcriptional regulators follows a common theme and is regulated in a glucose-dependent manner. This is not surprising given the fact that the levels of the substrate UDP-GlcNAc required for O-GlcNAc modification depends on the availability of glucose (Fig. 1). Production of UDP-GlcNAc by the hexosamine biosynthetic pathway (HBP) requires the glycolytic intermediate fructose-6-phosphate, which provides a link between glycolysis and HBP (Fig. 1). Thus, HBP together with O-GlcNAc modification serves as a glucose/nutrient sensor that links metabolism to activation of many signaling pathways in the cell.
Interestingly, many transcriptional regulators that participate in the regulation of the same target genes are modified by O-GlcNAc. Glucose induction of insulin gene expression in pancreatic beta cells requires the synergistic interaction of Pdx-1, MafA, and NeuroD1, and all three of these transcription factors are regulated by O-GlcNAc modification (Fig. 6). While O-GlcNAc modification of Pdx-1 regulates its ability to bind to the insulin promoter, modification of NeuroD1 causes its translocation into the nucleus (Fig. 6). Interestingly, expression of MafA by glucose requires the O-GlcNAc modification of an unknown transcriptional regulator. Another example of this type of regulation is the expression of gluconeogenic genes in liver by CREB and FoxO-1 (Fig. 7). Both of these transcription factors, as well as their transcriptional co-activators CRTC2 and PGC-1α, are also regulated by O-GlcNAc modification (Fig. 7). This type of regulation by O-GlcNAc modification may be essential to ensure the precise regulation of metabolic gene expression in response to changes in nutrient status.
Progress in understanding the detailed function of O-GlcNAc modification on proteins has been hindered by the fact that proteins can contain multiple O-GlcNAc modification sites with different functions, as illustrated for the transcription factor Sp1 (Fig. 8). There is also evidence that O-GlcNAc modification can modulate the function of transcription factors by multiple mechanisms in a tissue-specific manner. Like phosphorylation, O-GlcNAc modification occurs at serine and threonine residues, therefore mutating the O-GlcNAc modified sites may also abolish phosphorylation of these residues, making it difficult to reach conclusions on the function of O-GlcNAc modification on various proteins. Most of the O-GlcNAc modified transcription factors have been shown to undergo other posttranslational modifications, including phosphorylation, acetylation, and sumoylation, suggesting the idea that these modifications may exert a combinatorial effect on transcription factor function. Thus, it is difficult to assess the role O-GlcNAc modification without considering the combinatorial effect of other posttranslational modifications on the same transcription factor. It is also important to consider that in many cases O-GlcNAc modification of multiple residues may be required to regulate transcription factor function. Despite these hurdles, our understanding of O-GlcNAc modification on modulation of transcription factor function is steadily increasing with the development of new technologies to detect and analyze O-GlcNAc modifications on proteins.
Acknowledgements
We apologize to all authors, whose original publications could not be cited or discussed in this review due to space limitations. We thank the reviewers for their insightful comments. Research in the corresponding author's laboratory was supported by grants R01DK067581 from the NIH/NIDDK, P20RR020171 from NIH/NCRR, and 1-05-CD-15 from the American Diabetes Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem. 259(1984):3308–3317. [PubMed] [Google Scholar]
- 2.Davis LI, Blobel G. Identification and characterization of a nuclear pore complex protein. Cell. 45(1986):699–709. doi: 10.1016/0092-8674(86)90784-1. [DOI] [PubMed] [Google Scholar]
- 3.Holt GD, Hart GW. The subcellular distribution of terminal N-acetylglucosamine moieties. Localization of a novel protein-saccharide linkage, O-linked GlcNAc. J Biol Chem. 261(1986):8049–8057. [PubMed] [Google Scholar]
- 4.Holt GD, Snow CM, Senior A, Haltiwanger RS, Gerace L, Hart GW. Nuclear pore complex glycoproteins contain cytoplasmically disposed O-linked N-acetylglucosamine. J Cell Biol. 104(1987):1157–1164. doi: 10.1083/jcb.104.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shafi R, Iyer SP, Ellies LG, O'Donnell N, Marek KW, Chui D, Hart GW, Marth JD. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci U S A. 97(2000):5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Donnell N, Zachara NE, Hart GW, Marth JD. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol Cell Biol. 2004;24:1680–1690. doi: 10.1128/MCB.24.4.1680-1690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanover JA, Krause MW, Love DC. The hexosamine signaling pathway: OGlcNAc cycling in feast or famine. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbagen.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Love DC, Kochan J, Cathey RL, Shin SH, Hanover JA. Mitochondrial and nucleocytoplasmic targeting of O-linked GlcNAc transferase. J Cell Sci. 2003;116:647–654. doi: 10.1242/jcs.00246. [DOI] [PubMed] [Google Scholar]
- 9.Hanover JA, Yu S, Lubas WB, Shin SH, Ragano-Caracciola M, Kochran J, Love DC. Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene. Arch Biochem Biophys. 2003;409:287–297. doi: 10.1016/s0003-9861(02)00578-7. [DOI] [PubMed] [Google Scholar]
- 10.Traxinger RR, Marshall S. Coordinated regulation of glutamine:fructose-6-phosphate amidotransferase activity by insulin, glucose, and glutamine. Role of hexosamine biosynthesis in enzyme regulation. J Biol Chem. 1991;266:10148–10154. [PubMed] [Google Scholar]
- 11.Marshall S, Bacote V, Traxinger RR. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J Biol Chem. 1991;266:4706–4712. [PubMed] [Google Scholar]
- 12.Zachara NE, Hart GW. O-GlcNAc a sensor of cellular state: the role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim Biophys Acta. 2004;1673:13–28. doi: 10.1016/j.bbagen.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 13.Bouche C, Serdy S, Kahn CR, Goldfine AB. The cellular fate of glucose and its relevance in type 2 diabetes. Endocr Rev. 2004;25:807–830. doi: 10.1210/er.2003-0026. [DOI] [PubMed] [Google Scholar]
- 14.Taylor RP, Geisler TS, Chambers JH, McClain DA. Up-regulation of O-GlcNAc transferase with glucose deprivation in HepG2 cells is mediated by decreased hexosamine pathway flux. J Biol Chem. 2009;284:3425–3432. doi: 10.1074/jbc.M803198200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor RP, Parker GJ, Hazel MW, Soesanto Y, Fuller W, Yazzie MJ, McClain DA. Glucose deprivation stimulates O-GlcNAc modification of proteins through up-regulation of O-linked N-acetylglucosaminyltransferase. J Biol Chem. 2008;283:6050–6057. doi: 10.1074/jbc.M707328200. [DOI] [PubMed] [Google Scholar]
- 16.Cheung WD, Hart GW. AMP-activated protein kinase and p38 MAPK activate O-GlcNAcylation of neuronal proteins during glucose deprivation. J Biol Chem. 2008;283:13009–13020. doi: 10.1074/jbc.M801222200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slawson C, Hart GW. Dynamic interplay between O-GlcNAc and O-phosphate: the sweet side of protein regulation. Curr Opin Struct Biol. 2003;13:631–636. doi: 10.1016/j.sbi.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 18.Copeland RJ, Bullen JW, Hart GW. Cross-talk between GlcNAcylation and phosphorylation: roles in insulin resistance and glucose toxicity. Am J Physiol Endocrinol Metab. 2008;295:E17–28. doi: 10.1152/ajpendo.90281.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butkinaree C, Park K, Hart GW. O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim Biophys Acta. 1800:96–106. doi: 10.1016/j.bbagen.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laczy B, Hill BG, Wang K, Paterson AJ, White CR, Xing D, Chen YF, Darley-Usmar V, Oparil S, Chatham JC. Protein O-GlcNAcylation: a new signaling paradigm for the cardiovascular system. Am J Physiol Heart Circ Physiol. 2009;296:H13–28. doi: 10.1152/ajpheart.01056.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones SP, Zachara NE, Ngoh GA, Hill BG, Teshima Y, Bhatnagar A, Hart GW, Marban E. Cardioprotection by N-acetylglucosamine linkage to cellular proteins. Circulation. 2008;117:1172–1182. doi: 10.1161/CIRCULATIONAHA.107.730515. [DOI] [PubMed] [Google Scholar]
- 22.Lefebvre T, Guinez C, Dehennaut V, Beseme-Dekeyser O, Morelle W, Michalski JC. Does O-GlcNAc play a role in neurodegenerative diseases? Expert Rev Proteomics. 2005;2:265–275. doi: 10.1586/14789450.2.2.265. [DOI] [PubMed] [Google Scholar]
- 23.Lazarus BD, Love DC, Hanover JA. O-GlcNAc cycling: implications for neurodegenerative disorders. Int J Biochem Cell Biol. 2009;41:2134–2146. doi: 10.1016/j.biocel.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuzwa SA, Vocadlo DJ. O-GlcNAc modification and the tauopathies: insights from chemical biology. Curr Alzheimer Res. 2009;6:451–454. doi: 10.2174/156720509789207967. [DOI] [PubMed] [Google Scholar]
- 25.Akimoto Y, Hart GW, Hirano H, Kawakami H. O-GlcNAc modification of nucleocytoplasmic proteins and diabetes. Med Mol Morphol. 2005;38:84–91. doi: 10.1007/s00795-004-0264-1. [DOI] [PubMed] [Google Scholar]
- 26.Buse MG. Hexosamines, insulin resistance, and the complications of diabetes: current status. Am J Physiol Endocrinol Metab. 2006;290:E1–E8. doi: 10.1152/ajpendo.00329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dias WB, Hart GW. O-GlcNAc modification in diabetes and Alzheimer's disease. Mol Biosyst. 2007;3:766–772. doi: 10.1039/b704905f. [DOI] [PubMed] [Google Scholar]
- 28.Chou TY, Hart GW. O-linked N-acetylglucosamine and cancer: messages from the glycosylation of c-Myc. Adv Exp Med Biol. 2001;491:413–418. doi: 10.1007/978-1-4615-1267-7_26. [DOI] [PubMed] [Google Scholar]
- 29.Lehman DM, Fu DJ, Freeman AB, Hunt KJ, Leach RJ, Johnson-Pais T, Hamlington J, Dyer TD, Arya R, Abboud H, Goring HH, Duggirala R, Blangero J, Konrad RJ, Stern MP. A single nucleotide polymorphism in MGEA5 encoding OGlcNAc-selective N-acetyl-beta-D glucosaminidase is associated with type 2 diabetes in Mexican Americans. Diabetes. 2005;54:1214–1221. doi: 10.2337/diabetes.54.4.1214. [DOI] [PubMed] [Google Scholar]
- 30.Haberhausen G, Schmitt I, Kohler A, Peters U, Rider S, Chelly J, Terwilliger JD, Monaco AP, Muller U. Assignment of the dystonia-parkinsonism syndrome locus, DYT3, to a small region within a 1.8-Mb YAC contig of Xq13.1. Am J Hum Genet. 1995;57:644–650. [PMC free article] [PubMed] [Google Scholar]
- 31.Hanover JA, Krause MW, Love DC. The hexosamine signaling pathway: OGlcNAc cycling in feast or famine. Biochim Biophys Acta. 1800:80–95. doi: 10.1016/j.bbagen.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao Y, Miyazaki J, Hart GW. The transcription factor PDX-1 is post-translationally modified by O-linked N-acetylglucosamine and this modification is correlated with its DNA binding activity and insulin secretion in min6 beta-cells. Arch Biochem Biophys. 2003;415:155–163. doi: 10.1016/s0003-9861(03)00234-0. [DOI] [PubMed] [Google Scholar]
- 33.Akimoto Y, Hart GW, Wells L, Vosseller K, Yamamoto K, Munetomo E, Ohara-Imaizumi M, Nishiwaki C, Nagamatsu S, Hirano H, Kawakami H. Elevation of the post-translational modification of proteins by O-linked N-acetylglucosamine leads to deterioration of the glucose-stimulated insulin secretion in the pancreas of diabetic Goto-Kakizaki rats. Glycobiology. 2007;17:127–140. doi: 10.1093/glycob/cwl067. [DOI] [PubMed] [Google Scholar]
- 34.Han I, Kudlow JE. Reduced O glycosylation of Sp1 is associated with increased proteasome susceptibility. Mol Cell Biol. 1997;17:2550–2558. doi: 10.1128/mcb.17.5.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Majumdar G, Harmon A, Candelaria R, Martinez-Hernandez A, Raghow R, Solomon SS. O-glycosylation of Sp1 and transcriptional regulation of the calmodulin gene by insulin and glucagon. Am J Physiol Endocrinol Metab. 2003;285:E584–591. doi: 10.1152/ajpendo.00140.2003. [DOI] [PubMed] [Google Scholar]
- 36.Goldberg HJ, Whiteside CI, Hart GW, Fantus IG. Posttranslational, reversible O-glycosylation is stimulated by high glucose and mediates plasminogen activator inhibitor-1 gene expression and Sp1 transcriptional activity in glomerular mesangial cells. Endocrinology. 2006;147:222–231. doi: 10.1210/en.2005-0523. [DOI] [PubMed] [Google Scholar]
- 37.Chou TY, Dang CV, Hart GW. Glycosylation of the c-Myc transactivation domain. Proc Natl Acad Sci U S A. 1995;92:4417–4421. doi: 10.1073/pnas.92.10.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.James LR, Tang D, Ingram A, Ly H, Thai K, Cai L, Scholey JW. Flux through the hexosamine pathway is a determinant of nuclear factor kappaB- dependent promoter activation. Diabetes. 2002;51:1146–1156. doi: 10.2337/diabetes.51.4.1146. [DOI] [PubMed] [Google Scholar]
- 39.Golks A, Tran TT, Goetschy JF, Guerini D. Requirement for O-linked N-acetylglucosaminyltransferase in lymphocytes activation. Embo J. 2007;26:4368–4379. doi: 10.1038/sj.emboj.7601845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang WH, Park SY, Nam HW, Kim do H, Kang JG, Kang ES, Kim YS, Lee HC, Kim KS, Cho JW. NFkappaB activation is associated with its OGlcNAcylation state under hyperglycemic conditions. Proc Natl Acad Sci U S A. 2008;105:17345–17350. doi: 10.1073/pnas.0806198105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang WH, Kim JE, Nam HW, Ju JW, Kim HS, Kim YS, Cho JW. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat Cell Biol. 2006;8:1074–1083. doi: 10.1038/ncb1470. [DOI] [PubMed] [Google Scholar]
- 42.Shaw P, Freeman J, Bovey R, Iggo R. Regulation of specific DNA binding by p53: evidence for a role for O-glycosylation and charged residues at the carboxy-terminus. Oncogene. 1996;12:921–930. [PubMed] [Google Scholar]
- 43.Nanashima N, Asano J, Hayakari M, Nakamura T, Nakano H, Yamada T, Shimizu T, Akita M, Fan Y, Tsuchida S. Nuclear localization of STAT5A modified with O-linked N-acetylglucosamine and early involution in the mammary gland of Hirosaki hairless rat. J Biol Chem. 2005;280:43010–43016. doi: 10.1074/jbc.M509481200. [DOI] [PubMed] [Google Scholar]
- 44.Housley MP, Rodgers JT, Udeshi ND, Kelly TJ, Shabanowitz J, Hunt DF, Puigserver P, Hart GW. O-GlcNAc regulates FoxO activation in response to glucose. J Biol Chem. 2008;283:16283–16292. doi: 10.1074/jbc.M802240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuo M, Zilberfarb V, Gangneux N, Christeff N, Issad T. O-glycosylation of FoxO1 increases its transcriptional activity towards the glucose 6-phosphatase gene. FEBS Lett. 2008;582:829–834. doi: 10.1016/j.febslet.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 46.Dentin R, Hedrick S, Xie J, Yates J, 3rd, Montminy M. Hepatic glucose sensing via the CREB coactivator CRTC2. Science. 2008;319:1402–1405. doi: 10.1126/science.1151363. [DOI] [PubMed] [Google Scholar]
- 47.Housley MP, Udeshi ND, Rodgers JT, Shabanowitz J, Puigserver P, Hunt DF, Hart GW. A PGC-1alpha-O-GlcNAc transferase complex regulates FoxO transcription factor activity in response to glucose. J Biol Chem. 2009;284:5148–5157. doi: 10.1074/jbc.M808890200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Comer FI, Hart GW. O-GlcNAc and the control of gene expression. Biochim Biophys Acta. 1999;1473:161–171. doi: 10.1016/s0304-4165(99)00176-2. [DOI] [PubMed] [Google Scholar]
- 49.Jackson SP, Tjian R. O-glycosylation of eukaryotic transcription factors: implications for mechanisms of transcriptional regulation. Cell. 1988;55:125–133. doi: 10.1016/0092-8674(88)90015-3. [DOI] [PubMed] [Google Scholar]
- 50.Hanover JA. Glycan-dependent signaling: O-linked N-acetylglucosamine. FASEB J. 2001;15:1865–1876. doi: 10.1096/fj.01-0094rev. [DOI] [PubMed] [Google Scholar]
- 51.Wells L, Vosseller K, Hart GW. Glycosylation of nucleocytoplasmic proteins: signal transduction and O-GlcNAc. Science. 2001;291:2376–2378. doi: 10.1126/science.1058714. [DOI] [PubMed] [Google Scholar]
- 52.Zachara NE. Detecting the “O-GlcNAc-ome”; Detection, Purification, and Analysis of O-GlcNAc Modified Proteins. Methods Mol Biol. 2009;534:1–30. doi: 10.1007/978-1-59745-022-5_19. [DOI] [PubMed] [Google Scholar]
- 53.Rexach JE, Clark PM, Hsieh-Wilson LC. Chemical approaches to understanding O-GlcNAc glycosylation in the brain. Nat Chem Biol. 2008;4:97–106. doi: 10.1038/nchembio.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Z, Udeshi ND, O'Malley M, Shabanowitz J, Hunt DF, Hart GW. Enrichment and site-mapping of O-Linked N-Acetylglucosamine by a combination of chemical/enzymatic tagging, photochemical cleavage, and electron transfer dissociation (ETD) mass spectrometry. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M900268-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Butkinaree C, Park K, Hart GW. O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbagen.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Issad T, Kuo M. O-GlcNAc modification of transcription factors, glucose sensing and glucotoxicity. Trends Endocrinol Metab. 2008;19:380–389. doi: 10.1016/j.tem.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 57.Golks A, Guerini D. The O-linked N-acetylglucosamine modification in cellular signalling and the immune system. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:748–753. doi: 10.1038/embor.2008.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chatham JC, Not LG, Fulop N, Marchase RB. Hexosamine biosynthesis and protein O-glycosylation: the first line of defense against stress, ischemia, and trauma. Shock. 2008;29:431–440. doi: 10.1097/shk.0b013e3181598bad. [DOI] [PubMed] [Google Scholar]
- 59.Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 60.Lucas PC, McAllister-Lucas LM, Nunez G. NF-kappaB signaling in lymphocytes: a new cast of characters. Journal of cell science. 2004;117:31–39. doi: 10.1242/jcs.00904. [DOI] [PubMed] [Google Scholar]
- 61.Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109(Suppl):S81–96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 62.Schindler C, Levy DE, Decker T. JAK-STAT signaling: from interferons to cytokines. J Biol Chem. 2007;282:20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- 63.Levy DE, Darnell JE., Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 64.Bromberg J, Darnell JE., Jr. The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19:2468–2473. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- 65.Gewinner C, Hart G, Zachara N, Cole R, Beisenherz-Huss C, Groner B. The coactivator of transcription CREB-binding protein interacts preferentially with the glycosylated form of Stat5. J Biol Chem. 2004;279:3563–3572. doi: 10.1074/jbc.M306449200. [DOI] [PubMed] [Google Scholar]
- 66.Koo SH, Flechner L, Qi L, Zhang X, Screaton RA, Jeffries S, Hedrick S, Xu W, Boussouar F, Brindle P, Takemori H, Montminy M. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437:1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- 67.Barco A, Pittenger C, Kandel ER. CREB, memory enhancement and the treatment of memory disorders: promises, pitfalls and prospects. Expert Opin Ther Targets. 2003;7:101–114. doi: 10.1517/14728222.7.1.101. [DOI] [PubMed] [Google Scholar]
- 68.Conkright MD, Montminy M. CREB: the unindicted cancer co-conspirator. Trends Cell Biol. 2005;15:457–459. doi: 10.1016/j.tcb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 69.Rudolph D, Tafuri A, Gass P, Hammerling GJ, Arnold B, Schutz G. Impaired fetal T cell development and perinatal lethality in mice lacking the cAMP response element binding protein. Proc Natl Acad Sci U S A. 1998;95:4481–4486. doi: 10.1073/pnas.95.8.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lamarre-Vincent N, Hsieh-Wilson LC. Dynamic glycosylation of the transcription factor CREB: a potential role in gene regulation. J Am Chem Soc. 2003;125:6612–6613. doi: 10.1021/ja028200t. [DOI] [PubMed] [Google Scholar]
- 71.Shrivastava A, Calame K. An analysis of genes regulated by the multi-functional transcriptional regulator Yin Yang-1. Nucleic Acids Res. 1994;22:5151–5155. doi: 10.1093/nar/22.24.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shi Y, Lee JS, Galvin KM. Everything you have ever wanted to know about Yin Yang 1. Biochim Biophys Acta. 1997;1332:F49–66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 73.Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 74.Hiromura M, Choi CH, Sabourin NA, Jones H, Bachvarov D, Usheva A. YY1 is regulated by O-linked N-acetylglucosaminylation (O-glcNAcylation) J Biol Chem. 2003;278:14046–14052. doi: 10.1074/jbc.M300789200. [DOI] [PubMed] [Google Scholar]
- 75.Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 76.Puigserver P, Rhee J, Lin J, Wu Z, Yoon JC, Zhang CY, Krauss S, Mootha VK, Lowell BB, Spiegelman BM. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARgamma coactivator-1. Mol Cell. 2001;8:971–982. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- 77.Olson BL, Hock MB, Ekholm-Reed S, Wohlschlegel JA, Dev KK, Kralli A, Reed SI. SCFCdc4 acts antagonistically to the PGC-1alpha transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes Dev. 2008;22:252–264. doi: 10.1101/gad.1624208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Teyssier C, Ma H, Emter R, Kralli A, Stallcup MR. Activation of nuclear receptor coactivator PGC-1alpha by arginine methylation. Genes Dev. 2005;19:1466–1473. doi: 10.1101/gad.1295005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 80.Daitoku H, Yamagata K, Matsuzaki H, Hatta M, Fukamizu A. Regulation of PGC-1 promoter activity by protein kinase B and the forkhead transcription factor FKHR. Diabetes. 2003;52:642–649. doi: 10.2337/diabetes.52.3.642. [DOI] [PubMed] [Google Scholar]
- 81.Kuo M, Zilberfarb V, Gangneux N, Christeff N, Issad T. O-GlcNAc modification of FoxO1 increases its transcriptional activity: a role in the glucotoxicity phenomenon? Biochimie. 2008;90:679–685. doi: 10.1016/j.biochi.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 82.Bargonetti J, Manfredi JJ. Multiple roles of the tumor suppressor p53. Curr Opin Oncol. 2002;14:86–91. doi: 10.1097/00001622-200201000-00015. [DOI] [PubMed] [Google Scholar]
- 83.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 84.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 85.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 86.Bech-Otschir D, Kraft R, Huang X, Henklein P, Kapelari B, Pollmann C, Dubiel W. COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. Embo J. 2001;20:1630–1639. doi: 10.1093/emboj/20.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rodriguez MS, Desterro JM, Lain S, Lane DP, Hay RT. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol Cell Biol. 2000;20:8458–8467. doi: 10.1128/mcb.20.22.8458-8467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rodriguez MS, Desterro JM, Lain S, Midgley CA, Lane DP, Hay RT. SUMO-1 modification activates the transcriptional response of p53. Embo J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gostissa M, Hengstermann A, Fogal V, Sandy P, Schwarz SE, Scheffner M, Del Sal G. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. Embo J. 1999;18:6462–6471. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu K, Paterson AJ, Zhang F, McAndrew J, Fukuchi K, Wyss JM, Peng L, Hu Y, Kudlow JE. Accumulation of protein O-GlcNAc modification inhibits proteasomes in the brain and coincides with neuronal apoptosis in brain areas with high O-GlcNAc metabolism. J Neurochem. 2004;89:1044–1055. doi: 10.1111/j.1471-4159.2004.02389.x. [DOI] [PubMed] [Google Scholar]
- 91.Muramatsu M, Inoue S. Estrogen receptors: how do they control reproductive and nonreproductive functions? Biochem Biophys Res Commun. 2000;270:1–10. doi: 10.1006/bbrc.2000.2214. [DOI] [PubMed] [Google Scholar]
- 92.Parker MG. Structure and function of estrogen receptors. Vitam Horm. 1995;51:267–287. doi: 10.1016/s0083-6729(08)61041-9. [DOI] [PubMed] [Google Scholar]
- 93.Arnold SF, Obourn JD, Yudt MR, Carter TH, Notides AC. In vivo and in vitro phosphorylation of the human estrogen receptor. J Steroid Biochem Mol Biol. 1995;52:159–171. doi: 10.1016/0960-0760(94)00166-j. [DOI] [PubMed] [Google Scholar]
- 94.Atsriku C, Britton DJ, Held JM, Schilling B, Scott GK, Gibson BW, Benz CC, Baldwin MA. Systematic mapping of posttranslational modifications in human estrogen receptor-alpha with emphasis on novel phosphorylation sites. Mol Cell Proteomics. 2009;8:467–480. doi: 10.1074/mcp.M800282-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Callige M, Richard-Foy H. Ligand-induced estrogen receptor alpha degradation by the proteasome: new actors? Nucl Recept Signal. 2006;4:e004. doi: 10.1621/nrs.04004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jiang MS, Hart GW. A subpopulation of estrogen receptors are modified by O-linked N-acetylglucosamine. J Biol Chem. 1997;272:2421–2428. doi: 10.1074/jbc.272.4.2421. [DOI] [PubMed] [Google Scholar]
- 97.Cheng X, Hart GW. Glycosylation of the murine estrogen receptor-alpha. J Steroid Biochem Mol Biol. 2000;75:147–158. doi: 10.1016/s0960-0760(00)00167-9. [DOI] [PubMed] [Google Scholar]
- 98.Lonard DM, Nawaz Z, Smith CL, O'Malley BW. The 26S proteasome is required for estrogen receptor-alpha and coactivator turnover and for efficient estrogen receptor-alpha transactivation. Mol Cell. 2000;5:939–948. doi: 10.1016/s1097-2765(00)80259-2. [DOI] [PubMed] [Google Scholar]
- 99.Mosselman S, Polman J, Dijkema R. ER beta: identification and characterization of a novel human estrogen receptor. FEBS Lett. 1996;392:49–53. doi: 10.1016/0014-5793(96)00782-x. [DOI] [PubMed] [Google Scholar]
- 100.Tremblay GB, Tremblay A, Copeland NG, Gilbert DJ, Jenkins NA, Labrie F, Giguere V. Cloning, chromosomal localization, and functional analysis of the murine estrogen receptor beta. Mol Endocrinol. 1997;11:353–365. doi: 10.1210/mend.11.3.9902. [DOI] [PubMed] [Google Scholar]
- 101.Kuiper GG, Gustafsson JA. The novel estrogen receptor-beta subtype: potential role in the cell- and promoter-specific actions of estrogens and anti-estrogens. FEBS Lett. 1997;410:87–90. doi: 10.1016/s0014-5793(97)00413-4. [DOI] [PubMed] [Google Scholar]
- 102.Barkhem T, Carlsson B, Nilsson Y, Enmark E, Gustafsson J, Nilsson S. Differential response of estrogen receptor alpha and estrogen receptor beta to partial estrogen agonists/antagonists. Mol Pharmacol. 1998;54:105–112. doi: 10.1124/mol.54.1.105. [DOI] [PubMed] [Google Scholar]
- 103.Cheng X, Cole RN, Zaia J, Hart GW. Alternative O-glycosylation/O-phosphorylation of the murine estrogen receptor beta. Biochemistry. 2000;39:11609–11620. doi: 10.1021/bi000755i. [DOI] [PubMed] [Google Scholar]
- 104.Cheng X, Hart GW. Alternative O-glycosylation/O-phosphorylation of serine-16 in murine estrogen receptor beta: post-translational regulation of turnover and transactivation activity. J Biol Chem. 2001;276:10570–10575. doi: 10.1074/jbc.M010411200. [DOI] [PubMed] [Google Scholar]
- 105.Chen YX, Du JT, Zhou LX, Liu XH, Zhao YF, Nakanishi H, Li YM. Alternative O-GlcNAcylation/O-phosphorylation of Ser16 induce different conformational disturbances to the N terminus of murine estrogen receptor beta. Chem Biol. 2006;13:937–944. doi: 10.1016/j.chembiol.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 106.Hart GW, Sakabe K. Fine-tuning ER-beta structure with PTMs. Chem Biol. 2006;13:923–924. doi: 10.1016/j.chembiol.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 107.Cho JH, Tsai MJ. The role of BETA2/NeuroD1 in the development of the nervous system. Mol Neurobiol. 2004;30:35–47. doi: 10.1385/MN:30:1:035. [DOI] [PubMed] [Google Scholar]
- 108.Chu K, Nemoz-Gaillard E, Tsai MJ. BETA2 and pancreatic islet development. Recent Prog Horm Res. 2001;56:23–46. doi: 10.1210/rp.56.1.23. [DOI] [PubMed] [Google Scholar]
- 109.Andrali SS, Sampley ML, Vanderford NL, Ozcan S. Glucose regulation of insulin gene expression in pancreatic beta-cells. Biochem J. 2008;415:1–10. doi: 10.1042/BJ20081029. [DOI] [PubMed] [Google Scholar]
- 110.Bernardo AS, Hay CW, Docherty K. Pancreatic transcription factors and their role in the birth, life and survival of the pancreatic beta cell. Mol Cell Endocrinol. 2008;294:1–9. doi: 10.1016/j.mce.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 111.Andrali SS, Qian Q, Ozcan S. Glucose mediates the translocation of NeuroD1 by O-linked glycosylation. J Biol Chem. 2007;282:15589–15596. doi: 10.1074/jbc.M701762200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Petersen HV, Jensen JN, Stein R, Serup P. Glucose induced MAPK signalling influences NeuroD1-mediated activation and nuclear localization. FEBS Lett. 2002;528:241–245. doi: 10.1016/s0014-5793(02)03318-5. [DOI] [PubMed] [Google Scholar]
- 113.Dentin R, Liu Y, Koo SH, Hedrick S, Vargas T, Heredia J, Yates J, 3rd, Montminy M. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature. 2007;449:366–369. doi: 10.1038/nature06128. [DOI] [PubMed] [Google Scholar]
- 114.Al-Hakim AK, Goransson O, Deak M, Toth R, Campbell DG, Morrice NA, Prescott AR, Alessi DR. 14-3-3 cooperates with LKB1 to regulate the activity and localization of QSK and SIK. J Cell Sci. 2005;118:5661–5673. doi: 10.1242/jcs.02670. [DOI] [PubMed] [Google Scholar]
- 115.Screaton RA, Conkright MD, Katoh Y, Best JL, Canettieri G, Jeffries S, Guzman E, Niessen S, Yates JR, 3rd, Takemori H, Okamoto M, Montminy M. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 116.Monticelli S, Rao A. NFAT1 and NFAT2 are positive regulators of IL-4 gene transcription. Eur J Immunol. 2002;32:2971–2978. doi: 10.1002/1521-4141(2002010)32:10<2971::AID-IMMU2971>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 117.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 118.Wang CY, Petryniak B, Thompson CB, Kaelin WG, Leiden JM. Regulation of the Ets-related transcription factor Elf-1 by binding to the retinoblastoma protein. Science. 1993;260:1330–1335. doi: 10.1126/science.8493578. [DOI] [PubMed] [Google Scholar]
- 119.Juang YT, Solomou EE, Rellahan B, Tsokos GC. Phosphorylation and O-linked glycosylation of Elf-1 leads to its translocation to the nucleus and binding to the promoter of the TCR zeta-chain. J Immunol. 2002;168:2865–2871. doi: 10.4049/jimmunol.168.6.2865. [DOI] [PubMed] [Google Scholar]
- 120.Tsokos GC, Nambiar MP, Juang YT. Activation of the Ets transcription factor Elf-1 requires phosphorylation and glycosylation: defective expression of activated Elf-1 is involved in the decreased TCR zeta chain gene expression in patients with systemic lupus erythematosus. Ann N Y Acad Sci. 2003;987:240–245. doi: 10.1111/j.1749-6632.2003.tb06054.x. [DOI] [PubMed] [Google Scholar]
- 121.Kato GJ, Dang CV. Function of the c-Myc oncoprotein. FASEB J. 1992;6:3065–3072. doi: 10.1096/fasebj.6.12.1521738. [DOI] [PubMed] [Google Scholar]
- 122.Amati B, Land H. Myc-Max-Mad: a transcription factor network controlling cell cycle progression, differentiation and death. Curr Opin Genet Dev. 1994;4:102–108. doi: 10.1016/0959-437x(94)90098-1. [DOI] [PubMed] [Google Scholar]
- 123.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 124.Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 125.Luscher B, Kuenzel EA, Krebs EG, Eisenman RN. Myc oncoproteins are phosphorylated by casein kinase II. EMBO J. 1989;8:1111–1119. doi: 10.1002/j.1460-2075.1989.tb03481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Seth A, Alvarez E, Gupta S, Davis RJ. A phosphorylation site located in the NH2-terminal domain of c-Myc increases transactivation of gene expression. J Biol Chem. 1991;266:23521–23524. [PubMed] [Google Scholar]
- 127.Chou TY, Hart GW, Dang CV. c-Myc is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas. J Biol Chem. 1995;270:18961–18965. doi: 10.1074/jbc.270.32.18961. [DOI] [PubMed] [Google Scholar]
- 128.Gu W, Bhatia K, Magrath IT, Dang CV, Dalla-Favera R. Binding and suppression of the Myc transcriptional activation domain by p107. Science. 1994;264:251–254. doi: 10.1126/science.8146655. [DOI] [PubMed] [Google Scholar]
- 129.Fu Z, Tindall DJ. FOXOs, cancer and regulation of apoptosis. Oncogene. 2008;27:2312–2319. doi: 10.1038/onc.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120:2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 131.Biggs WH, 3rd, Meisenhelder J, Hunter T, Cavenee WK, Arden KC. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc Natl Acad Sci U S A. 1999;96:7421–7426. doi: 10.1073/pnas.96.13.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Matsuzaki H, Daitoku H, Hatta M, Tanaka K, Fukamizu A. Insulin-induced phosphorylation of FKHR (Foxo1) targets to proteasomal degradation. Proc Natl Acad Sci U S A. 2003;100:11285–11290. doi: 10.1073/pnas.1934283100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lefterova MI, Lazar MA. New developments in adipogenesis. Trends Endocrinol Metab. 2009;20:107–114. doi: 10.1016/j.tem.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 134.Yeh WC, Cao Z, Classon M, McKnight SL. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev. 1995;9:168–181. doi: 10.1101/gad.9.2.168. [DOI] [PubMed] [Google Scholar]
- 135.Johnson PF. Molecular stop signs: regulation of cell-cycle arrest by C/EBP transcription factors. J Cell Sci. 2005;118:2545–2555. doi: 10.1242/jcs.02459. [DOI] [PubMed] [Google Scholar]
- 136.Lane MD, Tang QQ. From multipotent stem cell to adipocyte. Birth Defects Res A Clin Mol Teratol. 2005;73:476–477. doi: 10.1002/bdra.20150. [DOI] [PubMed] [Google Scholar]
- 137.Timchenko NA. Aging and liver regeneration. Trends Endocrinol Metab. 2009;20:171–176. doi: 10.1016/j.tem.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 138.Robinson GW, Johnson PF, Hennighausen L, Sterneck E. The C/EBPbeta transcription factor regulates epithelial cell proliferation and differentiation in the mammary gland. Genes Dev. 1998;12:1907–1916. doi: 10.1101/gad.12.12.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tang QQ, Gronborg M, Huang H, Kim JW, Otto TC, Pandey A, Lane MD. Sequential phosphorylation of CCAAT enhancer-binding protein beta by MAPK and glycogen synthase kinase 3beta is required for adipogenesis. Proc Natl Acad Sci U S A. 2005;102:9766–9771. doi: 10.1073/pnas.0503891102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Li X, Kim JW, Gronborg M, Urlaub H, Lane MD, Tang QQ. Role of cdk2 in the sequential phosphorylation/activation of C/EBPbeta during adipocyte differentiation. Proc Natl Acad Sci U S A. 2007;104:11597–11602. doi: 10.1073/pnas.0703771104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kim JW, Tang QQ, Li X, Lane MD. Effect of phosphorylation and S-S bond-induced dimerization on DNA binding and transcriptional activation by C/EBPbeta. Proc Natl Acad Sci U S A. 2007;104:1800–1804. doi: 10.1073/pnas.0611137104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li X, Molina H, Huang H, Zhang YY, Liu M, Qian SW, Slawson C, Dias WB, Pandey A, Hart GW, Lane MD, Tang QQ. O-linked N-acetylglucosamine modification on CCAAT enhancer-binding protein {beta}: role during adipocyte differentiation. J Biol Chem. 2009;284:19248–19254. doi: 10.1074/jbc.M109.005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Vanderford NL, Andrali SS, Ozcan S. Glucose induces MafA expression in pancreatic beta cell lines via the hexosamine biosynthetic pathway. J Biol Chem. 2007;282:1577–1584. doi: 10.1074/jbc.M605064200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Norton JD. ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J Cell Sci. 2000;113(Pt 22):3897–3905. doi: 10.1242/jcs.113.22.3897. [DOI] [PubMed] [Google Scholar]
- 145.Yokota Y, Mori S, Narumi O, Kitajima K. In vivo function of a differentiation inhibitor, Id2. IUBMB Life. 2001;51:207–214. doi: 10.1080/152165401753311744. [DOI] [PubMed] [Google Scholar]
- 146.Wang S, Sdrulla A, Johnson JE, Yokota Y, Barres BA. A role for the helix-loop-helix protein Id2 in the control of oligodendrocyte development. Neuron. 2001;29:603–614. doi: 10.1016/s0896-6273(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 147.Gronning LM, Tingsabadh R, Hardy K, Dalen KT, Jat PS, Gnudi L, Shepherd PR. Glucose induces increases in levels of the transcriptional repressor Id2 via the hexosamine pathway. Am J Physiol Endocrinol Metab. 2006;290:E599–606. doi: 10.1152/ajpendo.00242.2005. [DOI] [PubMed] [Google Scholar]
- 148.Vicent D, Piper M, Gammeltoft S, Maratos-Flier E, Kahn CR. Alterations in skeletal muscle gene expression of ob/ob mice by mRNA differential display. Diabetes. 1998;47:1451–1458. doi: 10.2337/diabetes.47.9.1451. [DOI] [PubMed] [Google Scholar]
- 149.Weigert C, Brodbeck K, Sawadogo M, Haring HU, Schleicher ED. Upstream stimulatory factor (USF) proteins induce human TGF-beta1 gene activation via the glucose-response element-1013/-1002 in mesangial cells: up-regulation of USF activity by the hexosamine biosynthetic pathway. J Biol Chem. 2004;279:15908–15915. doi: 10.1074/jbc.M313524200. [DOI] [PubMed] [Google Scholar]
- 150.Kolm-Litty V, Sauer U, Nerlich A, Lehmann R, Schleicher ED. High glucose-induced transforming growth factor beta1 production is mediated by the hexosamine pathway in porcine glomerular mesangial cells. J Clin Invest. 1998;101:160–169. doi: 10.1172/JCI119875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Osterby R. Glomerular structural changes in type 1 (insulin-dependent) diabetes mellitus: causes, consequences, and prevention. Diabetologia. 1992;35:803–812. doi: 10.1007/BF00399925. [DOI] [PubMed] [Google Scholar]
- 152.Yamamoto T, Nakamura T, Noble NA, Ruoslahti E, Border WA. Expression of transforming growth factor beta is elevated in human and experimental diabetic nephropathy. Proc Natl Acad Sci U S A. 1993;90:1814–1818. doi: 10.1073/pnas.90.5.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Solomon SS, Majumdar G, Martinez-Hernandez A, Raghow R. A critical role of Sp1 transcription factor in regulating gene expression in response to insulin and other hormones. Life Sci. 2008;83:305–312. doi: 10.1016/j.lfs.2008.06.024. [DOI] [PubMed] [Google Scholar]
- 154.Majumdar G, Harrington A, Hungerford J, Martinez-Hernandez A, Gerling IC, Raghow R, Solomon S. Insulin dynamically regulates calmodulin gene expression by sequential o-glycosylation and phosphorylation of sp1 and its subcellular compartmentalization in liver cells. J Biol Chem. 2006;281:3642–3650. doi: 10.1074/jbc.M511223200. [DOI] [PubMed] [Google Scholar]
- 155.Dauphinee SM, Ma M, Too CK. Role of O-linked beta-N-acetylglucosamine modification in the subcellular distribution of alpha4 phosphoprotein and Sp1 in rat lymphoma cells. J Cell Biochem. 2005;96:579–588. doi: 10.1002/jcb.20508. [DOI] [PubMed] [Google Scholar]
- 156.Yang X, Su K, Roos MD, Chang Q, Paterson AJ, Kudlow JE. O-linkage of N-acetylglucosamine to Sp1 activation domain inhibits its transcriptional capability. Proc Natl Acad Sci U S A. 2001;98:6611–6616. doi: 10.1073/pnas.111099998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yang X, Zhang F, Kudlow JE. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell. 2002;110:69–80. doi: 10.1016/s0092-8674(02)00810-3. [DOI] [PubMed] [Google Scholar]
- 158.Roos MD, Su K, Baker JR, Kudlow JE. O glycosylation of an Sp1-derived peptide blocks known Sp1 protein interactions. Mol Cell Biol. 1997;17:6472–6480. doi: 10.1128/mcb.17.11.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lim K, Chang HI. O-GlcNAc modification of Sp1 inhibits the functional interaction between Sp1 and Oct1. FEBS Lett. 2009;583:512–520. doi: 10.1016/j.febslet.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 160.Lim K, Chang HI. O-GlcNAcylation of Sp1 interrupts Sp1 interaction with NF-Y. Biochem Biophys Res Commun. 2009 doi: 10.1016/j.bbrc.2009.03.075. [DOI] [PubMed] [Google Scholar]
- 161.Lim K, Chang HI. O-GlcNAc inhibits interaction between Sp1 and Elf-1 transcription factors. Biochem Biophys Res Commun. 2009;380:569–574. doi: 10.1016/j.bbrc.2009.01.121. [DOI] [PubMed] [Google Scholar]